Significance
p120-catenin, an armadillo-repeat protein, regulates the stability of classical cadherins, but its cellular functions in vivo remain unclear. Here, we show that genetic deletion of p120-catenin in the mouse embryo causes developmental arrest at midgestation with a bifurcation of the posterior body axis. This morphological defect is associated with an expanded primitive streak, p53-dependent cell death, and abnormal mesoderm migration. The expanded primitive streak in p120-catenin mutants is caused by ectopic activation of the canonical WNT signaling pathway, and the bifurcation of the axis is associated with accumulation of mesoderm cells that migrate without direction. The functions of p120-catenin in the early mouse embryo are likely to be important during other developmental epithelial-to-mesenchymal transitions (EMTs) and in tumor metastasis.
Keywords: WNT signaling, cell migration, gastrulation, epithelial–mesenchymal transition, p53-dependent cell death
Abstract
Epithelial-to-mesenchymal transitions (EMTs) require a complete reorganization of cadherin-based cell–cell junctions. p120-catenin binds to the cytoplasmic juxtamembrane domain of classical cadherins and regulates their stability, suggesting that p120-catenin may play an important role in EMTs. Here, we describe the role of p120-catenin in mouse gastrulation, an EMT that can be imaged at cellular resolution and is accessible to genetic manipulation. Mouse embryos that lack all p120-catenin, or that lack p120-catenin in the embryo proper, survive to midgestation. However, mutants have specific defects in gastrulation, including a high rate of p53-dependent cell death, a bifurcation of the posterior axis, and defects in the migration of mesoderm; all are associated with abnormalities in the primitive streak, the site of the EMT. In embryonic day 7.5 (E7.5) mutants, the domain of expression of the streak marker Brachyury (T) expands more than 3-fold, from a narrow strip of posterior cells to encompass more than one-quarter of the embryo. After E7.5, the enlarged T+ domain splits in 2, separated by a mass of mesoderm cells. Brachyury is a direct target of canonical WNT signaling, and the domain of WNT response in p120-catenin mutant embryos, like the T domain, is first expanded, and then split, and high levels of nuclear β-catenin levels are present in the cells of the posterior embryo that are exposed to high levels of WNT ligand. The data suggest that p120-catenin stabilizes the membrane association of β-catenin, thereby preventing accumulation of nuclear β-catenin and excessive activation of the WNT pathway during EMT.
p120-catenin (CTNND1) is a large cytoplasmic armadillo-repeat protein that binds to the juxtamembrane domain (JMD) of the cytoplasmic domain of classical cadherins, including both E-cadherin and N-cadherin (1). In the absence of p120-catenin at the JMD, the cadherins that mediate homophilic cell adhesion are susceptible to endocytosis and subsequent degradation (2).
Despite its role in the regulation of cadherin stability, the effects of deletion of p120-catenin in vivo can be relatively mild and vary between tissues. Drosophila and Caenorhabditis elegans have p120-catenin homologs that are not required for development or viability (3, 4). Nevertheless, Drosophila p120-catenin does affect cadherin endocytosis and thereby the kinetics of cell rearrangements and cell shape dynamics (5). In the mouse, p120-catenin is an essential gene, and tissue-specific deletion has shown that it is required for the morphogenesis of the salivary gland (6), the mammary gland (7), and the kidney (8), whereas p120-catenin deletion has no effect on the prostate (7). In addition to its roles in development, decreased expression of p120-catenin is observed frequently in human tumors (9), and p120-catenin acts as a tumor suppressor in mouse Kras-dependent pancreatic cancer (10, 11), although the mechanisms of action of p120 in tumorigenesis are unclear.
Dynamic changes in cadherin location and expression are a hallmark of epithelial-to-mesenchymal transitions (EMTs). Mouse gastrulation is an ideal context for the study of EMT, as it can be manipulated genetically relatively easily, and the process can be imaged at high resolution in vivo. In response to localized expression of WNT and NODAL ligands, gastrulation begins at a single position in the epiblast, the primitive streak, which marks the future posterior of the animal (12). At the primitive streak, individual cells move out of the epiblast epithelium and then migrate immediately away in the mesenchymal layer that gives rise to both the mesoderm and definitive endoderm (13, 14).
During the gastrulation EMT, cells switch from expression of E-cadherin (CDH1) in the epithelial epiblast to N-cadherin (CDH2) in the newly formed mesenchyme (15). The cytoplasmic domains of E-cadherin and N-cadherin bind the actin-binding protein α-catenin and the armadillo-repeat proteins β-catenin and p120-catenin. As p120-catenin regulates cadherin stability and thereby the stability of cell–cell adhesion, it seemed likely that it would play a role in the gastrulation EMT. p120-catenin is required for early mouse development (6), but its specific functions at that stage have not been examined.
Here, we show that p120-catenin has critical roles during the mouse gastrulation. p120-catenin is not required for cadherin switching but instead is a strong negative regulator of WNT signaling and a regulator of cell behavior during the EMT. In the absence of p120-catenin, the domain of the primitive streak expands severalfold and the level of nuclear β-catenin increases more than 2-fold in the cells near the primitive streak that are exposed to WNT ligands. At the center of the expanded mutant primitive streak, N-cadherin+, SNAIL+, KDR+ mesoderm cells accumulate and split the streak in 2, bifurcating the posterior body axis. These activities of p120-catenin in the regulation of WNT signaling and EMT are likely to be important in other EMTs in development and in tumorigenesis.
Results
Absence of p120-Catenin Causes Bifurcation of the Posterior Body Axis.
To study the role of p120-catenin in early development, we generated a null mutation from the conditional allele (6). Embryos homozygous for the p120-catenin–null allele arrested development at embryonic day 8.5 (E8.5), with a striking morphological phenotype: a splitting of the posterior body axis and a duplicated allantois.
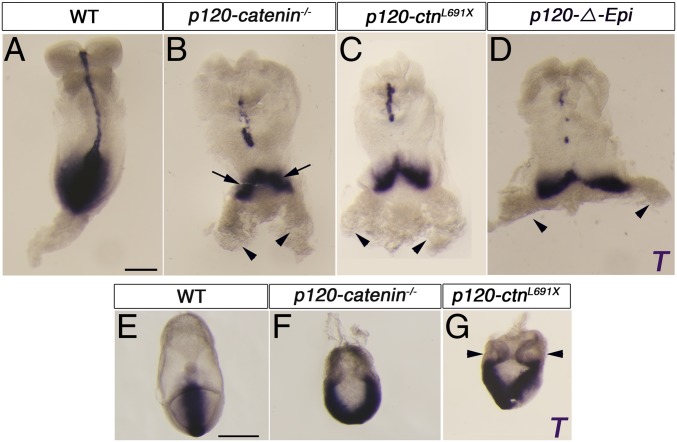
Wild-type embryos express Brachyury (T) in a single position that marks the posterior of the embryo (Fig. 1A). In contrast, 80% of E8.5 p120-null embryos (n > 30) had 2 separated posterior domains that expressed T (arrows in Fig. 1B). Wild-type embryos have a single posterior allantois, a posterior extraembryonic mesoderm structure; in contrast ∼80% of p120 mutant embryos had 2 separated allantoides (arrowheads in Fig. 1B). In the remaining 20% of mutants, there was a single allantois and an expanded and distorted T+ streak domain (SI Appendix, Fig. S1 A and B, arrow). We also isolated a nonsense allele of p120-catenin (L691X) based on its embryonic phenotype in an ENU-mutagenesis screen (16); L691X homozygotes showed the same splitting of the T expression as the targeted null allele (Fig. 1C), confirming the specificity of the phenotype. The bifurcation of the T+ primitive streak and duplication of the allantois was clear at E7.5 (Fig. 1 E–G), 1 d after the onset of gastrulation at E6.5. Other markers of the primitive streak, including Wnt3 and Mixl1, were also present in 2 separate domains in the mutants (SI Appendix, Fig. S1 D–K).
Fig. 1.
The posterior body axis is duplicated in p120-catenin mutant embryos. (A–D) Expression of T in E8.5 wild-type and p120-catenin mutant embryos, assayed by in situ hybridization, dorsal views. (A) Wild-type embryos express T in the primitive streak and the midline. (B) Approximately 80% of p120-catenin–null mutants have a posterior bifurcation of the Brachyury (T) expression domain in the primitive streak (arrows point to the 2 T-domains). Arrowheads point to 2 allantoides. (C) Embryos homozygous for a truncating point mutation of p120-catenin (LX169) recapitulate the null phenotype. The arrowheads point to 2 allantoides. (D) Conditional deletion of p120-catenin in the epiblast. Most p120-ΔEpi mutants develop 2 allantoides (arrowheads). (E–G) In situ hybridization for T in E7.5 wild-type and p120-catenin mutant embryos, posterior views. (E) T is expressed in the posterior of wild-type embryos. (F and G) T is expressed in 2 separated domains in the null (F) and mutant embryos with LX169 point mutation (G). The arrowheads point to 2 allantoides. (All scale bars, 50 μm.)
