Significance
We have previously demonstrated an increased autoantibody reactivity to Anoctamin 2 (ANO2), an ion channel expressed in the central nervous system (CNS), in multiple sclerosis (MS). We now show that ANO2 antibodies recognize a fragment of Epstein-Barr virus (EBV) nuclear antigen 1, thereby constituting an example of molecular mimicry. In this way, the immune response toward EBV may take part in and promote CNS inflammation, likely through T cells reactive with the same protein. In our very large case-control cohort, we demonstrate that the presence of ANO2 reactivity associates with a high MS risk, in particular together with HLA risk variants and high EBNA1 antibody titers, which we consider a strong argument for its relevance in MS ethiopathogenesis.
Keywords: Anoctamin 2, ANO2, multiple sclerosis, molecular mimicry, Epstein-Barr virus
Abstract
Multiple sclerosis (MS) is a chronic inflammatory, likely autoimmune disease of the central nervous system with a combination of genetic and environmental risk factors, among which Epstein-Barr virus (EBV) infection is a strong suspect. We have previously identified increased autoantibody levels toward the chloride-channel protein Anoctamin 2 (ANO2) in MS. Here, IgG antibody reactivity toward ANO2 and EBV nuclear antigen 1 (EBNA1) was measured using bead-based multiplex serology in plasma samples from 8,746 MS cases and 7,228 controls. We detected increased anti-ANO2 antibody levels in MS (P = 3.5 × 10−36) with 14.6% of cases and 7.8% of controls being ANO2 seropositive (odds ratio [OR] = 1.6; 95% confidence intervals [95%CI]: 1.5 to 1.8). The MS risk increase in ANO2-seropositive individuals was dramatic when also exposed to 3 known risk factors for MS: HLA-DRB1*15:01 carriage, absence of HLA-A*02:01, and high anti-EBNA1 antibody levels (OR = 24.9; 95%CI: 17.9 to 34.8). Reciprocal blocking experiments with ANO2 and EBNA1 peptides demonstrated antibody cross-reactivity, mapping to ANO2 [aa 140 to 149] and EBNA1 [aa 431 to 440]. HLA gene region was associated with anti-ANO2 antibody levels and HLA-DRB1*04:01 haplotype was negatively associated with ANO2 seropositivity (OR = 0.6; 95%CI: 0.5 to 0.7). Anti-ANO2 antibody levels were not increased in patients from 3 other inflammatory disease cohorts. The HLA influence and the fact that specific IgG production usually needs T cell help provides indirect evidence for a T cell ANO2 autoreactivity in MS. We propose a hypothesis where immune reactivity toward EBNA1 through molecular mimicry with ANO2 contributes to the etiopathogenesis of MS.
Multiple sclerosis (MS) is a chronic inflammatory disease of the central nervous system (CNS) characterized by damage to myelin and neurons/axons (1–3) often with onset during young adulthood. Etiology involves both genetic and environmental risk factors and several of these have been shown to jointly and interactively associate with increased risk for disease (4, 5). The strongest genetic association is with the HLA gene region on chromosome 6p21, which harbors a series of class II risk alleles (e.g., DRB1*15:01), as well as class I alleles (e.g., A*02:01) that have been found to affect the risk of MS (2, 6, 7). Among various lifestyle/environmental factors thought to affect the risk of MS, Epstein-Barr virus (EBV) infection is a strong candidate.
For the present study, it is of particular relevance that a combination of DRB1*15:01 carriage and high levels of Epstein-Barr virus nuclear antigen 1 (EBNA1) antibodies, primarily directed toward 2 EBNA1 peptide fragments [aa 385 to 420 and aa 402 to 502], increase the risk of developing MS 10-fold (8, 9). Since more than 95% of healthy individuals show an immune response to EBV, it cannot be the sole cause of MS. However, it could be a prerequisite for the disease and interact with other risk factors. The mechanisms are far from clear. One hypothesis is molecular mimicry (10). There are descriptions of T cell responses primarily against EBNA1 that cross-react with CNS/myelin components (11), but the mere existence of these does not inform us about their etiopathogenetic role. Well-known features of MS, such as the association with HLA class II alleles (6), similarly demyelinating disease in the CNS of antigen-induced rodent models (12), reduced disease activity with immunomodulatory treatments (13), and even increased numbers of T cells producing proinflammatory cytokines in response to CNS antigens (14, 15) strongly support, but do not prove, a role of an autoimmune response to self-antigens in the CNS. Defining reliable MS-specific autoantigens has proven difficult, which may partly be explained by epitope spreading (16) and the lack of validated assays for CNS antigen-specific T cells (17). It has been notoriously difficult to replicate findings of suggested autoantibodies in MS, despite the fact that demyelinating antibodies with unknown specificity are present (18). Nevertheless, the identification of MS-specific antigenic targets is essential for understanding MS pathogenesis.
We have previously identified increased autoantibody reactivity against Anoctamin 2 (ANO2) in an antibody screening of potential MS autoantigens with protein fragments representing ∼38% of all human proteins (19). This finding was later replicated where anti-ANO2 antibody levels were 5.3-fold higher in MS cases than in controls (20). ANO2 is a Ca2+ activated chloride channel important in, e.g., transepithelial ion transport, smooth muscle contraction, olfaction, phototransduction, nociception, and control of neuronal excitability (21). We have previously shown that neurons and glial cells from normal hippocampal and cortical regions express ANO2 and a clear increase in ANO2 staining intensity was detected near and inside MS plaques (20).
