Abstract
Background
Metastasis is responsible for the majority of deaths in a variety of cancer types, including breast cancer. Although several factors or biomarkers have been identified to predict the outcome of patients with breast cancer, few studies have been conducted to identify metastasis-associated biomarkers.
Methods
Quantitative iTRAQ proteomics analysis was used to detect differentially expressed proteins between lymph node metastases and their paired primary tumor tissues from 23 patients with metastatic breast cancer. Immunohistochemistry was performed to validate the expression of two upregulated (EpCAM, FADD) and two downregulated (NDRG1, αB-crystallin) proteins in 190 paraffin-embedded tissue samples. These four proteins were further analyzed for their correlation with clinicopathological features in 190 breast cancer patients.
Results
We identified 637 differentially regulated proteins (397 upregulated and 240 downregulated) in lymph node metastases compared with their paired primary tumor tissues. Data are available via ProteomeXchange with identifier PXD013931. Furthermore, bioinformatics analysis using GEO profiling confirmed the difference in the expression of EpCAM between metastases and primary tumors tissues. Two upregulated (EpCAM, FADD) and two downregulated (NDRG1, αB-crystallin) proteins were associated with the progression of breast cancer. Obviously, EpCAM plays a role in the metastasis of breast cancer cells to the lymph node. We further identified αB-crystallin as an independent biomarker to predict lymph node metastasis and the outcome of breast cancer patients.
Conclusion
We have identified that EpCAM plays a role in the metastasis of breast cancer cells to the lymph node. αB-crystallin, a stress-related protein that has recently been shown to be important for cell invasion and survival, was identified as a potential prognostic biomarker to predict the outcome of breast cancer patients.
Electronic supplementary material
The online version of this article (10.1186/s12885-019-6016-3) contains supplementary material, which is available to authorized users.
Keywords: Breast cancer, Metastasis, EpCAM, FADD, NDRG1, αB-crystallin, Biomarker, iTRAQ proteomic analysis
Background
Breast cancer is the most frequently diagnosed cancer and the leading cause of cancer death among females worldwide [1]. While the incidence rates are generally higher in more developed areas, such as North America and Australia, the incidence of breast cancer in developing countries has been increasing in recent years. In China, breast cancer has become the most common cancer in females and the leading cause of cancer-related death in younger women, especially in highly urbanized regions, which is possibly due to changes in lifestyle and reproductive behavior [2, 3]. With breast cancer, it is not the primary tumors but the metastasis that is responsible for the death of over 90% of breast cancer patients [4, 5]. Some breast cancer patients who initially present with distant metastases and resection are diagnosed with late-stage disease that is nearly incurable. It is possible that the seeds of metastasis are sown at a very early stage in the primary tumor development in the breast [5–8]. Other patients, who have no detectable metastases at the time of diagnosis, ultimately develop metastatic lesions, often months or years after the initial diagnosis [9, 10]. Therefore, the identification of metastasis-related factors warrants further investigation.
Enormous efforts have been made in identifying metastasis-related factors that can be used as prognostic markers to predict the transition from primary to systemic diseases [11–15]. Established prognostic factors that have been confirmed to be involved in breast cancer metastasis include tumor size, axillary lymph node status, and histological grade/subtype. New potential prognostic biomarkers of breast cancer metastasis are continuously being uncovered, which include uPA/PAI1, ER, PR, HER2/ErbB2, circulating tumor cells, the presence of epithelial cells in the bone marrow [12, 16], E-cadherin [17] and, more recently, nucleobindin-2 [18]. Unfortunately, each of these prognostic markers has limited prognostic value in only certain subgroups of patients with breast cancer. Moreover, metastasis to the lymph node, primarily the axillary nodes, is the earliest sign of the metastatic spread of breast cancer [19] and this process occurs at a higher rate than any single distant organ metastasis [20]. In addition to the well-known CXCL12/CXCR4 axis in directing the migration of breast cancer cells through the lymphatics [21, 22], very few studies have been conducted to identify biomarkers associated with the lymph metastasis of breast cancer.
Profiling the tumor tissue proteomics provides important information of biomarker discovery. This potentially useful strategy, however, is limited by the sensitivity of the currently available methods [16]. Isobaric tags for relative and absolute quantitation (iTRAQ) has been widely employed in quantitative proteomic studies in complex biological systems [23, 24] and has been successful in the characterization of protein bioindicators of diverse effects [25]. Recently, the combination of iTRAQ isobaric labeling, multidimensional liquid chromatography and ultrahigh resolution mass spectrometry has been used to identify tumor biomarkers in cancer, including breast cancer [26–30]. In this study, primary breast tumor tissues and paired lymph node metastases from breast cancer patients were analyzed in parallel by the quantitative iTRAQ proteomic method. Four differentially regulated proteins were validated by immunohistochemistry. Through further clinicopathological correlation and bioinformatic studies, we identified αB-crystallin as a potential prognostic biomarker to predict the occurrence of lymph metastasis and the clinical outcome of breast cancer patients.
Methods
Human subjects
This study was approved by the Research Ethics Committee of Central South University, China, and informed consent was obtained from all of the patients. All patients were diagnosed by two senior pathologists as invasive breast cancer (invasive ductal carcinoma or invasive lobular carcinoma) without radiotherapy or chemotherapy before surgery.
Mass collection methods for breast cancer
Select the cases with large lesions (> 1.5 cm × 1.5 cm × 1 cm) which were diagnosed as breast cancer by frozen section. Tissue samples were cut the tumors (> 0.5 cm × 0.5 cm × 0.5 cm) and preserved them in liquid nitrogen. We then decided whether to join the group according to routine diagnosis and lymph node metastasis.
Methods for collecting lymph node metastases
The lymph nodes with the largest diameter (> 1 cm) were selected, the adipose tissue around the lymph nodes was removed, the lymph nodes were cut along the largest diameter, and the color of the section was observed by naked eyes. The selected lymph nodes were divided into two parts, half of which were stored in liquid nitrogen, and the other half were stained with H&E and observed under a microscope to determine whether the lymph nodes really existed. In breast cancer metastasis, the criterion for admission was that metastatic cancer accounted for more than 90% of lymph nodes. The collected breast cancer tissues and matched metastatic lymph nodes were preserved in liquid nitrogen.
iTRAQ proteomics
Twenty-three paired fresh primary tumors and metastatic axillary LNs were collected from Hunan Cancer Hospital between November 2013 and March 2014. Each collected tissue sample was divided into two parts; one part was used for routine pathological examination, and the other part was stored in liquid nitrogen for the proteomic study. To minimize the influence of residual lymphoid tissues on protein identification, only the axillary LNs with > 95% neoplastic cells according to H&E examination were used for the proteomic study. Relative quantitative proteomics was performed using the Fitgene iTRAQ Proteomics Platform (http://www.fitgene.com) according to the standard procedure [28, 30]. Briefly, the prepared lysates (200 μg) were treated with 4 μL of reducing reagent for 1 h at 60 °C and then blocked with 2 μL of cysteine blocking reagent for 10 min at room temperature. After centrifugation, the supernatant was collected and incubated with trypsin and TEAB overnight at 37 °C. The samples were then mixed with the iTRAQ reagents and subjected to two-dimensional LC-MS/MS analysis and a database search. An expression change greater than 1.5-fold was considered a difference between the primary tumor tissues and the paired metastatic LN tissues.
