Significance
Marine unicellular cyanobacteria coexist in the oceans with lytic phages that infect and kill them. Yet, the cyanobacteria persist, fulfilling their role as important primary producers, due to population diversity that results in different sensitivity and resistance profiles to co-occurring cyanophages. Here, we report a surprising dichotomy in modes of resistance against specialist versus generalist phages: Resistance is primarily extracellular against specialist phages but intracellular against generalist phages. Known intracellular resistance mechanisms are absent from most marine Synechococcus and Prochlorococcus strains, suggesting that currently unknown defense mechanisms exist in marine cyanobacteria. Furthermore, phage DNA entry and replication, coupled with survival of the cyanobacterial cell, provide a means for horizontal transfer of genetic material into, and evolution of, marine cyanobacteria.
Keywords: cyanobacteria, virus, infection, resistance, polyploidy
Abstract
Long-term coexistence between unicellular cyanobacteria and their lytic viruses (cyanophages) in the oceans is thought to be due to the presence of sensitive cells in which cyanophages reproduce, ultimately killing the cell, while other cyanobacteria survive due to resistance to infection. Here, we investigated resistance in marine cyanobacteria from the genera Synechococcus and Prochlorococcus and compared modes of resistance against specialist and generalist cyanophages belonging to the T7-like and T4-like cyanophage families. Resistance was extracellular in most interactions against specialist cyanophages irrespective of the phage family, preventing entry into the cell. In contrast, resistance was intracellular in practically all interactions against generalist T4-like cyanophages. The stage of intracellular arrest was interaction-specific, halting at various stages of the infection cycle. Incomplete infection cycles proceeded to various degrees of phage genome transcription and translation as well as phage genome replication in numerous interactions. In a particularly intriguing case, intracellular capsid assembly was observed, but the phage genome was not packaged. The cyanobacteria survived the encounter despite late-stage infection and partial genome degradation. We hypothesize that this is tolerated due to genome polyploidy, which we found for certain strains of both Synechococcus and Prochlorococcus. Our findings unveil a heavy cost of promiscuous entry of generalist phages into nonhost cells that is rarely paid by specialist phages and suggests the presence of unknown mechanisms of intracellular resistance in the marine unicellular cyanobacteria. Furthermore, these findings indicate that the range for virus-mediated horizontal gene transfer extends beyond hosts to nonhost cyanobacterial cells.
The ability of a virus to reproduce requires a high degree of compatibility to its host. Viruses only reproduce in cells with recognizable cell surfaces, contain all of the machinery required for intracellular replication, and lack effective defenses against them. Thus, virus host range is intricately linked to its cellular requirements (extracellular and intracellular) as well as the cell’s resistance mechanisms against viral infection. Virus host range varies greatly, with both specialists and generalists commonly isolated from diverse environments (1–5). Specialists have a narrow host range, infecting a single or narrow set of similar hosts, whereas generalists have broader host ranges, infecting multiple distinct host types (3, 5). Cellular requirements for specialist and generalist phages may therefore vary, as might the defenses against them.
Unicellular cyanobacteria of the genera Synechococcus and Prochlorococcus are the most abundant photosynthetic organisms in the oceans and are estimated to perform 25% of oceanic primary production (6). They are diverse and belong to multiple phylogenetic lineages (7, 8). They coexist in the oceans with high numbers of cyanophages (9–11). The 2 major virus families infecting them are the T7-like cyanopodoviruses and the T4-like cyanomyoviruses (1, 4, 9, 11). T7-like cyanophages are specialists, whereas T4-like cyanophages consist of both specialists and generalists, some of which can infect members of both cyanobacterial genera (1, 4, 9, 11). Viruses from both families are lytic, killing the cell at the end of the infection cycle (1, 9, 11), despite the presence of an integrase gene in some T7-like cyanophages (12). Thus, cyanobacteria–cyanophage coexistence in the oceans is likely facilitated by the presence of both sensitive cells in which cyanophages reproduce and resistant cells that survive the encounter (9, 13).
Known active intracellular defense mechanisms, such as restriction modification, CRISPR-Cas, and a suite of recently discovered systems, are uncommon in marine Synechococcus and Prochlorococcus strains (14–16). Previous studies of experimentally evolved resistant cyanobacteria found that resistance is generally passive, being caused by mutations in cell-surface–related genes that prevent recognition and attachment of the virus to the cell (13, 17). Information is lacking, however, if an incompatible cell surface is the predominant mode of defense in intrinsically resistant nonhost cyanobacteria. This is despite the fact that many Synechococcus and Prochlorococcus cells are likely to be resistant to co-occurring viruses in the oceans (9, 13). By intrinsically resistant, we refer to cyanobacteria that were not experimentally evolved. We use the term resistant nonhosts for resistant bacteria that may not have been sensitive to the phage in question in their evolutionary past or that became resistant following coevolution or 1-sided evolution in nature (18, 19). Here, we set out to investigate resistance in a suite of Synechococcus and Prochlorococcus strains that are resistant nonhosts to different subsets of cyanophages and to determine whether modes of resistance are similar against specialist and generalist viruses.
Results and Discussion
Resistance to Generalist and Specialist Phages.
To determine viral host range and identify intrinsically resistant cyanobacteria, we challenged diverse cyanobacteria from our culture collection with T7-like and T4-like cyanophage isolates. This was done in liquid assays, as well as with plaque and spot assays (Methods). We define a cell as resistant when no viral progeny are produced, whereas a cell is sensitive when infective viruses result from the encounter. As found previously, T7-like cyanophages were specialists, infecting just a single host, whereas both specialist and generalist viruses were found among T4-like cyanophages (SI Appendix, Table S1). The generalists infected 7 to 9 strains from different lineages out of 17 Synechococcus and Prochlorococcus strains tested.
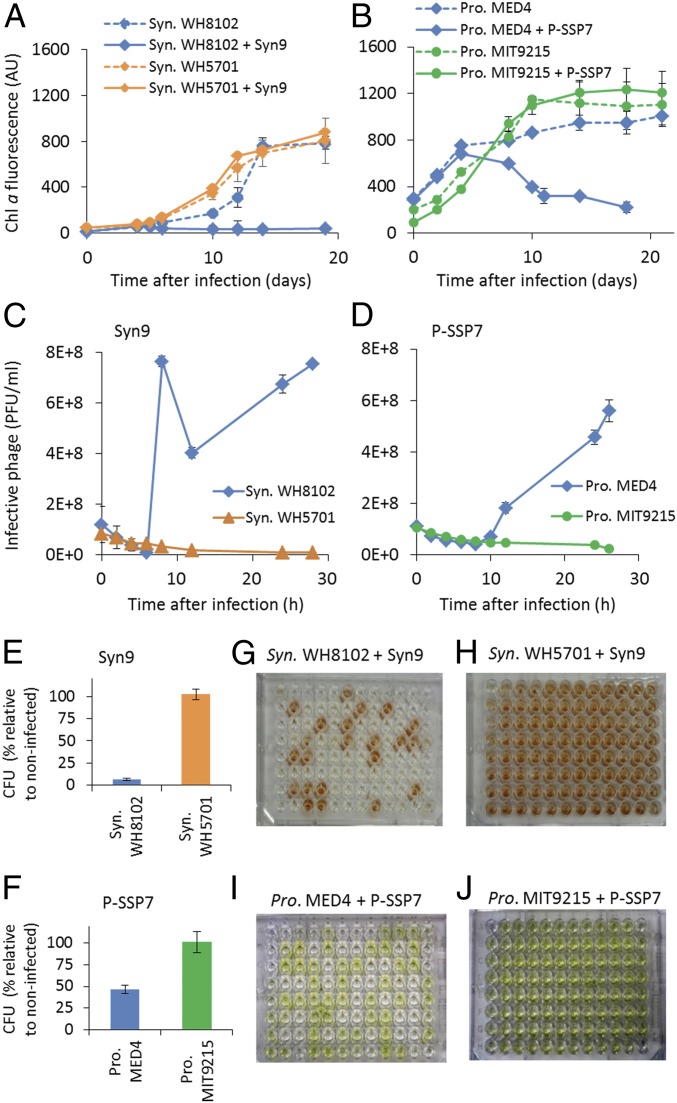
No production of infective virus progeny was found on resistant strains, even after many hours, as determined from virus growth experiments (Fig. 1 C and D). Furthermore, no sensitive subpopulations were detected, as seen from both viable colony assays (Fig. 1 E and F) and a single-cell lysis method (Methods) (Fig. 1 G–J). Interestingly, the resistant cyanobacteria survived the viral challenge with no observable effect on growth over periods of at least 2 to 3 wk (Fig. 1 A and B). In comparison, cultures of sensitive strains died rapidly and produced infective viral progeny (Fig. 1). These findings indicate that resistance was all-inclusive, with no sensitive subpopulations in the cultures, and that these are not cases of inefficient infection (20).
Fig. 1.
Determination of cyanobacterial resistance. Growth of cyanobacteria in the presence and absence of cyanophages (n = 6) (A and B) and production of infective cyanophages (n = 3) (C and D). These figures are representative of resistance and sensitivity for all interactions. Synechococcus (Syn.) WH8102 and Prochlorococcus (Pro.) MED4 are sensitive to the Syn9 and P-SSP7 phages, respectively, as seen by the decline in culture growth (A and B) and production of infective viral particles (C and D), whereas Syn. WH5701 and Pro. MIT9215 are resistant to these same cyanophages, respectively, as growth is unaffected by the cyanophages (A and B) and no infective particles were produced (C and D). Viable colony (E and F) and cell lysis (G–J) assays showed that all Syn. WH5701 and Pro. MIT9215 cells were resistant to Syn9 and P-SSP7, respectively (n = 3 for both assays, with a total of 576 cells analyzed in the cell lysis assay per interaction); Syn9 lysed 72% of sensitive Syn. WH8102 cells (G), and P-SSP7 lysed 52% of sensitive Pro. MED4 cells (I), while both phages lysed 0% of the respective resistant cells: Syn. WH5701 (H) and Pro. MIT9215 (J). AU, arbitrary units; Chl a, Chlorophyll a; CFU, colony-forming units; PFU, plaque-forming units.