In the current study, we have analyzed a large MS case-control cohort, to replicate and further evaluate anti-ANO2 antibody reactivity in MS. An observed interaction between anti-EBNA1 and anti-ANO2 antibody reactivity in the risk for MS prompted us to investigate the potential role of molecular mimicry. We found a sequence similarity between EBNA1 and ANO2, which overlaps with the defined minimal epitope of ANO2 and with one of the previously known EBNA1 peptide fragments associated with MS risk, and demonstrate cross-reactivity between the fragments. We show a genome-wide association of the HLA gene region, specifically the DRB1*04:01 haplotype, with anti-ANO2 antibody levels. We detect anti-ANO2 antibody reactivity at similar levels in controls as in 3 other inflammatory diseases.
Results
Anti-ANO2 IgG Reactivity in Multiple Sclerosis.
In support of our previous findings, we detected increased anti-ANO2 antibody reactivity in MS compared with controls (P = 3.5 × 10−36) and replicated our previous results in a validation cohort (P = 2.3 × 10−22; Table 1 and SI Appendix, Table S1). In a multivariate analysis, anti-EBNA1 antibody levels had the strongest influence on anti-ANO2 antibody levels (P = 7.1 × 10−95), followed by study type (P = 1.1 × 10−43), MS status (P = 7.7 × 10−28), and age (P = 4.6 × 10−21). A signal intensity threshold, set at the maximum change in association with MS (continuous association curve, CAC; SI Appendix, Material S5), resulted in 14.6% in cases and 7.8% in controls being positive (OR for MS = 1.6; 95%CI: 1.5 to 1.8; P = 3.5 × 10−19). The difference in proportion of ANO2 seropositivity in MS cases compared with controls was significant at various cutoffs (SI Appendix, Table S2). Anti-ANO2 antibodies were elevated in all stages of MS in comparison with controls. There was a higher proportion of ANO2 seropositivity among relapsing remitting (15.6%) compared with progressive cases (13.2% in secondary progressive and 11.7% in primary progressive); however, the effect was due to age confounding.
Table 1.
Association of anti-ANO2 antibody levels with disease
| Sample cohort | Cases (n) | Controls (n) | Beta | P | P* |
| Initial | 1,040 | 1,058 | 428 | 5.0 × 10−13 | 6.9 × 10−21 |
| Validation | 7,603 | 6,170 | 182 | 7.1 × 10−12 | 2.3 × 10−22 |
| Whole | 8,746 | 7,228 | 213 | 4.2 × 10−27 | 3.5 × 10−36 |
| Pre-MS | 476 | 478 | −80 | 0.42 | 0.23 |
| RA | 986 | 689 | −2 | 0.95 | 0.38 |
| IIM | 219 | 306** | 4 | 0.79 | 0.81 |
| SLE | 349 | 306** | −2 | 0.85 | 0.36 |
Association analyses were adjusted for age, sex, and EBNA1 status. Analyses were also adjusted for study type for the validation and whole cohorts. *Log10 transformed signal intensities. **Same control set (selected to match SLE cases) were used for both IIM and SLE cohorts. MS, multiple sclerosis; RA, rheumatoid arthritis; IIM, idiopathic inflammatory myopathy; SLE, systemic lupus erythematosus.
ANO2 Seropositivity in Relation to Other Multiple Sclerosis Risk Factors.
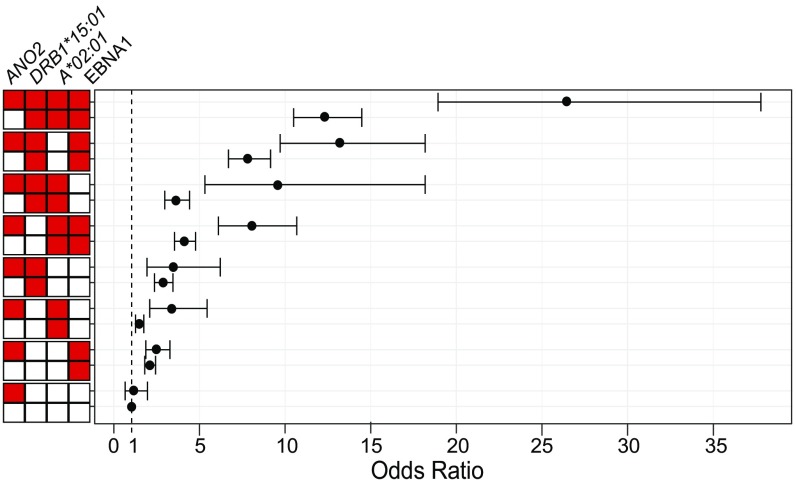
In individuals exposed to 3 other known risk factors for MS: DRB1*15:01 carriage, absence of A*02:01, and EBNA1-high (IgG signal intensity above median in controls), the risk for developing MS was higher in ANO2-seropositive subjects. Setting the group with none of the risk factors as reference, the risk for MS was significantly higher in the group with all 4 risk factors (OR = 26.4; 95%CI: 18.9 to 37.8) compared with the group carrying all risk factors except ANO2 seropositivity (OR = 12.3; 95%CI: 10.5 to 14.5). We also detected a significant risk increase among ANO2-seropositive individuals carrying 2 other risk factors compared with those carrying the same risk factors except ANO2 seropositivity (Fig. 1 and SI Appendix, Table S3). We detected interaction on the additive scale between ANO2 seropositivity and carriage of DRB1*15:01 (AP = 0.37; 95%CI: 0.24 to 0.50; P = 1.6 × 10−8), and between ANO2 seropositivity and EBNA1-high (AP = 0.24; 95%CI: 0.10 to 0.37; P = 7.0 × 10−4) as we reported previously (20). We now also detected interaction between ANO2 seropositivity and absence of A*02:01 in the risk for MS (AP = 0.43; 95%CI: 0.31 to 0.55; P = 1.2 × 10−12). Thus, the increased MS risk when ANO2 seropositivity was present can be due to an interaction between these risk factors.