At the time of developmental arrest at ∼E8.5, mesodermal tissues that arise from the primitive streak were specified but were organized abnormally in the mutant embryos. Cardiac mesoderm, marked by expression of Nkx2.5, was specified in a single anterior domain (SI Appendix, Fig. S1 L and M). Paraxial mesoderm, marked by expression of Meox1, was present and flanked the midline, but segmented somites were not formed (SI Appendix, Fig. S1 N and O). The axial mesoderm of the wild-type midline expresses T, and T was expressed in a small discontinuous midline domain in the mutants. Consistent with the midline T expression pattern, the axial domain of FOXA2 was also interrupted and there was a small abnormal-shaped FOXA2+ node in the p120-catenin mutants (SI Appendix, Fig. S2 A–D).
p120-Catenin Acts Autonomously in the Epiblast.
Partial or complete axis duplication in the mouse embryo can be caused by defects in collective migration of a population of extraembryonic organizer cells, the anterior visceral endoderm (AVE) (17). Although p120-catenin has been reported to regulate collective cell migration (7), Hex-GFP+ AVE cells (18) in the p120 mutants migrated to their normal destination at the anterior border between the embryonic and extraembryonic regions (SI Appendix, Fig. S3 A and B). Other markers of the AVE, Dkk1 and Cer1, were also expressed in the correct domain in the AVE of the mutant embryos (SI Appendix, Fig. S3 C–F).
To test directly whether p120-catenin acts within cells of the embryo proper to control posterior axis specification, we deleted p120-catenin in the epiblast by crossing animals carrying the p120-catenin conditional allele with mice expressing the epiblast-specific Sox2-Cre transgene (19). The p120-catenin epiblast-deleted (p120-ΔEpi) embryos recapitulated the features of the p120-catenin–null phenotype: >70% of the mutant embryos (n = 17/24) showed duplicated expression of the streak marker T and formed 2 allantoides (Fig. 1D). As in the null mutants, the remaining mutant embryos accumulated a bulge of T+ cells near the primitive streak (n = 7/24; SI Appendix, Fig. S1C, arrow). Thus, p120-catenin is required in cells of the epiblast for specification of a single normal posterior body axis.
The Cadherin Switch Occurs in the Absence of p120-Catenin.
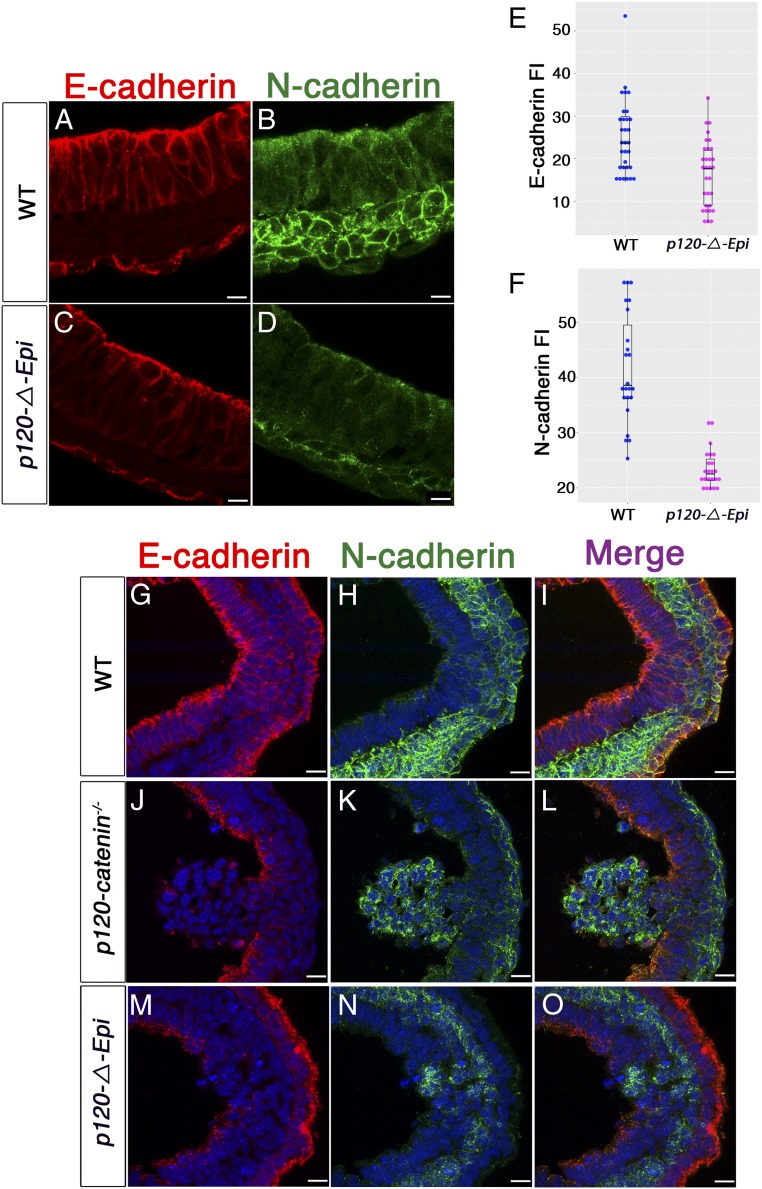
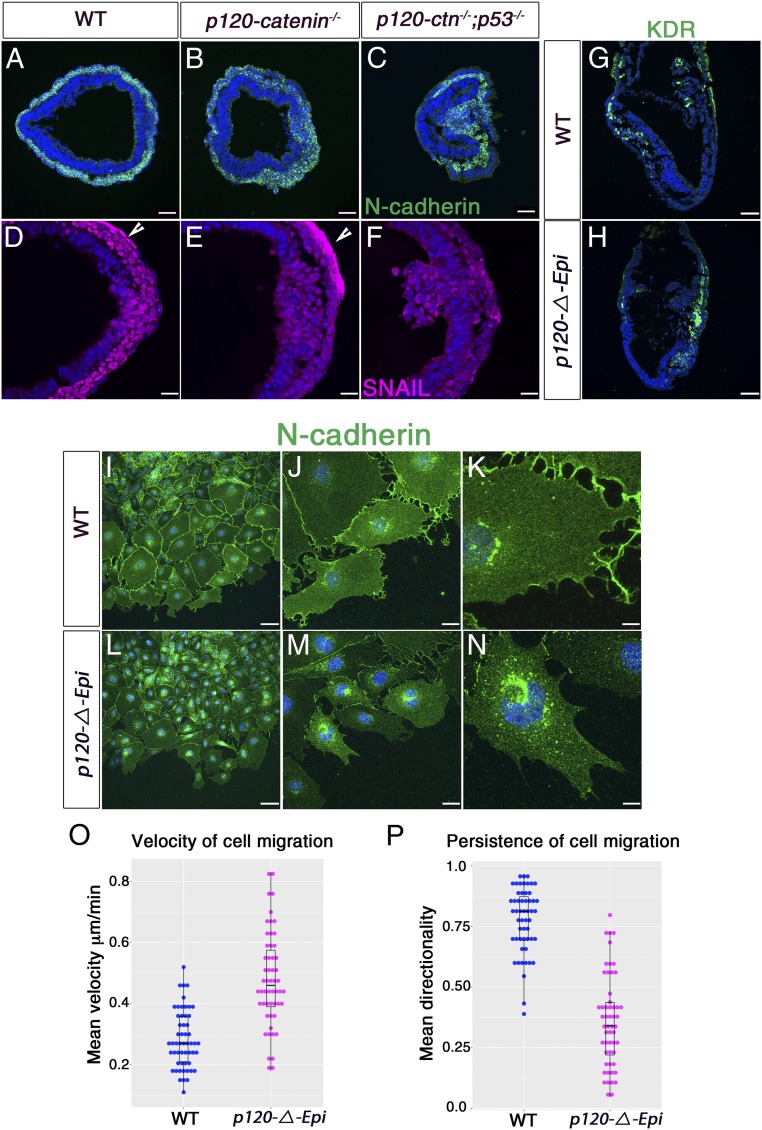
p120-catenin stabilizes membrane-associated classical cadherins (20, 21) and was expressed uniformly in the epiblast, primitive streak, and mesoderm of wild-type embryos; its expression was not detected in these cells types in the p120-ΔEpi mutant (SI Appendix, Fig. S4 A–D). To quantitate the effect of p120-catenin on cadherin levels, we measured signal intensity of plasma membrane-associated cadherins in individual embryos. E-cadherin and N-cadherin were both expressed in the correct locations of p120-ΔEpi mutant embryos: E-cadherin in lateral plasma membranes of the epiblast and N-cadherin in the plasma membrane in the mesoderm layer (Fig. 2 A–D). Although expressed in the correct tissue layers, quantitative analysis showed that the levels of both cadherins were lower in p120 mutant embryos than in wild type. There was about 1.5-fold more E-cadherin in the epiblast and 1.8-fold more N-cadherin in the mesoderm in wild type than in mutants (31 cells scored for E-cadherin in both genotypes; P < 0.0002; 23 cells of each genotype scored for N-cadherin; P < 0.0001) (Fig. 2 E and F). These data were supported by Western blot analysis (SI Appendix, Fig. S5 A–D), although there was more variability in the Western data, presumably because of pooling of embryos of slightly different ages for Westerns. Thus, p120-catenin promotes normal levels of E-cadherin and N-cadherin in the early embryo.
