Abstract
Aims: Asthma, characterized by airway obstruction and hyper-responsiveness, is more severe and less responsive to treatment in obese subjects. While alterations in mitochondrial function and redox signaling have been implicated in asthma pathogenesis, it is unclear whether these mechanisms differ in lean versus obese asthmatics. In addition, we previously demonstrated that circulating platelets from asthmatic individuals have altered bioenergetics; however, it is unknown whether platelet mitochondrial changes reflect those observed in airway epithelial cells. Herein we hypothesized that lean and obese asthmatics show differential bioenergetics and redox signaling in airway cells and that these alterations could be measured in platelets from the same individual.
Results: Using freshly isolated bronchial airway epithelial cells and platelets from lean and obese asthmatics and healthy individuals, we show that both cell types from obese asthmatics have significantly increased glycolysis, basal and maximal respiration, and oxidative stress compared with lean asthmatics and healthy controls. This increased respiration was associated with enhanced arginine metabolism by arginase, which has previously been shown to drive respiration. Inducible nitric oxide synthase (iNOS) was also upregulated in cells from all asthmatics. However, due to nitric oxide synthase uncoupling in obese asthmatics, overall nitric oxide (NO) bioavailability was decreased, preventing NO-dependent inhibition in obese asthmatic cells that was observed in lean asthmatics.
Innovation and Conclusion: These data demonstrate bioenergetic differences between lean and obese asthmatics that are, in part, due to differences in NO signaling. They also suggest that the platelet may serve as a useful surrogate to understand redox, oxidative stress and bioenergetic changes in the asthmatic airway.
Keywords: asthma, mitochondria, obesity, glycolysis, metabolism
Introduction
Asthma is one of the most common chronic diseases in the United States and is characterized by reversible airflow limitation, bronchial hyper-responsiveness, and recruitment and activation of inflammatory cells, ultimately leading to airway remodeling. Over the last decade, it has been realized that asthma is a heterogeneous disease with variability among patients in characteristics, including lung function, weight, steroid responsiveness, and inflammatory markers (28, 37). Cluster analyses have advanced our understanding of how to treat various groups of asthmatics, including a subset of obese asthmatics (body mass index [BMI] above 30 kg/m2) who report increased asthma severity, increased exacerbations and hospital stays, and are generally refractory to steroid therapy as they do not display common T2 inflammatory markers associated with atopic inflammation (15, 42).
Innovation
This study begins to elucidate bioenergetic and redox mechanisms unique to obese versus lean asthmatics, which may contribute to differential pathogenesis of these two clinical phenotypes. Furthermore, data demonstrating that platelet bioenergetics reflect airway epithelial bioenergetics raise the possibility of utilizing platelets as a surrogate to measure and study airway bioenergetics more easily and less invasively in human asthma cohorts.
On a molecular level, pathogenesis of asthma is multifaceted, but studies of both lean and obese asthmatics show exacerbated production of reactive oxygen species (ROS) in airway cells accompanied by decreased activity of antioxidants (13, 17, 44), and increased metabolism of arginine to ornithine due to the upregulation of arginase (5, 16, 52). Airway inflammation is also characterized by increased expression of inducible nitric oxide synthase (iNOS), which results in enhanced fractional exhaled nitric oxide (NO) (FENO) in lean asthmatics (20, 36, 51). However, FENO has been observed to be much lower in some asthmatic phenotypes despite iNOS upregulation, and this is thought to be due to deficiency of l-arginine, a critical substrate for nitric oxide synthase (NOS), and accumulation of asymmetric dimethylarginine (ADMA), leading to decreased iNOS activity and uncoupling of the enzyme, which prevents NO synthesis (2, 5, 16, 30, 43, 49).
Accumulating studies also demonstrate that alterations in mitochondrial structure and function are present in asthmatic patients and have been implicated in disease development and progression. For example, changes in mitochondrial shape and increased number have been observed in the airways of asthmatic subjects (6, 25, 26, 45, 52). In addition, airway epithelial cells from asthmatic subjects show increased mitochondrial respiration (45, 52). This increase in respiration has been attributed to enhanced arginine metabolism to ornithine by upregulated arginase, which ultimately generates glutamate (through catalysis by ornithine amino transferase) to drive the production of substrates for the tricarboxylic acid (TCA) cycle (52). These studies and others link altered mitochondrial function to enhanced arginine metabolism as well as changes in NO signaling in the asthmatic airway. However, it remains unclear whether bioenergetics is different in lean versus obese asthmatics.
Notably, mitochondrial and metabolic changes do not appear to be confined to the airways of asthmatics, but instead present systemically. For example, studies investigating whole-body metabolism with control of dietary intake suggest mild asthmatics are metabolically more efficient than healthy subjects (34), and childhood asthma has been linked with abnormal lipid and glucose metabolism (9). At the cellular level, we have previously shown that circulating platelets from asthmatics show less reliance on glycolysis and greater TCA cycle activity than platelets from healthy subjects (50). While these data suggest that measurement of bioenergetics in a circulating cell type, such as the platelet, may provide information about metabolic changes in the airways, it remains unknown whether platelets accurately reflect bioenergetic changes in airway cells.
We hypothesized that obese and lean asthmatics have differential systemic cellular bioenergetics and that these mitochondrial alterations can be detected in the airway epithelium as well as circulating platelets. Here we show that airway epithelial cells from obese asthmatics show higher rates of glycolysis and oxidative phosphorylation in comparison with lean asthmatics. Our data demonstrate that iNOS uncoupling and decreased NO bioavailability in obese asthmatics contribute to increased ROS production and enhanced maximal respiratory capacity, respectively. Notably, these bioenergetic and redox changes are present in both platelets and airway epithelial cells from the same subjects. These data further demonstrate that mitochondrial alterations are present in obese asthmatics and suggest that platelets may serve as a surrogate to measure bioenergetic and redox alterations in asthmatic subjects.
Results
Obese asthmatics show greater glycolytic and respiratory rate than lean asthmatics
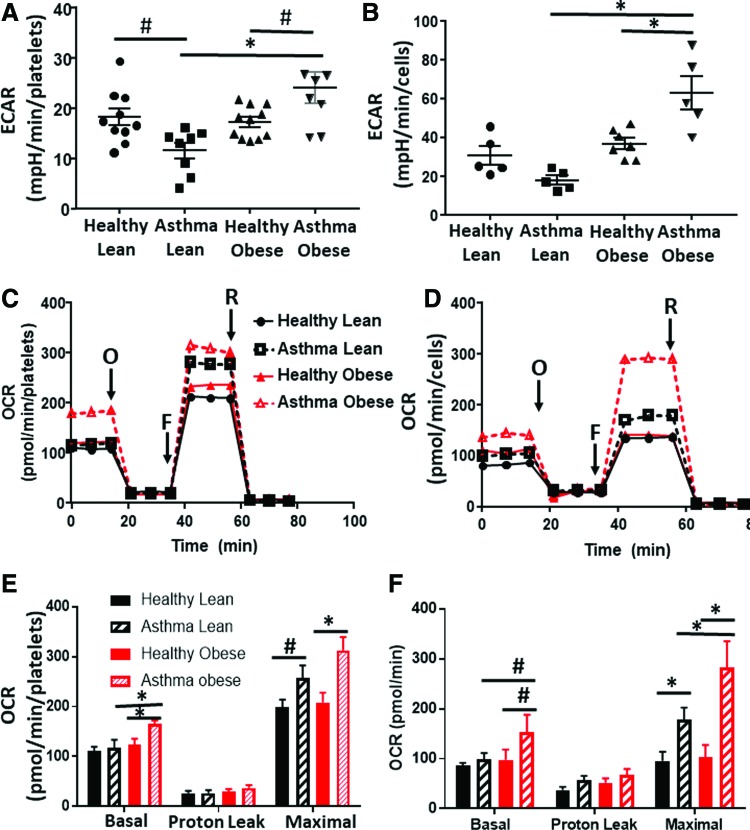
In the first series of experiments, we measured cellular bioenergetics in freshly isolated intact platelets from lean asthmatics, obese asthmatics, healthy lean subjects, and healthy obese subjects (see Table 1 for demographics). Fresh airway epithelial cells were obtained from a subgroup of subjects in each of these cohorts and bioenergetic measurements were made in parallel. Levels of basal platelet activation were measured and found not to differ between groups (Supplementary Fig. S1). Basal glycolytic rate was assessed in both cell types by measuring the extracellular acidification rate (ECAR) that was sensitive to the glycolytic inhibitor 2-deoxyglucose (2-DG) using extracellular flux analysis. Consistent with prior studies (50), platelets from lean asthmatics showed a significant decrease in the basal glycolytic rate compared with healthy subjects (Fig. 1A) and this effect was paralleled in airway epithelial cells (Fig. 1B). While there was no significant change in the glycolytic rate of platelets and airway epithelial cells from healthy obese subjects compared with healthy lean subjects, platelets and epithelial cells from obese asthmatics showed a significant increase in glycolysis in comparison with all other groups (Fig. 1A, B).
Table 1.