Our investigation focused on cyanobacterial interactions with 3 generalist T4-like cyanophages and 5 specialist viruses, 3 from the T7-like and 2 from the T4-like cyanophage families (SI Appendix, Table S1). First, we characterized whether the mode of resistance was extracellular or intracellular in a total of 63 cyanobacteria–cyanophage interactions. Extracellular resistance is manifested as either lack of attachment to the cell surface or lack of subsequent entry of genetic material into the cell. We tested for attachment using adsorption assays (Methods). Specialist phages, from both the T7-like and T4-like families of cyanophages, did not attach to the vast majority of resistant cyanobacterial strains (Fig. 2 A–E), indicating that resistance was extracellular in most cases (79%, 34 of 43 interactions). In stark contrast, generalist cyanophages attached to nearly all resistant cyanobacteria, indicating that resistance was rarely extracellular against generalists (10%, 2 of 20 interactions) (Fig. 2 F–H). However, attachment was not indiscriminant, as none of the cyanophages attached to 3 heterotrophic marine bacteria tested (Fig. 2).
Fig. 2.
Attachment of specialist and generalist cyanophages to resistant cyanobacteria. Attachment of specialist T7-like cyanopodoviruses (A–C), specialist T4-like cyanomyoviruses (D and E), and generalist T4-like cyanomyoviruses (F–H) to resistant Prochlorococcus (green), Synechococcus (orange), and heterotrophic marine bacteria (gray) compared with sensitive cyanobacterial strains (blue) and growth medium (red) controls. Phage names are shown above each graph (n = 6). Attachment significantly greater than the negative growth medium control as determined from the Mann–Whitney U test, **P < 0.01.
To assess whether cyanophages that attached to resistant cells also inserted their DNA into the cytoplasm and initiated infection, we assessed transcription of early expressed genes in the viral transcriptional program. Generalist cyanophages expressed early genes inside resistant cells in all 12 interactions tested (Fig. 3 A–C), indicating that their DNA was inserted into the cell and that resistance was intracellular. Gene transcription, and thus genome entry, was also found for 5 of the 7 interactions tested with specialist viruses that had attached to cells (SI Appendix, Fig. S1), indicating that in a minor number of cases, resistance was also intracellular against specialist viruses.
Fig. 3.
Phage gene expression in resistant cyanobacteria. (A–C) Early gene transcription of generalist T4-like cyanophages in resistant Prochlorococcus (Pro.; green) and Synechococcus (Syn.; orange) strains relative to a sensitive Synechococcus strain (blue) (n = 3). Results with specialist cyanophages are shown in SI Appendix, Fig. S1. (D–H) Phage transcript levels of 2 genes in each transcription cluster: early (red), middle (green), and late (blue) genes are shown for resistant cyanobacteria relative to those in a sensitive cyanobacterium (Cyanobact.) (n = 3). The sensitive strain was Synechococcus WH8102 for the 3 generalist phages (Syn9, P-TIM40, and S-TIM4) and Prochlorococcus MED4 and MIT9515 for the specialist P-SSP7 and P-TIM68 phages, respectively. (I and J) Numbers of phage proteins detected by MS for 2 generalist phages are shown for interactions with a sensitive and multiple resistant cyanobacteria (n = 3). Unclassified, proteins for S-TIM4 without sequence homology to Syn9. Color-coding of strains is as in A–C. Fewer phage proteins were found in all expression classes and interactions in resistant cyanobacteria relative to the sensitive strain (P ≤ 0.05), except for Syn9 early proteins in Synechococcus WH5701 (P = 0.5), as determined from the Mann–Whitney U test.
These findings indicate that resistance to specialist cyanophages was primarily extracellular (at least 79% of interactions). In contrast, resistance against generalist cyanophages was predominantly intracellular (90% of interactions). This holds for resistance against cyanophages from both T7-like and T4-like families and for resistant cyanobacteria from both the Synechococcus and Prochlorococcus genera.
These findings raise the intriguing question of why different modes of resistance are found against specialist versus generalist cyanophages. Clearly, resistance will be extracellular against a specialist phage if the resistant nonhost never had the specific cell-surface receptor molecule that the specialist virus attaches to. Indeed, cell-surface genes are often present in only a subset of marine unicellular cyanobacteria (13, 21, 22). In addition, extracellular defenses could have developed through coevolution between host and phage, with continuous reshaping of the gene repertoire coding for cell-surface molecules used by phages as receptors (13, 19, 22, 23). This process of continuous mutation and loss and gain of genes from the genome during host–phage coevolution would have occurred in a “Red Queen”-like manner, and ultimately resulted in a phage that did not regain the ability to attach to the host cell surface (13, 19, 23, 24). In both cases, the end result is specialist phages that attach to cell-surface molecules encoded by lineage-specific genes (13, 25). Such passive extracellular defenses against specialist phages are highly efficient and metabolically cost-effective for the host as the phage does not even enter the cell.
We consider it probable that generalist phages have evolved to attach to essential cell-surface molecules coded by highly conserved genes across the cyanobacteria, as known for some bacteria/phage systems (25, 26). Thus, it would be considerably more costly, even lethal, to mutate or exchange such genes, although it is also possible that generalism results from phages having evolved the ability to attach to different cell-surface molecules in distinct hosts (25, 27). Either way, generalist phages enter the cell. Thus, the only way for the cell to avoid being killed and selected against is development of intracellular resistance. This is consistent with model predictions that resistance will develop after entry into the cell when the cost of resistance at the stage of recognition and attachment is high (28).
Resistance can also be considered a lack of adaptation of the phage to the cyanobacterial cell. For specialists, this is manifested as the absence of changes in the phage that facilitate attachment to a cell-surface molecule on the resistant nonhost. For generalists, this could be due to the lack of ability of the phage to overcome active intracellular defenses or, alternatively, to its inability to utilize the cell’s intracellular machinery for its replication. This intracellular incompatibility could arise through a passive mechanism of adaptive loss from the cell or be due to an intrinsic lack of the machinery required for phage replication in the first place. We consider it unlikely that nonhost cells lacked the machinery required for phage replication by generalists. This is because generalist phages successfully infect cells belonging to multiple distinct lineages, and even across genera, within the monophyletic clade of the marine unicellular cyanobacteria. Thus, for this to be the case, each sensitive cyanobacterial lineage would have had to independently gain the machinery needed by the phage, which we consider less probable than adaptive losses in the resistant lineages.
Let us attempt to understand the advantages and disadvantages of specialism versus generalism from the perspective of the phages. Specialism has a distinct advantage as no loss occurs with irreversible attachment to nonhosts. Thus, the range of attachment is the same as the range of reproduction (29). For this to be a true advantage, abundances of the specific host must be sufficiently high so that chances of host encounter prior to loss of infectivity are relatively high (1, 4, 30). Indeed, observations and models indicate that phage specialism is favored when hosts are common (31, 32). Furthermore, specialism allows optimization of the infection cycle to a particular host, whereas adaptation of a generalist to new hosts can lead to reduced fitness on other hosts (20, 33, 34).
In comparison, a clear trade-off to promiscuous entry for generalist phages exists as irreversible attachment and entry into nonhost cells lead to the loss of the phage. Nonetheless, generalism would be advantageous when abundances of an optimal host are low or fluctuate greatly in the environment (31, 32). In this case, generalism provides the phage with reasonable alternatives for reproduction and increases the effective number of hosts, although a certain degree of hedge-betting is employed: It is better to enter cells that may not be suitable for reproduction rather than to lose infectivity before meeting a rare specific host. Indeed, previous findings have shown that specialist cyanophages tend to infect abundant high-light–adapted Prochlorococcus ecotypes, while generalist cyanophages tend to infect Synechococcus and low-light–adapted Prochlorococcus ecotypes (1, 4), which are less abundant in vast oligotrophic regions of the oceans and fluctuate seasonally (6, 7).
Intracellular Prevention of Virus Production.
The finding that resistance was largely intracellular against generalist cyanophages was surprising since known intracellular defense mechanisms are found in only a quarter of the marine Synechococcus and Prochlorococcus strains used in this study (14–16) (SI Appendix, Table S2). We followed the progression of the infection cycle of the generalist Syn9 phage in multiple resistant cyanobacteria to determine the stage at which infection was arrested. We also examined certain stages of infection in other interactions to assess how general our findings are. Overall, we investigated resistance in Synechococcus and Prochlorococcus strains against 3 generalist and 5 specialist cyanophages for which attachment and entry were observed (Methods). Below, we present the data by stage of infection for all interactions investigated.