Fig. 1.
Odds ratios for MS with different combinations of risk factors. Risk factors include ANO2 seropositivity, DRB1*15:01 carrier, A*02:01 noncarrier, and EBNA1-high (anti-EBNA1 antibody levels above median in controls). Red box indicates that the group of individuals is exposed to the risk factor and white box that they are not. The ORs were calculated by comparison with a reference group, carrying none of the risk factors. Total numbers of MS cases and controls in each group and P values are presented in SI Appendix, Table S3. Bars in the graph indicate 95% confidence intervals.
ANO2 Antibody Epitope Mapping.
In a fine-tuned epitope mapping of ANO2 using 15 peptides (15 mer) with a 14 amino acid overlap covering the region [aa 128 to 156] (SI Appendix, Fig. S1), we confirmed the minimal epitope of 12 amino acids HAGGPGDIELGP [aa 136 to 147], as shown previously using different fragment structures and assays (20). The minimal epitope was represented by a series of overlapping peptides where ANO2 [aa 135 to 149] was one of the most highly reactive ones. ANO2 [aa 135 to 149] and ANO2 [aa 128 to 142], as a negative control, were used in the consecutive experiments.
Anti-ANO2 Antibodies Cross-React with EBNA1.
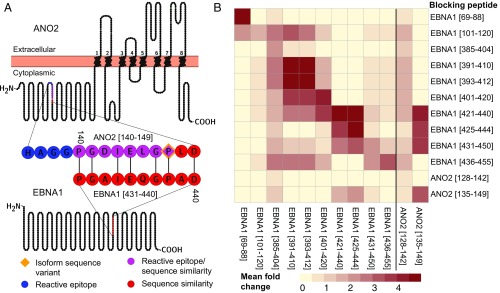
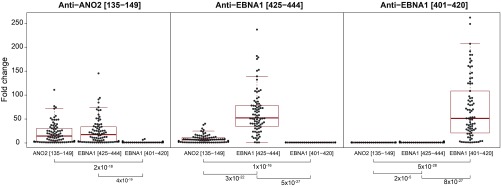
A high sequence similarity between ANO2 [aa 140 to 149] and EBNA1 [aa 431 to 440] with 7/10 identical amino acids and no gaps with BLAST score 18.1 bits, was detected when the full-length sequences of ANO2 and EBNA1 were aligned. The amino acid sequence with high similarity in ANO2 overlaps with 6 of the 12 amino acids of the ANO2-reactive epitope [aa 13 to 147] (Fig. 2A). We therefore set out to investigate if anti-ANO2 antibodies also recognize and bind to EBNA1 and vice versa. In a competitive assay with protein fragments, EBNA1 specifically reduced the signal from the ANO2 fragment [aa 79 to 167] but not fragments representing other proteins (SI Appendix, Fig. S1). In a peptide competition assay, EBNA1 peptides overlapping with the sequence of high similarity to ANO2, i.e., EBNA1 [aa 421 to 440], [aa 425 to 444], and [aa 431 to 450], reduced the signal from the reactive ANO2 peptide [aa 135 to 149] (Fig. 2B). The effect of EBNA1 [aa 425 to 444] on ANO2 [aa 135 to 149] was confirmed in a follow-up competition assay in a larger sample set (Fig. 3). Here, the signal from ANO2 [aa 135 to 149] was affected to the same extent by blocking with EBNA1 [aa 425 to 444] as by its own peptide. Similarly, the signal from EBNA1 [aa 425 to 444] was decreased when the samples were blocked with ANO2 [aa 135 to 149], although not to the same degree as with its own peptide. The negative control, EBNA1 [aa 401 to 420], had no effect on EBNA1 [aa 425 to 444] or ANO2 [aa 135 to 149] reactivity.
Fig. 2.
Competition assay screening. The amino acid stretch showing high sequence similarity between ANO2 and EBNA1 overlaps with the ANO2 antibody epitope as marked in a schematic overview of the ANO2 and EBNA1 full-length sequences. Illustration was generated with the Protter tool (43), and subsequently modified (A). A heatmap with the mean fold change between not blocking and blocking with peptides, representing ANO2 and EBNA1 (rows), in plasma samples from 7 MS cases across the same peptides (columns). The signal intensity from ANO2 [aa 135 to 149] was affected by 3 EBNA1 peptides [aa 421 to 440], [aa 425 to 444], and [aa 431 to 450] (B). The sequence in common between these 3 EBNA1 sequences is EBNA1 [aa 431 to 440] PGAIEQGPAD, which is the sequence with similarity to ANO2 [aa 140 to 147] PGDIELGPLD.
Fig. 3.
Effects on antibody reactivity to ANO2 and EBNA1 peptides after peptide competition. Fold change between signal intensity without blocking and signal intensity when blocked with each peptide are presented on the y axis. ANO2 [aa 135 to 149], EBNA1 [aa 401 to 420], and EBNA1 [aa 425 to 444] were included in the assay with 82 MS case samples, selected to represent both high and low anti-ANO2 and anti-EBNA1 antibody levels. The x axis presents blocking peptides and P values of the differences between fold changes (Wilcoxon rank-sum test), demonstrating that the signal intensity from ANO2 [aa 135 to 149] is equally affected by ANO2 [aa 135 to 149] and EBNA1 [aa 425 to 444].