Demographics
| Lean healthy (n = 10) | Lean asthmatic (n = 8) | Obese healthy (n = 11) | Obese asthmatic (n = 8) | |
|---|---|---|---|---|
| Age | 33 (21–52) | 31 (19–51) | 30 (19–43) | 36 (21–58) |
| Sex, female, % | 66 | 71 | 71 | 56 |
| BMI | 24 (21–25) | 22 (19–22) | 33 (31–37) | 36 (30–44) |
| FEV1% predicted | 99 (81–124) | 93 (80–107) | 102 (88–115) | 82 (61–109) |
| FVC (L) | 4.6 (3.6–6.1) | 3.9 (2.4–5.8) | 4.5 (3.7–6.3) | 3.6 (2.0–5.7) |
| FVC % predicted | 101 (89–121) | 98 (77–124) | 101 (89–115) | 89 (73–117) |
| FEV1/FVC | 0.81 (0.71–0.91) | 0.80 (0.64–1.0) | 0.83 (0.73–0.89) | 0.75 (0.67–0.81) |
| eNO ppb | 16 (7–32) | 33 (9–100) | 17 (8–31) | 16 (7–30) |
| ICS% | N/A | 50 | N/A | 78 |
BMI, body mass index; eNO, exhaled nitric oxide; FEV1%, forced exhalation percent predicted; FVC, forced vital capacity; ICS%, percent inhaled corticosteroids.
FIG. 1.
Platelet and airway epithelial cell bioenergetics differs in lean and obese asthmatics. (A, B) Measured basal ECAR of (A) platelets and (B) airway epithelial cells isolated from healthy lean, asthmatic lean, healthy obese, and asthmatic obese individuals. (C, D) Representative oxygen consumption traces of (C) platelets and (D) airway epithelial cells from all four experimental groups. Basal respiration rate was measured followed by proton leak after the addition of oligomycin (O), maximal respiration after the addition of FCCP (F), and nonmitochondrial respiration after the addition of rotenone (R). (E, F) Quantification of several traces in (E) platelets and (F) epithelial cells such as those shown in (C, D), respectively. For platelet studies: healthy lean (n = 10), lean asthmatic (n = 8), healthy obese (n = 11), and asthmatic obese (n = 8). For airway epithelial cells: all groups n = 5. All data are mean ± SEM. Statistical significance was tested by one-way ANOVA. #p < 0.05; *p < 0.01. ECAR, extracellular acidification rate; FCCP, carbonyl cyanide-ρ-trifluoromethoxyphenylhydrazone; SEM, standard error of the mean. Color images are available online.
To test whether rates of oxidative phosphorylation were also altered, we next measured oxygen consumption rates (OCRs) of the platelets and epithelial cells (Fig. 1C, D). Basal OCR did not differ between platelets from lean healthy and lean asthmatic subjects, although maximal OCR was increased in lean asthmatic platelets (Fig. 1E). This increase in maximal OCR was also significant in epithelial cells from lean asthmatics (Fig. 1F). Notably, in both cell types, cells from obese asthmatic subjects showed an even greater increase in both basal and maximal OCRs, significant compared with all healthy controls and lean asthmatic cells (Fig. 1E, F). Measurement of basal ATP production rate showed that both airway epithelial cells and platelets from lean and obese asthmatics showed increased rates of ATP production compared with healthy controls, consistent with prior studies (50, 52). While cells from obese asthmatics showed a trend to even greater ATP production compared with lean asthmatic cells, this did not reach statistical significance (Table 2).
Table 2.
ATP Production
| Platelets (pmol/min/106 cells) | Epithelial cells (nmol/min/50,000 cells) | p | |
|---|---|---|---|
| Lean healthy (n = 10) | 72 ± 6 | 56 ± 11 | |
| Lean asthmatic (n = 8) | 89 ± 8 | 73 ± 8 | <0.05 versus lean healthy, obese healthy |
| Obese healthy (n = 11) | 67 ± 10 | 50 ± 8 | |
| Obese asthmatic (n = 8) | 98 ± 7 | 81 ± 6 | <0.01 versus lean healthy, obese healthy |
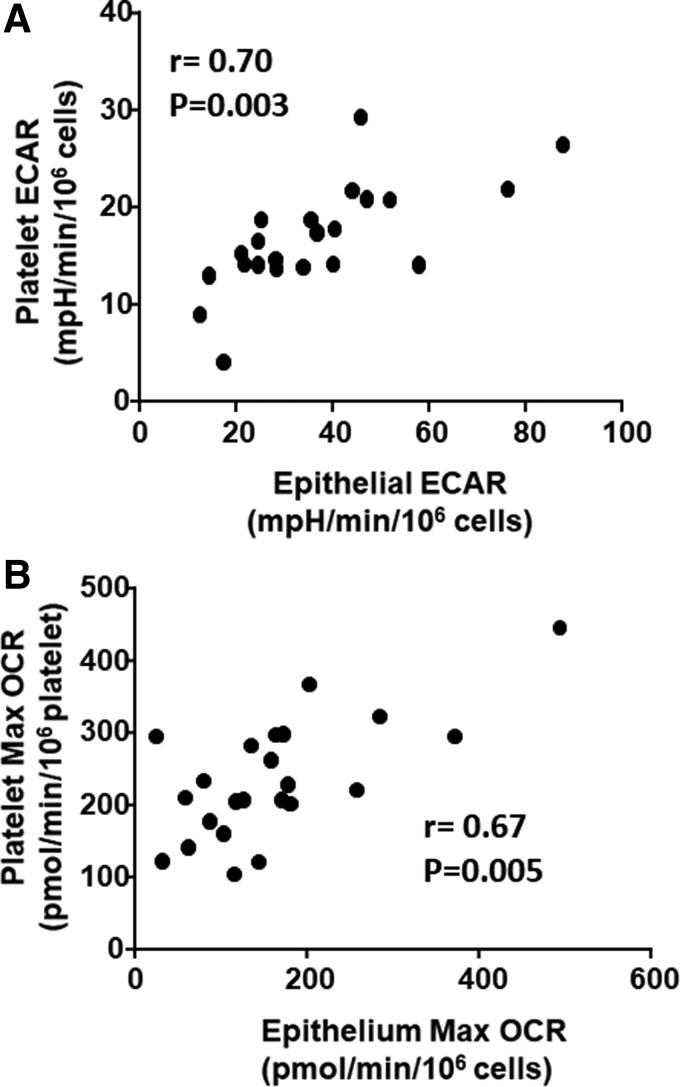
Given that bioenergetic parameters showed similar effects in both platelets and epithelial cells, we next sought to assess whether platelet bioenergetics accurately reflects airway epithelial bioenergetics. We performed a correlation between platelet and airway epithelial cell basal ECAR (Fig. 2A) and maximal OCR (Fig. 2B) in each subject. We found a significant correlation between ECAR (r = 0.70; p = 0.03) and maximal OCR (r = 0.67; p = 0.005) in these two cell types, demonstrating that circulating platelet bioenergetics reflects airway epithelial bioenergetics in healthy and asthmatic humans.
FIG. 2.
Platelet ECAR and maximal OCR correlate with airway epithelial ECAR and maximal OCR. The correlation between (A) basal ECAR and (B) maximal OCR in platelets and airway epithelial cells in the same individuals. Pearson R and p-value are reported for each graph. n = 5 for each experimental group (20 total). OCR, oxygen consumption rate.
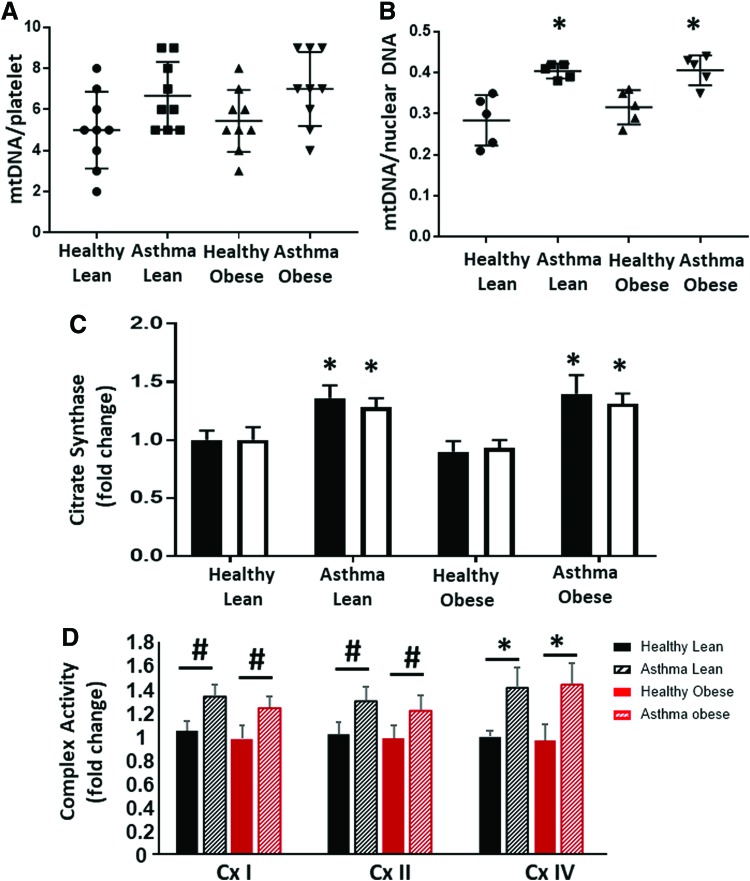
To determine whether the difference between obese and lean asthmatic bioenergetics was due to changes in mitochondrial number, we next measured mitochondrial DNA as a measure of mitochondrial number. Platelets from lean and obese asthmatic subjects showed a small increase in mitochondrial number that did not reach significance compared with platelets from lean and obese healthy subjects (Fig. 3A). Airway epithelial cells isolated from lean and obese asthmatics also showed an ∼25% increase in mitochondrial DNA copy number, and this reached statistical significance compared with healthy controls (Fig. 3B). Both platelets and airway epithelial cells from lean and obese asthmatics had significantly increased activity of the matrix TCA cycle enzyme citrate synthase, consistent with an increase in mitochondrial number as well as increased TCA cycle activity (Fig. 3C). The specific activities of electron transport chain complexes I, II, and IV were increased in airway epithelial cells from lean and obese asthmatics compared with healthy controls, consistent with previous studies (Fig. 3D).