Fig. 2.
p120-catenin is required for normal Cadherin levels, but not for the E-cadherin to N-cadherin switch. (A–D) High-magnification images of immunostained E7.5 transverse wild-type and mutant embryo sections stained for E-cadherin (red) and N-cadherin (green). (A and B) Wild type. (C and D) p120-ΔEpi mutants. (A) E-cadherin is concentrated in both apical and lateral membranes of the wild-type epiblast; (C) E-cadherin levels are reduced in the mutant, especially in lateral epiblast membranes. N-cadherin is expressed in the mesoderm layer of both (B) wild-type and (D) p120 mutant embryos, but the staining appears more diffuse and punctate in the mutant. (Scale bars, 16 μm.) (E) The E-cadherin fluorescence intensity (FI) of wild-type and mutant epiblast cells. The mean E-cadherin FI for the wild-type epiblast cells was 25.06 1.57, whereas in the mutant FI = 16.50 1.40; P < 0.0002 (n = 31 cells each for wild type and mutant). (F) N-cadherin FI for wild-type mesoderm cells was 41.67 2.06; mesoderm mutant cells have a mean FI of 23.52 0.71, P < 0.0001 (n = 23 cells each for wild type and mutant). Points are values for individual cells; bars represent the mean and SD. (G–O) Higher-magnification views of cadherin expression in the streak region of E7.5 embryos. (G–I) Wild-type embryo. (J–L) A cluster of cells protruding from the mutant streak into the amniotic cavity expresses N-cadherin, and not E-cadherin. (M–O) In other mutants, cells contiguous with the epiblast layer express N-cadherin. (Scale bars, 16 μm.)
A hallmark of the gastrulation EMT, in which cells change identity from epithelial to mesenchymal is a switch in cadherin expression. E-cadherin is present at high levels in cell membranes of epiblast cells; when cells move through the primitive streak, they down-regulate E-cadherin and up-regulate N-cadherin (13). Although it has been proposed that p120-catenin might be required for cadherin switching (22), the switch from E-cadherin to N-cadherin took place during the EMT in the p120 mutants, and an N-cadherin+ mesoderm layer spread around the embryonic circumference of E7.5 mutants, although the mesoderm layer was thinner in the mutant than in wild type (SI Appendix, Fig. S5 E–M). Thus, p120-catenin is not essential for cadherin switching during mouse gastrulation. Nevertheless, the EMT was not normal in the mutants. Epithelial cells of the wild-type primitive streak that are preparing to undergo EMT express E-cadherin, whereas cells at that position in mutant embryos expressed N-cadherin and not E-cadherin and appeared to be mesenchymal rather than epithelial (Fig. 2 G–O).
p120-Catenin Regulates the Size and Organization of the Primitive Streak.
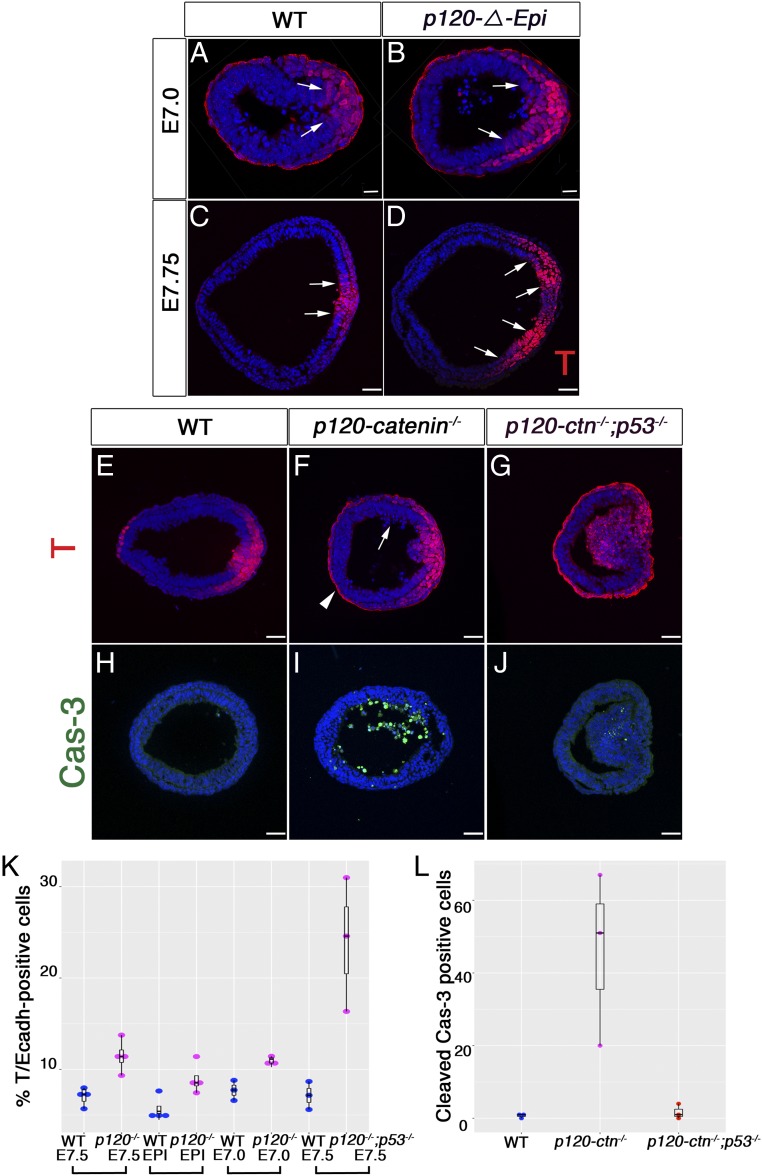
In wild-type embryos, the Brachyury (T) transcription factor is expressed in the cells of the epiblast that are competent to undergo the gastrulation EMT. In immunostained cross-sections of wild-type embryos, where single-cell resolution is possible, T expression was detected in cells of the posterior epiblast and in the newly generated mesoderm cells adjacent to the streak (Fig. 3 A, C, and E). In mutant embryos at E7.0, a single T domain was observed in the posterior epiblast layer (Fig. 3B). In the majority (∼80%) of E7.75 null and p120-ΔEpi mutant embryos (n = 12/15 and n = 8/10, respectively), the T domain was split into 2 domains separated by T-negative nonepithelial cells (Fig. 3D). In the remaining 20% of null and p120-ΔEpi mutants (n = 3/15 and n = 2/10), a ball of T+ cells protruded into the central amniotic cavity of the embryo (Fig. 3F).
Fig. 3.
p120-catenin limits the size of the primitive streak and inhibits apoptosis during the EMT. (A and B) Immunostaining for T in E7.0 transverse section; posterior to the right. (A) Wild-type embryos show T in the posterior. (B) In the mutant epiblast, the single domain of T is expanded. (C–G) Cryosections of E7.5 and E7.75 embryos stained for T. (A–D) The arrows mark the edges of T-expression domain in the epiblast layer. (C and E) Wild-type embryos express T in the streak region of the posterior epiblast and in the nascent mesoderm derived from the streak. (D) In 80% of p120-catenin–null embryos (n = 12/15) and p120-ΔEpi (n = 8/10) mutant embryos, the domain of T expression in the posterior epiblast is expanded and split into 2 domains by T-negative cells. (F) In the remaining null and p120-ΔEpi mutants (n = 3/15 and n = 2/10), a cluster of T+ cells protrudes into the central amniotic cavity. The arrow points to pyknotic nuclei. The arrowhead points to nonspecific binding of T antibody to the visceral endoderm; compare with D using a different T antibody. (G) T expression in p120−/− p53−/− mutant embryos. (Scale bar, 50 μm.) (H–J) Sections of E7.5 embryos stained for Cleaved Caspase-3 (green). (H) Wild-type embryos do not have cell death. (J) p120−/− p53−/− double mutants have fewer apoptotic bodies than (I) p120 single mutants; speckles of Cleaved Caspase-3 were observed in p120 p53 double-mutant embryos (J). (All scale bars, 50 μm.) (K) Fraction of T-positive cells in the epiblast layer of wild-type and p120-catenin mutant embryos. Each dot represents the value from 1 section from 1 embryo; black bars represent the mean and SD. EPI, epiblast deleted; WT, wild type. In E7.5 wild-type embryos, 7.0% 0.52 of the cells in a single optical section were T+, whereas 11.4% 0.90 of the cells were T+ in E7.5 p120-ctn−/−, stained in the same experiment. In an independent experiment, 5.7 0.67% of epiblast cells were T+ in E7.5 wild-type, whereas 8.9 0.85% of cells were T+ in E7.5 p120-ΔEpi. At E7.0, 7.7% 0.63 of cells were T+ in wild type, whereas 10.9% 0.35 of cells were T+ cells in p120-ΔEpi embryos. In E7.5 p120 p53 double-mutant embryos 24 4.20 were T+ cells, whereas 7.1 0.89 were T+ in wild-type sections stained in parallel. The brackets indicate experiments stained in the same batch. (L) Quantitation of Cleaved Caspase-3 apoptotic bodies per section. Data are the mean ± SD (black bars). The mean number of Caspase-3+ apoptotic bodies detected per wild-type embryo was 0.66 0.33; the mean was 46 14 in p120−/− mutants; this number was reduced to 1.70 1.2 in p120−/− p53−/− double mutants (n = 3 for each genotype; P < 0.0053).
The size of the T-expression domain in the epiblast was expanded in both null and p120-ΔEpi E7.75 mutant embryos. To measure the size of the epiblast T domain, we counted the number of cells positive for both T and E-cadherin in the epiblast layer in a single transverse plane of a Z-stack per embryo, compared with the total DAPI+ nuclei in that transverse plane. Shortly after the initiation of gastrulation, at E7.0, p120 mutant embryos had ∼40% more T-positive cells than wild-type embryos (Fig. 3K). At E7.5, the number of T+ cells in the epiblast of both null and epiblast-deleted mutants was ∼60% greater than in wild type (Fig. 3K) and T expression was split into 2 domains, suggesting that the T+ streak domain in the mutant epiblast expanded before it split in 2.