The raw data acquired from LC-MS/MS was processed with AB Sciex ProteinPilot 4.0 (AB Sciex, Concord, Ontario, Canada), and protein identification and quantification were achieved by searching the UniProt database (Release 2014.5.14). Proteomics profiling and database searching based on the TripleTOF® 5600+ System (AB Sciex) and ProteinPilot 4.0 (AB Sciex) were performed following the manufacturer’s recommendations. The parameters were set as follows: Unused ≥1.3; Credibility ≥95%; C.V. ≤ 0.5; AVG. ≥ 1.5 or ≤ 0.67; T.TEST < 0.05; Peptides (95%) ≥ 4. To ensure the reliability and stability of the reported data, we performed the following steps for data quality control. First, before database searching, we selected “Run False Discovery Rate Analysis” in the software AB Sciex ProteinPilot for FDR control. Second, we removed the results identified by the reverse database. Third, we removed those proteins with extremely high or low ratios. Finally, we removed those proteins with abnormal quantification between technical repetition and biological repetition.
The coefficients of variation (CV) of biological repetition were analyzed for data from different groups of samples. By observing the experimental data, when the coefficient of variation is within (+ 50%), 60% of the identified proteins can be covered. Most of the data exceeding the coefficient of variation are caused by individual differences of organisms. In subsequent analysis, this part of data will be excluded from the scope of analysis. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [31] partner repository with the dataset identifier PXD013931.
Immunohistochemical analysis
A total of 106 paired paraffin-embedded tissue samples with lymph node metastasis were obtained from female patients with breast disease who were operated on in Hunan Cancer Hospital between May 1996 and May 2008. None of the patients underwent preoperative chemotherapy or radiotherapy. The tissue samples were fixed with 10% formaldehyde in PBS, embedded in paraffin and cut into consecutive 4-μm sections. Breast cancer was staged according to the Nottingham modified program of Bloom-Richardson scoring system.
For immunohistochemistry, a two-step polymer-based detection method (EnVison™) was used according to our recently published protocol [18]. The primary antibodies (all diluted 1:200) were rabbit monoclonal antibodies obtained from Abcam (Cambridge, MA, USA) (EpCAM [ab124825], FADD [ab108601], αB-crystallin [ab76467]) or CST (Danvers, MA, USA) (NDRG1 [#9485]). The staining was examined by two senior pathologists, and the total immunostaining score (TIS) was calculated as described.
Clinicopathological correlation study
A total of 190 breast cancer patients admitted to Hunan Cancer Hospital between May 1996 and March 2005 were followed up for over 10 years, and the clinicopathological parameters, including age at diagnosis, tumor size, axillary node status, clinical stage, histological type/grade, ER/PR/HER2 status, and menstruation history, were recorded. These parameters were correlated with the expression levels of the four metastasis-associated proteins.
GEO analysis
The difference in the expression levels of αB-crystallin between normal breast tissues and breast cancers was analyzed online in the Gene Expression Omnibus (GEO) profile (https://www.ncbi.nlm.nih.gov/geo/) using the search terms of “invasive breast cancer” and “CRYAB”.
Statistical analysis
The statistical analysis was performed using SPSS 2.0 Software. A Wilcoxon signed-rank test was used to compare the expression of the metastasis-associated proteins between the paired primary tumors and the metastatic lesions of breast cancer on immunohistochemistry. A chi-square (χ2) test was used to evaluate the metastasis-associated proteins with the clinicopathological parameters. Survival analysis was performed using the Kaplan-Meier method. The Student’s t test was used to compare the mRNA expression of FADD and αB-crystallin between normal breast and breast cancer tissues from the GEO profile. A p value of less than 0.05 was considered statistically significant.
Results
Identification of lymph metastasis-associated proteins in breast cancer patients
To identify the proteins associated with lymph metastasis of breast cancer, we first analyzed 23 paired primary tumors and axillary lymph node metastases from patients with metastatic breast cancer using iTRAQ-based proteomic analysis. The quantitative data are presented in Additional file 3: Table S1. A total of 637 differentially regulated proteins (397 upregulated and 240 downregulated) between the primary sites and the lymph node metastases of breast cancer were identified based on a 95% confidence interval and a difference ratio of ≥1.5 for up-regulated protein, and ratio ≤ 0.67 for down-regulated. The top 30 upregulated and downregulated proteins are presented in Additional file 4: Table S2 and Table S3, respectively.
To gain insights into the biological and molecular characteristics of these proteins, gene ontology (GO) analysis was performed on the differentially regulated proteins. An analysis of the biological process annotations of the 397 proteins that were upregulated in metastatic sites is shown in Additional file 1: Figure S1A. These proteins were predominantly involved in cellular nitrogen compound metabolism and biosynthesis, followed by signal transduction, small molecule metabolism, and stress responses. The GO enrichment analysis of cellular components indicated that these upregulated proteins were primarily distributed in the nucleus and the cytoplasm (Additional file 1: Figure S1B). In terms of molecular functions, the majority of these upregulated proteins were involved in binding activities, such as RNA binding and ion binding (Additional file 1: Figure S1C). The 240 proteins that were downregulated in lymph node metastases were primarily associated with signal transduction, anatomical structure development, stress response, and cell differentiation (Additional file 1: Figure S1D). For cellular distribution, the downregulated proteins were predominantly localized in the extracellular region, the organelles, and the cytoplasm (Additional file 1: Figure S1E). The most significant molecular function of these downregulated proteins was ion binding (Additional file 1: Figure S1F).
Validation of differentially regulated proteins
We filtered out four proteins (two upregulated proteins and two downregulated proteins) for further validation. These proteins were chosen based on the following criteria: 1) they had a fold-change of greater than 1.5 (for the upregulated proteins) or less than 0.67 (for the downregulated proteins); 2) they had a peptide number of greater than 3 in the iTRAQ identification; and 3) they are known to be related to cancer cell invasion/metastasis based on previous studies. These four proteins were EpCAM (epithelial cell adhesion molecule) [32], FADD (Fas-associated death domain protein) [33], NDRG1 (N-myc downstream-regulated gene 1) [34] and αB-crystallin (Alpha-crystallin B chain) [35], and their ratios of metastatic vs. primary tumor sites were 1.85, 1.51, 0.33, and 0.34, respectively. The mass annotated product ion spectra of these four proteins were obtained (data not shown). The biological processes, cellular locations, and molecular functions of these four individual proteins (Additional file 4: Table S4) were analyzed using the UniProt knowledgebase (http://www.uniprot.org/), which was in agreement with the abovementioned GO analysis results.