FIG. 3.
Mitochondrial number and enzymatic activity are increased in cells from lean and obese asthmatic subjects. (A) Mitochondrial DNA copy number (normalized to platelet number) in platelets and from healthy lean, asthmatic lean, healthy obese, and asthmatic obese individuals. (B) Mitochondrial DNA copy number normalized to corresponding nuclear DNA copy number in airway epithelial cells from the same groups. Bars in (A, B) represent mean ± SEM. (C) Citrate synthase activity (represented as a fold change of the respective healthy lean group) in airway epithelial cells (black bars) and platelets (white bars). (D) Enzymatic activity of complexes I, II, and IV measured in airway epithelial cells and represented as a fold change of the healthy lean group. n = 10 for platelets and n = 5 for airway epithelial cells. Statistical significance determined by one-way ANOVA. #p < 0.05, *p < 0.01 versus respective healthy control. Color images are available online.
Platelet and airway epithelium from obese asthmatics show increased oxidant production
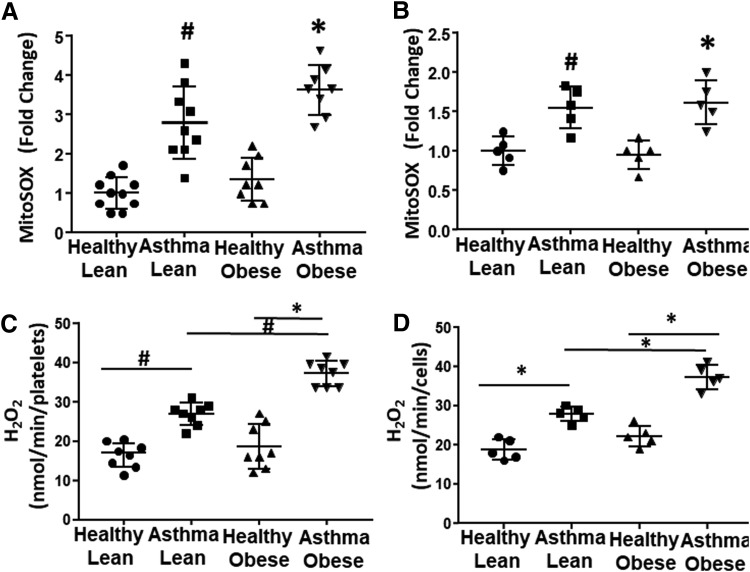
Mitochondrial superoxide production is regulated by the activity of the electron transport chain. To determine whether the observed alterations in bioenergetics change mitochondrial oxidant production in asthmatics, we next used MitoSOX to measure mitochondrial superoxide production in both platelets and airway epithelial cells. There was no statistically significant difference in mitochondrial oxidant production between lean and obese healthy subjects in platelet or epithelial cells (Fig. 4A, B). In contrast, lean asthmatics showed a significant increase in mitochondrial superoxide production in platelets (Fig. 4A) and this increase was recapitulated in airway epithelial cells (Fig. 4B). Obese asthmatics also showed increases in mitochondrial oxidant production in both platelets and airway epithelial cells (Fig. 4A, B).
FIG. 4.
Platelets and airway epithelial cells show increased mitochondrial and cellular oxidant production. (A, B) Mitochondrial superoxide production in (A) platelets and (B) airway epithelial cells as well as (C, D) total cellular H2O2 production measured in (C) platelets and (D) airway epithelial cells from healthy lean, asthmatic lean, healthy obese, and asthmatic obese individuals. All data are individual values of each subject with bars representing the mean and standard deviation. Statistical significance determined by one-way ANOVA. #p < 0.05 and *p < 0.01 versus respective healthy control if not otherwise noted. n = 10 for platelets and n = 5 for airway epithelial cells. H2O2, hydrogen peroxide.
Prior studies have established that antioxidant defenses are inactivated in asthmatics and that cellular sources of ROS beyond the mitochondrion are increased in asthmatic airway cells (13, 17, 22). Thus, we next measured total cellular hydrogen peroxide (H2O2) generation in both platelets and airway epithelial cells. Both platelets and airway epithelial cells from lean asthmatics showed a significant increase in H2O2 production compared with lean and obese healthy controls. In addition, both cell types isolated from obese asthmatics showed a significantly greater increase in H2O2 compared with lean asthmatics (Fig. 4C, D).
NOS and arginase activities are increased in cells from asthmatics
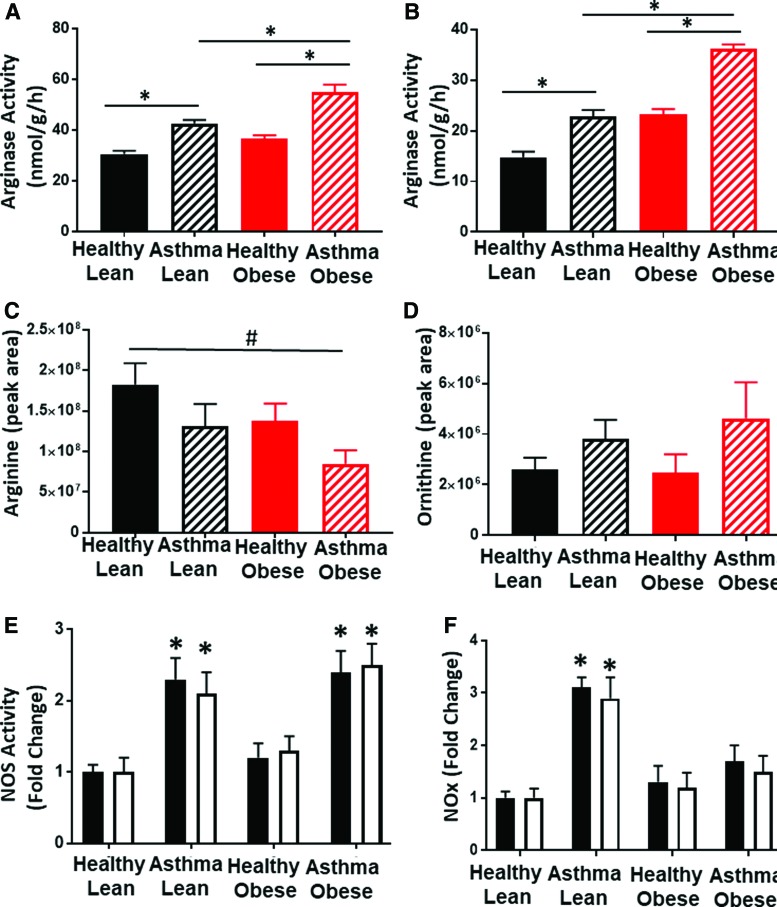
Alterations in arginine metabolism through the upregulation of arginase and iNOS have been shown to modulate both bioenergetics and oxidant production in airway epithelial cells from asthmatics (5, 51, 52). Measurement of arginase activity in the airway epithelial cells showed that cells isolated from lean asthmatics had significantly increased arginase activity compared with lean and obese healthy controls, consistent with prior studies (16, 52). In addition, arginase activity was further increased in cells isolated from obese asthmatics (Fig. 5A). Notably, although absolute arginase activity was much lower in platelets than epithelial cells, a similar increase in activity was also observed in platelets from lean and obese asthmatics compared with lean and obese healthy subjects (Fig. 5B). Arginase catalyzes the conversion of arginine to ornithine, and consistent with an upregulation of cellular arginase, plasma levels of l-arginine were decreased in both lean and obese asthmatics and healthy obese compared with healthy lean subjects. l-Arginine levels in obese asthmatics were significantly lower when compared with healthy lean subjects. Ornithine levels trended higher in both lean and obese asthmatics compared with both lean and obese controls (Fig. 5C, D) although these values did not reach statistical significance.
FIG. 5.
Arginine metabolism and NO production are altered in cells from asthmatics. (A, B) Arginase activity in (A) airway epithelial cells and (B) platelets from all four experimental groups. (C) Arginine and (D) ornithine levels in the plasma of all four experimental groups (n = X for each group). (E) Activity of NOS (expressed as fold change of healthy lean group) in airway epithelial cells (black bars) and platelets (white bars). (F) Nitrite and nitrate levels combined (expressed as fold change of healthy lean group) in airway epithelial cells (black bars) and platelets (white bars). All data expressed from healthy lean, asthmatic lean, healthy obese, and asthmatic obese groups as mean ± SEM. Statistical significance was determined by one-way ANOVA. For cell data, n = 5 for all groups. #p < 0.05 and *p < 0.01. NOS, nitric oxide synthase. Color images are available online.
l-Arginine is also the substrate for iNOS, which is known to be upregulated in asthma. Thus, we next assessed NOS content by measuring the conversion of exogenously added l-arginine to l-citrulline. These experiments demonstrated that in both airway epithelial cells and platelets, NOS activity was significantly increased in cells isolated from both lean and obese asthmatics compared with healthy controls (Fig. 5E). However, when nitrite and nitrate concentrations were assessed (as a measure of total NO production), lean asthmatics showed significantly greater levels of nitrite and nitrate than healthy controls or obese asthmatics (Fig. 5F). Consistent with the measurements of nitrite and nitrate, the plasma levels of the nitro-fatty acid, NO2-conjugated linoleic acid (NO2-cLA), were also increased in lean asthmatics (but not obese asthmatics), indicative of increased NO bioavailability in lean asthmatics (data not shown). This difference in NO production was recapitulated in the measurements of exhaled NO in these subjects, as exhaled NO was significantly increased in lean asthmatics, but not in obese asthmatics (Table 1). Taken together, these data suggest that while total NOS levels are equivalent and upregulated in lean and obese asthmatics, NO bioavailability is significantly decreased in obese asthmatics.