After entry into the cell, cyanophage DNA is transcribed in 3 transcription clusters (35, 36): Early genes involved in host takeover are transcribed first, followed by middle genes for DNA metabolism and replication, and ending with late genes for virion morphogenesis and exit. We measured transcript levels of 2 phage genes from each transcriptional cluster by real-time qPCR in resistant strains and compared them with those measured during infection of a sensitive host. A variety of transcriptional results were found for Syn9 in different resistant cyanobacteria, ranging from transcript levels highly similar to those in the sensitive strain down to 1% or less (Fig. 3D). Similar ranges in transcription were found for other generalist phages and a specialist phage (Fig. 3 E–H), with overall levels being somewhat lower than in the sensitive strain (10 to 60%). Thus, transcription levels were not dictated by the identity of the phage, and they were not related solely to the identity of the cyanobacterium, as seen from dramatic variation of transcript levels in 2 strains (Prochlorococcus MIT9215 and Synechococcus CC9311). However, Synechococcus WH5701 had transcript levels similar to those in the sensitive host for all 3 generalist phages. Thus, even though the phages’ transcriptional programs moved beyond early-phase transcription in all resistant strains, transcription levels varied greatly and were interaction-specific.
Next, we assessed de novo phage protein production by comparing the number of phage proteins detected in resistant strains with those in a sensitive strain using mass spectrometry (MS) (Methods). Seventy-five percent of Syn9 proteins were detected in resistant Synechococcus WH5701 (Fig. 3I and SI Appendix, Table S3A). In 4 other interactions with Syn9, 35% or fewer phage proteins were detected. Investigation of the S-TIM4 phage showed that Synechococcus WH5701 also produced the most phage proteins (30%), while no phage proteins were found in the other 2 resistant cyanobacteria (Fig. 3J). Thus, Synechococcus WH5701 displayed both consistently high phage transcript levels and the highest number of phage proteins among resistant strains. In contrast, Prochlorococcus MIT9215 produced the least proteins for both phages (Fig. 3 I and J and SI Appendix, Table S3), even though S-TIM4 transcript levels were high and similar to those in Synechococcus WH5701 (Fig. 3F). This indicates that transcript levels alone do not explain the presence or absence of phage proteins.
Even though 75% of Syn9 proteins were produced in Synechococcus WH5701, a number of key phage proteins were lacking, whose combined absence would prevent the formation of an infective phage particle (SI Appendix, Table S3A). These include the small terminase subunit (TerS; T4 gp16 homolog) and protease (T4 gp21 homolog), both of which are required for DNA packaging and capsid maturation (37), and proteins required for tail termination and attachment to the capsid (homologs of T4 gp3, gp15, and gp13) (38). The absence of these proteins was not due to their lack of transcription as messenger RNA (mRNA) was found for all 4 genes (SI Appendix, Fig. S2). This suggests that certain phage proteins were not translated or were specifically targeted for proteolysis after translation in Synechococcus WH5701.
In summary, the protein repertoire for both the Syn9 and S-TIM4 phages was incomplete in all resistant strains investigated, ranging from 0 to 75% of those detected in the sensitive cyanobacterium. In some interactions, this was likely due to insufficient transcription. In many other interactions, resistance appears to have been due to posttranscriptional processes that led to the absence of phage proteins. These caused a general lack of translation in some interactions, while to the absence of only a select set of key proteins in others.
Next, we assessed whether infection proceeded to phage genome DNA (gDNA) replication. Surprisingly, Syn9 genome replication was just as high and nearly as rapid in resistant Synechococcus WH5701 as in the sensitive strain (Fig. 4A). In contrast, no phage genome replication was found in the other 5 resistant cyanobacteria (Fig. 4A). Investigation of other interactions, those with both generalist (Fig. 4) and specialist (SI Appendix, Fig. S3) phages, revealed that phage genome replication was fairly common, if entry occurred, with more than half of all interactions (11 of 17) displaying some degree of genome replication. However, differences in the extent and rapidity of DNA replication were obvious (SI Appendix, Supplementary Results and Discussion and Table S4). In some interactions, replication was similar or even higher than in the sensitive strain, including for multiple generalist phages in Synechococcus WH5701 (Fig. 4) and multiple specialist phages in Prochlorococcus MIT9215 (SI Appendix, Fig. S3).
Fig. 4.
Phage DNA replication and cyanobacterial DNA degradation in resistant cyanobacteria. Intracellular phage DNA (A, C, and E) and cyanobacterial DNA (B, D, and F) levels were monitored for 10 or 12 h after infection with 3 generalist phages: Syn9 (A and B), P-TIM40 (C and D), and S-TIM4 (E and F) (n = 3). Longer term assessment (144 h) is shown after infection by Syn9 of sensitive Synechococcus WH8102 (G) and resistant Synechococcus WH5701 (H). Results in G and H are representative of 4 independent experiments. Phage DNA was measured by qPCR for g20 (portal protein gene) and is relative to maximal levels in the sensitive strain. Cyanobacterial DNA was measured by qPCR for rbcL (RuBisco gene) and is relative to levels immediately after infection (at t0). Levels during interactions with resistant Synechococcus (Syn.; orange), resistant Prochlorococcus (Pro.; green), and sensitive Synechococcus (blue) strains are shown. Results with specialist phages are shown in SI Appendix, Fig. S3.
Host genome degradation is common during virus infection, including that by cyanophages (35, 36, 39–41), and provides nucleotides for phage genome replication (39). Cyanobacterial genome degradation occurred in all resistant strains in which phage gDNA was replicated, including Synechococcus WH5701 with the Syn9 phage (Fig. 4). In contrast, no degradation was observed in cyanobacterial strains for which the phage genome was not replicated (compare left and right panels in Fig. 4 and SI Appendix, Fig. S3). Investigation of the long-term fate of Syn9 phage DNA in Synechococcus WH5701 showed that it was degraded by 14 to 24 h after infection, at which time cyanobacterial genome levels also began increasing (Fig. 4H).
The concomitant degradation of cyanobacterial DNA with phage DNA replication is particularly intriguing as no detrimental effect on cyanobacterial growth was observed in the resistant strains (compare Fig. 1A with Fig. 4A and Fig. 1B with SI Appendix, Fig. S3B). We hypothesized that this may be due to the presence of multiple copies of the genome (polyploidy) in a single cell, a known phenomenon in many organisms, including many cyanobacteria (42, 43). An investigation of ploidy in 21 cyanobacteria revealed 6 Synechococcus and 3 Prochlorococcus strains with more than 1 genome copy (SI Appendix, Fig. S4). Most interestingly, multiple genome copies were detected in the 2 strains for which concomitant phage genome replication and cyanobacterial genome degradation was a common occurrence: Synechococcus WH5701 and Prochlorococcus MIT9215 (SI Appendix, Fig. S4). This is consistent with polyploidy providing genome copy redundancy that would allow cells to withstand partial genome degradation without negatively affecting growth. It further points to a potential benefit of polyploidy in bacteria and eukaryotic cells not considered previously, similar to a recent report suggesting that polyploidy may help cells compensate for ultraviolet damage to the chromosome (44).
Our findings of some resistant cyanobacteria in which phage genomes are expressed and replicated raised the question of whether infective phage particles are produced intracellularly and resistance is due to lack of release from the cell. We investigated this for Syn9 in Synechococcus WH5701, the interaction for which prior stages of the infection process were most similar to the sensitive Synechococcus WH8102.
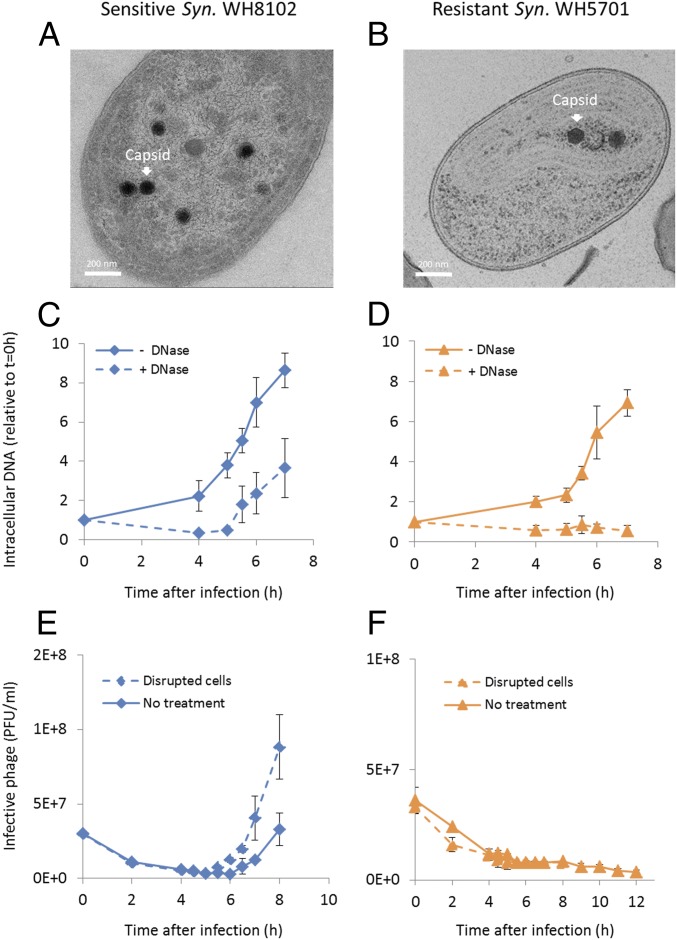
First, we assessed whether virus particles are formed intracellularly through direct observation in thin cell sections by transmission electron microscopy (TEM). To our surprise, capsids were assembled in the resistant strain (Fig. 5 A and B). However, fewer were present in each thin section in the resistant (1.2 ± 0.9, n = 28 sections) relative to the sensitive (3.9 ± 1.7, n = 18 sections) cyanobacterium (P < 0.001). Capsid sizes were similar in the resistant and sensitive strains (Fig. 5 A and B and SI Appendix, Fig. S5), as well as to those reported previously for Syn9 (45), and are significantly smaller than carboxysomes (SI Appendix, Fig. S5). Phage tails were not observed in the sensitive or resistant strain, so no conclusions about their assembly and attachment can be reached.