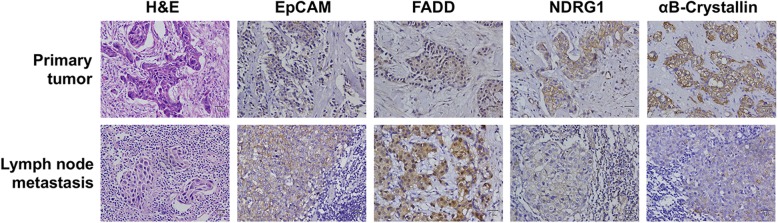
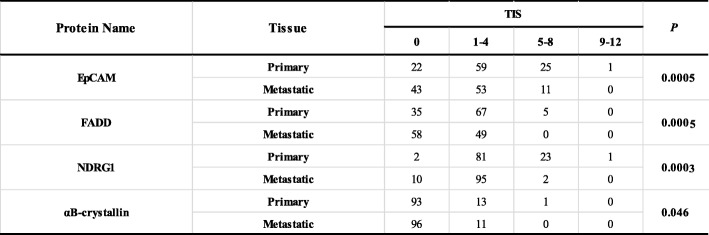
Next, we used immunohistochemistry to verify the expression of the four breast cancer lymph metastasis-associated proteins in 106 cases of paraffin-embedded paired primary tumors and lymph metastasis tissues obtained from metastatic breast cancer patients. The representative staining images are presented in Fig. 1, and the quantitatively analyzed results, which are presented as total immunostaining score (TIS), are summarized in Table 1. As shown in Fig. 1, most of the EpCAM was localized on the plasma membrane, which is in agreement with its known cellular localization. FADD was primarily localized in the cytoplasm and the nucleus. NDRG1 was located in the plasma membrane and the cytoplasm. The αB-crystallin protein was primarily expressed on the plasma membrane and in the cytoplasm. Consistent with the iTRAQ data, NDRG1 and αB-crystallin were downregulated at the metastatic sites compared with the primary tumors in terms of TIS (Table 1) (P = 0.0003 [NDRG1] or P = 0.046 [αB-crystallin]). However, the expression levels of EpCAM and FADD were also lower at the metastatic sites compared with the primary tumors (P = 0.0005).
Fig. 1.
Immunohistochemical analysis of the expression of four breast cancer metastasis-associated proteins. The expression levels of EpCAM, FADD, NDRG1, and αB-crystallin were evaluated by the immunohistochemical staining of paraffin-embedded paired primary and metastatic tissue sections that were obtained from patients with metastatic breast cancer
Table 1.
Summary of the expression of the four metastasis-associated proteins in the paired primary and metastatic tissues of breast cancer
Correlation of metastasis-associated proteins with the clinicopathological features of breast cancer patients
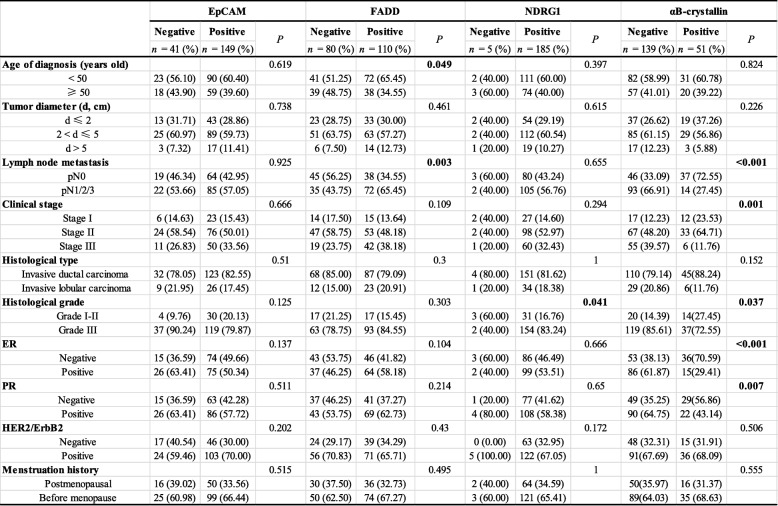
To clarify the clinical relevance of the proteins identified from iTRAQ proteomics that were associated with lymph metastasis, we analyzed the relationship between these four proteins and the clinicopathological parameters of 190 cases of breast cancer patients. We showed that EpCAM was not correlated with any of the clinicopathological parameters examined (Table 2). However, FADD expression was positively correlated with a younger age at diagnosis (P = 0.049) and lymph node metastasis (P = 0.003). NDRG1 expression was correlated with worse histological grade (P = 0.041) but not with lymph node metastasis (P = 0.655). αB-crystallin expression was inversely correlated with lymph node metastasis (P < 0.001), clinical stage (P = 0.001), histological grade (P = 0.037), ER (P < 0.001), and PR status (P = 0.007).
Table 2.
The association between the four metastasis-associated proteins and the clinicopathological features of 190 breast cancer patients
Association of metastasis-associated proteins with overall survival of breast cancer patients
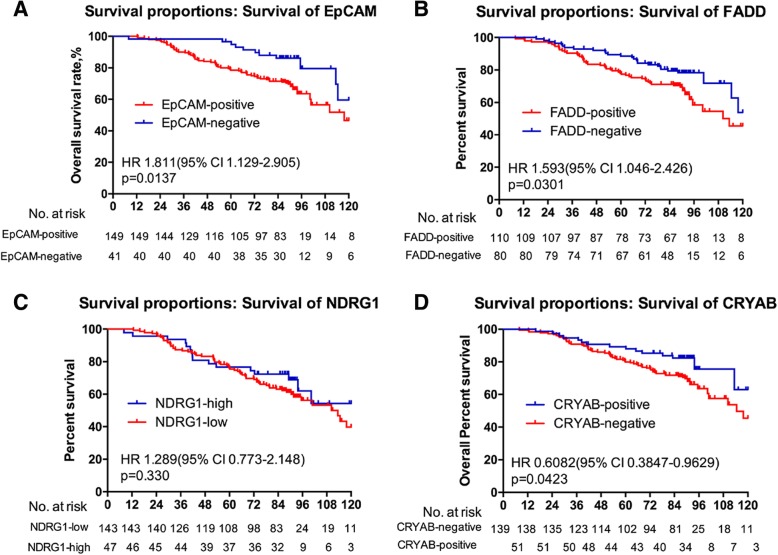
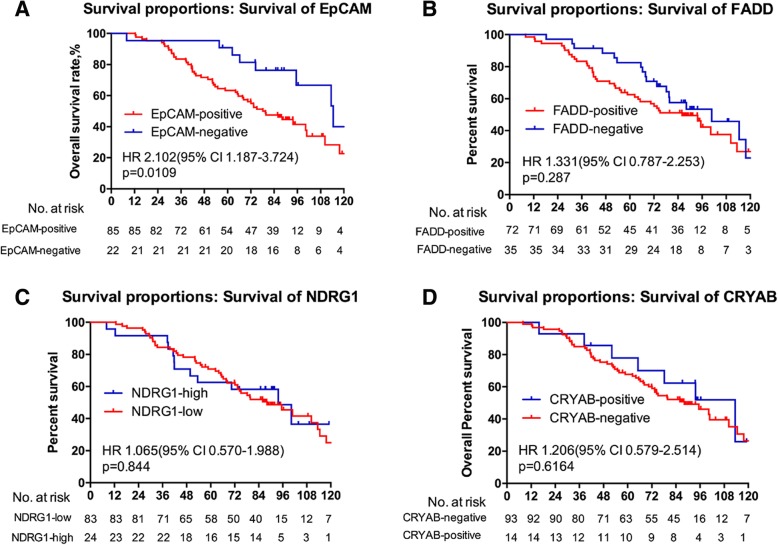
In addition, we followed up 190 breast cancer patients for over 10 years and conducted a survival analysis for the positivity of expression (EpCAM, FADD, and αB-crystallin) or the level of expression (NDRG1) in the primary tumor sites. The results revealed that the patients who had positive expression of EpCAM or FADD survived for a shorter time compared with those with negative expression (Fig. 2a-b). Those who had positive expression of αB-crystallin survived longer than those with negative expression (Fig. 2d). However, the expression level of NDRG1 had no prognostic value for breast cancer patients (Fig. 2c). Moreover, the prognostic value of EpCAM only applied to patients with lymph node metastasis (Fig. 3a-d). Univariable analysis linked with tumor diameter, TNM stage and histology stage and type, but multivariable analysis assigned significance only to histology type (lobular carcinoma vs. duct carcinoma) (Table 3).