HLA Gene Region Is Genome-Wide Associated with Anti-ANO2 Antibody Levels.
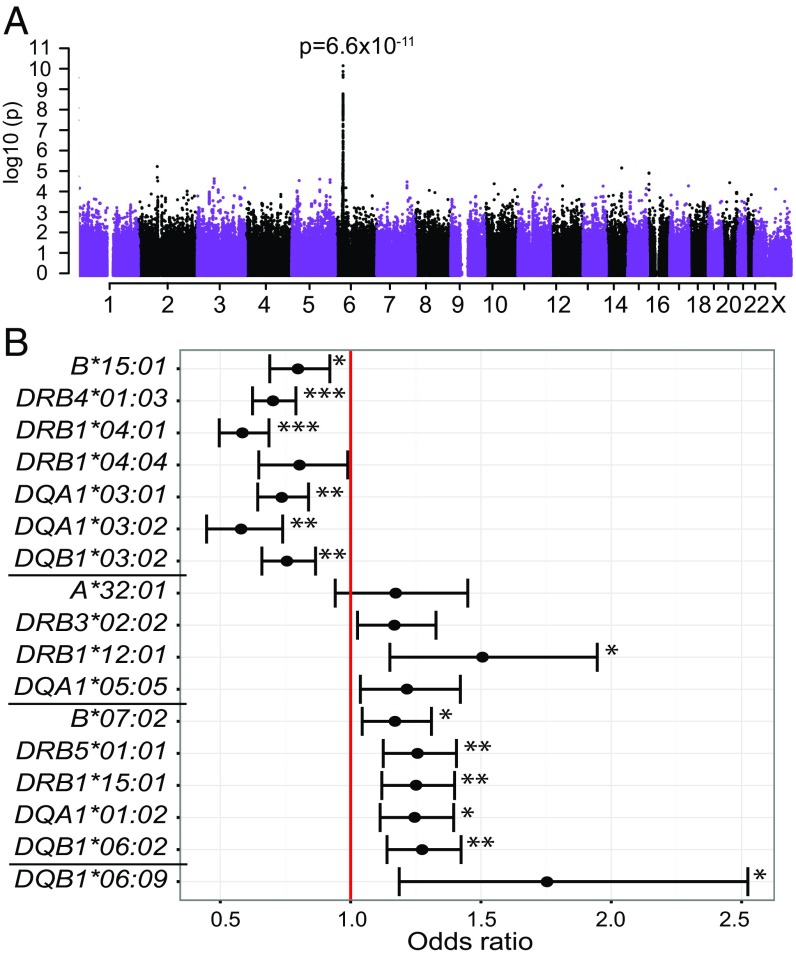
We detected a genome-wide significant association with anti-ANO2 antibody levels (Fig. 4A) consisting of 46 significant SNPs with the top SNP rs2516049 (P = 6.6 × 10−11) being in linkage disequilibrium (LD) with 41 SNPs (r2 > 0.6) covering ∼316 kb of the HLA gene region on chromosome (chr) 6: 32.34 to 32.66 Mb (6,406 cases and 5,530 controls; SI Appendix, Material S1 and Table S4). The region encompassed 8 genes: HCG23, BTNL2, HLA-DRA, HLA-DRB5, HLA-DRB1, HLA-DRB6, HLA-DQA1, and HLA-DQB1. No other region in the genome was significantly associated with anti-ANO2 antibody levels. To analyze the HLA gene region in detail, genetic association analysis was also performed using SNP data from the MS replication chip (22). While this SNP chip does not cover the whole genome, it includes a denser marker map across the HLA region (chr 6: 29.45 to 33.20 Mb), with 7,027 SNPs compared with 4,101 SNPs on the OmniExpress chip. In addition, more individuals from the whole cohort were genotyped with this chip (7,062 cases and 6,098 controls; SI Appendix, Material S1). The association analysis of anti-ANO2 antibody levels resulted in 119 genome-wide significant SNPs, where the top SNP rs6916742 (P = 1.4 × 10−12) was in LD with 18 SNPs covering ∼329 kb (chr 6: 32.34 to 32.67 Mb) of the HLA gene region and second top SNP rs477515, not in LD with top SNP but with 102 SNPs defining the region ∼336 kb (chr 6: 32.34 to 32.68 Mb). The associated region encompassed the same 8 genes as the region defined in genome-wide association study (GWAS) (SI Appendix, Fig. S2).
Fig. 4.
HLA gene region is genome-wide associated with anti-ANO2 antibody levels. A genome-wide significant signal was detected in the HLA gene region using OmniExpress genotype data (A). Imputed HLA allele data, generated from SNP genotypes from MS replication chip, defined 16 HLA alleles significantly associated with anti-ANO2 antibody levels. The associated alleles represented 3 haplotypes (separated by black lines, SI Appendix, Table S5) (23–26). DRB4*01:03 and DRB1*04:01, conferred with the most significant ORs for anti-ANO2 seropositivity (B). Significant P values are marked with asterisks next to error bars (<10−8 ***, <10−4 **, and <10−2 *).
DRB1*04:01 Haplotype Associates with Reduced Anti-ANO2 Antibody Levels.