Fig. 5.
Formation of Syn9 phage particles in resistant Synechococcus WH5701. Capsid assembly is observed from TEM images of thin cell sections (A and B), packaging of DNA into capsids determined from DNase protection (C and D), and intracellular production of infective phage determined from plaque-forming units (E and F) during Syn9 infection of sensitive Synechococcus (Syn.) WH8102 (A, C, and E) and resistant Synechococcus WH5701 (B, D, and F) strains. TEM images are representative of 2 independent experiments (n = 18 sections for Synechococcus WH8102 and n = 28 sections for Synechococcus WH5701). Size distribution of phage particles is shown in SI Appendix, Fig. S5. (C and D) “− DNase” treatment indicates total intracellular DNA, while “+ DNase” treatment indicates intracellular DNA that is protected from DNase digestion after cell disruption (n = 3). Phage DNA levels were determined by qPCR for the g20 portal gene. (E and F) “Disrupted cells” treatment shows the number of infective phages present intracellularly and in the extracellular medium, while “No treatment” shows the number of infective phages in the extracellular medium (n = 3). PFU, plaque-forming units.
Next, we determined whether the replicated phage genome in the resistant strain (Fig. 4 A and H) was packaged into capsids. This was done by testing for protection of intracellular DNA from deoxyribonuclease (DNase) digestion (Methods). While phage DNA was protected in the sensitive strain (Fig. 5C), it was not protected in the resistant strain (Fig. 5D), indicating that phage DNA was not packaged. These findings are in line with expectations based on the proteomic data since 2 key packaging proteins (TerS and the protease) were not present in the resistant strain (discussed above).
Finally, we tested whether phage particles inside resistant cells were infective, even though this is not expected if DNA is not packaged. We disrupted cells and tested for infective phages in the supernatant (Methods). As expected, no infective phages were found in the resistant strain either in disrupted cells or released from untreated cells (Fig. 5F), whereas infective phages were found in the sensitive strain in both conditions (Fig. 5E). Thus, only partial noninfective virus particles were formed, and the stage at which resistance occurred was prior to release from the cell. Taken together, our findings suggest that resistance to Syn9 in Synechococcus WH5701 occurred due to the lack of key proteins that prevented DNA packaging (and perhaps tail attachment), thus thwarting the formation of the next generation of infective phage progeny.
Our combined results for all interactions indicate that the stage at which intracellular resistance acts is interaction-specific, with the infection process of the same generalist phage being arrested at different stages in distinct cyanobacteria (Fig. 6 and SI Appendix, Table S5). Furthermore, the infection cycle of diverse phages ended at different stages in a single cyanobacterial strain (SI Appendix, Table S5), suggesting that multiple means of defense are present in 1 strain. Two strains (Synechococcus WH5701 and Prochlorococcus MIT9215) were more prone than others, however, to allow phages to reach later stages of the infection cycle (SI Appendix, Table S5).
Fig. 6.
Infection by generalist cyanophages is arrested at different stages. A schematic diagram of the progression of infection of a single generalist phage, Syn9, in different resistant cyanobacteria is shown. (Left) Progression of infection in the sensitive strain. (Right) Resistant strains are separated into 3 groups depending on the progress of intracellular infection after entry. Details of the stage of infection reached for all phages and interactions investigated in this study appear in SI Appendix, Table S5. Pro., Prochlorococcus; Syn., Synechococcus.
Of the 17 cyanobacterial strains investigated in this study, only 4 have recognizable active defense systems, and these are nuclease-based (16, 46) (SI Appendix, Supplementary Results and Discussion and Table S2). The progression of the infection cycle and the cellular processes we observed in most cyanobacteria–cyanophage interactions are not in line with these known defenses. Such observations include phage genome replication, partial phage particle formation, bacterial genome degradation, and survival of the cell. Indeed, many intracellular defense mechanisms lead to rapid phage genome degradation, including CRISPR-Cas and restriction modification-like systems (46–48). Others cause death of the bacterium, such as inducible chromosomal islands and abortive infection systems, including those that inhibit the translation machinery (16, 46, 49). Resistance due to lysogeny also does not match the profile we report here of lytic gene expression and phage genome replication (46). Thus, novel means of intracellular defenses likely exist in the marine unicellular cyanobacteria. Whether these are currently unknown active mechanisms of resistance or are more passive mechanisms due to adaptive losses of cellular components essential for phage reproduction remains to be seen.
Over the past decade, it has become increasingly apparent that both specialist and generalist phages exist in nature (2, 3, 5). How much intrinsic resistance is cell surface-related against specialists and intracellular against generalists in other bacterial phyla is currently unknown. Nonetheless, entry by generalists into more bacteria than specialists seems likely. This requires cells to have different modes of resistance against specialists versus generalists, even in the same cell lineage, as cells in nature are not exclusively infected by either specialist or generalist viruses.
Widespread intracellular resistance with virus DNA replication in nonhosts has important implications for horizontal gene transfer. This extends the range of effective DNA transfer by generalist viruses: not only from one host to another but also to a suite of nonhosts that survive the encounter. This, coupled with persistence of phage DNA in the cell for hours, provides considerable opportunity for transfer of phage DNA into bacterial genomes and for transmission of that DNA to subsequent generations. The pervasive entry of phage DNA into nonhosts reported here is thus likely to be a major process driving genome diversification in the abundant marine unicellular cyanobacteria Synechococcus and Prochlorococcus and possibly for a suite of other bacterial phyla as well.
Methods
Bacterial Growth and Phage Propagation.
Cyanobacterial cultures were grown in liquid medium or in agarose pour-plates. Prochlorococcus strains MED4, MIT9215, MIT9515, MIT9312, MIT9313, and NATL2A were grown in the Pro99 seawater-based medium (50), and Synechococcus strains WH8102, WH8109, WH5701, WH7803, WH7805, CC9311, CC9605, RS9916, RS9917, BL107, and MITS9902 were grown in an artificial seawater (ASW) medium (51) with modifications as described by Lindell et al. (52). Cyanobacterial cultures were grown at a temperature of 21 °C and at a light intensity of 10 μmol of photons per m−2⋅s−1 under a 14:10 light/dark regime. Growth of the cultures was monitored using chlorophyll a autofluorescence (excitation/emission: 440/680 nm) as a proxy for biomass (50) using a Synergy 2 microplate reader (BioTek). Typical growth rates of these cyanobacteria under these conditions are a doubling every 1 to 2 d.
Cells were enumerated by flow cytometry prior to experimentation using either an LSR-II flow cytometer (BD Biosciences) equipped with a 488-nm laser or an Influx flow cytometer (BD Biosciences) equipped with 488-nm and 457-nm lasers. Populations were determined based on their forward scatter and autofluorescence. Prochlorococcus cells were detected by their red fluorescence (emission at 692/640 nm), and Synechococcus cells were detected by their orange fluorescence (emission at 580/30 nm). Yellow-green 1-μm-diameter microspheres (Fluoresbrite) were added to each sample as an internal standard for size and fluorescence.
Growth of Prochlorococcus and Synechococcus strains in pour-plates was used for colony formation and to produce lawns for plaque formation and for spot assays as described by Moore et al. (50), with modifications as described by Lindell (53). Ultrapure low-melting-point agarose (Invitrogen) at 0.28% was added to the growth medium, which was also supplemented with 1 mM sodium sulfite after cooling to ∼30 °C. For growth of colonies, the cultures were serially diluted and the helper bacterium Alteromonas sp. EZ55 was added to the plates to obtain higher plating efficiency of the cyanobacteria (54). In pour-plating, the cyanobacteria are added to the low-melting-point agarose mix after cooling to ∼30 °C and are poured into the Petri dish together with the agarose mix and allowed to solidify. As such, the cyanobacteria grow within the agarose and are not spread on top of the semisolid medium.
Heterotrophic bacteria were grown overnight at 37 °C with shaking at 200 rpm. Alteromonas sp. strain EZ55 was grown in YTSS medium (4 g of yeast extract [Becton Dickinson], 2.5 g of tryptone [Becton Dickinson], and 20 g of sea salts [Sigma-Aldrich] per liter) (55). Vibrio harvei and Photobacterium leiognathi were grown in marine broth (Becton Dickinson) (56). Heterotrophic bacteria were enumerated by plating on YTSS agar plates with 1.5% Bacto Agar (Becton Dickinson) or Marine Agar (Becton Dickinson). Before use in adsorption assays, these heterotrophic bacteria were concentrated by centrifugation to 5⋅107 cells per milliliter (6,000 × g at room temperature for 2 min), washed 3 times, and resuspended in ASW medium to remove the rich growth medium.