Fig. 2.
The association between four metastasis-associated proteins and the overall survival of breast cancer patients. Kaplan-Meier plots of the association between the expression of EpCAM (a), FADD (b), NDRG1 (c), and αB-crystallin (d) and the overall survival probability of breast cancer patients
Fig. 3.
The association between four metastasis-associated proteins and the overall survival in breast cancer patients with metastasis. Kaplan-Meier plots of the association between the expression of EpCAM (a), FADD (b), NDRG1 (c), and αB-crystallin (d) and the overall survival probability in breast cancer patients with metastasis
Table 3.
Univariate and Multivariate Analysis by a Cox Proportional Hazards Regression Model in Cohort
| Variable | OS | |||
|---|---|---|---|---|
| Univariate | Multivariate | |||
| HR (95% CI) | P Value | HR (95% CI) | P Value | |
| Age, years (> 45 vs. ≤ 45) | 0.898 (0.581–1.390) | 0.630 | NA | |
| ER (positive vs. negative) | 1.114 (0.711–1.745) | 0.637 | NA | |
| PR (positive vs. negative) | 1.213 (0.775–1.899) | 0.398 | NA | |
| CrebB-2 (positive vs. negative) | 1.128 (0.705–1.806) | 0.615 | NA | |
| Menstrual history (presence vs. absence) | 1.381 (0.851–2.241) | 0.191 | NA | |
| Operation | 0.649 | NA | ||
| Modified radical mastectomy vs. radical correction | 1.150 (0.727–1.820) | 0.550 | NA | |
| Other operation vs. radical correction | 0.642 (0.155–2.663) | 0.542 | NA | |
| FADD (positive vs. negative) | 1.580 (0.995–2.509) | 0.053 | NA | |
| NDRG1 (low vs. high) | 1.302 (0.762–2.226) | 0.335 | NA | |
| CRYAB (positive vs. negative) | 1.561 (0.902–2.701) | 0.112 | NA | |
| Tumor diameter, cm | 0.072 | NS | ||
| > 5 vs. > 2 and ≤ 5 | 1.923 (1.019–3.636) | 0.043 | ||
| > 5 vs. ≤2 | 2.230 (1.093–4.549) | 0.027 | ||
| TNM stage | < 0.0001 | NS | ||
| III vs. I | 4.329 (1.824–10.273) | 0.001 | ||
| III vs. II | 2.101 (1.333–3.311) | 0.001 | ||
| Histology stage (poorly differentiation vs. high-middle differentiation) | 2.286 (1.100–4.751) | 0.027 | NS | |
| Histology type (lobular carcinoma vs. duct carcinoma) | 1.720 (1.025–2.886) | 0.040 | 1.846 (1.093–3.118) | 0.022 |
| Lymph node metastasis (presence vs. absence) | 2.810 (1.694–4.662) | < 0.0001 | 2.801 (1.688–4.649) | < 0.0001 |
| EpCAM (positive vs. negative) | 2.306 (1.218–4.367) | 0.010 | 2.585 (1.351–4.944) | 0.004 |
Data in bold are P values < 0.05
Downregulation of αB-crystallin mRNA expression in breast cancer
Finally, to examine whether αB-crystallin (gene name: CRYAB) was also involved in human breast cancer development, using the public database, we reviewed the mRNA expression of CRYAB in normal breast and invasive breast cancer tissues in Gene Expression Omnibus (GEO) (Expression Profile GDS3324). The results are presented in Additional file 2: Figure S2. The expression of CRYAB was significantly lower in breast cancer tissues compared with normal breast tissues (P = 0.001). We further found that the level of expression of αB-crystallin was indeed lower in breast cancer tissues compared with benign breast lesions, with metastatic breast cancer having the lowest expression (Table 4). These findings support the tumor-suppressive role of αB-crystallin in the development of breast cancer.
Table 4.
Summary of the expression of CRYAB in different stages of breast tissues
| Tissue | TIS | P | |||
|---|---|---|---|---|---|
| 0 | 1–4 | 5–8 | 9–12 | ||
| Benign | 1 | 24 | 16 | 6 | 0.0003 |
| Non-metastatic | 46 | 28 | 6 | 3 | |
| Metastatic | 189 | 24 | 1 | 0 | |
Discussion
Metastasis is one of the most important factors that causes the death of patients with breast cancer. Detection of breast cancer metastasis at the earliest possible stage is critical for the successful management of breast cancer progression. Therefore, it is very important to search for effective biomarkers for breast cancer metastasis and prognosis. In proteomic comparative studies of breast cancer metastasis, with tumor tissue as the research object, the commonly used method is based on the comparison of lymph node metastasis or other organ metastases, gene expression or protein expression of primary breast cancer with metastasis and without metastasis. In this study, we used the iTRAQ proteomic technique to analyze the differentially regulated proteins between the primary tumor sites and their corresponding lymph node metastases in metastatic breast cancer patients, and this comparison method can more accurately compare the differences in protein expression of breast cancer cells with varying metastatic capacity. Four proteins (EpCAM, FADD, NDRG1, and αB-crystallin) were chosen for validation by immunohistochemistry. Specially, αB-crystallin could potentially be addressed as a potential prognostic biomarker to predict the lymph node metastasis and clinical outcomes of breast cancer patients.
αB-crystallin, also called HspB5, is a member of the α-crystallin family small heat shock proteins and is an important component of the vertebrate lens [36]. In nonlens tissues, αB-crystallin is an integral part of the cellular proteostasis system, which is associated with a broad spectrum of human diseases, including cancer [37]. αB-crystallin plays an important role in stress responses, such as heat shock and radiation poisoning. As a molecular chaperone, αB-crystallin is expressed in human cells at a higher level under pathological conditions. The expression of αB-crystallin in human renal carcinogenesis, triple-negative (basal-like) breast cancer, hepatocellular carcinoma, and squamous cell carcinoma of the head and neck is related to poor prognosis [36, 37], suggesting an oncogenic role for αB-crystallin in promoting tumorigenesis. In breast cancer, αB-crystallin has been shown to be an oncoprotein that predicts poor prognosis [38–41] and resistance to neoadjuvant chemotherapy, especially for triple-negative breast cancer [40, 42]. However, the role of αB-crystallin as a tumor suppressor has also been reported [43]. These contradictory findings indicate that the role of αB-crystallin in carcinogenesis is complicated. The present study demonstrated that αB-crystallin was downregulated in the lymph metastases compared with the primary breast tumors. This finding is inconsistent with the previous finding that αB-crystallin expression promotes the brain metastasis of breast cancer [38, 44]. Recently, the majority of lymphatic and distant metastases were shown to originate differently in human colorectal cancer [45]. This phenomenon is also true for breast cancer metastasis, in which approximately 1/3 of breast cancer patients without lymph metastasis develop distant metastasis [46]. These observations suggest that the two routes of cancer spreading may occur independently and may use different sets of molecular routers to drive the metastatic spread of cancer cells through either the lymphatics or the blood vessels. Reconciling our data with the previous reports, it is possible that αB-crystallin plays a role of router to switch between lymphatic and hematogenous spreading. That is, the role of αB-crystallin in breast cancer progression needs to be reevaluated. It is speculated that αB-crystallin may function as a tumor promoter in hematogenous metastasis – to the brain, for example, but αB-crystallin may function as a tumor suppressor in lymph node metastasis. However, this speculation should be validated experimentally through in vitro and in vivo studies. Clearly, our findings further support a tumor-suppressor role for αB-crystallin in breast cancer development.