To clarify the association in the HLA region with anti-ANO2 antibody levels, we tested association of imputed HLA alleles with anti-ANO2 antibody levels. In total, 16 alleles were significantly associated with anti-ANO2 antibody levels representing primarily 3 haplotypes (23–26) (Fig. 4B and SI Appendix, Table S5). The top associated alleles were DRB4*01:03 (P = 4.0 × 10−12) and DRB1*04:01 (P = 1.7 × 10−11), located on the same haplotype, showed protective effects on ANO2 seropositivity (OR = 0.7; 95%CI 0.6 to 0.8 and OR = 0.6; 95%CI 0.5 to 0.7; respectively). One of the associated haplotypes is a known MS risk haplotype (24) and 4 alleles on that haplotype (DRB5*01:01, DRB1*15:01, DQB*06:02, and DQA1*01:02) were the top significant alleles in a HLA allele association analysis with anti-EBNA1 antibody levels and associated with increased risk for high anti-EBNA1 antibody levels (P < 10−38). The HLA alleles most strongly associated with anti-ANO2 antibody levels also associated with anti-EBNA1 antibody levels in the same direction. When conditioning on DRB1*04:01 and DRB4*01:03 in GWAS of anti-ANO2 antibody levels, there was a complete loss of association (SI Appendix, Fig. S2). The same correction using the MS replication chip resulted in complete loss of association to the second top SNP rs6916742 and associated SNPs in LD, whereas the association to the top SNP rs477515 was decreased but not lost (SI Appendix, Fig. S2). The remaining association signal can likely be explained by additional associated HLA alleles. When conditioning on alleles representing the 2 other associated haplotypes, DRB1*12:01 or DRB1*15:01 in GWAS, there was no major impact on the association (P = 1.4 × 10−10 and P = 5.3 × 10−9, respectively).
Anti-ANO2 Antibodies Present before Multiple Sclerosis Onset.
In the pre-MS cohort, which consisted of MS cases sampled before symptom onset and age/sex matched controls, the frequency of ANO2 seropositivity was 10.3% among cases and 6.1% among controls. Neither anti-ANO2 antibody levels (Table 1 and SI Appendix, Table S1) nor ANO2 seropositivity were significantly associated with MS when adjusting for EBNA1 status. Mean time to first symptom was 7.8 y (median 7.0; range 0.6 to 23.7 y) in ANO2-seropositive MS cases (n = 49) and there was no correlation between anti-ANO2 antibody levels and years to symptom onset. The frequency of EBNA1-high individuals was significantly different in ANO2 seropositives (91.0%) compared with ANO2 seronegatives (58.0%; P = 7.3 × 10−10). The difference was also significant in MS cases (P = 8.3 × 10−5) and controls (P = 6.4 × 10−5). Using the definition of EBNA1 seronegativity at a signal intensity below 200 to define individuals with absence of an immune response toward EBNA1, 9.9% (n = 78) of the individuals in the pre-MS cohort were EBNA1 seronegative and out of these, 1 individual was ANO2 seropositive. In the whole cohort, 3.5% (n = 573) were EBNA1 seronegative out of which 1.2% (n = 7) were ANO2 seropositive (P = 6.6 × 10−22). Thus, it is extremely rare for individuals to be ANO2 seropositive in absence of an immune response toward EBNA1.
No Association of ANO2 IgG Reactivity with Other Autoimmune Diseases.
We found no association between rheumatoid arthritis (RA) and anti-ANO2 antibody levels (Table 1). Frequencies of ANO2 seropositivity were similar in RA cases (4.8%) and controls (4.9%). EBNA1 status had a significant effect on anti-ANO2 antibody levels in this dataset (P = 3.8 × 10−5). In a separate serology analysis, anti-ANO2 IgG reactivity was not increased in patients with idiopathic inflammatory myopathy (IIM) nor systemic lupus erythematosus (SLE) compared with population controls (n = 306, age and sex matched to SLE; Table 1). EBNA1 status had significant effects on anti-ANO2 antibody levels also in these disease cohorts.
Multiple Antigen Representations of ANO2 Show Antibody Reactivity.
We observed a high correlation between anti-ANO2 antibodies detected in our previous study (20) and the current measurement of the initial cohort, despite methodological differences (SI Appendix, Fig. S3A). In the present study, antibody reactivities toward the longer fragment of ANO2 [aa 1 to 365] showed higher levels in MS cases compared with controls in the initial, validation, whole, and pre-MS cohorts (SI Appendix, Table S6). The signal intensity was overall lower for ANO2 [aa 1 to 365] compared with ANO2 [aa 79 to 165], but the levels correlated well (SI Appendix, Fig. S3B).
Anti-ANO2 and Anti-EBNA1 Antibodies Are Detected in Cerebrospinal Fluid.
Reanalysis of data generated previously on planar microarrays (19) demonstrated high correlation (Pearson’s correlation = 0.83; Spearman’s rho = 0.75) between anti-ANO2 [aa 79 to 165] measured in plasma and cerebrospinal fluid (CSF) (SI Appendix, Fig. S5). In concordance with these results, reactivity toward ANO2 [aa 79 to 165] and ANO2 [aa 136 to 150] showed a high correlation in a separate, matched set of plasma and CSF samples from 118 individuals analyzed using protein and peptide bead arrays (SI Appendix, Fig. S6). Moreover, ANO2 reactivity also correlated with anti-EBNA1 reactivity in both plasma and CSF in this dataset (SI Appendix, Fig. S6).