The cyanophages used in this study are double-stranded DNA viruses belonging to the Caudovirales and included 3 generalist T4-like myoviruses: Syn9, P-TIM40, and S-TIM4; 2 specialist T4-like myoviruses: P-TIM68 and P-TIM75; and 3 specialist T7-like podoviruses: P-SSP7, Syn5, and S-TIP37. They were isolated from various oceanic regions on a variety of hosts (SI Appendix, Table S6). All of the T4-like myoviruses belong to clade III based on g20 portal protein phylogeny, while the T7-like podoviruses belong to clade A (Syn5) or clade B (P-SSP7 and S-TIP37) based on DNApol phylogeny. The genomes of these phages have been sequenced. Phages from the 2 families are very different from each other (12, 57), and each phage within a family is distinct, with at least 40 to 50% of its genome content being different from each of the other phages within the same phage family. The length of the lytic cycle of these cyanophages ranges from 1 to 12 h (SI Appendix, Table S6), and is considerably shorter than the generation time of the cyanobacterial strains. They were propagated by infecting a sensitive strain (SI Appendix, Table S6) at a low multiplicity of infection (MOI) of <0.01 and allowing the culture to clear under cyanobacterial growth conditions. Cells were removed from the lysates by filtration through a 0.2-μm pore-sized filter. For small volumes (<20 mL), an Acrodisc syringe filter (Pall Corp.) was used, whereas Nalgene Rapid Flow 50-mm filter units (Thermo Fisher Scientific) were used for larger volumes.
The concentration of infective phages was determined by the plaque assay. Lysates were serially diluted and plated in pour-plates with a sensitive cyanobacterium at a concentration of 2 × 106 cells to form a cyanobacterial lawn. On occasion, spot assays were conducted to test for infectivity by pipetting 15 μL of undiluted phage on an existing lawn of cyanobacteria and monitoring for clearings (plaques).
Cyanobacterial Resistance and Sensitivity and Cyanophage Host Range.
To determine which strains of cyanobacteria were sensitive or resistant to each phage, as well as the phage’s host range, the 17 cyanobacterial strains mentioned above were challenged with each of the 8 phages in liquid culture and the growth of the culture was followed for a period of 2 to 4 wk. Sensitivity was determined by a decline in cyanobacterial growth in the phage treatments relative to uninfected controls. If no decline of growth was detected in liquid, the ability to infect was tested either by the plaque assay or a spot assay (discussed above). These assays show the ability of the phage to infect over multiple potential infection cycles over the period of 1 d to a week. A strain was considered sensitive if there was either a decline in growth of the liquid culture or if clearings formed in plaque or spot assays. A strain was considered resistant if there was no decline in growth of the liquid culture and there was no formation of plaques or spots on cyanobacterial lawns. The host range of the phage consists of strains that are sensitive to the phage. Phages were considered specialists if they had 1 sensitive host and generalists if they had more than 4 sensitive hosts out of the 17 cyanobacteria tested.
To verify that lack of viral production was not due to inefficient infections (20), we tested for production of infective phages using phage growth curves (58). This was done for the Syn9 phage on the resistant strain Synechococcus WH5701 and for the P-SSP7 phage on the resistant strain Prochlorococcus MIT9215. Experiments were initiated by adding phage at an MOI of 0.1 to cell cultures (108 cells per milliliter), diluted 100-fold after 1 h, and monitored for phage release into the extracellular medium using the plaque assay after filtering out cells. This was done for a period of 26 h, which is longer than the time of phage release (the latent period) on sensitive strains by more than 5-fold for Syn9 (36) and 3-fold for P-SSP7 (35).
To assess if a small subpopulation of resistant cultures was sensitive to the phages, we investigated the same 2 interactions with resistant cyanobacteria described above and assessed sensitivity at the single-cell level. Two different assays were used. In the first, the number of viable cells remaining after challenge with phage was determined by colony counts (viable cells assay). Phages were added to the resistant culture at an MOI of 3 and allowed to adsorb to the cells. Phages that did not adsorb were removed by centrifugation (7,500 × g for 5 min at room temperature for Synechococcus-infected cells and 9,500 × g for 15 min at room temperature for Prochlorococcus-infected cells), and serial dilutions of the cells were plated for colony formation. The percentage of viable cells was determined from the number of colonies formed in the infected cultures compared with those in the same, but uninfected, control cultures.
The second assay measures cell lysis (59). Phages were added to the resistant culture at an MOI of 1 and allowed to adsorb to the cells. Cells from the infected resistant culture were diluted 1,000-fold prior to sorting into wells containing a culture of a sensitive strain, with each well receiving a single cell. This dilution prevents cosorting of free phages into the wells with the cells due to the greater separation between cells and phages (59). Thus, a clearing of the culture in the well indicates that the sorted cell produced infective phage that caused the collapse of the sensitive strain. A total of 576 cells were tested per interaction. Sorting was conducted with a FACSAria-IIIu (BD Biosciences) flow cytometer and was triggered based on autofluorescence and size of the cyanobacterium in the culture, such that free phages are not sorted into the wells.
A decline in colony number (viable cells assay) or lysis of the sensitive strain in the wells (lysis assay) would indicate that the culture contains sensitive cells. Adsorption was 4 h for Syn9 and 7 h for P-SSP7, periods that correspond to maximal adsorption to sensitive strains but prior to phage release. Positive controls for infection were conducted with a sensitive strain for the 2 phages in both assays.
Adsorption Assays.
To determine if resistance to the phage was due to lack of the ability to attach to the cell surface, we carried out adsorption assays following the method of Avrani et al. (13), but without cell concentration. Phages were added to exponentially growing resistant cyanobacteria (108 cells per milliliter) at an MOI of 3. Phage abundance in the extracellular medium was measured by the plaque assay with a sensitive strain immediately after phage addition (at time 0 [t0]) and at a period of time corresponding to maximal adsorption (t1). Cells were removed by filtration prior to the plaque assay. A decline in the concentration of phages in the extracellular medium over this time period indicates attachment. The period of maximal adsorption ranged from 15 min to 8 h, depending on the phage (SI Appendix, Table S6), and was determined from phage growth curves (discussed above). Positive control experiments were carried out with a sensitive cyanobacterium for each phage. Note that for S-TIP37, the lengths of the latent period and maximal adsorption were the same (3 h). Therefore, t1 was sampled at 2.5 h to prevent sampling after cell lysis. Negative control experiments for attachment to the experimental vessel, a 30-mL glass tube, were carried out in the absence of cells (growth medium controls) for each phage.
For ease of interpretation, the results are presented as the percentage of phages that attached to cells: The difference in the number of phages between t0 and t1 was divided by the number of initial phages at t0. In cases where the concentration of free phages at t1 was slightly (but not significantly) higher than the initial concentration at t0, values were converted to 0 rather than presenting negative attachment values. Attachment was considered to be positive when significantly more cyanophages attached to cells than to the experimental vessel, as determined from the negative control experiments. Significance was determined by the nonparametric Mann–Whitney U test (Statistical Analyses).
Infection Experiments.
Infection experiments were carried out to determine whether resistance was intracellular and to determine the intracellular stage at which the viral infection cycle was arrested in a subset of the interactions for which adsorption occurred. The following stages of the infection cycle were investigated: 1) entry of phage DNA into the cell, which was assessed from the transcription of early genes as it is the first stage of infection and phage RNA can only be detected inside an infected cell; 2) gene expression (both transcription and translation of early, middle, and late genes); 3) phage DNA replication and cyanobacterial DNA degradation; 4) capsid assembly; 5) DNA packaging; and 6) formation of intracellular infective phages.
Exponentially growing cyanobacteria (108 cells per milliliter) were infected with phages at an MOI of 3 to ensure infection of nearly all cells in the culture. Infected cultures were incubated in growth conditions for the duration of the experiments. Experiments with sensitive cyanobacteria were carried out as positive controls in all cases to verify infectivity of the phage and to ensure that infection progressed as expected for sensitive strains. Infection experiments were carried out in triplicate as a minimum, each replicate with independent cultures, except for capsid assembly, which was carried out twice. The number of independent experiments performed (n) is mentioned in the figure legends.
Phage gene transcription.
Early phage gene transcription was investigated for 12 of 18 interactions with generalist phages and 7 of 9 interactions with specialist phages for which adsorption was observed. Progression of the transcriptional program was investigated for the same 12 interactions with generalist phages and for 3 of the interactions with specialist phages.
Cells (1 to 1.5 mL) were collected at 0, 0.5, 1, 2, and 4 h postinfection and were centrifuged at 15,000 × g and 4 °C for 10 min. Cells were flash-frozen in liquid nitrogen and stored at −80 °C prior to RNA extraction. Phage entry was assessed from transcription of an early gene for all cyanophages, except for P-TIM75, for which a late gene was analyzed as its genome has yet to be sequenced. Transcription profiles are presented as the number of mRNA copies per nanogram of RNA. The progression of the phage transcriptional program and relative transcript levels were assessed for 2 early, middle, and late genes for the Syn9, S-TIM4, P-TIM40, P-TIM68, and P-SSP7 phages. Transcript levels are presented relative to those in the sensitive strain by comparing maximal levels in the resistant strain with maximal levels in the sensitive strain, irrespective of the time point at which maximal transcript levels were found. Genes from the different transcriptional clusters for Syn9 and P-TIM40 were based on data reported in a study by Doron et al. (36), for S-TIM4 based on homology to Syn9, for P-TIM68 as per Fridman et al. (60), for P-SSP7 based on a study by Lindell et al. (35), for Syn5 based on studies by Raytcheva et al. (61) and Kirzner (62), and for S-TIP37 as per Kirzner (62). The genes used to assess phage transcription in each interaction are shown in SI Appendix, Table S7.