Many studies have shown that there is close link between FADD and many cancers, such as nonsmall cell lung cancer [47], gastric cancer [48] and hepatocellular carcinoma (HCC) [49]. In the first two of these cancers, the expression of FADD was correlated with lymph node metastasis and the poor prognosis of patients, and the loss of FADD expression plays an important role in HCC carcinogenesis. FADD expression is associated with T stage and perineural invasion [50]. An increase in FADD expression was shown to be associated with a higher incidence of lymph node metastasis at presentation and with a shorter DMFI when lymph node metastases are present [33]. These studies only involved the comparison between cancer and the surrounding normal tissues, whereas we focused on the differences in FADD expression between primary tumors and metastases. Using proteomic results, we determined that the expression of FADD was upregulated in metastasis. Furthermore, the IHC results revealed that there were significant differences in FADD expression between the primary tumors and metastases, but the rate of FADD-positive tumors decreased, which is inconsistent with the proteomic results. The possible reason for this inconsistency is that proteomics analyzes the relative quantity of protein expression, whereas immunohistochemistry analyzes the positive rate of protein expression, and thus results from these two methods are not always consistent. In addition, we also investigated potential correlations between FADD expression and the clinical pathological characteristics of 190 patients with breast cancer. We performed a 120-months survival analysis and found that FADD expression was associated with lymph node metastasis. Furthermore, higher expression levels of FADD were identified in patients with breast cancer, which were also correlated with a shorter survival time. These finding suggest that there is a close relationship between FADD expression and the lymph node metastasis and poor prognosis of breast cancer. Moreover, the regulatory mechanism of FADD in breast cancer metastasis warrants further investigation.
NDRG1 has been reported to function as a metastasis suppressor gene, and it is downregulated in gastric cancer [34], prostate [51, 52], pancreatic cancer [53] and breast cancers [45]. However, compared with normal tissue, NDRG1 expression was shown to be upregulated in homologous hepatocellular carcinoma [54] and oral squamous cell carcinoma [55]. In this study, all of the proteomics and IHC results revealed that NDRG1 expression was downregulated in metastases compared to the primary tumors. The expression of NDRG1 in various tissues may be affected by many factors, such as metal ions, oxygen, proto-oncogenes, tumor suppressor genes, hormones or vitamins. For example, NDRG1 expression in prostate cancer cells was shown to be affected by androgens, whereas NDRG1 expression in breast cancer cells is mainly associated with estradiol. Thus, the expression of NDRG1 is variable. In the clinical pathology and survival analysis, significant differences in NDRG1 expression were not detected in this study.
EpCAM is a transmembrane glycoprotein and appears to play a role in tumorigenesis and metastasis of carcinomas [56]. EpCAM is frequently upregulated in carcinomas but is not expressed in cancers of non-epithelial origin. At present, the FDA approves the automated cell detection method for EpCAM as biomarker, and this method has been used to detect circulating tumor cells in patients with breast [57], prostate [32, 58] and esophageal cancer [59]. The expression of EpCAM was shown to be high in laryngeal carcinoma but low in bone marrow as a metastatic niche for disseminated cancer cells [60]. These findings are consistent with our IHC results. However, EpCAM expression was increased in the metastatic group compared to the nonmetastatic group according to both iTRAQ and the proteomics analysis. Furthermore, the survival analysis showed that the survival rate was lower in the EpCAM-positive group. Therefore, the expression of EpCAM should be further clarified in breast cancer metastasis. Taken together, these data suggest that EpCAM plays a critical role in the metastatic process of breast cancer.
Conclusions
In summary, we discovered differentially regulated proteins between the primary breast tumors and their lymph node metastatic sites using the iTRAQ proteomics analysis. Through further immunohistochemical study, clinicopathological correlation analysis, and GEO profiling, we identified αB-crystallin as an independent biomarker to predict the outcome of breast cancer patients in the lymph node. Obviously, αB-crystallin plays a role in the metastasis of breast cancer cells to the lymph node, but its exact role in each step of breast cancer metastasis and the underlying signaling mechanism remain to be fully clarified. EpCAM, FADD and NDRG1 expression were shown to be associated with the progression of breast cancer, but the questions of how certain oncogenes may initiate dissemination before triggering aggressive proliferation and how tumor-suppressor pathways suppress metastasis in breast cancer warrant further investigation.
Additional files
Figure S1. GO analysis of the differentially regulated proteins in lymph node metastases vs. primary breast tumor tissues. The upregulated (A-C) and downregulated (D-F) proteins identified by the iTRAQ proteomics were analyzed by the GO Consortium and categorized according to their biological processes, cellular locations, and molecular functions. (TIF 5559 kb)
Figure S2. GEO analysis of CRYAB mRNA expression in normal breast and breast cancer tissues. (A) The mRNA expression of CRYAB in normal breast tissues (n =5) and breast cancer tissues (n = 28) was analyzed from the Affymetrix Human Genome Microarray at the GEO website (https://www.ncbi.nlm.nih.gov/geoprofiles/54408377 for αB-crystallin). (B) Quantification of the mRNA expression of CRYAB in normal breast tissues and breast cancer tissues. (TIF 4929 kb)
Table S1. Identification of differentially expressed proteins between primary breast cancer tissues and metastatic lymph node tissues by the iTRAQ technique. (XLS 1215 kb)
Table S2. Partial up-regulated proteins in metastatic lymph node compared with primary tumor in breast cancer. Table S3. Partial down-regulated proteins in metastatic lymph node compared with primary tumor in breast cancer. Table S4. UniProt analysis of the biological processes, cellular locations, and molecular functions of the four metastasis-associated proteins. (DOCX 29 kb)
Acknowledgements
We thank the Proteomic technique platform from Lei Xue by FitGene Biotechnology Co., Ltd. (Guangzhou, P. R. China, http://www.fitgene.com) for the iTRAQ proteomics analysis and the PRIDE partner repository. The clerical assistance from Kassey Deng, Lu Lu, Chao Chen and Huimei Yi is highly appreciated.
Abbreviations
- EpCAM
Epithelial cell adhesion molecule
- FADD
Fas-associated death domain
- GEO
Gene expression omnibus
- GO
Gene ontology
- HCC
Hepatocellular carcinoma
- iTRAQ
Isobaric tags for relative and absolute quantitation
- NDRG1
N-myc downstream-regulated gene 1
- TIS
Total immunostaining score
Authors’ contributions
LZ, XD, JZ, YZ, GH, PT and YT conceived of the study. LZ, XD, LY, XT, JZ, YZ, GH, SZ, PT and YT analyzed and interpreted the data. LZ, XD, LY, XT, JZ, YZ, GH, and YT performed the histological examination of the tumor tissue. LZ, JZ, YZ, LY, XT, GH, SZ, PT and YT participated in the study design and coordination, helped to interpret the data, and helped to draft the manuscript. LZ, SZ, PT and YT helped to interpret the data and to draft the manuscript. LZ, XD, JZ, and YT drafted the manuscript, and LZ, XD, and YT were major contributors in writing the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the Hunan Province Science and Technology Project (2014FJ6090 to LZ) and the National Natural Science Foundation of China (81472496 to XD) in the design of this study and collection, analysis, and interpretation of data.