Cell Death in the Absence of p120-Catenin Masks the Extent of Streak Expansion.
Pyknotic nuclei were detected in the mesenchymal cells at the mutant streak (E-cadherin negative, N-cadherin positive) (Fig. 3F, n = 8/10). Staining for Cleaved Caspase-3 confirmed that the number of apoptotic bodies was elevated ∼50-fold in the mutant embryos and showed that dying cells accumulated in the amniotic cavity of the mutants, suggesting that they had been apically extruded from the epiblast layer (Fig. 3 I and L). Elevated rates of cell detachment and subsequent cell death were also observed after conditional deletion of p120 in the terminal end buds from the mouse mammary gland (7) and after apical extrusion of p120 mutant cells in the pancreas (11), suggesting that p120-catenin has a general prosurvival role in epithelia.
Several apoptotic pathways act through up-regulation of expression of the tumor suppressor p53 (23, 24). No p53 expression was detected in wild-type embryos (SI Appendix, Fig. S6 E–G) or in the anterior epiblast of mutant embryos; however, p53 was detected in some cells in the posterior epiblast of E7.0 mutant embryos (SI Appendix, Fig. S6I, arrowheads). We generated double mutants to test whether the apoptosis in p120 embryos depended on p53. p120 p53 double-homozygous E7.5 embryos (either double-null mutants or embryos that lacked both genes in the epiblast) had >20-fold fewer Cleaved Caspase-3+ apoptotic bodies than p120 single mutants (0.66 0.33 apoptotic bodies per wild-type embryo; 46 14 cells in p120−/− mutants and 1.7 1.2 in p120−/− p53−/− double mutants) (n = 3 for each genotype; P < 0.0053; Fig. 3 I, J, and L).
The decrease in apoptosis in the p120 p53 double mutants correlated with an increase in the number of T+ cells. Nearly 25% of the cells in the epiblast of the double-mutant embryos expressed T, compared with ∼7% in the wild-type controls (Fig. 3 C and E). The dramatic increase of the T+ domain in the epiblast of the double mutants compared with the single mutants (Fig. 3 E–G and SI Appendix, Fig. S4 K–M) suggested that the majority of the T+ cells in the p120 single mutants undergo cell death. We also observed T+ cells detaching and floating in the amniotic cavity of p120-catenin mutant embryos (SI Appendix, Fig. S6B, arrow). Thus, in the absence of p120-catenin and p53, the size of the streak domain is expanded more than 3-fold, a phenotype that has not been reported in other mouse mutants.
p120-Catenin Is a Negative Regulator of WNT Signaling at the Primitive Streak.
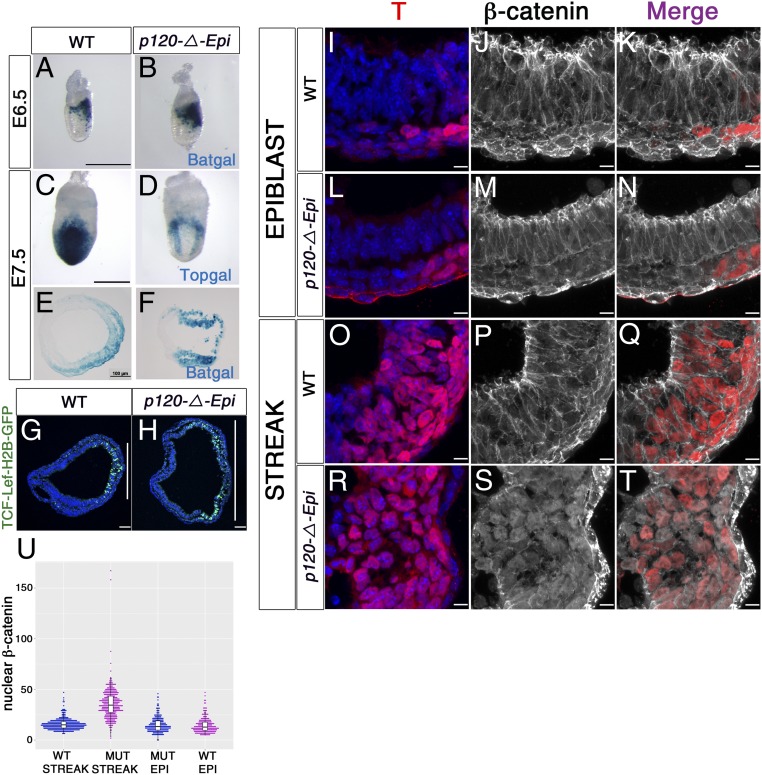
The data show that p120-catenin is a strong negative regulator of the Brachyury expression domain, and it is known that transcription of the T gene is directly activated by β-catenin, the transcriptional effector of the canonical WNT pathway (25–28). We used 3 different transgenic reporters for canonical WNT signaling to test whether WNT signaling was affected in p120-catenin mutant embryos. The Batgal WNT reporter (29) was expressed in a single domain in the posterior of both E6.5 wild-type and p120-catenin mutant embryos, but the Batgal domain was broader in the mutant (Fig. 4 A and B). Two other WNT reporters that are expressed later, at E7.5, Topgal and TCF-Lef:H2B-GFP (30, 31), were expressed in a single posterior domain in wild type and in 2 separated domains in the posterior of mutant embryos (Fig. 4 C and D). The expression of TCF-Lef:H2B-GFP, which can be imaged at single-cell resolution, encompassed nearly all of the posterior half of the embryo, split by a central reporter-negative domain (n = 3; Fig. 4 G and H). Thus, like the T-expression domain, the WNT response domain in p120 embryos was expanded at E6.5 to E7.0 and split into 2 discrete domains by E7.5.
Fig. 4.
p120-catenin restricts the domain and level of WNT response in the posterior epiblast. (A–H) WNT reporter gene expression. (A and B) β-Galactosidase staining for the Batgal WNT reporter in E6.5 (A) wild-type and (B) mutant embryos. Posterior to the right. (C and D) Topgal expression in E7.5 (C) wild-type and (D) p120-catenin–null embryos. Posterior view. (E and F) Transverse sections of E7.5 embryos stained for Batgal, (E) wild-type, and (F) p120-catenin mutants; posterior to the right. (G and H) Immunostaining for TCF-Lef:H2B-GFP-reporter expression in transverse sections of E7.5 embryos. The vertical white lines represent the extent of the GFP+ domain. (Scale bars, 50 μm.) (I–U) Nuclear β-catenin. (I–T) High-magnification views of immunostained E7.5 embryo sections, showing localization of β-catenin (white) and T (red). Upper panels correspond to the lateral epiblast of (I–K) wild type and (L–N) mutant; Lower panels show the streak of (O–Q) wild type and (R–T) mutant. Note colocalization β-catenin and T in nuclei of cells of the mutant streak. (Scale bar, 8 μm.) (U) Quantitation of fluorescence intensity (FI) of β-catenin in the nuclei of streak and epiblast cells (EPI). In wild-type embryos, the β-catenin FI in the nuclei at the streak was 15.65 5.57; in contrast, in the mutant streak, the nuclear β-catenin FI was 35.86 14.94 (P < 0.0001). The mean β-catenin FI in wild-type epiblast nuclei was 15.09 7.49, whereas in the mutant epiblast was 14.45 7.30 (P < 0.351). The black bars in the graphic represent the mean + SD.
p120-Catenin Negatively Regulates Nuclear Localization of β-Catenin in WNT-Responding Cells.
β-Catenin, the key transcriptional effector of canonical WNT signaling, is also a core protein of adherens junctions (AJs), where, like p120-catenin, it is tethered to the cytoplasmic domain of E-cadherin (32). In wild-type embryos, β-catenin was highly enriched at cell–cell junctions throughout the epiblast, including at the primitive streak (Fig. 4 I–K, O–Q and SI Appendix, Fig. S7 A–F). β-Catenin was also present at junctions in anterior and lateral epiblast cells of p120-catenin mutants (Fig. 4 L–N). The mean nuclear β-catenin fluorescence intensity (FI) in the wild-type epiblast nuclei was 15.09 7.49, which was not significantly different from the level in the mutant epiblast (14.45 7.30; P < 0.351).
In contrast, β-catenin in the streak region of p120 mutant embryos was depleted from junctions and instead could easily be seen to be enriched in nuclei, where it colocalized with the T transcription factor (Fig. 4 R–T and SI Appendix, Fig. S7 A–I). The mean FI of nuclear β-catenin adjacent to the mutant streak was more than twice that in wild-type streak cells (Fig. 4U): in wild-type embryos, the β-catenin FI in the nuclei at the streak was 15.65 5.57, whereas the nuclear β-catenin FI in the mutant streak was 35.86 14.94 (P < 0.0001). Thus, p120-catenin limits the amount of nuclear β-catenin, but only in the posterior of the embryo, the region exposed to WNT ligands (26).
p120 Mutants Fail to Complete the EMT.