Discussion
We here validate increased antibody response to ANO2 in MS in a large case-control cohort. The correlation between anti-ANO2 antibody levels between the previous (20) and the current study was high, which was reassuring considering the methodological differences. The fact that we also detect reactivity to the longer ANO2 [aa 1 to 365] and that it associates with MS further supports our findings. The lower signal intensity acquired with the longer fragment is likely explained by a lower antibody accessibility of the epitope. We also detected anti-ANO2 antibodies in CSF, and ANO2 antibody reactivity in plasma and CSF correlated well. This further supports our results that ANO2 is an important autoantigen in MS. Moreover, we show anti-EBNA1 antibody reactivity in CSF, as has been described previously (27), with a high correlation with anti-ANO2 reactivity in CSF.
There are several lifestyle and environmental factors associated with an increased risk of MS (4, 28, 29), including various aspects of virus infections. Many different infections have been claimed to be associated with MS but have in most cases been refuted. However, EBV infection remains a strong suspect for the following reasons: 1) in a nested case-control study, MS did not occur until persons had developed an immune response to EBV (30); 2) there is an increased antibody response to EBNA1 in persons affected by MS (31, 32); 3) a history of infectious mononucleosis doubles the risk for MS (33); and 4) persons with clinically isolated syndrome (CIS) at risk for MS displayed much higher antibody responses to EBNA1, but to no other EBV antigens, compared with controls; and the anti-EBNA1 antibody levels predicted conversion to definite MS as well as new CNS lesions (34).
Still, definite evidence for a causal link is lacking and the different aspects of EBV-related phenomena in MS such as increased anti-EBNA1 antibody levels and infectious mononucleosis, apart from the mere EBV infections, remain unexplained. For example, it cannot be completely excluded that an EBV immune response in MS could be secondary to the immunogenetics of MS. There are several mutually nonexclusive mechanisms by which EBV could trigger MS; for example, direct antiviral immunity could lead to bystander damage in CNS (35). Furthermore, the viral infection might initiate an autoimmune response through molecular mimicry in which the infectious agent displays epitopes mimicking CNS self-epitopes. Such a mechanism has been demonstrated for Theiler’s murine encephalomyelitis virus infection (36). In humans, causality of any autoimmunity related to infections is much more difficult to prove. Nevertheless, molecular mimicry is one out of several possible mechanisms by which EBV could trigger MS (37, 38). As circumstantial evidence for such a mechanism, EBNA1-specific T cells can cross-react with myelin antigens (11). Still, the etiopathogenetic role of these phenomena remains speculative. We here add a further dimension to the molecular mimicry hypothesis by demonstrating that a precise peptide stretch of EBNA1, one of the most associated to MS (8), cross-reacts with a precise ANO2 antibody epitope. Importantly, this antibody reactivity is associated with a drastic increased risk for MS, which has not been demonstrated before, giving strong support for an etiologic role of this autoimmunity. EBNA1 protein fragments blocked anti-ANO2 reactivity, but not nonrelated antibody reactivities. Furthermore, anti-ANO2 reactivity was rarely detected in individuals who did not yet have serological evidence of having had an EBV infection. Hence, an immune response toward certain EBNA1 protein fragments may be a prerequisite for the development of anti-ANO2 antibodies.
The mechanisms of how autoimmune response against ANO2 might act in MS is unclear. First, the antibodies are directed against an intracellular part of the ion channel ANO2, and it is difficult to assume direct antibody-mediated damage to the CNS. However, there are numerous examples in endocrine autoimmunity with antibodies to intracellular targets such as GAD65, proinsulin, and IA-2 in diabetes (39) or 21-hydroxylase in adrenal disease (40, 41). For 21-hydroxylase and proinsulin, the antibodies are markers of a T cell response to the same antigen, and these may be the real effectors (41, 42). We speculate that the anti-ANO2 antibodies are markers of a T cell response, which may have pathogenic consequences. According to immunological dogma, an IgG antibody response depends on T cell help. Furthermore, we have demonstrated a striking influence of certain HLA class II alleles on the anti-ANO2 antibody response. The class II molecules present antigens to T cells, thus the data provide indirect evidence of a role for ANO2-reactive T cells. Several HLA class II alleles were associated with anti-ANO2 antibody levels but the strongest effect was observed for DRB1*04:01 haplotype with a potential protective influence on the presence of anti-ANO2 antibodies. This could potentially depend on more efficient elimination of ANO2-specific T cells in the thymus or down-regulation in the systemic compartment. Interestingly, the associated HLA alleles also affected anti-EBNA1 antibody levels with the same direction of effect, again suggesting cross-reactivity. While obtaining T cell data remains a valuable next step, it is however, due to the extensive work and time necessary to produce proper data, beyond the scope of this study.
We have not been able to find any specific clinical features, which differ between ANO2-seropositive and ANO2-seronegative persons with MS. How the anti-ANO2 antibody levels differ with age in the same individual is not possible to conclude from the current study. It would therefore be interesting to study sequential samples from the same individual especially before and after a documented immune response to EBNA1, as well as before and after MS onset. We detect anti-ANO2 antibodies in all phases of MS; before symptom onset, CIS, relapsing-remitting MS stage, and progressive phase of MS. The proportion of anti-ANO2 seropositive individuals is specifically increased in MS, although anti-ANO2 antibody reactivity is still detected—at lower frequencies—in population controls and other inflammatory conditions. Thus, this autoimmune reaction is present in the general population but is specifically acting in MS to increase risk together with other risk factors.