Availability of data and materials
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [1] partner repository with the dataset identifier PXD013931.
Ethics approval and consent to participate
All participants signed informed consent forms, and the Research Ethics Committee of Central South University, China approved this study, reference number is EC20101220005.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Liang Zeng, Email: zlxx03@126.com.
Xiyun Deng, Email: dengxiyunmed@hunnu.edu.cn.
Jingmin Zhong, Email: zhongjingmin123@126.com.
Li Yuan, Email: lizzyklarck@126.com.
Xiaojun Tao, Email: xiaojtao@126.com.
Sai Zhang, Email: zhangsai2000@csu.edu.cn.
Yong Zeng, Email: 13118408@bjtu.edu.cn.
Guangchun He, Email: 18874093428@163.com.
Pingping Tan, Email: tanpingping@hnszlyy.com.
Yongguang Tao, Email: taoyong@csu.edu.cn.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Chen W, Zheng R, Zuo T, Zeng H, Zhang S, He J. National cancer incidence and mortality in China, 2012. Chin J Cancer Res. 2016;28(1):1–11. doi: 10.3978/j.issn.1000-9604.2016.02.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jin X, Mu P. Targeting breast Cancer metastasis. Breast Cancer (Auckl) 2015;9(Suppl 1):23–34. doi: 10.4137/BCBCR.S25460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schwartz RS, Erban JK. Timing of metastasis in breast Cancer. N Engl J Med. 2017;376(25):2486–2488. doi: 10.1056/NEJMcibr1701388. [DOI] [PubMed] [Google Scholar]
- 6.DeMichele A, Yee D, Esserman L. Mechanisms of resistance to neoadjuvant chemotherapy in breast Cancer. N Engl J Med. 2017;377(23):2287–2289. doi: 10.1056/NEJMcibr1711545. [DOI] [PubMed] [Google Scholar]
- 7.Harper KL, Sosa MS, Entenberg D, Hosseini H, Cheung JF, Nobre R, Avivar-Valderas A, Nagi C, Girnius N, Davis RJ, et al. Mechanism of early dissemination and metastasis in Her2(+) mammary cancer. Nature. 2016;540(7634):588. doi: 10.1038/nature20609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hosseini H, Obradovic MM, Hoffmann M, Harper KL, Sosa MS, Werner-Klein M, Nanduri LK, Werno C, Ehrl C, Maneck M, et al. Early dissemination seeds metastasis in breast cancer. Nature. 2016;540(7634):552. doi: 10.1038/nature20785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O'Shaughnessy J. Extending survival with chemotherapy in metastatic breast cancer. Oncologist. 2005;10(Suppl 3):20–29. doi: 10.1634/theoncologist.10-90003-20. [DOI] [PubMed] [Google Scholar]
- 10.Spolverato G, Vitale A, Bagante F, Connolly R, Pawlik TM. Liver resection for breast Cancer liver metastases: a cost-utility analysis. Ann Surg. 2017;265(4):792–799. doi: 10.1097/SLA.0000000000001715. [DOI] [PubMed] [Google Scholar]
- 11.Ohsfeldt RL, Ward MM, Schneider JE, Jaana M, Miller TR, Lei Y, Wakefield DS. Implementation of hospital computerized physician order entry systems in a rural state: feasibility and financial impact. J Am Med Inform Assoc. 2005;12(1):20–27. doi: 10.1197/jamia.M1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347(25):1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 13.Chandran UR, Ma C, Dhir R, Bisceglia M, Lyons-Weiler M, Liang W, Michalopoulos G, Becich M, Monzon FA. Gene expression profiles of prostate cancer reveal involvement of multiple molecular pathways in the metastatic process. BMC Cancer. 2007;7:64. doi: 10.1186/1471-2407-7-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Appierto V, Di Cosimo S, Reduzzi C, Pala V, Cappelletti V, Daidone MG. How to study and overcome tumor heterogeneity with circulating biomarkers: the breast cancer case. Semin Cancer Biol. 2017;44:106–116. doi: 10.1016/j.semcancer.2017.04.007. [DOI] [PubMed] [Google Scholar]
- 15.Ramaswamy S, Ross KN, Lander ES, Golub TR. A molecular signature of metastasis in primary solid tumors. Nat Genet. 2003;33(1):49–54. doi: 10.1038/ng1060. [DOI] [PubMed] [Google Scholar]
- 16.Lorusso G, Ruegg C. New insights into the mechanisms of organ-specific breast cancer metastasis. Semin Cancer Biol. 2012;22(3):226–233. doi: 10.1016/j.semcancer.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 17.Fry SA, Sinclair J, Timms JF, Leathem AJ, Dwek MV. A targeted glycoproteomic approach identifies cadherin-5 as a novel biomarker of metastatic breast cancer. Cancer Lett. 2013;328(2):335–344. doi: 10.1016/j.canlet.2012.10.011. [DOI] [PubMed] [Google Scholar]
- 18.Zeng Liang, Zhong Jingmin, He Guangchun, Li Fangjun, Li Jing, Zhou Wen, Liu Wenbin, Zhang Yun, Huang Sanqian, Liu Zhihong, Deng Xiyun. Identification of Nucleobindin-2 as a Potential Biomarker for Breast Cancer Metastasis Using iTRAQ-based Quantitative Proteomic Analysis. Journal of Cancer. 2017;8(15):3062–3069. doi: 10.7150/jca.19619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Castle J, Shaker H, Morris K, Tugwood JD, Kirwan CC. The significance of circulating tumour cells in breast cancer: a review. Breast. 2014;23(5):552–560. doi: 10.1016/j.breast.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 20.Lu X, Kang Y. Organotropism of breast cancer metastasis. J Mammary Gland Biol Neoplasia. 2007;12(2–3):153–162. doi: 10.1007/s10911-007-9047-3. [DOI] [PubMed] [Google Scholar]
- 21.Patsialou A, Wang Y, Lin J, Whitney K, Goswami S, Kenny PA, Condeelis JS. Selective gene-expression profiling of migratory tumor cells in vivo predicts clinical outcome in breast cancer patients. Breast Cancer Res. 2012;14(5):R139. doi: 10.1186/bcr3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van’t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415(6871):530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 23.Zieske LR. A perspective on the use of iTRAQ reagent technology for protein complex and profiling studies. J Exp Bot. 2006;57(7):1501–1508. doi: 10.1093/jxb/erj168. [DOI] [PubMed] [Google Scholar]
- 24.Evans C, Noirel J, Ow SY, Salim M, Pereira-Medrano AG, Couto N, Pandhal J, Smith D, Pham TK, Karunakaran E, et al. An insight into iTRAQ: where do we stand now? Anal Bioanal Chem. 2012;404(4):1011–1027. doi: 10.1007/s00216-012-5918-6. [DOI] [PubMed] [Google Scholar]