NOS contributes to oxidant production in cells from obese asthmatics
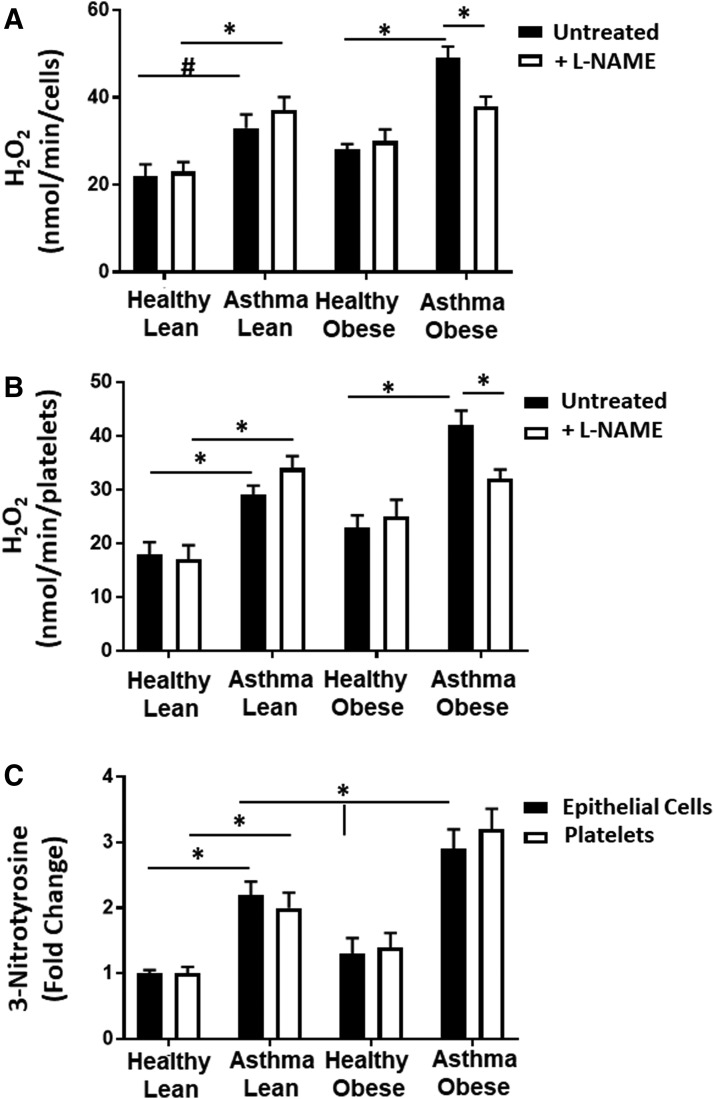
Decreases in l-arginine and increased ADMA are known to cause NOS uncoupling. Furthermore, our results are consistent with prior studies that implicate uncoupling of NOS in cellular oxidant generation in asthmatic epithelial cells and suggest that NOS uncoupling is increased with obesity in asthmatics (16, 49). To determine whether NOS uncoupling contributes to increased cellular oxidant production in cells from obese asthmatic subjects, we next measured cellular H2O2 production in the presence of L-NG-Nitroarginine methyl ester (L-NAME) (100 μM), a pharmacological inhibitor of NOS (Fig. 6A, B). Consistent with NOS uncoupling, inhibition of NOS by L-NAME significantly decreased H2O2 production in the airway epithelial cells and platelets of obese asthmatics (Fig. 6A, B). Furthermore, measurement of nitrotyrosine, an indicator of the reaction of NO with superoxide, which has been associated with NOS uncoupling, showed that nitrotyrosine was significantly increased in lean asthmatics compared with healthy controls and this was further significantly elevated in obese asthmatics in both airway epithelial cells and platelets (Fig. 6C).
FIG. 6.
NOS contributes to oxidant production in cells from obese asthmatic subjects. (A, B) Total cellular H2O2 production measured in the absence (black bars) and presence (white bars) of L-NAME (100 μM) in (A) airway epithelial cells and (B) platelets. (C) The 3-nitrotyrosine concentration measured in epithelial cells (black bars) and platelets (white bars) from healthy lean, asthmatic lean, healthy obese, and asthmatic obese individuals. n = 5 in each group. Data are mean ± SEM. Statistical significance determined by one-way ANOVA. #p < 0.05 and *p < 0.01. L-NAME, L-NG-nitroarginine methyl ester.
Decreased NO bioavailability regulates respiration in obese asthmatic cells
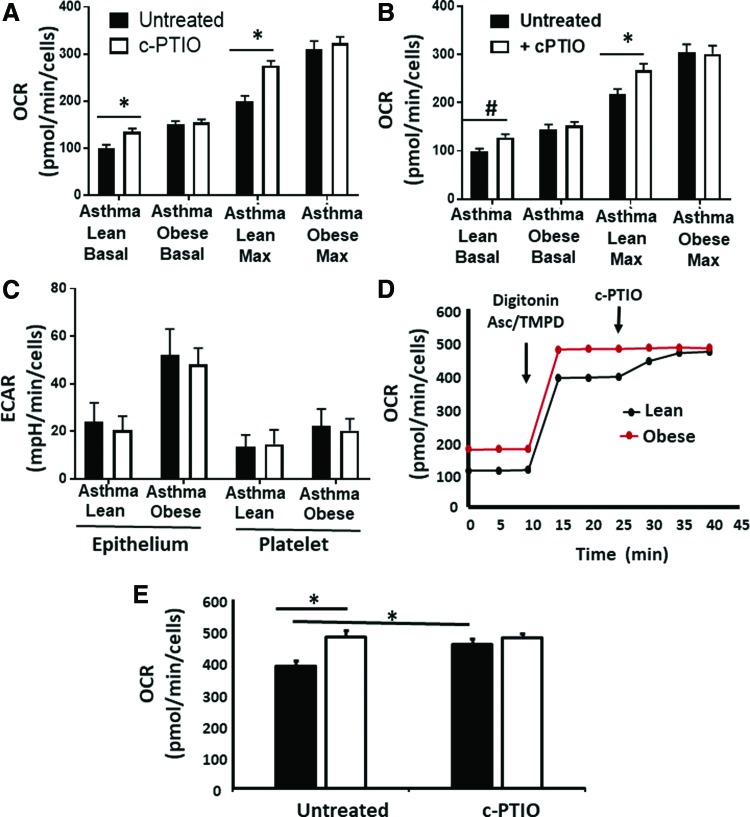
Prior studies demonstrate that increased arginine metabolism, resulting in elevated ornithine levels, enhances mitochondrial respiration in airway epithelial cells from subjects with asthma (52). Conversely, NO is a potent inhibitor of mitochondrial oxygen consumption through its reversible binding to cytochrome c oxidase (complex IV) (29, 40, 41). Given the increase in ornithine in both asthmatic groups, but difference in NO production, we next tested whether differences in respiratory rate between cells isolated from lean and obese asthmatics were due to increased NO bioavailability in the lean asthmatic group. Respiration was measured in airway epithelial cells isolated from lean and obese asthmatics in the presence and absence of the NO scavenger 2-phenyl-4,4,5,5,-tetramethylimidazoline-1-oxyl 3-oxide (c-PTIO) (100 μM; Fig. 7A). While no significant change in respiration was observed in cells from obese asthmatics, epithelial cells from lean asthmatics showed an increased rate of both basal and maximal respiration on exposure to c-PTIO (Fig. 7A). Similar results were observed in platelets from lean and obese asthmatics (Fig. 7B). These data are consistent with NO-dependent inhibition of respiration, which is absent in obese asthmatic epithelial cells. Notably, no change in basal ECAR was observed in either obese or lean platelets or airway epithelial cells in the presence of c-PTIO (Fig. 7C).
FIG. 7.
NO inhibits respiration and complex IV but not glycolysis in cells from lean asthmatic subjects. (A, B) Basal and maximal respiratory rates in (A) airway epithelial cells and (B) platelets isolated from lean and obese asthmatic individuals and treated with (white bars) or without (black bars) c-PTIO (100 μM). (C) Basal ECAR in airway epithelial cells and platelets isolated from lean and obese asthmatic individuals and treated with or without c-PTIO. (D) Representative trace of platelets permeabilized with digitonin (20 μg/mL) and treated with TMPD (0.5 mM) and ascorbate (2 mM) to measure oxygen consumption by complex IV in the presence and absence of c-PTIO. (E) Quantification of eight traces similar to those shown in (D). Statistical significance determined by one-way ANOVA. n = 4; Data are mean ± SEM #p < 0.05 and *p < 0.01. c-PTIO, 2-phenyl-4,4,5,5,-tetramethylimidazoline-1-oxyl 3-oxide; TMPD, tetramethyl-p-phenylenediamine. Color images are available online.