Cells for RNA extraction were thawed on ice and resuspended in 100 μL of 10 mM Tris⋅HCl (pH 8) with 100 units of RNase inhibitor (Applied Biosystems). Cyanobacterial cells were then treated twice with lysozyme (15,000 units of lysozyme [Sigma-Aldrich] at 37 °C for 30 min) prior to the addition of the cell lysis buffer and the isolation of RNA using a Quick RNA Mini Prep Kit (Zymo). Residual DNA was removed using a TURBO DNA-free Kit (Invitrogen) by incubating with DNase at 37 °C for 1 h. Reverse transcription (RT) was conducted with random hexamer primers using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). The cyanobacterial rnpB gene was used as a positive control for RT for all samples. No RT controls were performed on all samples to ensure that reported transcript levels were not from remaining phage DNA. The RT procedure consisted of primer annealing at 25 °C for 10 min, followed by cDNA synthesis at 37 °C for 120 min and reaction termination at 85 °C for 5 min. Samples were diluted 5-fold in 10 mM Tris⋅HCl (pH 8) and stored at −20 °C prior to real-time qPCR (discussed below). The primers used to assess phage transcription in each interaction are shown in SI Appendix, Table S7.
Phage protein expression.
Phage protein presence was investigated for 8 of 18 interactions with generalist phages for which adsorption was observed. Cells (50 mL per sample) were collected by centrifugation at 12,000 × g and 4 °C for 25 min, flash-frozen in liquid nitrogen, and stored at −80 °C prior to protein extraction. The presence of viral proteins was assessed for the Syn9 and S-TIM4 phages in a sensitive strain and in 5 and 3 resistant strains for Syn9 and S-TIM4, respectively. Only proteins detected in the sensitive strain were investigated for their presence or absence in resistant strains. Because we were interested in assessing de novo protein synthesis during the infection process, structural proteins making up the phage particle were largely excluded from the analyses, since such high-copy proteins are detected in the cultures from the phages used for initiating the infection and would confound interpretation of de novo protein synthesis. However, some cases of the absence of low-copy phage structural proteins from infections of resistant strains are reported. Proteins were separated into early, middle, or late expression clusters as determined for Syn9 by Doron et al. (36) and based on Syn9 homology for S-TIM4.
Proteins were extracted from cells in 2% sodium deoxycholate (SDC) and 50 mM ammonium bicarbonate in 2 cycles of sonication. They were then reduced with 3 mM dithiothreitol at 54 °C for 45 min, modified with 10 mM iodoacetamide in the dark at room temperature for 30 min, and digested at 37 °C overnight with modified trypsin (Promega), at a ratio of 1:50 enzyme to protein, in 1% SDC and 50 mM ammonium bicarbonate. A second trypsin digestion was done for 4 h under the same conditions. The deoxycholate was removed by centrifugation at 10,000 × g at room temperature for 10 min; 1% formic acid was added, and the samples were centrifuged again as described above. The tryptic peptides in the supernatant were desalted using C18 tips (Ultra-Micro), dried, and resuspended in 0.1% formic acid. The peptides were resolved by reverse-phase chromatography on 0.075 × 250-mm fused silica capillaries (J&W; Agilent) packed with ReproSil reverse-phase material (Dr. Maisch GmbH). The peptides were eluted in 0.1% formic acid at a flow rate of 0.15 μL⋅min−1 with linear gradients of 5 to 28% acetonitrile for 180 min and 28 to 95% acetonitrile for 15 min, followed by 95% acetonitrile for 25 min. MS was performed with a Q Exactive Plus Mass Spectrometer (Thermo Fisher Scientific) in positive mode with repetitive full MS scans followed by collision-induced dissociation of the 10 most dominant ions selected from the first MS scan. MS data were analyzed using either Proteome Discoverer (Thermo Fisher Scientific) or MaxQuant 1.5.2.8 software against the relevant proteomes from the UniProt database. The MS proteomics data have been deposited in the ProteomeXchange Consortium (63) via the PRIDE (64) partner repository with the dataset identifier PXD013030. Peptide false discovery rates were filtered to 1% using the target-decoy strategy (65). A protein was considered present when at least 1 peptide was detected in 2 of 3 independent experiments. A Mann–Whitney U test was performed to determine if the number of distinct phage proteins identified in each expression cluster in the resistant strain was significantly different from the number identified in the sensitive strain.
Phage DNA replication and cyanobacterial DNA degradation.
Phage DNA replication and cyanobacterial DNA degradation were investigated for 12 of 18 interactions with generalist phages and 7 of 9 interactions with specialist phages for which adsorption was observed. Samples for phage and cyanobacterial DNA quantification were collected at various time points after infection (ranging from 10 to 144 h, depending on the experiment). A quantitative procedure was used to extract DNA from cyanobacterial cells (66). Cells from 200 μL were collected on 25-mm, 0.2-μm pore-sized polycarbonate filters (General Electric) by filtration (at a vacuum of 20 mmHg). Cells were washed 3 times with 3 mL of ASW medium and once with 3 mL of preservation solution (10 mM Tris⋅HCl [pH 8], 100 mM ethylenediaminetetraacetic acid [EDTA], 0.5 M NaCl). The cells were then frozen in liquid nitrogen and stored at −80 °C prior to DNA extraction.
Cells were removed from filters by immersion in 10 mM Tris⋅HCl (pH 8) and agitation in a minibead beater for 2 min at 5,000 rpm without beads. The cells were removed from the shredded filter and heated at 95 °C for 15 min to lyse the cells and extract the DNA (66). DNA was quantified by real-time qPCR (discussed below). Phage DNA was quantified using the portal protein gene, g20, for the T4-like myoviruses and the DNA polymerase gene, DNApol, for the T7-like podoviruses. Cyanobacterial DNA was quantified using the gene for the large subunit of RubisCo, rbcL. The primers used are shown in SI Appendix, Table S7. Phage DNA levels at each time point are presented relative to the maximal concentration of phage DNA in the sensitive strain. Cyanobacterial genome levels are presented relative to measurements at t0, immediately after phage addition, in each interaction.
Capsid assembly.
Intracellular capsid assembly was investigated for the Syn9 phage in the resistant Synechococcus strain WH5701 and compared with that in the sensitive Synechococcus strain WH8102. Thin sections of infected cyanobacterial cells were imaged using TEM in the sensitive and resistant Synechococcus strain, as well as uninfected controls of the resistant Synechococcus WH5701 strain. Two independent experiments were conducted. Imaging was performed on infected cells collected at 5.5 h and 7 h postinfection for the sensitive WH8102 and resistant WH5701 strains, respectively. A longer period of time after phage addition was allowed for the resistant strain in case assembly took longer than in the sensitive strain. Cells from 10-mL cultures were harvested by centrifugation at 12,000 × g and 4 °C for 25 min, fixed with 4% formaldehyde and 2% glutaraldehyde in 0.1 M cacodylate buffer pH 7.4 (Sigma-Aldrich), and stored at 4 °C in the dark for up to 2 wk.
Cells were washed with 0.1 M cacodylate buffer and concentrated by centrifugation, as described above. The cell pellet was embedded in 3.4% agarose and fixed with 1% osmium tetroxide, 0.5% potassium dichromate, and 0.5% potassium hexacyanoferrate in 0.1 M cacodylate buffer. After washing 3 times in 0.1 M cacodylate buffer, samples were incubated for 1 h in an aqueous solution of 2% uranyl acetate and then washed 3 times with water (Becton Dickinson). The cells were dehydrated by 3 washes in dehydrated ethanol. Cells were then embedded in agar 100 epoxy resin (Agar Scientific). Alternatively, cell pellets were mounted on an aluminum disk with a depth of 100 μm (Engineering Office M. Wohlwend GmbH) and covered with a flat disk. The sandwiched sample was frozen in an EM ICE high-pressure freezing machine (Leica Microsystems). Cells were subsequently freeze-substituted in an AFS2 freeze substitution device (Leica Microsystems) in anhydrous acetone containing 0.2% uranyl acetate and 0.2% osmium tetroxide for 2 d at −90 °C and then warmed to −30 °C over 24 h. They were washed 3 times with acetone, brought to room temperature, and infiltrated with resin for 5 d in a series of increasing concentrations of agar 100 epoxy resin in acetone. After polymerization at 60 °C, 60- to 80-nm sections were stained with 2% uranyl acetate and 0.1% lead citrate (67). Samples were imaged using a Tecnai Spirit T12 or Tecnai T12 transmission electron microscope (FEI) operating at 120 kV. Images were acquired using a 2 k × 2 k charge-coupled device camera (Eagle; FEI) or a Gatan 4 k × 4 k OneView complementary metal-oxide semiconductor camera (Gatan).
The number and diameter of capsids and carboxysomes were measured using ImageJ 1.4g software. Two-tailed Student’s t tests were performed to test if the number of phage capsids per cell and the average capsid diameter were different between the sensitive and resistant strains and if there was a significant difference in the diameter of phage capsids and cyanobacterial carboxysomes in each strain.
DNA packaging and formation of intracellular infective phages.
DNA packaging into capsids and the formation of infective phages intracellularly were investigated for the Syn9 phage in the resistant Synechococcus strain WH5701 and compared with that in the sensitive Synechococcus strain WH8102. At different time points postinfection, cells were disrupted using the following procedure. Cells were incubated with 30,000 units (4 mg/mL) lysozyme (Sigma–Aldrich) at 37 °C for 30 min and subjected to 4 temperature change cycles of 2 min at −5 °C followed by 2 min at 37 °C. The weakened cells were then disrupted during incubation in 1% chloroform (vol/vol) at room temperature for 1 h with continuous inversion at 20 × g. Disrupted cultures were then filtered through 0.2-μm pore-sized syringe filters (Acrodisc Syringe filter; Pall Corp.) to remove remaining intact cyanobacteria and large cellular debris. This cell disruption procedure was first tested on Syn9 phage lysates and shown not to negatively affect infectivity.