- 25.Putz SM, Boehm AM, Stiewe T, Sickmann A. iTRAQ analysis of a cell culture model for malignant transformation, including comparison with 2D-PAGE and SILAC. J Proteome Res. 2012;11(4):2140–2153. doi: 10.1021/pr200881c. [DOI] [PubMed] [Google Scholar]
- 26.Zeidan B, Manousopoulou A, Garay-Baquero DJ, White CH, Larkin SET, Potter KN, Roumeliotis TI, Papachristou EK, Copson E, Cutress RI, et al. Increased circulating resistin levels in early-onset breast cancer patients of normal body mass index correlate with lymph node negative involvement and longer disease free survival: a multi-center POSH cohort serum proteomics study. Breast Cancer Res. 2018;20(1):19. doi: 10.1186/s13058-018-0938-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bouchal P, Dvorakova M, Roumeliotis T, Bortlicek Z, Ihnatova I, Prochazkova I, Ho JT, Maryas J, Imrichova H, Budinska E, et al. Combined proteomics and transcriptomics identifies carboxypeptidase B1 and nuclear factor kappaB (NF-kappaB) associated proteins as putative biomarkers of metastasis in low grade breast Cancer. Mol Cell Proteomics. 2015;14(7):1814–1830. doi: 10.1074/mcp.M114.041335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bouchal P, Roumeliotis T, Hrstka R, Nenutil R, Vojtesek B, Garbis SD. Biomarker discovery in low-grade breast cancer using isobaric stable isotope tags and two-dimensional liquid chromatography-tandem mass spectrometry (iTRAQ-2DLC-MS/MS) based quantitative proteomic analysis. J Proteome Res. 2009;8(1):362–373. doi: 10.1021/pr800622b. [DOI] [PubMed] [Google Scholar]
- 29.Jesneck JL, Mukherjee S, Yurkovetsky Z, Clyde M, Marks JR, Lokshin AE, Lo JY. Do serum biomarkers really measure breast cancer? BMC Cancer. 2009;9:164. doi: 10.1186/1471-2407-9-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruppen I, Grau L, Orenes-Pinero E, Ashman K, Gil M, Algaba F, Bellmunt J, Sanchez-Carbayo M. Differential protein expression profiling by iTRAQ-two-dimensional LC-MS/MS of human bladder cancer EJ138 cells transfected with the metastasis suppressor KiSS-1 gene. Mol Cell Proteomics. 2010;9(10):2276–2291. doi: 10.1074/mcp.M900255-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M, et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019;47(D1):D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ni J, Cozzi PJ, Duan W, Shigdar S, Graham PH, John KH, Li Y. Role of the EpCAM (CD326) in prostate cancer metastasis and progression. Cancer Metastasis Rev. 2012;31(3–4):779–791. doi: 10.1007/s10555-012-9389-1. [DOI] [PubMed] [Google Scholar]
- 33.Pattje WJ, Melchers LJ, Slagter-Menkema L, Mastik MF, Schrijvers ML, Gibcus JH, Kluin PM, Hoegen-Chouvalova O, van der Laan BF, Roodenburg JL, et al. FADD expression is associated with regional and distant metastasis in squamous cell carcinoma of the head and neck. Histopathology. 2013;63(2):263–270. doi: 10.1111/his.12174. [DOI] [PubMed] [Google Scholar]
- 34.Chang X, Xu X, Ma J, Xue X, Li Z, Deng P, Zhang S, Zhi Y, Chen J, Dai D. NDRG1 expression is related to the progression and prognosis of gastric cancer patients through modulating proliferation, invasion and cell cycle of gastric cancer cells. Mol Biol Rep. 2014;41(9):6215–6223. doi: 10.1007/s11033-014-3501-2. [DOI] [PubMed] [Google Scholar]
- 35.Liu S, Yan B, Lai W, Chen L, Xiao D, Xi S, Jiang Y, Dong X, An J, Chen X, et al. As a novel p53 direct target, bidirectional gene HspB2/alphaB-crystallin regulates the ROS level and Warburg effect. Biochim Biophys Acta. 2014;1839(7):592–603. doi: 10.1016/j.bbagrm.2014.05.017. [DOI] [PubMed] [Google Scholar]
- 36.Cvekl A, McGreal R, Liu W. Lens development and Crystallin gene expression. Prog Mol Biol Transl Sci. 2015;134:129–167. doi: 10.1016/bs.pmbts.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 37.Haslbeck M, Peschek J, Buchner J, Weinkauf S. Structure and function of alpha-crystallins: Traversing from in vitro to in vivo. Biochim Biophys Acta. 2016;1860(1 Pt B):149–166. doi: 10.1016/j.bbagen.2015.06.008. [DOI] [PubMed] [Google Scholar]
- 38.Malin D, Strekalova E, Petrovic V, Deal AM, Al Ahmad A, Adamo B, Miller CR, Ugolkov A, Livasy C, Fritchie K, et al. alphaB-crystallin: a novel regulator of breast cancer metastasis to the brain. Clin Cancer Res. 2014;20(1):56–67. doi: 10.1158/1078-0432.CCR-13-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malin D, Strekalova E, Petrovic V, Rajanala H, Sharma B, Ugolkov A, Gradishar WJ, Cryns VL. ERK-regulated alphaB-crystallin induction by matrix detachment inhibits anoikis and promotes lung metastasis in vivo. Oncogene. 2015;34(45):5626–5634. doi: 10.1038/onc.2015.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Petrovic V, Malin D, Cryns VL. alphaB-crystallin promotes oncogenic transformation and inhibits caspase activation in cells primed for apoptosis by Rb inactivation. Breast Cancer Res Treat. 2013;138(2):415–425. doi: 10.1007/s10549-013-2465-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang F, Chen X, Li C, Sun Q, Chen Y, Wang Y, Peng H, Liu Z, Chen R, Liu K, et al. Pivotal role of augmented alphaB-crystallin in tumor development induced by deficient TSC1/2 complex. Oncogene. 2014;33(34):4352–4358. doi: 10.1038/onc.2013.401. [DOI] [PubMed] [Google Scholar]
- 42.Chen Z, Ruan Q, Han S, Xi L, Jiang W, Jiang H, Ostrov DA, Cai J. Discovery of structure-based small molecular inhibitor of alphaB-crystallin against basal-like/triple-negative breast cancer development in vitro and in vivo. Breast Cancer Res Treat. 2014;145(1):45–59. doi: 10.1007/s10549-014-2940-8. [DOI] [PubMed] [Google Scholar]
- 43.Huang Z, Cheng Y, Chiu PM, Cheung FM, Nicholls JM, Kwong DL, Lee AW, Zabarovsky ER, Stanbridge EJ, Lung HL, et al. Tumor suppressor alpha B-crystallin (CRYAB) associates with the cadherin/catenin adherens junction and impairs NPC progression-associated properties. Oncogene. 2012;31(32):3709–3720. doi: 10.1038/onc.2011.529. [DOI] [PubMed] [Google Scholar]
- 44.Voduc KD, Nielsen TO. Perou CM, Harrell JC, Fan C, Kennecke H, Minn AJ, Cryns VL, Cheang MCU. alphaB-crystallin expression in breast Cancer is associated with brain metastasis. NPJ Breast Cancer. 2015;1:15014. doi: 10.1038/npjbcancer.2015.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bandyopadhyay S, Pai SK, Hirota S, Hosobe S, Takano Y, Saito K, Piquemal D, Commes T, Watabe M, Gross SC, et al. Role of the putative tumor metastasis suppressor gene Drg-1 in breast cancer progression. Oncogene. 2004;23(33):5675–5681. doi: 10.1038/sj.onc.1207734. [DOI] [PubMed] [Google Scholar]