Irrespective of any etiopathogenetic role, the mere presence of anti-ANO2 antibodies in MS provides an additional association to MS risk along with a whole series of genetic and lifestyle/environmental factors. Our results strongly support the role of ANO2 as a MS-specific autoantigen in a subset of MS patients, and that ANO2 immune reactivity is potentially induced by an immune response toward EBNA1 through molecular mimicry, in part providing a mechanistic link between EBV and MS.
Materials and Methods
All experiments were approved by the Regional Ethical Review Board in Stockholm, and Umeå, Regional Ethical Review Board, Sweden. All study participants, except those in the pre-MS cohort, gave written informed consent. Pre-MS cohort participants were informed about the study and given the opportunity to withdraw. Description of study cohorts and details regarding experimental procedures for serological analysis, epitope mapping, analysis of cross-reactivity, genotyping, and HLA imputation as well as data and statistical analyses are available in SI Appendix. The data that support the findings of this study are available from the corresponding author, upon request.
Supplementary Material
Acknowledgments
We thank Ingileif Jónsdóttir and Kári Stefánsson at deCODE for providing the OmniExpress genotypes. The study was supported by grants from the Swedish Research Council (grant 2015-02419 to P. Sundström and 2017-00777 to T.O.), the Swedish Brain Foundation, the Knut and Alice Wallenberg Foundation, the Margareta af Ugglas Foundation, Astra Zeneca (SciLifeLab collaboration), and the Swedish Rheumatism Association. The KTH Center for Applied Precision Medicine (KCAP) was funded by the Erling-Persson Family Foundation. K.L. was supported by the King Gustaf Vs 80th Birthday Fund. I.K., P. Stridh, and J. Huang were supported by Horizon 2020 MultipleMS grant 733161. K.T. was supported by the Swedish Brain Foundation, MS Forskningsfond, and the Swedish Association of Persons with Neurological Disabilities (Neuroförbundet).
Footnotes
Conflict of interest statement: Outside this work, T.O. has received unrestricted MS research grants, lecture and/or advisory board honoraria from: Biogen, Novartis, Merck, Sanofi, and Roche. Outside this work, P.K. is working at Roche Diagnostics in unrelated projects.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1902623116/-/DCSupplemental.
References
- 1.Nylander A., Hafler D. A., Multiple sclerosis. J. Clin. Invest. 122, 1180–1188 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hollenbach J. A., Oksenberg J. R., The immunogenetics of multiple sclerosis: A comprehensive review. J. Autoimmun. 64, 13–25 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Olsson T., Piehl F., “The immunobiology of multiple sclerosis” in Encyclopedia of Immunobiology, Ratcliffe M. J. H., Ed. (Academic, Oxford, 2016), vol. 5, pp. 180–191. [Google Scholar]
- 4.Olsson T., Barcellos L. F., Alfredsson L., Interactions between genetic, lifestyle and environmental risk factors for multiple sclerosis. Nat. Rev. Neurol. 13, 25–36 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Kockum I., Alfredsson L., Olsson T., “Genetic and environmental risk factors for multiple sclerosis—A role for interaction analysis” in Between the Lines of Genetic Code, Genetic Interactions in Understanding Disease and Complex Phenotypes, Padyukov L., Ed. (Academic Press, 2014), pp. 101–114. [Google Scholar]
- 6.Moutsianas L., et al. ; International IBD Genetics Consortium (IIBDGC) , Class II HLA interactions modulate genetic risk for multiple sclerosis. Nat. Genet. 47, 1107–1113 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fogdell-Hahn A., Ligers A., Grønning M., Hillert J., Olerup O., Multiple sclerosis: A modifying influence of HLA class I genes in an HLA class II associated autoimmune disease. Tissue Antigens 55, 140–148 (2000). [DOI] [PubMed] [Google Scholar]
- 8.Sundqvist E., et al. , Epstein-Barr virus and multiple sclerosis: Interaction with HLA. Genes Immun. 13, 14–20 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Sundström P., Nyström M., Ruuth K., Lundgren E., Antibodies to specific EBNA-1 domains and HLA DRB1*1501 interact as risk factors for multiple sclerosis. J. Neuroimmunol. 215, 102–107 (2009). [DOI] [PubMed] [Google Scholar]
- 10.Libbey J. E., McCoy L. L., Fujinami R. S., Molecular mimicry in multiple sclerosis. Int. Rev. Neurobiol. 79, 127–147 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lünemann J. D., et al. , EBNA1-specific T cells from patients with multiple sclerosis cross react with myelin antigens and co-produce IFN-gamma and IL-2. J. Exp. Med. 205, 1763–1773 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weissert R., et al. , MHC haplotype-dependent regulation of MOG-induced EAE in rats. J. Clin. Invest. 102, 1265–1273 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olsson T., The new era of multiple sclerosis therapy. J. Intern. Med. 275, 382–386 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Olsson T., Cytokine-producing cells in experimental autoimmune encephalomyelitis and multiple sclerosis. Neurology 45(suppl. 6), S11–S15 (1995). [DOI] [PubMed] [Google Scholar]
- 15.Olsson T., et al. , Autoreactive T lymphocytes in multiple sclerosis determined by antigen-induced secretion of interferon-gamma. J. Clin. Invest. 86, 981–985 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vanderlugt C. J., Miller S. D., Epitope spreading. Curr. Opin. Immunol. 8, 831–836 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hohlfeld R., Dornmair K., Meinl E., Wekerle H., The search for the target antigens of multiple sclerosis, part 1: Autoreactive CD4+ T lymphocytes as pathogenic effectors and therapeutic targets. Lancet Neurol. 15, 198–209 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Elliott C., et al. , Functional identification of pathogenic autoantibody responses in patients with multiple sclerosis. Brain 135, 1819–1833 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ayoglu B., et al. , Autoantibody profiling in multiple sclerosis using arrays of human protein fragments. Mol. Cell. Proteomics 12, 2657–2672 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ayoglu B., et al. , Anoctamin 2 identified as an autoimmune target in multiple sclerosis. Proc. Natl. Acad. Sci. U.S.A. 113, 2188–2193 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pedemonte N., Galietta L. J., Structure and function of TMEM16 proteins (anoctamins). Physiol. Rev. 94, 419–459 (2014). [DOI] [PubMed] [Google Scholar]
- 22.International Multiple Sclerosis Genetics Consortium; N. A. Patsopoulos et al.; Australia and New Zealand IBDGC; Belgium Genetic Consortium; Initiative on Crohn and Colitis, NIDDK IBDGC; United Kingdom IBDGC; Wellcome Trust Case Control Consortium, The Multiple Sclerosis Genomic Map: Role of peripheral immune cells and resident microglia in susceptibility. bioRxiv:10.1101/143933 (13 July 2017).