Packaging of phage DNA into capsids was assessed in the filtrate of the disrupted cells by testing for protection from DNase. DNase treatment was carried out in conditions known to fully digest free DNA in seawater without detrimentally affecting phage DNA present in capsids (10). Samples were incubated with 5 units/mL DNase I (Sigma-Aldrich) at 37 °C for 1 h. The enzyme was inactivated by adding 50 mM EDTA (pH 8). The sample was diluted 100-fold in 10 mM Tris⋅HCl (pH 8). Phage DNA that had been protected from DNase digestion was then quantified using qPCR with primers for the g20 gene (discussed below).
The plaque assay was used to determine the number of infective phages in the filtrate of the disrupted cells at each time point. This measures the number of infective phages present both intracellularly and that had been released to the extracellular medium. At each time point, this was compared with the number of infective phages in untreated cells (without cell disruption), which provides the number of infective phages in the extracellular medium alone. Thus, the timing and number of infective phages produced intracellularly are determined from the difference between the disrupted and untreated samples.
Determination of Ploidy in Marine Cyanobacteria.
The number of genome copies per cyanobacterial cell (ploidy) was determined for 21 cyanobacterial strains. DNA was extracted from 200-μL cultures after collection on 25-mm, 0.2-μm pore-sized polycarbonate filters (General Electric) using the quantitative extraction procedure described above. qPCR was carried out for the rbcL and rnpB genes, which are hundreds of thousands of base pairs apart on nearly all of the cyanobacterial genomes. Gene copy numbers were determined using standard curves (discussed below). The number of genome copies per cell was inferred after quantifying cyanobacterial cell abundances in each culture using an Influx flow cytometer (discussed above).
Real-Time qPCR.
Each qPCR reaction contained 1X LightCycler 480 SYBR Green I Master mix (Roche), 200 nM desalted primers (SI Appendix, Table S7), and 5 μL of template (in 10 mM Tris⋅HCl [pH 8]) in a total reaction volume of 25 μL. Reactions were carried out on a LightCycler 480 Real-Time PCR System (Roche). The cycling program began with a denaturation step of 95 °C for 5 to 10 min, followed by 35 to 45 cycles of amplification. Each cycle consisted of denaturation at 95 °C for 10 s, annealing at 55 to 65 °C (5 °C below the primers’ calculated melting temperature [Tm]) for 10 s, and elongation at 72 °C for 10 s, at the end of which plate fluorescence was read (6-Carboxyfluorescein [6-FAM]; excitation/emission: 465/510 nm). The point at which the fluorescence of a sample rose above the background fluorescence was calculated using LightCycler 480 software (release 1.5.0) using the absolute quantification/second-derivative maximum analysis package (68). Specificity of the amplified PCR product was verified by performing melting curve analysis on the LightCycler 480 instrument.
Standard curves were generated from genomic DNA for each amplified gene and were used to calculate the number of gene copies in the samples. Genomic DNA for standard curves from cyanobacterial cells was extracted using a DNeasy Blood & Tissue Kit (Qiagen) and from phages using a phenol/chloroform-based method (69). DNA concentrations in nanograms per milliliter were measured by absorbance at 260 nm using a Synergy 2 microplate reader (BioTek). DNA concentrations were converted to genome copies per milliliter by entering the genome length of the cyanobacterium or phage into the URI Genomics & Sequencing Center calculator for determining the number of copies of a template (http://cels.uri.edu/gsc/cndna.html).
Homology-Based Searches for Defense Systems.
We investigated the presence of known active defense systems in the marine cyanobacteria that enable the host cell to remain viable. We searched for restriction enzymes using the Restriction Enzyme Database (http://rebase.neb.com/rebase/rebase.html) using each cyanobacterial strain’s name. We also performed key word searches in the genome entry of each of the 17 strains used here for the following words: restriction, endonuclease, BREX, cas, and CRISPR. The Pfam database (https://pfam.xfam.org/) was used to confirm all annotations by searching for protein domains and domain families belonging to known active defense systems. When discrepancies arose, the annotation selected was that in Pfam. Furthermore, since it has been suggested that defense systems are clustered in specific genomic locations (70), once we found a defense related protein, we investigated the neighboring protein sequences for defense genes using Pfam. We also performed protein BLAST searches using the National Center for Biotechnology Information for all proteins identified to assess if homologous proteins are present in other marine cyanobacteria. It should be noted that we did not search for known inducible chromosome islands or abortive infection systems, as these cause the death of the cell.
Statistical Analysis.
All statistical analysis was performed using IBM SPSS Statistics software, version 23/24. Either the Student’s t test (parametric) or Mann–Whitney U test (nonparametric) was used depending on the distribution of the data and equality of variance between the datasets. Normality was tested using the Shapiro–Wilk test for normality. Equality of variance was tested using Levene’s test using SPSS software.
Supplementary Material
Acknowledgments
We thank Ayalla Fedida for her mentorship at the early stages of this project, Dvora Rywka Sehtman and Shira Shankshalil for work on resistance to Syn5 and S-TIP37, Naama Dekel-Bird and Daniel Schwartz for the isolation of the P-TIM75 phage, and D.L. laboratory members Oded Beja and Sarit Avrani for discussions and suggestions throughout the study and for comments on the manuscript. Protein analyses were carried out at the Technion Smoler Proteomics Center. Flow cytometry was done at the Infrastructure unit of the Technion’s Lokey Interdisciplinary Center for Life Sciences & Engineering. Electron microscopy was conducted by Eyal Shimoni at the Weizmann Institute’s Irving and Cherna Moskowitz Center for Nano and Bio-Nano Imaging. This work was supported by grants from the Simons Foundation (Grants 329108 and 529554) and the European Research Council (Starting Grant 203406) to D.L. This is a contribution of the Simons Collaboration on Ocean Processes and Ecology (SCOPE).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Proteome data were submitted to the ProteomeXchange Consortium with dataset identifier PXD013030.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1906897116/-/DCSupplemental.
References
- 1.Sullivan M. B., Waterbury J. B., Chisholm S. W., Cyanophages infecting the oceanic cyanobacterium Prochlorococcus. Nature 424, 1047–1051 (2003). Erratum in: Nature 426, 584 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Flores C. O., Meyer J. R., Valverde S., Farr L., Weitz J. S., Statistical structure of host-phage interactions. Proc. Natl. Acad. Sci. U.S.A. 108, E288–E297 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koskella B., Meaden S., Understanding bacteriophage specificity in natural microbial communities. Viruses 5, 806–823 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dekel-Bird N. P., Sabehi G., Mosevitzky B., Lindell D., Host-dependent differences in abundance, composition and host range of cyanophages from the Red Sea. Environ. Microbiol. 17, 1286–1299 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Ross A., Ward S., Hyman P., More is better: Selecting for broad host range bacteriophages. Front. Microbiol. 7, 1352 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flombaum P., et al. , Present and future global distributions of the marine Cyanobacteria Prochlorococcus and Synechococcus. Proc. Natl. Acad. Sci. U.S.A. 110, 9824–9829 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scanlan D. J., et al. , Ecological genomics of marine picocyanobacteria. Microbiol. Mol. Biol. Rev. 73, 249–299 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biller S. J., Berube P. M., Lindell D., Chisholm S. W., Prochlorococcus: The structure and function of collective diversity. Nat. Rev. Microbiol. 13, 13–27 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Waterbury J. B., Valois F. W., Resistance to co-occurring phages enables marine synechococcus communities to coexist with cyanophages abundant in seawater. Appl. Environ. Microbiol. 59, 3393–3399 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baran N., Goldin S., Maidanik I., Lindell D., Quantification of diverse virus populations in the environment using the polony method. Nat. Microbiol. 3, 62–72 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suttle C. A., Chan A. M., Marine cyanophages infecting oceanic and coastal strains of Synechococcus–Abundance, morphology, cross-infectivity and growth-characteristics. Mar. Ecol. Prog. Ser. 92, 99–109 (1993). [Google Scholar]
- 12.Labrie S. J., et al. , Genomes of marine cyanopodoviruses reveal multiple origins of diversity. Environ. Microbiol. 15, 1356–1376 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Avrani S., Wurtzel O., Sharon I., Sorek R., Lindell D., Genomic island variability facilitates Prochlorococcus-virus coexistence. Nature 474, 604–608 (2011). [DOI] [PubMed] [Google Scholar]