The final step of the EMT is the movement of mesoderm cells away from their epithelium of origin. The epithelial cells in the WT primitive streak are a well-organized columnar E-cadherin+ epithelium, from which individual cells ingress to join the mesenchymal layer (14). In contrast, at the position of the primitive streak in E7.5 p120-catenin mutants, clusters of E-cadherin–negative, nonepithelial cells accumulated (Fig. 2 G–O). These cells expressed N-cadherin and SNAIL and therefore appeared to have mesodermal identity, although most of these cells remained at the streak region in contrast to the relatively small number of N-cad+SNAIL+ cells found in the mutant mesodermal layer (Fig. 5 A, B, D, and E).
Fig. 5.
p120-catenin promotes persistent directional mesoderm migration. (A–C) Immunostaining of E7.5 transverse embryo sections for N-cadherin (green) in (A) wild type, (B) p120-catenin, and (C) p53−/− p120−/− double mutants. (D–F) Transverse sections of E7.5 embryos stained for SNAIL. (E) p120−/− and (F) p53−/− p120−/− double-mutant embryos have more SNAIL-expressing cells (magenta) in the epiblast layer than seen in (D) wild type. (D and E) The arrowheads point to nonspecific binding of SNAIL antibody to the visceral endoderm. (G and H) Immunofluorescent staining of transverse sections of E7.5 wild-type and p120-catenin mutant embryos for KDR (green). (Scale bars, 50 μm.) N-cadherin expression in mesoderm explants of (I–K) wild-type and (L–N) p120-ΔEpi mutant embryos. (Scale bars: I and L, 80 μm; J and M, 24 μm; K and N, 8 μm.) (O) The average velocity of migration of wild-type and mutant mesoderm cell explants over a 7-h period of culture. Points are values for individual cells; bars represent the mean and SD. The mean velocity for the mutant explants was 0.48 0.02 μm/min, 70% faster than wild type (0.28 0.01 μm/min; P < 0.0001) (n = 54 cells each for wild type and mutant). (P) Comparison of the cell migration directionality (persistence) in wild-type and mutant mesoderm cell explants over a 7-h period of culture. Data are shown as the mean persistence and SD for wild-type (0.78 0.02) and mutant (0.36 0.03) mesoderm cells (P < 0.0001). A value of 1.0 is a constant direction.
Cells of the allantois and other extraembryonic mesoderm structures arise from the proximal epiblast (33); we therefore examined expression of markers of extraembryonic mesoderm in the mutants. In wild-type embryos, extraembryonic mesoderm cells migrated to the proximal region of the embryo; the extraembryonic mesoderm marker KDR was expressed in cells of the extraembryonic mesoderm and the anterior mesoderm (Fig. 5G). In contrast, in the p120-catenin mutants, most of the KDR+ cells accumulated near the streak and the adjacent posterior mesoderm (Fig. 5H and SI Appendix, Fig. S6 C and D), indicating that posterior mesoderm failed to migrate away from the streak.
To test whether the failure of the EMT reflected a defect in migration of the nascent mesodermal out of the epithelial layer, we analyzed the behavior of cells in mesoderm explants from E7.5 embryos (ref. 34; Methods). Cells in wild-type mesoderm explants were loosely connected through cell-to-cell contacts at sites enriched with membrane N-cadherin (Fig. 5 I–K). In contrast, the intensity of N-cadherin at contacts between cells was less intense in the mutant explants (Fig. 5 L–N).
Live imaging of cultured migrating mesoderm cells showed that the mutant cells migrated ∼2-fold faster than the wild-type mesoderm cells (Fig. 5O), consistent with decreased cell–cell adhesion. In addition, mutant cells had lost normal directional migration. WT cells migrated in the same direction ∼75% of the time between frames taken at every 15 min. In contrast, mutant cells changed direction between frames in the majority of cases (∼60% of the time) (P < 0.0001) (Fig. 5P, SI Appendix, Fig. S8 A and B, and Movies S1–S4).
The Split Posterior Streaks of E8.5 p120 Mutants Are Separated by Nonmigrating Mesoderm.
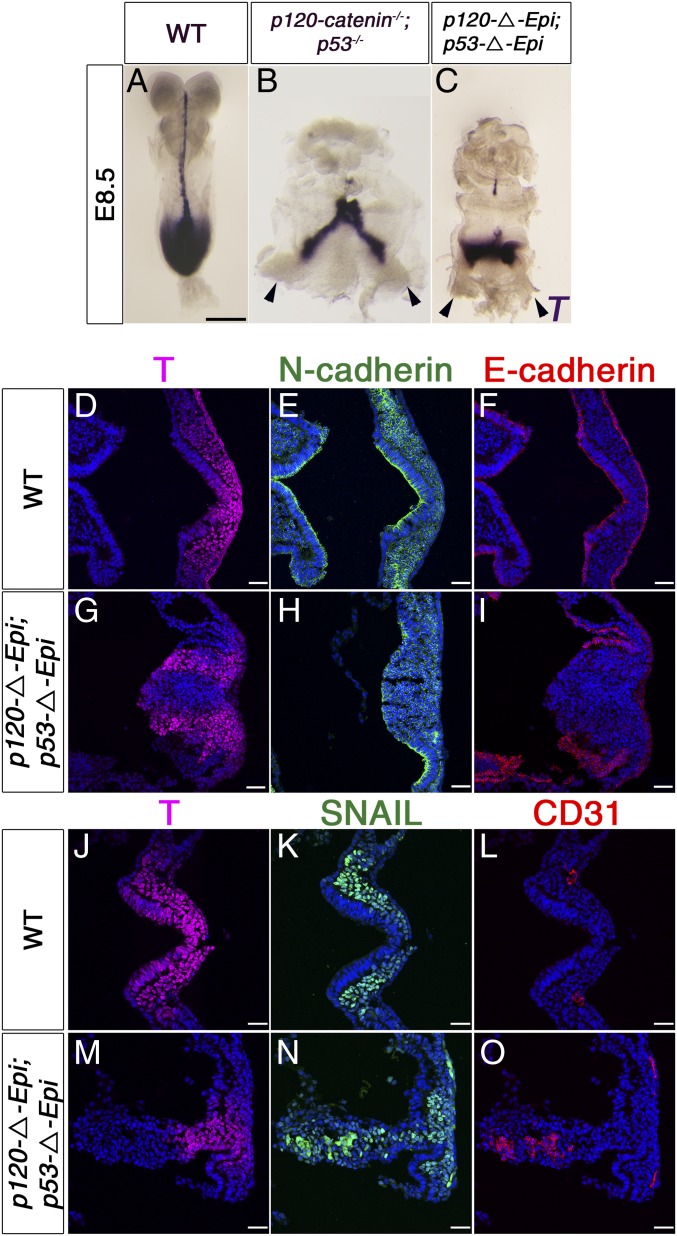
As many cells in the region of the mutant streak undergo cell death, we examined p120 mutant embryos in which cell death was blocked by the removal of p53 more closely. The overall body plan of the double mutants was the same as in the single mutants: Whole-mount in situ hybridization showed that individual E8.5 p120 p53 double-mutant embryos had 2 stripes of T expression each just anterior to an independent allantois as well as T-expressing cells connecting the independent primitive streaks (n = 5; Fig. 6 A–C). Thus, it was not the domain of cell death in the primitive streak that led to the bifurcation of the posterior axis.
Fig. 6.
Posterior axis splitting and tissue disorganization in p120 p53 double-mutant embryos. (A–C) Whole-mount in situ hybridization of E8.5 embryos for T expression. (A) T expression in the wild-type embryo. (B) The 2-T main domains seem connected by T-expressing cells, and there are 2 allantoides in p120−/−p53−/− and (C) p120-ΔEpi p53-ΔEpi mutant embryos. The arrowheads point to 2 allantoides. (Scale bar, 50 μm.) (D–O) Primitive streak of E8.5 wild-type and p120 p53 double-mutant embryos. (D and J) In wild-type, T (magenta) is expressed in the streak and in the nascent mesoderm. (E) N-cadherin (green) marks mesoderm cells and cadherin switching has started in the apical epiblast. (F) E-cadherin (red) is expressed throughout the epiblast. In the majority of p120 p53 double-mutant embryos examined at this stage (3/5), (G) T is expressed in 2 domains, (H) separated by a mass of cells expressing punctate N-cadherin and not covered by (I) E-cadherin–expressing epiblast. In the other E8.5 double mutants (M–O), the tissue in the posterior of the double mutants is disorganized, with regions expressing SNAIL (green) and CD31 (red) (N and O). (Scale bar, 50 μm.)
At E7.5, T+, SNAIL+, N-cadherin+ cells accumulated at the center of the streak region in all p120 p53 double-homozygous embryos examined (Fig. 5 C and F; n = 6), with larger numbers of cells accumulated in the streak region than in p120-catenin single mutants. In one-half (3/6) of the E7.5 double-mutant embryos, a group of SNAIL+ cells appeared to be expelled apically from the posterior epiblast, spilling into the amniotic cavity (Fig. 5F), suggesting that these cells could move, but in a random manner. In the other E7.5 double-mutant embryos, a large ball of cells protruded into the amniotic cavity; this ball of cells had a mesoderm identity, based on N-cadherin expression (Fig. 5C).