To determine whether the difference in respiration observed between lean and obese cells was due to inhibition of complex IV, we next directly measured complex IV activity in platelets from obese and lean asthmatic individuals. Although our data showed increased complex IV enzymatic activity in both lean and obese cells compared with healthy controls (Fig. 3D), spectrophotometric measurement of this activity was performed in lysed cells, and reversible posttranslational modifications that may be present in the environment of the cell were not taken into account. To take into account the cellular milieu, the plasma membrane of platelets was permeabilized with digitonin such that the cytoskeletal structure and mitochondrial membranes remained intact. Electrons were then directly passed to complex IV by the addition of tetramethyl-p-phenylenediamine (TMPD) and ascorbate, and then, the OCR was measured. In these conditions, complex IV activity was significantly lower in platelets from lean asthmatic individuals and this activity was increased to the rate of platelets from obese asthmatics by the addition of c-PTIO to scavenge NO (Fig. 7D, E). These data confirm that NO-dependent inhibition of complex IV accounts for the decreased respiration rate observed in platelets from lean asthmatics compared with obese asthmatics.
Discussion
The major finding of this study is that lean and obese asthmatics display differential bioenergetics in both their airway epithelial cells and platelets. These data confirm prior studies demonstrating that platelets from lean asthmatics have a lower basal glycolytic rate compared with healthy lean subjects (50). We have now extended these observations to show that cells from obese asthmatics have significantly enhanced basal glycolysis, as well as basal and maximal respiratory rates compared with lean asthmatics and healthy individuals. Recent studies report that the upregulation of arginase 2 within the mitochondrion, resulting in greater ornithine levels, drives the production of TCA cycle intermediates to increase mitochondrial respiration in airway epithelial cells from asthmatic subjects (52). The data presented herein, showing increased respiration associated with upregulation of arginase and increased ornithine levels, are consistent with this pathway, but further elucidate an NO-dependent mechanism that contributes to the enhanced respiratory rate observed in cells from obese asthmatics compared with their lean counterparts.
While it is well established that iNOS is upregulated in the airways of asthmatics, increasing BMI has been associated in some studies with decreased NO bioavailability as measured by FENO (4, 22). Consistent with this, we show equivalent upregulation of NOS activity in lean and obese asthmatic subjects, but decreased NO bioavailability in cells from obese asthmatic subjects in this study. This difference in bioavailability appears to be at least partially due to NOS uncoupling and is evidenced by increased oxidant production in cells from obese asthmatics that is attenuated by NOS inhibition. Nitric oxide potently inhibits mitochondrial respiration through its reversible binding to cytochrome c oxidase (29, 40, 41). Our data suggest that while both lean and obese asthmatics show increased respiratory rate compared with healthy controls, respiration in lean asthmatics is significantly suppressed by NO. This inhibition plays a lesser role in obese asthmatics since NO bioavailability is decreased, and thus leads to the apparent increased respiration rate observed in obese asthmatic cells compared with those from lean asthmatics.
It is unclear what role NO-dependent inhibition of respiration plays in airway inflammation and hyper-responsiveness. The inhibition of cytochrome c oxidase has been implicated in numerous signaling pathways (7). For example, NO-dependent inhibition of respiration has been shown to redirect oxygen signaling within the cell, preventing the stabilization of hypoxia-inducible factor 1 alpha (HIF-1α) (14, 27). Notably, HIF-1α is stabilized in several cell types from animal models of asthma and propagates the inflammatory response (1, 11). Similarly, cytochrome c oxidase inhibition can also enhance mitochondrial superoxide production, leading to downstream signaling, including the activation of the proinflammatory transcription factor NF-κB (13, 33). Interestingly, Hüttemann et al. showed that mice deficient in cytochrome c oxidase activity in the lung demonstrated airway hypo-responsiveness (18), raising the question of whether the lack of NO-mediated inhibition in airway cells from obese asthmatics contributes to the exacerbated hyper-responsiveness observed in some of these subjects, compared with lean asthmatics. Further study is required to determine which downstream inflammatory pathways are initiated by NO-dependent inhibition of respiration, and whether this inhibition contributes to the differences in pathogenesis observed between lean and obese asthmatics.
Beyond differences in NO bioavailability, our data demonstrate that NOS uncoupling contributes to enhanced oxidant production observed in cells from obese asthmatics. Prior studies have observed that ADMA levels are increased in asthma (16, 30, 38), and decreases in the ratio of l-arginine/ADMA have been associated with decreased FENO (16). Our data show that arginase activity is significantly higher in cells from healthy obese subjects compared with lean controls and this effect is even greater in obese asthmatics, which likely contributes to the decreased l-arginine levels observed. Although we did not see a significant difference in ADMA levels in obese asthmatics, it is important to note that this species is highly compartmentalized and plasma levels may not reflect intracellular concentrations.
In addition to oxidant production by uncoupled NOS, our data demonstrate an increase in mitochondrial oxidant production in cells from asthmatics compared with healthy controls. While we did not elucidate the exact source of mitochondrial oxidant production in this study, complexes I and III of the electron transport chain likely contribute. A limitation of this study is that the mitochondrial membrane potential was not measured. Measurement of this parameter would be important in understanding whether increased membrane potential contributes to electron transport chain-dependent oxidant production. Another limitation of the study is the lack of measurement of mitochondrial biogenesis, which could also contribute to increases in respiration and oxidant production observed in obese asthmatics.
Our data demonstrate a significant increase in glycolysis in both platelets and airway epithelial cells from obese asthmatics, while glycolysis is decreased (compared with healthy controls) in lean asthmatic cells. Prior studies have reported both decreased and increased lactate levels and glycolysis in patients with asthma (32, 35, 50). Xu et al. reported a wide range of lactate levels in the bronchial alveolar lavage from asthmatics, with the highest lactate concentration present in severe asthmatics (compared with mild and moderate asthmatics) (51). Notably, in that study, BMI was highest (>30) in the severe asthmatic group, making this effect consistent with the results, we report here, of increased glycolytic rate in obese versus lean asthmatics. However, in our cohort, obese asthmatics had lower FENO than lean asthmatics, which is in contrast to Xu et al. data showing that glycolysis was highest in patients with the highest FENO (51). The reason for this discrepancy is unclear but suggests that perhaps NO is not the predominant mechanistic regulator of glycolytic rate in these patients. Our data showing no change in ECAR with c-PTIO treatment confirm that the acute, reversible actions of NO are not responsible for the enhanced glycolytic rate observed, although longer term studies with NOS inhibition are required to rule out the role of NO completely. Deeper mechanistic studies are required to investigate the mechanism of upregulated glycolysis, including the roles of glycolytic enzyme expression, changes in substrate utility, or increased oxidant production that has previously been shown to enhance glycolysis through HIF-1α (39, 53).
Our data demonstrating a significant correlation between platelet and airway epithelial bioenergetics are novel. The correlation between respiration and glycolytic rate of these two cell types in healthy individuals is consistent with the growing concept that bioenergetic profiling of circulating cells can serve as a marker of systemic mitochondrial function, and potentially as a surrogate for these measures in other cell types. Molina and colleagues recently showed in nonhuman primates that monocytes and platelets reflect respiratory patterns of skeletal and cardiac muscle (46) and that platelets mimic glucose metabolism measured in the brain by positron emission tomography imaging (47). Furthermore, we and others have shown changes in bioenergetics in circulating cells from individuals with different pathologies and these changes can be correlated with clinical and mechanistic parameters of disease progression (3, 8, 23, 31, 47, 48, 50). Notably, the data herein also demonstrate that redox changes within the platelet, particularly iNOS upregulation and uncoupling as well as enhanced arginase activity, reflect those observed in the airway epithelial cell. These data support accumulating studies suggesting that metabolic and redox dysfunction in asthma is systemic rather than confined to the airway (34, 50). More study is required to determine whether the reflection of airway epithelial bioenergetics is unique to the platelet or common to all circulating cells. This relationship may be unique to platelets as they have been shown to play a role in asthma pathogenesis through their secretion of mitogenic factors, modulation of leukocyte chemotactic factors, and the formation of eosinophil complexes (12, 19, 21). In addition, a recent study showing that platelets are produced in the lung raises the possibility that these cells uniquely reflect lung physiology more closely than other circulating cell types (24). Whether or not these systemic alterations are confined to platelets, our data suggest that platelets could serve as a useful and readily measurable surrogate to assess and understand changes in airway epithelial metabolism with the progression and/or treatment of asthma.
Materials and Methods
Human subjects
All studies were performed in accordance with the University of Pittsburgh Medical Center Institutional Review Board and measurements were performed only after informed consent. All asthmatics had mild to moderate disease, were previously diagnosed by a physician, and had either a significant (>12%) bronchodilator response or a ≥20% drop from baseline in the forced exhalation volume in the first second (FEV1) after a methacholine challenge. Participants were recruited from the Asthma Institute Registry, advertising, other research studies, and the general population. Demographics of human subjects are shown in Table 1.
Human airway epithelial cell isolation
Human airway epithelial cell samples were obtained during bronchoscopies (in subjects described in Table 1), using a standard sterile single-sheathed nylon cytology brush (ConMed, REF 129R; ConMed Corporation, New York). A total of four to six brushings were obtained from each subject and cells were placed into 10 mL of ice-cold phosphate-buffered saline (PBS), centrifuged 10 min at 600gmax room temperature, washed, and resuspended in serum-free hormonally supplemented bronchial epithelial growth medium (Clonetics, San Diego, CA) containing 50 μg of gentamicin and 50 μg of amphotericin. After counting, cell density was adjusted at 1.0 × 106 cells/mL in bronchial epithelial growth medium. For Seahorse experiments, cells were immediately plated on Seahorse plates. After 48 h in submerged conditions, the cells were measured by Seahorse extracellular flux analysis as described below. Cells used for other analyses were also primary cells measured 2 days after isolation and plating.