- 14.Zhao F., et al. , Genome-wide analysis of restriction-modification system in unicellular and filamentous cyanobacteria. Physiol. Genomics 24, 181–190 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Cai F., Axen S. D., Kerfeld C. A., Evidence for the widespread distribution of CRISPR-Cas system in the Phylum Cyanobacteria. RNA Biol. 10, 687–693 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doron S, et al. (2018) Systematic discovery of antiphage defense systems in the microbial pangenome. Science 359, eaar4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stoddard L. I., Martiny J. B. H., Marston M. F., Selection and characterization of cyanophage resistance in marine Synechococcus strains. Appl. Environ. Microbiol. 73, 5516–5522 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Antonovics J., et al. , The origin of specificity by means of natural selection: Evolved and nonhost resistance in host-pathogen interactions. Evolution 67, 1–9 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Schwartz D. A., Lindell D., Genetic hurdles limit the arms race between Prochlorococcus and the T7-like podoviruses infecting them. ISME J. 11, 1836–1851 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Howard-Varona C., et al. , Multiple mechanisms drive phage infection efficiency in nearly identical hosts. ISME J. 12, 1605–1618 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kettler G. C., et al. , Patterns and implications of gene gain and loss in the evolution of Prochlorococcus. PLoS Genet. 3, e231 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodriguez-Valera F., et al. , Explaining microbial population genomics through phage predation. Nat. Rev. Microbiol. 7, 828–836 (2009). [DOI] [PubMed] [Google Scholar]
- 23.Martiny J. B. H., Riemann L., Marston M. F., Middelboe M., Antagonistic coevolution of marine planktonic viruses and their hosts. Annu. Rev. Mar. Sci. 6, 393–414 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Avrani S., Schwartz D. A., Lindell D., Virus-host swinging party in the oceans: Incorporating biological complexity into paradigms of antagonistic coexistence. Mob. Genet. Elements 2, 88–95 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bertozzi Silva J., Storms Z., Sauvageau D., Host receptors for bacteriophage adsorption. FEMS Microbiol. Lett. 363, fnw002 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Inoue T., Matsuzaki S., Tanaka S., A 26-kDa outer membrane protein, OmpK, common to Vibrio species is the receptor for a broad-host-range vibriophage, KVP40. FEMS Microbiol. Lett. 125, 101–105 (1995). [DOI] [PubMed] [Google Scholar]
- 27.Scholl D., Rogers S., Adhya S., Merril C. R., Bacteriophage K1-5 encodes two different tail fiber proteins, allowing it to infect and replicate on both K1 and K5 strains of Escherichia coli. J. Virol. 75, 2509–2515 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fenton A., Antonovics J., Brockhurst M. A., Two-step infection processes can lead to coevolution between functionally independent infection and resistance pathways. Evolution 66, 2030–2041 (2012). [DOI] [PubMed] [Google Scholar]
- 29.Sieber M., Gudelj I., Do-or-die life cycles and diverse post-infection resistance mechanisms limit the evolution of parasite host ranges. Ecol. Lett. 17, 491–498 (2014). [DOI] [PubMed] [Google Scholar]
- 30.Chibani-Chennoufi S., Bruttin A., Dillmann M. L., Brüssow H., Phage-host interaction: An ecological perspective. J. Bacteriol. 186, 3677–3686 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guyader S., Burch C. L., Optimal foraging predicts the ecology but not the evolution of host specialization in bacteriophages. PLoS One 3, e1946 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heineman R. H., Springman R., Bull J. J., Optimal foraging by bacteriophages through host avoidance. Am. Nat. 171, E149–E157 (2008). [DOI] [PubMed] [Google Scholar]
- 33.Duffy S., Turner P. E., Burch C. L., Pleiotropic costs of niche expansion in the RNA bacteriophage phi 6. Genetics 172, 751–757 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Enav H., Kirzner S., Lindell D., Mandel-Gutfreund Y., Beja O., Adaptation to sub-optimal hosts is a driver of viral diversification in the ocean. Nat. Commun. 9, 4698 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lindell D., et al. , Genome-wide expression dynamics of a marine virus and host reveal features of co-evolution. Nature 449, 83–86 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Doron S., et al. , Transcriptome dynamics of a broad host-range cyanophage and its hosts. ISME J. 10, 1437–1455 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Black L. W., Rao V. B., Structure, assembly, and DNA packaging of the bacteriophage T4 head. Adv. Virus Res. 82, 119–153 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fokine A., et al. , The molecular architecture of the bacteriophage T4 neck. J. Mol. Biol. 425, 1731–1744 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weed L. L., Cohen S. S., The utilization of host pyrimidines in the synthesis of bacterial viruses. J. Biol. Chem. 192, 693–700 (1951). [PubMed] [Google Scholar]
- 40.Zeng Q., Chisholm S. W., Marine viruses exploit their host’s two-component regulatory system in response to resource limitation. Curr. Biol. 22, 124–128 (2012). [DOI] [PubMed] [Google Scholar]
- 41.Thompson L. R., et al. , Phage auxiliary metabolic genes and the redirection of cyanobacterial host carbon metabolism. Proc. Natl. Acad. Sci. U.S.A. 108, E757–E764 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Griese M., Lange C., Soppa J., Ploidy in cyanobacteria. FEMS Microbiol. Lett. 323, 124–131 (2011). [DOI] [PubMed] [Google Scholar]
- 43.Binder B. J., Chisholm S. W., Cell cycle regulation in marine Synechococcus sp. strains. Appl. Environ. Microbiol. 61, 708–717 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ohbayashi R., et al. , Coordination of polyploid chromosome replication with cell size and growth in a cyanobacterium. MBio 10, e00510–e00519 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weigele P. R., et al. , Genomic and structural analysis of Syn9, a cyanophage infecting marine Prochlorococcus and Synechococcus. Environ. Microbiol. 9, 1675–1695 (2007). [DOI] [PubMed] [Google Scholar]
- 46.van Houte S., Buckling A., Westra E. R., Evolutionary ecology of prokaryotic immune mechanisms. Microbiol. Mol. Biol. Rev. 80, 745–763 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Simmon V. F., Lederberg S., Degradation of bacteriophage lambda deoxyribonucleic acid after restriction by Escherichia coli K-12. J. Bacteriol. 112, 161–169 (1972). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Garneau J. E., et al. , The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468, 67–71 (2010). [DOI] [PubMed] [Google Scholar]
- 49.Labrie S. J., Samson J. E., Moineau S., Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 8, 317–327 (2010). [DOI] [PubMed] [Google Scholar]
- 50.Moore L. R., et al. , Culturing the marine cyanobacterium Prochlorococcus. Limnol. Oceanogr. Methods 5, 353–362 (2007). [Google Scholar]
- 51.Wyman M., Gregory R. P. F., Carr N. G., Novel role for phycoerythrin in a marine cyanobacterium, Synechococcus strain DC2. Science 230, 818–820 (1985). [DOI] [PubMed] [Google Scholar]
- 52.Lindell D., Padan E., Post A. F., Regulation of ntcA expression and nitrite uptake in the marine Synechococcus sp. strain WH 7803. J. Bacteriol. 180, 1878–1886 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lindell D., “The genus Prochlorococcus, phylum cyanobacteria” in The Prokaryotes–Other Major Lineages of Bacteria and the Archaea, Rosenberg E., Stackebrandt E., DeLong E., Lory S., Thompson F. L., Eds. (Springer, ed. 4, 2014), vol. 11, pp. 829–845. [Google Scholar]
- 54.Morris J. J., Kirkegaard R., Szul M. J., Johnson Z. I., Zinser E. R., Facilitation of robust growth of Prochlorococcus colonies and dilute liquid cultures by “helper” heterotrophic bacteria. Appl. Environ. Microbiol. 74, 4530–4534 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morris J. J., Johnson Z. I., Szul M. J., Keller M., Zinser E. R., Dependence of the cyanobacterium Prochlorococcus on hydrogen peroxide scavenging microbes for growth at the ocean’s surface. PLoS One 6, e16805 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zobell C. E., Studies on marine bacteria. I. The cultural requirements of heterotrophic aerobes. J. Mar. Res. 4, 41–75 (1941). [Google Scholar]
- 57.Sullivan M. B., et al. , Genomic analysis of oceanic cyanobacterial myoviruses compared with T4-like myoviruses from diverse hosts and environments. Environ. Microbiol. 12, 3035–3056 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ellis E. L., Delbrück M., The growth of bacteriophage. J. Gen. Physiol. 22, 365–384 (1939). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kirzner S., Barak E., Lindell D., Variability in progeny production and virulence of cyanophages determined at the single-cell level. Environ. Microbiol. Rep. 8, 605–613 (2016). [DOI] [PubMed] [Google Scholar]
- 60.Fridman S., et al. , A myovirus encoding both photosystem I and II proteins enhances cyclic electron flow in infected Prochlorococcus cells. Nat. Microbiol. 2, 1350–1357 (2017). [DOI] [PubMed] [Google Scholar]
- 61.Raytcheva D. A., Haase-Pettingell C., Piret J. M., King J. A., Intracellular assembly of cyanophage Syn5 proceeds through a scaffold-containing procapsid. J. Virol. 85, 2406–2415 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kirzner S., “Different infection properties of two distinct T7-like podoviruses infecting the marine Synechococcus strain WH8109,” PhD thesis, Technion – Israel Institute of Technology, Haifa, Israel (2015).
- 63.Deutsch E. W., et al. , The ProteomeXchange consortium in 2017: Supporting the cultural change in proteomics public data deposition. Nucleic Acids Res. 45, D1100–D1106 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Perez-Riverol Y., et al. , The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 47, D442–D450 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Elias J. E., Gygi S. P., Target-decoy search strategy for mass spectrometry-based proteomics. Methods Mol. Biol. 604, 55–71 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zinser E. R., et al. , Prochlorococcus ecotype abundances in the North Atlantic Ocean as revealed by an improved quantitative PCR method. Appl. Environ. Microbiol. 72, 723–732 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Reynolds E. S., The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J. Cell Biol. 17, 208–212 (1963). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Heid C. A., Stevens J., Livak K. J., Williams P. M., Real time quantitative PCR. Genome Res. 6, 986–994 (1996). [DOI] [PubMed] [Google Scholar]
- 69.Pickard D. J., Preparation of bacteriophage lysates and pure DNA. Methods Mol. Biol. 502, 3–9 (2009). [DOI] [PubMed] [Google Scholar]
- 70.Makarova K. S., Wolf Y. I., Snir S., Koonin E. V., Defense islands in bacterial and archaeal genomes and prediction of novel defense systems. J. Bacteriol. 193, 6039–6056 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.