At E8.5, 2 physically separated domains of T+ mesenchymal cells were apparent in the majority of the immunostained sections of p120 p53 double-mutant embryos (n = 5; Fig. 6 G–I), corresponding to the 2 stripes of T expression seen in whole-mount embryos (Fig. 6 A–C). Between the 2 mesenchymal T+ domains, there was a large mass of mesenchymal cells that expressed N-cadherin, SNAIL, and CD31 (PECAM), a marker of extraembryonic mesoderm, indicating that these cells had a more mature mesodermal identity but had failed to move away from their site of origin (Fig. 6 D–O). We conclude that the absence of p120-catenin causes an early expansion of the primitive streak domain due to elevated WNT signaling and that the expanded domain resolves into 2 independent mesoderm-producing primitive streaks separated by differentiating nonmigratory mesoderm.
Discussion
Here, we define several essential roles of p120-catenin in the gastrulation EMT: p120 limits the activity of canonical WNT signals that promote EMT; p120 prevents EMT-associated cell death; and p120 is required for the organized delamination of cells from the epiblast to generate the mesoderm layer.
The domain of canonical WNT signaling is expanded in p120 mutant embryos as soon as it can be detected in the gastrulating embryo. The increased WNT response in p120 mutant embryos is seen at E6.5 in the expansion of the domain of expression of the WNT target genes T and Wnt3 and the increased number of cells expressing WNT reporter genes. High levels of WNT signaling are detected only in the posterior region of the embryo where WNT ligands are expressed, suggesting that the absence of p120-catenin amplifies the response to localized WNT ligands. The heightened WNT response in the p120-catenin mutants leads to high levels of nuclear β-catenin mutant near the primitive streak, the region of the embryo exposed to high levels of WNT ligands. These posterior cells have low levels of membrane-associated β-catenin, suggesting that, in the absence of p120-catenin, WNT drives relocalization of β-catenin from the membrane to nuclei.
The role of p120-catenin as a negative regulator of the nuclear localization of β-catenin seen here contrasts with previous work that suggested p120-catenin inhibits canonical WNT signaling by blocking the transcriptional repressive activity of Kaiso, a Zinc finger-BTB protein (35). While this could be a context-dependent effect, Kaiso-null mutant mice are viable and fertile, which argues against an essential role for Kaiso in canonical WNT signaling in mouse development (36). As seen here, loss of p120-catenin leads to elevated activated β-catenin in some tumors: For example, mosaic deletion of p120 in the intestine leads to clusters of cells with elevated levels of nonmembrane associated β-catenin, which promotes the formation of adenomas (37). Thus, we propose that p120-catenin has a general role as a negative regulator of WNT signaling in epithelia.
In the standard view of canonical WNT signaling, the β-catenin that is activated in response to WNT ligand comes from a cytoplasmic pool regulated by the APC-containing destruction complex, and there is no exchange between the pools of β-catenin regulated by the destruction complex and AJs (38). In contrast, our data argue that p120-catenin limits the amount of β-catenin that can be released from the AJ complex to the nucleus in response to WNT signals. Several studies suggest that WNT signaling at the embryonic primitive streak may depend on β-catenin derived from dissolving AJs, but this topic has been controversial (39). One set of experiments, based on differentiating embryonic stem cells, suggested that p120-catenin from AJs is a positive regulator of signaling and helps activate WNT target gene expression (40). In contrast, work in the mouse embryo suggested that the junctional cadherin complex acts as a negative regulator of signaling by sequestering β-catenin until it is released from AJ by WNT ligand in a process regulated by fibroblast growth factor signaling (41).
The WNT-dependent removal of E-cadherin and β-catenin from junctions during the EMT is likely to be a complex process that requires posttranscriptional mechanisms that depend on known and unknown proteins. For example, the FERM domain protein LULU/EPB41l5 is required for down-regulation of E-cadherin at the streak (42), and LULU/EPB41l5 binds specifically to p120-catenin (43). Similarly, the apical protein CRB2 is also required for down-regulation of E-cadherin at the streak (14).
After the initiation of a broad primitive streak domain in p120-catenin mutants, the streak domain is further expanded, but it becomes physically split into 2 domains by a central region with mesoderm identity to create 2 distinct primitive streaks. In the wild-type embryo, cells ingress into the mesoderm layer from the primitive streak epithelium as individuals in an apparently stochastic pattern, while retaining the integrity of the epiblast epithelium (14). In contrast, cells in the central region of the p120 mutant primitive streak region become mesenchymal but fail to move out of their position in the epithelial layer. These central-streak N-cad+SNA+KDR+ cells have a mesoderm identity but do not migrate efficiently into the mesoderm layer. This cluster of abnormal mesoderm cells appears to physically block communication between cells of the 2 epiblast T+ domains; as a result, each T+ domain acts as an autonomous primitive streak to produce mesoderm in 2 separated tails.
The results suggest that p120-catenin is required for nascent mesoderm cells to migrate out of the epithelial layer in response to directional signals. p120-catenin can regulate the actin cytoskeleton through inhibition of the small GTPase RhoA (44). However, we did not observe an increase in stress fibers, a hallmark of increased RhoA activity (SI Appendix, Fig. S8 C–H). The loss of directional migration of cells from the primitive streak in p120 mutants is consistent with findings showing that knockdown of N-cadherin in cultured mammalian cells and in zebrafish embryos leads to more rapid and less directional migration (45, 46). However, mouse N-cadherin–null mutants make mesoderm and somites (47), indicating that p120-catenin plays roles beyond N-cadherin stabilization in the mesoderm, perhaps through stabilization of other classical cadherins such as CDH3 (P-cadherin) (48). Alternatively, elevated nuclear β-catenin in nascent mesoderm cells could prevent them from migrating away from their origin, as has been seen in Xenopus neural crest cells (49).
Loss or altered expression of p120-catenin has been observed frequently in human tumors of the colon, prostate, lung, stomach, breast, and pancreas (9). The relationship between EMT and tumor progression is provocative and controversial (50, 51). Nevertheless, the combination of cellular phenotypes seen in cells undergoing the gastrulation EMT in p120 p53 mutant embryos are strikingly similar to the characteristics of metastatic tumor cells. We see that the absence of p120-catenin amplifies responses to WNT ligands, and it has been shown that WNT signaling can act in a positive-feedback loop to promote further EMT, as well as promote transcription of genes that drive proliferation (52, 53). p120-catenin mutant cells that have just completed the gastrulation EMT have increased motility but lose directionality, and most of these cells die. When p53 is deleted in p120 mutant cells, embryonic cells that have completed the EMT survive and migrate rapidly without proper direction, a hallmark of metastatic cancer.
Methods
Mouse Strains.
The p120-catenin conditional allele was a gift from Elaine Fuchs (Rockefeller University, New York, NY) and Albert Reynolds (Vanderbilt University, Nashville, TN). To generate the null allele, the conditional allele of p120-catenin was crossed to CAG-Cre transgenic animals (The Jackson Laboratory). Mice expressing the p120-catenin floxed allele were crossed with Sox2-Cre–expressing mice (19) to generate p120-ΔEpi embryos. Null and conditional p53 alleles were a gift from Scott Lowe (Memorial Sloan Kettering Cancer Center [MSKCC], New York, NY). Mice that were 8 to 16 weeks old were used to generate E6.5 to E8.5 embryos. Analysis of the p120 mutant phenotype was performed in the FVB genetic background, with the exception of p120 p53 double-mutant mice, which were maintained in a mixed background. The TCF-LEF-H2B-GFP strain was a gift from Anna-Katarina Hadjantonakis (31) (MSKCC). For imaging of mesoderm explants, mT/mG mice (54) were crossed to CAG-Cre mice to generate mice expressing GFP at the membrane. Mice were housed and bred under standard conditions in accordance with Institutional Animal Care and Use Committee (IACUC) guidelines. The MSKCC IACUC approved the experiments. Primers for genotyping the p120-catenin and p53 alleles have been published (6, 55).
ENU Allele Isolation, Sequencing, Genotyping.
The p120-catenin point mutation (L691X) was generated by ENU mutagenesis of C57BL/6J males and identified based on its embryonic phenotype, as described previously (16). The L691X allele has a single T-to-A transversion in the 2555 nucleotide position of p120-catenin coding sequence, leading to a nonsense mutation (in the amino acid position 691 changes Leu to Stop). The L691X mutation created a SpeI restriction site used for genotyping.
Immunostaining.
Immunofluorescent staining of embryo sections and whole-mount embryos was performed according to standard protocols. Briefly, embryos were fixed for 120 min in 4% paraformaldehyde (PFA) on ice followed by 4 washes with PBS, 10 min each. For sectioning, embryos were incubated in 30% sucrose in PBS on ice until they sank. Embryos were embedded in OCT (Tissue-Tek), frozen, and cryosectioned at 10 to 12 μm. Immunostaining was performed in blocking buffer: PBS containing 4% heat-inactivated donkey serum (Gemini; Bio-products) and 0.1% Triton X-100. The following primary antibodies were incubated overnight in blocking buffer: p120-catenin (1:300; Sigma-Aldrich), β-catenin (1:50; EMD Millipore), E-cadherin (1:300; Sigma-Aldrich), N-cadherin (1:300; Santa Cruz), N-cadherin (1:300; Cell Signaling Technology), SNAIL (1:100; kindly donated by Antonio García de Herreros, Institut Hospital del Mar d’Investigacions Mèdiques, Barcelona, Spain), Cleaved Caspase-3 (1:400; Promega), T (1:300; EMD Millipore), p53 (1:400; Cell Signaling), T (1:400; Cell Signaling), KDR (1:300; BD Pharmingen), CD31 (1:300; Dianova), and FOXA2 (1:500; Abcam). Fluorescent secondary antibodies (Invitrogen) were diluted at 1:400 in blocking buffer and incubated for 1 h at room temperature (RT). The mouse embryo sections were mounted in ProLong Gold (Thermo Fisher) and imaged using a Leica-upright SP5 laser point-scanning microscope. Confocal images were reconstructed and analyzed by using Volocity software package (PerkinElmer).