- 46.Weigelt B, Peterse JL, van’t Veer LJ. Breast cancer metastasis: markers and models. Nat Rev Cancer. 2005;5(8):591–602. doi: 10.1038/nrc1670. [DOI] [PubMed] [Google Scholar]
- 47.Cimino Y, Costes A, Damotte D, Validire P, Mistou S, Cagnard N, Alifano M, Regnard JF, Chiocchia G, Sautes-Fridman C, et al. FADD protein release mirrors the development and aggressiveness of human non-small cell lung cancer. Br J Cancer. 2012;106(12):1989–1996. doi: 10.1038/bjc.2012.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoo NJ, Lee SH, Jeong EG, Lee JW, Soung YH, Nam SW, Kim SH, Lee JY, Lee SH. Expression of nuclear and cytoplasmic phosphorylated FADD in gastric cancers. Pathol Res Pract. 2007;203(2):73–78. doi: 10.1016/j.prp.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 49.Tu W, Luo M, Wang Z, Yan W, Xia Y, Deng H, He J, Han P, Tian D. Upregulation of SATB1 promotes tumor growth and metastasis in liver cancer. Liver Int. 2012;32(7):1064–1078. doi: 10.1111/j.1478-3231.2012.02815.x. [DOI] [PubMed] [Google Scholar]
- 50.Choi EJ, Yun JA, Jabeen S, Jeon EK, Won HS, Ko YH, Kim SY. Prognostic significance of TMEM16A, PPFIA1, and FADD expression in invasive ductal carcinoma of the breast. World J Surg Oncol. 2014;12:137. doi: 10.1186/1477-7819-12-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bandyopadhyay S, Pai SK, Gross SC, Hirota S, Hosobe S, Miura K, Saito K, Commes T, Hayashi S, Watabe M, et al. The Drg-1 gene suppresses tumor metastasis in prostate cancer. Cancer Res. 2003;63(8):1731–1736. [PubMed] [Google Scholar]
- 52.Bandyopadhyay S, Wang Y, Zhan R, Pai SK, Watabe M, Iiizumi M, Furuta E, Mohinta S, Liu W, Hirota S, et al. The tumor metastasis suppressor gene Drg-1 down-regulates the expression of activating transcription factor 3 in prostate cancer. Cancer Res. 2006;66(24):11983–11990. doi: 10.1158/0008-5472.CAN-06-0943. [DOI] [PubMed] [Google Scholar]
- 53.Maruyama Y, Ono M, Kawahara A, Yokoyama T, Basaki Y, Kage M, Aoyagi S, Kinoshita H, Kuwano M. Tumor growth suppression in pancreatic cancer by a putative metastasis suppressor gene Cap43/NDRG1/Drg-1 through modulation of angiogenesis. Cancer Res. 2006;66(12):6233–6242. doi: 10.1158/0008-5472.CAN-06-0183. [DOI] [PubMed] [Google Scholar]
- 54.Cheng J, Xie HY, Xu X, Wu J, Wei X, Su R, Zhang W, Lv Z, Zheng S, Zhou L. NDRG1 as a biomarker for metastasis, recurrence and of poor prognosis in hepatocellular carcinoma. Cancer Lett. 2011;310(1):35–45. doi: 10.1016/j.canlet.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 55.Chang JT, Wang HM, Chang KW, Chen WH, Wen MC, Hsu YM, Yung BY, Chen IH, Liao CT, Hsieh LL, et al. Identification of differentially expressed genes in oral squamous cell carcinoma (OSCC): overexpression of NPM, CDK1 and NDRG1 and underexpression of CHES1. Int J Cancer. 2005;114(6):942–949. doi: 10.1002/ijc.20663. [DOI] [PubMed] [Google Scholar]
- 56.Grover PK, Cummins AG, Price TJ, Roberts-Thomson IC, Hardingham JE. Circulating tumour cells: the evolving concept and the inadequacy of their enrichment by EpCAM-based methodology for basic and clinical cancer research. Ann Oncol. 2014;25(8):1506–1516. doi: 10.1093/annonc/mdu018. [DOI] [PubMed] [Google Scholar]
- 57.Gastl G, Spizzo G, Obrist P, Dunser M, Mikuz G. Ep-CAM overexpression in breast cancer as a predictor of survival. Lancet. 2000;356(9246):1981–1982. doi: 10.1016/S0140-6736(00)03312-2. [DOI] [PubMed] [Google Scholar]
- 58.Ni J, Cozzi P, Hao J, Beretov J, Chang L, Duan W, Shigdar S, Delprado W, Graham P, Bucci J, et al. Epithelial cell adhesion molecule (EpCAM) is associated with prostate cancer metastasis and chemo/radioresistance via the PI3K/Akt/mTOR signaling pathway. Int J Biochem Cell Biol. 2013;45(12):2736–2748. doi: 10.1016/j.biocel.2013.09.008. [DOI] [PubMed] [Google Scholar]
- 59.Driemel C, Kremling H, Schumacher S, Will D, Wolters J, Lindenlauf N, Mack B, Baldus SA, Hoya V, Pietsch JM, et al. Context-dependent adaption of EpCAM expression in early systemic esophageal cancer. Oncogene. 2014;33(41):4904–4915. doi: 10.1038/onc.2013.441. [DOI] [PubMed] [Google Scholar]
- 60.Romeu C, Farre X, Cardesa A, Nadal A. Expression of ep-CAM, but not of E48, associates with nodal involvement in advanced squamous cell carcinomas of the larynx. Histopathology. 2013;62(6):954–961. doi: 10.1111/his.12108. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. GO analysis of the differentially regulated proteins in lymph node metastases vs. primary breast tumor tissues. The upregulated (A-C) and downregulated (D-F) proteins identified by the iTRAQ proteomics were analyzed by the GO Consortium and categorized according to their biological processes, cellular locations, and molecular functions. (TIF 5559 kb)
Figure S2. GEO analysis of CRYAB mRNA expression in normal breast and breast cancer tissues. (A) The mRNA expression of CRYAB in normal breast tissues (n =5) and breast cancer tissues (n = 28) was analyzed from the Affymetrix Human Genome Microarray at the GEO website (https://www.ncbi.nlm.nih.gov/geoprofiles/54408377 for αB-crystallin). (B) Quantification of the mRNA expression of CRYAB in normal breast tissues and breast cancer tissues. (TIF 4929 kb)
Table S1. Identification of differentially expressed proteins between primary breast cancer tissues and metastatic lymph node tissues by the iTRAQ technique. (XLS 1215 kb)
Table S2. Partial up-regulated proteins in metastatic lymph node compared with primary tumor in breast cancer. Table S3. Partial down-regulated proteins in metastatic lymph node compared with primary tumor in breast cancer. Table S4. UniProt analysis of the biological processes, cellular locations, and molecular functions of the four metastasis-associated proteins. (DOCX 29 kb)
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [1] partner repository with the dataset identifier PXD013931.