Human platelet isolation
Platelets were isolated by differential centrifugation from human venous blood collected in citrate containing tubes as previously described (8). Briefly, whole blood was centrifuged (150 g, 10 min) in the presence of prostaglandin I2 (PGI2, 1 μg/mL; Sigma-Aldrich, St. Louis, MO) to obtain platelet-rich plasma. Platelets were subsequently pelleted from the platelet-rich plasma by centrifugation at 1500 g for 10 min. The platelet pellets were washed with erythrocyte lysis buffer (Qiagen, Valencia, CA) and PGI2. The final samples were resuspended in modified Tyrode's buffer (20 mM HEPES, 128 mM NaCl, 12 mM bicarbonate, 0.4 mM NaH2PO4, 5 mM glucose, 1 mM MgCl2, 2.8 mM KCl, pH 7.4) before study.
Measurement of platelet activation
Isolated platelets were incubated with phycoerythrin-labeled mouse anti-human CD41a antibody and APC-labeled mouse anti-human CD62P antibody (30 min; 25°C) to measure surface p-selectin expression by flow cytometry (LSRFortessa with FACSDiva software; Becton Dickinson). Platelets were identified by their characteristic light scatter and CD41a antibody binding. Activated platelets are reported as the percentage of 10,000 CD41+ platelets exhibiting APC-CD62P fluorescence.
Measurement of OCR and ECAR in platelets and epithelial cells
OCR and ECAR were measured in isolated platelets and epithelial cells using the Seahorse Extracellular Flux (XF96) Analyzer (Seahorse Bioscience, Inc., North Billerica, MA) as previously described (8, 50). Isolated platelets were diluted in unbuffered Dulbecco's modified Eagle's media (pH 7.4 at 37°C) to 50 × 106 cells/mL and 500 μL of sample loaded per well in standard XF24 plates. Epithelial cells (once isolated) were seeded and grown overnight at the density of 5 × 104 cells/well. Once in the XF24, the platelets or epithelial cells were consecutively treated with oligomycin A (2.5 μM), carbonyl cyanide-ρ-trifluoromethoxyphenylhydrazone (FCCP) (0.7 μM), 2-DG (100 μM), and rotenone (2 μM). Three measurements of OCR and ECAR were made over 1.5 min after addition of the agents and a 3-min mix step. OCR and ECAR readings were normalized to cell number.
Measurement of complex IV activity in permeabilized cells
Platelets (50 × 106) were seeded and equilibrated in standard XF24 plates as described above. Once a stable baseline rate was established, cells were treated with digitonin (20 μg/mL) and TMPD (500 μM). Ascorbate (2 mM) was also injected to maintain TMPD in its reduced form. OCR was measured by extracellular flux analysis as described above.
Measurement of electron transport complex and TCA cycle enzyme activities
All enzymatic activities were measured as previously described (8, 50) in cells (50–100 μg of protein) after undergoing three cycles of freeze/thaw. Briefly, citrate synthase activity was measured by monitoring the rate of conversion of acetyl coenzyme A (CoA) (100 μM) and oxaloacetate (200 μM) to citrate spectrophotometrically at 412 nm by coupling CoA production with the colorimetric indicator dithionitrobenzoic acid (200 μM). Complex I activity was measured using NADH (100 μM) and ubiquinone (100 μM) as substrates and monitoring the rotenone (10 μM)-sensitive decrease in absorbance of NADH at 340 nm. Complex II activity was measured using succinate (1.2 mM) and ubiquinone (50 μM) as substrates, coupling the (i) complex II catalyzed transfer of an electron from succinate to ubiquinone and the (ii) ubiquinol reduction of the dye 2,6-dichlorophenolindophenol (150 μM), which is monitored by a decrease in its absorbance at 600 nm and inhibited by thenoyltrifluoroacetone (50 μM). Complex IV activity was measured using reduced cytochrome c (50 μM) as the substrate and monitoring the oxidation of cytochrome c at 550 nm that was inhibitable by potassium cyanide (50 μM). All complex activities were normalized to protein concentration as determined by the Bradford protein assay method.
Measurement of ATP production
ATP production was measured by using a luciferase-based luminescence assay kit (PerkinElmer, Waltham, MA) and measuring the linear rate of luminescence over 5 min as previously described (8, 50). ATP content was normalized to platelet or airway epithelial cell number.
Measurement of mtDNA
Mitochondrial DNA was isolated from airway epithelial cells and platelets and DNA copy number quantified by real time-PCR (RT-PCR) using the primer for ND1 (R: 5′ GGC GTC TGC AAA TGG TTG TAA; F: 5′ AAT CGC CAT AGC CTT CCT AAC AT). These results were expressed as a ratio of nuclear DNA quantified by RT-PCR using the primer for histone 19 (F: 5′ GTA CCC ACC TGT CGT CC; R: 5′ GTC CAC GAG ACC AAT GAC TG) in airway epithelial cells. Since platelets do not contain nuclear DNA, mtDNA was normalized to platelet number.
Measurement of ROS production
Mitochondrial superoxide production was measured by MitoSOX as previously described (8). Briefly, cells were incubated with MitoSOX reagent (5 μM, 10 min; Invitrogen, Carlsbad, CA) and washed with PBS. Fluorescent intensity (510/580 nm) was measured kinetically for 10 min and normalized to cell number. Total H2O2 production was measured by spectrophotometrically monitoring the oxidation of Amplex Red by platelets or epithelial cells over 10 min to calculate the rate of H2O2 production. Rates are normalized for cell number.
Nitrotyrosine content
Nitrotyrosine concentration was assessed in platelets and airway epithelial cells by enzyme-linked immunosorbent assay as per the manufacturer's instructions (Ab113848; Abcam).
NOS activity
Total cellular NOS activity was assessed in lysed platelet or airway epithelial cells by quantifying the conversion of [3H]-arginine to citrulline in the presence and absence of L-NAME according to the kit manufacturer's instructions (catalog No. 781001; Cayman Chemicals).
NOx measurement
NOx encompasses total NO species, including nitrite and nitrate. NOx was measured by vanadium chloride-based reductive chemiluminescence as previously described (10). Briefly, cells were placed in fresh PBS for 10 min, and then, the PBS was removed and injected into a solution of vanadium chloride kept at 90°C. The vessel was purged with helium gas and connected inline to a nitric oxide analyzer (Seivers). The area under the curve of the resulting signal was measured and compared with known concentrations of nitrite/nitrate to calculate NOx concentration. NOx concentration was normalized to cell number.
Arginase activity
Total cellular arginase activity was quantified in frozen/thawed airway epithelial cells or platelets utilizing a colorimetric arginase activity kit (MAK 112; Sigma-Aldrich). Arginase activity yields ornithine and urea. In this assay, urea reacts with kit components to generate a colored compound, which was measured at 430 nm.
High-resolution liquid chromatography mass spectrometry to measure arginine and ornithine
Sample preparation
One hundred microliters of plasma was spiked with an isotopically labeled standard (Taurine-1,1,2,2-d4; Sigma-Aldrich, St. Louis, MO) to a final concentration of 100 nM. Simultaneous protein precipitation and polar amino acid extraction were performed using 300 μL ice-cold 1:1 (v:v) methanol:ethanol as previously described (1). Briefly, samples were cleared by centrifugation at 16,000 g and the supernatant dried under nitrogen gas. Samples were resuspended in 5% ACN in water and 5 μL subjected to online liquid chromatography tandem mass spectrometry (LC-MS/MS) analysis.
LC-MS/MS method
Analyses were performed by untargeted LC-MS/MS. Briefly, samples were separated over a reversed-phase Phenomenex Kinetex C18+ column (2.1 × 100 mm, 1.7 μm particle size) maintained at 40°C. For the 20-min LC gradient, the mobile phase consisted of the following: solvent A (1.5 mM ammonium fluoride) and solvent B (100% acetonitrile). The gradient was the following: 0–12.0 min 5% B to 1000% B, 12.0–15.0 min hold at 100% B, 15.0–15.1 100% to 5% B, 15.1–20.0 min 5% B. The Q Exactive mass spectrometer was operated in positive ion mode, scanning in full MS mode (2 microscans) from 66.7 to 1000 m/z at 70,000 resolution with an AGC target of 3e6, dd-ms2 fragmentation was selected for the top 4 ions per full scan using an AGC target of 1e5, NCE of 35, using an isolation window of 1 AMU. Source settings were 4.5 kV spray voltage, 20 sheath gas, 10 auxiliary gas at 320°C, and 4 sweep gas. Calibration was performed before analysis using the Pierce™ negative ion calibration solutions (Thermo Fisher Scientific). Integrated peak areas were then extracted manually using Quan Browser (Thermo Fisher Xcalibur version 2.7). Graphs and statistical analyses (either t-test or ANOVA) were prepared with GraphPad Prism 7.0 (GraphPad Software, Inc., La Jolla, CA).