In Situ Hybridization.
In situ hybridization was performed according to protocols previously described (56). Briefly, the embryos were fixed overnight in 4% PFA. The next day, the embryos were washed in PBS with 0.1% Tween and dehydrated in graduated methanol series. The following day, the embryos were hydrated in graduated methanol series, incubated in 10 μg/mL PK/PBS solution for 3 to 7 min, and hybridized in hybridization solution plus RNA probe at 70 °C overnight. A series of washes in 2XSSC and MAB solutions (100 mM maleic acid and 150 mM NaCl in PBS) was performed the next day. The embryos were incubated in 1:1,000 anti-DIG antibody (Roche) in 10% goat serum/0.1% Tween-20 in PBS overnight and then washed extensively in PBT (0.1% BSA; 0.1% Tween 20 in PBS). The embryos were incubated in BM purple solution (Roche) until the development of purple color.
Batgal and Topgal Staining.
For β-galactosidase staining, we used previously described protocols (57). Briefly, the embryos were dissected in cold 1% BSA-PBS and fixed in fixing solution (1% PFA, 0.2% glutaraldehyde, 0.02% Nonidet P-40 in PBS) for 10 min at RT. The embryos were washed in cold PBS 3 times and incubated in staining solution (2 mM MgCl2, 5 mM potassium ferricyanide, 1 mg/mL X-gal in PBS) at 37 °C until the development of the blue color.
Mesoderm Explants.
Mesoderm explants were performed as previously described (57). Briefly, E7.5 membrane-GFP–expressing embryos (54) were dissected in cold DMEM/F12 media; after dissection, embryos were transferred to a 2.5% pancreatin/0.5% trypsin in Tyrode Ringer’s saline solution on ice for 10 min. After digestion, the embryos were washed 3 times in culture media (DMEM/F12; 10% FBS and 1% penicillin–streptomycin). Visceral endoderm and epiblast were removed using sharp forceps, and then the mesodermal wings were allowed to attach to cover-glass bottom chambers coated with Fibronectin (50 μg/mL PBS) and cultured for 24 h at 37 °C with 5% of CO2 before live imaging or immunostaining. Live imaging was performed for 7 h at 37 °C in a culture chamber with 5% CO2 using an inverted ZEN Imaging system (Zeiss).
The velocity of mesoderm cell movement was determined using the Cell Tracking macro from Fiji. This macro generates a text document with the distance and velocity per minute for each cell. We tracked 13 different mesoderm cells randomly chosen at the leading edge of 4 wild-type and 4 p120-catenin mutant explants every 15 min for 7 h (54 cells in total for each genotype). The data obtained from the Manual Cell tracking was used in the Chemotaxis plug-in from Fiji to generate the mesoderm cell directionality values and plots. Persistence was calculated by comparing the Euclidean distance (length of straight line between cell start and end point) to the accumulated distance. We obtained the mean and SD of velocity and persistence/directionality using Prism 7.
Quantitation of Nuclear β-Catenin and Cadherin FI.
Quantitation was performed on high-magnification images of cryosections stained for β-catenin and T (high-magnification images similar to the ones in Fig. 4 I–T). We used the mean gray value tool from Fiji to obtain the FI values from the nuclei. For cells at the streak, we used 1 Z-confocal plane chosen in the middle of the stack with T-positive cells that we overlapped to the same Z-plane on the β-catenin immunostained image; epiblast cells were defined as T-negative cells. We used DAPI to confirm the specific staining in the nuclei and to delimit the signal by using the Fiji Binary tool in this channel that we copy and overlap to the β-catenin signal. The mean FI values from streak and epiblast (from either wild-type or p120-mutant nuclei) were used for the statistical analysis. Quantitation of Cadherin intensity was performed on high-magnification images (63×) of cryosections stained for E-cadherin and N-cadherin (high-magnification images similar to the ones in Fig. 2 A–D). We used the Mean gray value tool from Fiji to obtain the FI values from either the epiblast or mesoderm. We used 1 Z-confocal plane chosen in the middle of the stack to measure the FI for 31 epiblast cells and 23 mesoderm cells from both the wild-type and the mutant embryos.
Quantitation of Cleaved Caspase-3 and T-Positive Cells.
We used Fiji to manually count the number of either T-positive cell nuclei or Cleaved Caspase-3 apoptotic bodies in a single Z-plane per embryo from a stack of the wild-type and the mutant sections. The percentage of T-positive cells was determined from the number of T-positive cells located within the epiblast (T+ and E-cadherin+) in a single Z-plane (SI Appendix, Fig. S4 E–M). We selected a section approximately from the same proximo-distal level, which is at about 30% of the distance from the extraembryonic ectoderm to the node, usually corresponding with the widest section of the embryo. For Cleaved Caspase-3+ cells, only cell-sized signals were counted; speckles were not considered as a positive signal. The mutant and the wild-type embryo sections were stained at the same time.
Western Blot Analysis.
E7.5 and E8.5 wild-type and p120-catenin mutant embryos were dissected in cold PBS and lysed by pipetting up and down in the lysis buffer (1% Nonidet P-40, 150 mM NaCl, 50 mM Tris⋅HCl, phosphatase inhibitor mixture I and II [Calbiochem]; and one tablet of Minicomplete [Roche] per 10 mL). The following antibodies were used at a 1:1,000 dilution in 5% BSA in TBST: E-cadherin (Cell Signaling), N-cadherin (Cell Signaling), and GAPDH (Santa Cruz). Secondary antibodies for each specific antigen were incubated in 5% BSA in TBST for 1 h at RT with rocking. We measured the intensity of the protein bands by using the Gel Analyzer macro from Fiji. The relative E-cadherin and N-cadherin values were calculated by dividing the area obtained from Gel Analyzer for the cadherin by the total area of loading control GAPDH in the wild-type and the mutant embryos. Averages represent the arithmetic mean of 3 to 4 different experiments with different embryos.
Statistics and Graphs.
Embryo images n > 3 for all of the experiments. We used Prism 7 to perform 2-tailed Student’s t tests and one-way ANOVA to evaluate the significance of all measurements. Graphs were plotted using the geom_dotplot function of the ggplot2 R library (https://ggplot2.tidyverse.org/reference/geom_dotplot.html). Each dot represents 1 observation. Dot plots convey information about a distribution in a similar way to a box plot or histogram but have the advantage that clusters and outliers in the data are more easily visualized.
Supplementary Material
Acknowledgments
We thank Elaine Fuchs (Rockefeller University) and Albert Reynolds (Vanderbilt University) for the p120-catenin conditional mice. We are grateful to Ed Espinoza for identification of the p120-L691X mutation in p120-catenin. We thank the Mouse Genetics and Molecular Cytology core facilities at MSKCC for their support. The SNAIL antibody was a gift from Antonio García de Herreros (Institut Hospital del Mar d’Investigacions Mèdiques, Barcelona, Spain). We thank members of the K.V.A. laboratory and Eric Brooks for their comments on the manuscript. The work was supported by NIH Grant R01/R37 HD03455 (to K.V.A.); MSKCC Cancer Center Support Grant P30 CA008748; and grants from the Pew Latin American Fellows Program and Consejo Nacional de Ciencia y Tecnología, Mexico (to R.H.-M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1902843116/-/DCSupplemental.