- 23.Farh K. K., et al. , Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature 518, 337–343 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patsopoulos N. A., et al. ; IMSGC; ANZgene , Fine-mapping the genetic association of the major histocompatibility complex in multiple sclerosis: HLA and non-HLA effects. PLoS Genet. 9, e1003926 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Erlich H. A., et al. ; Type 1 Diabetes Genetics Consortium (T1DGC) , Next generation sequencing reveals the association of DRB3*02:02 with type 1 diabetes. Diabetes 62, 2618–2622 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maiers M., Gragert L., Klitz W., High-resolution HLA alleles and haplotypes in the United States population. Hum. Immunol. 68, 779–788 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Nociti V., et al. , Epstein-Barr virus antibodies in serum and cerebrospinal fluid from multiple sclerosis, chronic inflammatory demyelinating polyradiculoneuropathy and amyotrophic lateral sclerosis. J. Neuroimmunol. 225, 149–152 (2010). [DOI] [PubMed] [Google Scholar]
- 28.Hedström A. K., Alfredsson L., Olsson T., Environmental factors and their interactions with risk genotypes in MS susceptibility. Curr. Opin. Neurol. 29, 293–298 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Montgomery S., et al. , Concussion in adolescence and risk of multiple sclerosis. Ann. Neurol. 82, 554–561 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Munger K. L., Levin L. I., O’Reilly E. J., Falk K. I., Ascherio A., Anti-Epstein-Barr virus antibodies as serological markers of multiple sclerosis: A prospective study among United States military personnel. Mult. Scler. 17, 1185–1193 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ascherio A., Munger K. L., Epstein-barr virus infection and multiple sclerosis: A review. J. Neuroimmune Pharmacol. 5, 271–277 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Levin L. I., Munger K. L., O’Reilly E. J., Falk K. I., Ascherio A., Primary infection with the Epstein-Barr virus and risk of multiple sclerosis. Ann. Neurol. 67, 824–830 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Belbasis L., Bellou V., Evangelou E., Ioannidis J. P., Tzoulaki I., Environmental risk factors and multiple sclerosis: An umbrella review of systematic reviews and meta-analyses. Lancet Neurol. 14, 263–273 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Lünemann J. D., et al. , Elevated Epstein-Barr virus-encoded nuclear antigen-1 immune responses predict conversion to multiple sclerosis. Ann. Neurol. 67, 159–169 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Serafini B., et al. , Dysregulated Epstein-Barr virus infection in the multiple sclerosis brain. J. Exp. Med. 204, 2899–2912 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Croxford J. L., Olson J. K., Anger H. A., Miller S. D., Initiation and exacerbation of autoimmune demyelination of the central nervous system via virus-induced molecular mimicry: Implications for the pathogenesis of multiple sclerosis. J. Virol. 79, 8581–8590 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Münz C., Lünemann J. D., Getts M. T., Miller S. D., Antiviral immune responses: Triggers of or triggered by autoimmunity? Nat. Rev. Immunol. 9, 246–258 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pender M. P., Infection of autoreactive B lymphocytes with EBV, causing chronic autoimmune diseases. Trends Immunol. 24, 584–588 (2003). [DOI] [PubMed] [Google Scholar]
- 39.Pihoker C., Gilliam L. K., Hampe C. S., Lernmark A., Autoantibodies in diabetes. Diabetes 54 (suppl. 2), S52–S61 (2005). [DOI] [PubMed] [Google Scholar]
- 40.Napier C., Pearce S. H., Autoimmune addison’s disease. Presse Med. 41, e626–e635 (2012). [DOI] [PubMed] [Google Scholar]
- 41.Blahnik G., et al. , Analysis of pancreatic beta cell specific CD4+ T cells reveals a predominance of proinsulin specific cells. Cell. Immunol. 335, 68–75 (2019). [DOI] [PubMed] [Google Scholar]
- 42.Dawoodji A., et al. , High frequency of cytolytic 21-hydroxylase-specific CD8+ T cells in autoimmune Addison’s disease patients. J. Immunol. 193, 2118–2126 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Omasits U., Ahrens C. H., Müller S., Wollscheid B., Protter: Interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics 30, 884–886 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.