NO2-cLA sample preparation and LC-MS/MS
Plasma (0.5 mL) samples containing 10 nM [13C18]-NO2-OA internal standard were incubated with 10 mM HgCl2 for 30 min at 37°C to release adducted NO2-cLA before extraction with 1 mL hexane:isopropanol:1 M formic acid (30:20:2) and an additional 1 mL of hexanes. The samples were vortexed and centrifuged at 2800 rpm for 10 min at 4°C and the organic phase was transferred to a new glass vial and dried under N2. Samples were stored at −80°C until analysis.
A CTC PAL autosampler (Leap Technologies, Carrboro, NC) and a Shimadzu LC-20AD pump (Columbia, MD) coupled to a Sciex 5000 triple quadrupole mass spectrometer (Framingham, MA) were used for the quantification of NO2-cLA. Samples were reconstituted in 100 μL of methanol and 10 μL was injected onto a reversed-phase column [Phenomenex Luna C18(2), 2 × 100 mm] at a flow rate of 0.75 mL/min. The gradient used for separation consisted of solvent A: water and 0.1% acetic acid and solvent B: acetonitrile and 0.1% acetic acid. The 15-min method started at 40% B and increased to 100% B over 10 min and was held for 2 min, following 3 min of equilibration at initial conditions. MS analyses was conducted using negative ion mode electrospray ionization with the following source parameters: collision gas 5 units, curtain gas 50 units, ion source gas (ISG)#1 55 units and ISG#2 50 units, ion spray voltage −4500 V, and source temperature 650°C. The declustering potential was set at −60, entrance potential −10, collision energy −35, and the collision exit potential −3. NO2-cLA was monitored at m/z 324→46 (loss of nitro group) and the internal standard [13C18]-NO2-OA at m/z 344→46. Endogenous NO2-cLA was quantified using a standard curve developed with synthetic NO2-cLA and [13C18]-NO2-OA internal standard.
Statistical analyses
Data are shown as mean ± standard error. All statistical comparisons are performed using the Student's t-test or one-way ANOVA as appropriate. The level of significance for p was chosen at 0.05. All data were analyzed with statistical program JMP Pro 10 (SAS Institute, Cary, NC).
Supplementary Material
Acknowledgments
This work was supported by the Hemophilia Center of Western Pennsylvania and NIH grant 1 R01 HL133003-01A1 to S.S.
Abbreviations Used
- 2-DG
2-deoxyglucose
- ADMA
asymmetric dimethylarginine
- BMI
body mass index
- c-PTIO
2-phenyl-4,4,5,5-tetramethylimidazoline-1-oxyl 3-oxide
- CoA
Coenzyme A
- ECAR
extracellular acidification rate
- FCCP
carbonyl cyanide-ρ-trifluoromethoxyphenylhydrazone
- FENO
fractional exhaled NO
- H2O2
hydrogen peroxide
- HIF-1α
hypoxia-inducible factor 1 alpha
- iNOS
inducible nitric oxide synthase
- ISG
ion source gas
- LM-MS/MS
liquid chromatography tandem mass spectrometry
- L-NAME
L-NG-Nitroarginine methyl ester
- NO
nitric oxide
- NO2-cLA
NO2-conjugated linoleic acid
- NOS
nitric oxide synthase
- OCR
oxygen consumption rate
- PBS
phosphate-buffered saline
- PGI2
prostaglandin I2
- ROS
reactive oxygen species
- SEM
standard error of the mean
- TCA
tricarboxylic acid
- TMPD
tetramethyl-p-phenylenediamine
Author Disclosure Statement
No competing financial interests exist.
Supplementary Material
References
- 1. Ahmad T, Kumar M, Mabalirajan U, Pattnaik B, Aggarwal S, Singh R, Singh S, Mukerji M, Ghosh B, and Agrawal A. Hypoxia response in asthma: differential modulation on inflammation and epithelial injury. Am J Respir Cell Mol Biol 47: 1–10, 2012 [DOI] [PubMed] [Google Scholar]
- 2. Ahmad T, Mabalirajan U, Ghosh B, and Agrawal A. Altered asymmetric dimethyl arginine metabolism in allergically inflamed mouse lungs. Am J Respir Cell Mol Biol 42: 3–8, 2010 [DOI] [PubMed] [Google Scholar]
- 3. Avila C, Huang RJ, Stevens MV, Aponte AM, Tripodi D, Kim KY, and Sack MN. Platelet mitochondrial dysfunction is evident in type 2 diabetes in association with modifications of mitochondrial anti-oxidant stress proteins. Exp Clin Endocrinol Diabetes 120: 248–251, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Barros R, Moreira A, Fonseca J, Moreira P, Fernandes L, de Oliveira JF, Delgado L, and Castel-Branco MG. Obesity and airway inflammation in asthma. J Allergy Clin Immunol 117: 1501–1502, 2006 [DOI] [PubMed] [Google Scholar]
- 5. Benson RC, Hardy KA, and Morris CR. Arginase and arginine dysregulation in asthma. J Allergy (Cairo) 2011: 736319, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bhatraju NK. and Agrawal A. Mitochondrial dysfunction linking obesity and asthma. Ann Am Thorac Soc 14: S368–S373, 2017 [DOI] [PubMed] [Google Scholar]
- 7. Brookes P. and Darley-Usmar VM. Hypothesis: the mitochondrial NO(*) signaling pathway, and the transduction of nitrosative to oxidative cell signals: an alternative function for cytochrome C oxidase. Free Radic Biol Med 32: 370–374, 2002 [DOI] [PubMed] [Google Scholar]
- 8. Cardenes N, Corey C, Geary L, Jain S, Zharikov S, Barge S, Novelli EM, and Shiva S. Platelet bioenergetic screen in sickle cell patients reveals mitochondrial complex V inhibition, which contributes to platelet activation. Blood 123: 2864–2872, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cottrell L, Neal WA, Ice C, Perez MK, and Piedimonte G. Metabolic abnormalities in children with asthma. Am J Respir Crit Care Med 183: 441–448, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Curtis E, Hsu LL, Noguchi AC, Geary L, and Shiva S. Oxygen regulates tissue nitrite metabolism. Antioxid Redox Signal 17: 951–961, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dewitz C, McEachern E, Shin S, Akong K, Nagle DG, Broide DH, Akuthota P, and Crotty Alexander LE. Hypoxia-inducible factor-1alpha inhibition modulates airway hyperresponsiveness and nitric oxide levels in a BALB/c mouse model of asthma. Clin Immunol 176: 94–99, 2017 [DOI] [PubMed] [Google Scholar]
- 12. Dudek AZ, Nesmelova I, Mayo K, Verfaillie CM, Pitchford S, and Slungaard A. Platelet factor 4 promotes adhesion of hematopoietic progenitor cells and binds IL-8: novel mechanisms for modulation of hematopoiesis. Blood 101: 4687–4694, 2003 [DOI] [PubMed] [Google Scholar]
- 13. Ghosh S, Willard B, Comhair SA, Dibello P, Xu W, Shiva S, Aulak KS, Kinter M, and Erzurum SC. Disulfide bond as a switch for copper–zinc superoxide dismutase activity in asthma. Antioxid Redox Signal 18: 412–423, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hagen T, Taylor CT, Lam F, and Moncada S. Redistribution of intracellular oxygen in hypoxia by nitric oxide: effect on HIF1alpha. Science 302: 1975–1978, 2003 [DOI] [PubMed] [Google Scholar]
- 15. Haldar P, Pavord ID, Shaw DE, Berry MA, Thomas M, Brightling CE, Wardlaw AJ, and Green RH. Cluster analysis and clinical asthma phenotypes. Am J Respir Crit Care Med 178: 218–224, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Holguin F, Comhair SA, Hazen SL, Powers RW, Khatri SS, Bleecker ER, Busse WW, Calhoun WJ, Castro M, Fitzpatrick AM, Gaston B, Israel E, Jarjour NN, Moore WC, Peters SP, Teague WG, Chung KF, Erzurum SC, and Wenzel SE. An association between L-arginine/asymmetric dimethyl arginine balance, obesity, and the age of asthma onset phenotype. Am J Respir Crit Care Med 187: 153–159, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Holguin F. and Fitzpatrick A. Obesity, asthma, and oxidative stress. J Appl Physiol (1985) 108: 754–759, 2010 [DOI] [PubMed] [Google Scholar]
- 18. Hüttemann M, Lee I, Gao X, Pecina P, Pecinova A, Liu J, Aras S, Sommer N, Sanderson TH, Tost M, Neff F, Aguilar-Pimentel JA, Becker L, Naton B, Rathkolb B, Rozman J, Favor J, Hans W, Prehn C, Puk O, Schrewe A, Sun M, Höfler H, Adamski J, Bekeredjian R, Graw J, Adler T, Busch DH, Klingenspor M, Klopstock T, Ollert M, Wolf E, Fuchs H, Gailus-Durner V, Hrabě de Angelis M, Weissmann N, Doan JW, Bassett DJ, and Grossman LI. Cytochrome c oxidase subunit 4 isoform 2-knockout mice show reduced enzyme activity, airway hyporeactivity, and lung pathology. FASEB J 26: 3916–3930, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kameyoshi Y, Dörschner A, Mallet AI, Christophers E, and Schröder JM. Cytokine RANTES released by thrombin-stimulated platelets is a potent attractant for human eosinophils. J Exp Med 176: 587–592, 1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kasteleyn MJ, Bonten TN, de Mutsert R, Thijs W, Hiemstra PS, le Cessie S, Rosendaal FR, Chavannes NH, and Taube C. Pulmonary function, exhaled nitric oxide and symptoms in asthma patients with obesity: a cross-sectional study. Respir Res 18: 205, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Klinger MH. Platelets and inflammation. Anat Embryol (Berl) 196: 1–11, 1997 [DOI] [PubMed] [Google Scholar]