References
- 1.Ishiyama N., et al. , Dynamic and static interactions between p120 catenin and E-cadherin regulate the stability of cell-cell adhesion. Cell 141, 117–128 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Nanes B. A., et al. , p120-catenin binding masks an endocytic signal conserved in classical cadherins. J. Cell Biol. 199, 365–380 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Myster S. H., Cavallo R., Anderson C. T., Fox D. T., Peifer M., Drosophila p120catenin plays a supporting role in cell adhesion but is not an essential adherens junction component. J. Cell Biol. 160, 433–449 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pettitt J., Cox E. A., Broadbent I. D., Flett A., Hardin J., The Caenorhabditis elegans p120 catenin homologue, JAC-1, modulates cadherin-catenin function during epidermal morphogenesis. J. Cell Biol. 162, 15–22 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bulgakova N. A., Brown N. H., Drosophila p120-catenin is crucial for endocytosis of the dynamic E-cadherin-Bazooka complex. J. Cell Sci. 129, 477–482 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis M. A., Reynolds A. B., Blocked acinar development, E-cadherin reduction, and intraepithelial neoplasia upon ablation of p120-catenin in the mouse salivary gland. Dev. Cell 10, 21–31 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Kurley S. J., et al. , p120-catenin is essential for terminal end bud function and mammary morphogenesis. Development 139, 1754–1764 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marciano D. K., et al. , p120 catenin is required for normal renal tubulogenesis and glomerulogenesis. Development 138, 2099–2109 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thoreson M. A., Reynolds A. B., Altered expression of the catenin p120 in human cancer: Implications for tumor progression. Differentiation 70, 583–589 (2002). [DOI] [PubMed] [Google Scholar]
- 10.Mann K. M., et al. ; Australian Pancreatic Cancer Genome Initiative , Sleeping beauty mutagenesis reveals cooperating mutations and pathways in pancreatic adenocarcinoma. Proc. Natl. Acad. Sci. U.S.A. 109, 5934–5941 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hendley A. M., et al. , p120 catenin suppresses basal epithelial cell extrusion in invasive pancreatic neoplasia. Cancer Res. 76, 3351–3363 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ben-Haim N., et al. , The nodal precursor acting via activin receptors induces mesoderm by maintaining a source of its convertases and BMP4. Dev. Cell 11, 313–323 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Ferrer-Vaquer A., Viotti M., Hadjantonakis A. K., Transitions between epithelial and mesenchymal states and the morphogenesis of the early mouse embryo. Cell Adhes. Migr. 4, 447–457 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramkumar N., et al. , Crumbs2 promotes cell ingression during the epithelial-to-mesenchymal transition at gastrulation. Nat. Cell Biol. 18, 1281–1291 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Acloque H., Adams M. S., Fishwick K., Bronner-Fraser M., Nieto M. A., Epithelial-mesenchymal transitions: The importance of changing cell state in development and disease. J. Clin. Invest. 119, 1438–1449 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.García-García M. J., et al. , Analysis of mouse embryonic patterning and morphogenesis by forward genetics. Proc. Natl. Acad. Sci. U.S.A. 102, 5913–5919 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Omelchenko T., et al. , β-Pix directs collective migration of anterior visceral endoderm cells in the early mouse embryo. Genes Dev. 28, 2764–2777 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Srinivas S., Rodriguez T., Clements M., Smith J. C., Beddington R. S., Active cell migration drives the unilateral movements of the anterior visceral endoderm. Development 131, 1157–1164 (2004). [DOI] [PubMed] [Google Scholar]
- 19.Hayashi S., Lewis P., Pevny L., McMahon A. P., Efficient gene modulation in mouse epiblast using a Sox2Cre transgenic mouse strain. Mech. Dev. 119 (suppl. 1), S97–S101 (2002). [DOI] [PubMed] [Google Scholar]
- 20.Ireton R. C., et al. , A novel role for p120 catenin in E-cadherin function. J. Cell Biol. 159, 465–476 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davis M. A., Ireton R. C., Reynolds A. B., A core function for p120-catenin in cadherin turnover. J. Cell Biol. 163, 525–534 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wheelock M. J., Shintani Y., Maeda M., Fukumoto Y., Johnson K. R., Cadherin switching. J. Cell Sci. 121, 727–735 (2008). [DOI] [PubMed] [Google Scholar]
- 23.Bieging K. T., Attardi L. D., Deconstructing p53 transcriptional networks in tumor suppression. Trends Cell Biol. 22, 97–106 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghosh A., Chen T. C., Kapila Y. L., Anoikis triggers Mdm2-dependent p53 degradation. Mol. Cell. Biochem. 343, 201–209 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu P., et al. , Requirement for Wnt3 in vertebrate axis formation. Nat. Genet. 22, 361–365 (1999). [DOI] [PubMed] [Google Scholar]
- 26.Tortelote G. G., et al. , Wnt3 function in the epiblast is required for the maintenance but not the initiation of gastrulation in mice. Dev. Biol. 374, 164–173 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arnold S. J., et al. , Brachyury is a target gene of the Wnt/beta-catenin signaling pathway. Mech. Dev. 91, 249–258 (2000). [DOI] [PubMed] [Google Scholar]
- 28.Yamaguchi T. P., Takada S., Yoshikawa Y., Wu N., McMahon A. P., T (Brachyury) is a direct target of Wnt3a during paraxial mesoderm specification. Genes Dev. 13, 3185–3190 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maretto S., et al. , Mapping Wnt/beta-catenin signaling during mouse development and in colorectal tumors. Proc. Natl. Acad. Sci. U.S.A. 100, 3299–3304 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DasGupta R., Fuchs E., Multiple roles for activated LEF/TCF transcription complexes during hair follicle development and differentiation. Development 126, 4557–4568 (1999). [DOI] [PubMed] [Google Scholar]
- 31.Ferrer-Vaquer A., et al. , A sensitive and bright single-cell resolution live imaging reporter of Wnt/ß-catenin signaling in the mouse. BMC Dev. Biol. 10, 121 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nelson W. J., Dickinson D. J., Weis W. I., Roles of cadherins and catenins in cell-cell adhesion and epithelial cell polarity. Prog. Mol. Biol. Transl. Sci. 116, 3–23 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Lawson K. A., Meneses J. J., Pedersen R. A., Cell fate and cell lineage in the endoderm of the presomite mouse embryo, studied with an intracellular tracer. Dev. Biol. 115, 325–339 (1986). [DOI] [PubMed] [Google Scholar]
- 34.Burdsal C. A., Damsky C. H., Pedersen R. A., The role of E-cadherin and integrins in mesoderm differentiation and migration at the mammalian primitive streak. Development 118, 829–844 (1993). [DOI] [PubMed] [Google Scholar]
- 35.Park J. I., et al. , Kaiso/p120-catenin and TCF/beta-catenin complexes coordinately regulate canonical Wnt gene targets. Dev. Cell 8, 843–854 (2005). Erratum in: Dev. Cell.9, 305 (2005). [DOI] [PubMed] [Google Scholar]
- 36.Prokhortchouk A., et al. , Kaiso-deficient mice show resistance to intestinal cancer. Mol. Cell. Biol. 26, 199–208 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smalley-Freed W. G., et al. , Adenoma formation following limited ablation of p120-catenin in the mouse intestine. PLoS One 6, e19880 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nusse R., Clevers H., Wnt/β-catenin signaling, disease, and emerging therapeutic modalities. Cell 169, 985–999 (2017). [DOI] [PubMed] [Google Scholar]
- 39.McEwen A. E., Escobar D. E., Gottardi C. J., Signaling from the adherens junction. Subcell. Biochem. 60, 171–196 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Przybyla L., Lakins J. N., Weaver V. M., Tissue mechanics orchestrate Wnt-dependent human embryonic stem cell differentiation. Cell Stem Cell 19, 462–475 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ciruna B., Rossant J., FGF signaling regulates mesoderm cell fate specification and morphogenetic movement at the primitive streak. Dev. Cell 1, 37–49 (2001). [DOI] [PubMed] [Google Scholar]
- 42.Lee J. D., Silva-Gagliardi N. F., Tepass U., McGlade C. J., Anderson K. V., The FERM protein Epb4.1l5 is required for organization of the neural plate and for the epithelial-mesenchymal transition at the primitive streak of the mouse embryo. Development 134, 2007–2016 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Hirano M., Hashimoto S., Yonemura S., Sabe H., Aizawa S., EPB41L5 functions to post-transcriptionally regulate cadherin and integrin during epithelial-mesenchymal transition. J. Cell Biol. 182, 1217–1230 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Anastasiadis P. Z., p120-ctn: A nexus for contextual signaling via Rho GTPases. Biochim. Biophys. Acta 1773, 34–46 (2007). [DOI] [PubMed] [Google Scholar]
- 45.Camand E., Peglion F., Osmani N., Sanson M., Etienne-Manneville S., N-cadherin expression level modulates integrin-mediated polarity and strongly impacts on the speed and directionality of glial cell migration. J. Cell Sci. 125, 844–857 (2012). [DOI] [PubMed] [Google Scholar]
- 46.Warga R. M., Kane D. A., A role for N-cadherin in mesodermal morphogenesis during gastrulation. Dev. Biol. 310, 211–225 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Radice G. L., et al. , Developmental defects in mouse embryos lacking N-cadherin. Dev. Biol. 181, 64–78 (1997). [DOI] [PubMed] [Google Scholar]
- 48.Saito M., Tucker D. K., Kohlhorst D., Niessen C. M., Kowalczyk A. P., Classical and desmosomal cadherins at a glance. J. Cell Sci. 125, 2547–2552 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maj E., et al. , Controlled levels of canonical Wnt signaling are required for neural crest migration. Dev. Biol. 417, 77–90 (2016). [DOI] [PubMed] [Google Scholar]
- 50.Li W., Kang Y., Probing the fifty shades of EMT in metastasis. Trends Cancer 2, 65–67 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ye X., Weinberg R. A., Epithelial-mesenchymal plasticity: A central regulator of cancer progression. Trends Cell Biol. 25, 675–686 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tetsu O., McCormick F., Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature 398, 422–426 (1999). [DOI] [PubMed] [Google Scholar]
- 53.He T. C., et al. , Identification of c-MYC as a target of the APC pathway. Science 281, 1509–1512 (1998). [DOI] [PubMed] [Google Scholar]
- 54.Muzumdar M. D., Tasic B., Miyamichi K., Li L., Luo L., A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605 (2007). [DOI] [PubMed] [Google Scholar]
- 55.Jacks T., et al. , Tumor spectrum analysis in p53-mutant mice. Curr. Biol. 4, 1–7 (1994). [DOI] [PubMed] [Google Scholar]
- 56.Eggenschwiler J. T., Anderson K. V., Dorsal and lateral fates in the mouse neural tube require the cell-autonomous activity of the open brain gene. Dev. Biol. 227, 648–660 (2000). [DOI] [PubMed] [Google Scholar]
- 57.Behringer R. G. M., Vintersten K., Nagy A., Manipulating the Mouse Embryo: A Laboratory Manual (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, ed. 3, 2003). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.