- 22. Komakula S, Khatri S, Mermis J, Savill S, Haque S, Rojas M, Brown L, Teague GW, and Holguin F. Body mass index is associated with reduced exhaled nitric oxide and higher exhaled 8-isoprostanes in asthmatics. Respir Res 8: 32, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kramer PA, Chacko BK, George DJ, Zhi D, Wei CC, Dell'Italia LJ, Melby SJ, George JF, and Darley-Usmar VM. Decreased Bioenergetic Health Index in monocytes isolated from the pericardial fluid and blood of post-operative cardiac surgery patients. Biosci Rep 35: e00237, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lefrançais E, Ortiz-Muñoz G, Caudrillier A, Mallavia B, Liu F, Sayah DM, Thornton EE, Headley MB, David T, Coughlin SR, Krummel MF, Leavitt AD, Passegué E, and Looney MR. The lung is a site of platelet biogenesis and a reservoir for haematopoietic progenitors. Nature 544: 105–109, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mabalirajan U, Dinda AK, Kumar S, Roshan R, Gupta P, Sharma SK, and Ghosh B. Mitochondrial structural changes and dysfunction are associated with experimental allergic asthma. J Immunol 181: 3540–3548, 2008 [DOI] [PubMed] [Google Scholar]
- 26. Mabalirajan U, Dinda AK, Sharma SK, and Ghosh B. Esculetin restores mitochondrial dysfunction and reduces allergic asthma features in experimental murine model. J Immunol 183: 2059–2067, 2009 [DOI] [PubMed] [Google Scholar]
- 27. Mateo J, García-Lecea M, Cadenas S, Hernández C, and Moncada S. Regulation of hypoxia-inducible factor-1alpha by nitric oxide through mitochondria-dependent and -independent pathways. Biochem J 376: 537–544, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Modena BD, Bleecker ER, Busse WW, Erzurum SC, Gaston BM, Jarjour NN, Meyers DA, Milosevic J, Tedrow JR, Wu W, Kaminski N, and Wenzel SE. Gene expression correlated with severe asthma characteristics reveals heterogeneous mechanisms of severe disease. Am J Respir Crit Care Med 195: 1449–1463, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Moncada S. and Erusalimsky JD. Does nitric oxide modulate mitochondrial energy generation and apoptosis? Nat Rev Mol Cell Biol 3: 214–220, 2002 [DOI] [PubMed] [Google Scholar]
- 30. Morris CR. Arginine and asthma. Nestle Nutr Inst Workshop Ser 77: 1–15, 2013 [DOI] [PubMed] [Google Scholar]
- 31. Nguyen QL, Corey C, White P, Watson A, Gladwin MT, Simon MA, and Shiva S. Platelets from pulmonary hypertension patients show increased mitochondrial reserve capacity. JCI Insight 2: e91415, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ostroukhova M, Goplen N, Karim MZ, Michalec L, Guo L, Liang Q, and Alam R. The role of low-level lactate production in airway inflammation in asthma. Am J Physiol Lung Cell Mol Physiol 302: L300–30L307, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Palacios-Callender M, Quintero M, Hollis VS, Springett RJ, and Moncada S. Endogenous NO regulates superoxide production at low oxygen concentrations by modifying the redox state of cytochrome c oxidase. Proc Natl Acad Sci U S A 101: 7630–7635, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Picado C, Deulofeu R, Lleonart R, Agustí M, Casals E, Quintó L, and Mullol J. Lipid and protein metabolism in asthma. Effects of diet and corticosteroid therapy. Allergy 54: 569–575, 1999 [DOI] [PubMed] [Google Scholar]
- 35. Qian X, Aboushousha R, van de Wetering C, Chia SB, Amiel E, Schneider RW, van der Velden JLJ, Lahue KG, Hoagland DA, Casey DT, Daphtary N, Ather JL, Randall MJ, Aliyeva M, Black KE, Chapman DG, Lundblad LKA, McMillan DH, Dixon AE, Anathy V, Irvin CG, Poynter ME, Wouters EFM, Vacek PM, Henket M, Schleich F, Louis R, van der Vliet A, and Janssen-Heininger YMW. IL-1/inhibitory kappaB kinase epsilon-induced glycolysis augment epithelial effector function and promote allergic airways disease. J Allergy Clin Immunol 142: 435–450.e10, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Roos AB, Mori M, Grönneberg R, Österlund C, Claesson HE, Wahlström J, Grunewald J, Eklund A, Erjefält JS, Lundberg JO, and Nord M. Elevated exhaled nitric oxide in allergen-provoked asthma is associated with airway epithelial iNOS. PLoS One 9: e90018, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Scherzer R. and Grayson MH. Heterogeneity and the origins of asthma. Ann Allergy Asthma Immunol 121: 400–405, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Scott JA, North ML, Rafii M, Huang H, Pencharz P, Subbarao P, Belik J, and Grasemann H. Asymmetric dimethylarginine is increased in asthma. Am J Respir Crit Care Med 184: 779–785, 2011 [DOI] [PubMed] [Google Scholar]
- 39. Shi DY, Xie FZ, Zhai C, Stern JS, Liu Y, and Liu SL. The role of cellular oxidative stress in regulating glycolysis energy metabolism in hepatoma cells. Mol Cancer 8: 32, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shiva S, Brookes PS, Patel RP, Anderson PG, and Darley-Usmar VM. Nitric oxide partitioning into mitochondrial membranes and the control of respiration at cytochrome c oxidase. Proc Natl Acad Sci U S A 98: 7212–7217, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shiva S. and Darley-Usmar VM. Control of the nitric oxide-cytochrome c oxidase signaling pathway under pathological and physiological conditions. IUBMB Life 55: 585–590, 2003 [DOI] [PubMed] [Google Scholar]
- 42. Sutherland ER, Goleva E, King TS, Lehman E, Stevens AD, Jackson LP, Stream AR, Fahy JV, Leung DY; Asthma Clinical Research Network. Cluster analysis of obesity and asthma phenotypes. PLoS One 7: e36631, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tajti G, Papp C, Kardos L, Keki S, Pak K, Szilasi ME, Gesztelyi R, Mikaczo A, Fodor A, Szilasi M, and Zsuga J. Positive correlation of airway resistance and serum asymmetric dimethylarginine (ADMA) in bronchial asthma patients lacking evidence for systemic inflammation. Allergy Asthma Clin Immunol 14: 2, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Teng Y, Sun P, Zhang J, Yu R, Bai J, Yao X, Huang M, Adcock IM, and Barnes PJ. Hydrogen peroxide in exhaled breath condensate in patients with asthma: a promising biomarker? Chest 140: 108–116, 2011 [DOI] [PubMed] [Google Scholar]
- 45. Trian T, Benard G, Begueret H, Rossignol R, Girodet PO, Ghosh D, Ousova O, Vernejoux JM, Marthan R, Tunon-de-Lara JM, and Berger P. Bronchial smooth muscle remodeling involves calcium-dependent enhanced mitochondrial biogenesis in asthma. J Exp Med 204: 3173–3181, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tyrrell DJ, Bharadwaj MS, Jorgensen MJ, Register TC, and Molina AJ. Blood cell respirometry is associated with skeletal and cardiac muscle bioenergetics: implications for a minimally invasive biomarker of mitochondrial health. Redox Biol 10: 65–77, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tyrrell DJ, Bharadwaj MS, Jorgensen MJ, Register TC, Shively C, Andrews RN, Neth B, Keene CD, Mintz A, Craft S, and Molina AJA. Blood-based bioenergetic profiling reflects differences in brain bioenergetics and metabolism. Oxid Med Cell Longev 2017: 7317251, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Willig AL, Kramer PA, Chacko BK, Darley-Usmar VM, Heath SL, and Overton ET. Monocyte bioenergetic function is associated with body composition in virologically suppressed HIV-infected women. Redox Biol 12: 648–656, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Winnica D, Que LG, Baffi C, Grasemann H, Fiedler K, Yang Z, Etling E, Wasil K, Wenzel SE, Freeman B, and Holguin F. l-Citrulline prevents asymmetric dimethylarginine-mediated reductions in nitric oxide and nitrosative stress in primary human airway epithelial cells. Clin Exp Allergy 47: 190–199, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Xu W, Cardenes N, Corey C, Erzurum SC, and Shiva S. Platelets from asthmatic individuals show less reliance on glycolysis. PLoS One 10: e0132007, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Xu W, Comhair SAA, Janocha AJ, Lara A, Mavrakis LA, Bennett CD, Kalhan SC, and Erzurum SC. Arginine metabolic endotypes related to asthma severity. PLoS One 12: e0183066, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Xu W, Ghosh S, Comhair SA, Asosingh K, Janocha AJ, Mavrakis DA, Bennett CD, Gruca LL, Graham BB, Queisser KA, Kao CC, Wedes SH, Petrich JM, Tuder RM, Kalhan SC, and Erzurum SC. Increased mitochondrial arginine metabolism supports bioenergetics in asthma. J Clin Invest 126: 2465–2481, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yu L, Lu M, Jia D, Ma J, Ben-Jacob E, Levine H, Kaipparettu BA, and Onuchic JN. Modeling the genetic regulation of cancer metabolism: interplay between glycolysis and oxidative phosphorylation. Cancer Res 77: 1564–1574, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.