Significance
The intestinal epithelium is a protective barrier against ingested pharmaceuticals and microbiota. Delamination of intestinal epithelial cells (IEC) is a common feature of pharmaceutical-induced enteropathies, but physiological functions and underlying mechanisms remain unknown. Using zebrafish, we define the mechanisms underlying intestinal toxicity of a human pharmaceutical, the NSAID Glafenine. Glafenine induced IEC delamination independent of microbiota colonization, yet Glafenine treatment in colonized animals caused inflammation and microbiota dysbiosis. Glafenine-induced IEC delamination was mediated by the unfolded protein response and protected from excessive inflammation and mortality. Glafenine toxicity resulted not from NSAID activity but from off-target inhibition of multidrug-resistance efflux pumps. These results reveal the mechanisms of Glafenine toxicity, and implicate IEC delamination as a protective response to pharmaceutical-induced enteropathies.
Keywords: NSAID, MDR efflux pump, zebrafish, microbiota, intestine
Abstract
Intestinal epithelial cell (IEC) shedding is a fundamental response to intestinal damage, yet underlying mechanisms and functions have been difficult to define. Here we model chronic intestinal damage in zebrafish larvae using the nonsteroidal antiinflammatory drug (NSAID) Glafenine. Glafenine induced the unfolded protein response (UPR) and inflammatory pathways in IECs, leading to delamination. Glafenine-induced inflammation was augmented by microbial colonization and associated with changes in intestinal and environmental microbiotas. IEC shedding was a UPR-dependent protective response to Glafenine that restricts inflammation and promotes animal survival. Other NSAIDs did not induce IEC delamination; however, Glafenine also displays off-target inhibition of multidrug resistance (MDR) efflux pumps. We found a subset of MDR inhibitors also induced IEC delamination, implicating MDR efflux pumps as cellular targets underlying Glafenine-induced enteropathy. These results implicate IEC delamination as a protective UPR-mediated response to chemical injury, and uncover an essential role for MDR efflux pumps in intestinal homeostasis.
The intestine is lined with a single layer of polarized intestinal epithelial cells (IECs), which function both to absorb dietary nutrients and to provide a physical barrier against microbiota residing in the intestinal lumen. The diversity of physical, chemical, and biological signals present in the intestinal environment can exert stresses on IECs, and coordinated epithelial renewal and barrier function is essential for maintenance of intestinal homeostasis. The intestinal epithelium is one of the most rapidly proliferating tissues and cellular turnover is tightly controlled. Dysregulation of IEC renewal is associated with a number of pathological conditions and diseases (1), yet rigorous investigation of these processes in vivo has remained challenging with mammalian models.
Xenobiotics are introduced into the intestine through the oral route as dietary components or pharmaceuticals. IECs play essential roles in orally delivered pharmaceutical biology, functioning as primary sites of absorption, metabolism, and excretion (2–4). As the front line of the host–xenobiotic interface, the intestinal epithelium is also a primary site of action for xenobiotic toxicity. Multidrug resistance (MDR) efflux pumps are 1 mechanism by which eukaryotes minimize intestinal toxicity of xenobiotics. MDR efflux pumps are ancient, evolutionarily conserved active transporters expressed on both apical and basolateral surfaces of IECs, which act on a variety of substrates (5). A growing number of xenobiotics (including many pharmaceuticals) have been identified as MDR efflux pump substrates; however, the impact of these transporters and their ligands on intestinal homeostasis remains unclear (6, 7). IECs are known to possess additional means for mitigating xenobiotic and other environmental stressors, including phase I metabolism (8) and activation of the unfolded protein response (UPR) (9) and inflammatory pathways (10). However, the role of these pathways in mediating intestinal responses to injury remains poorly understood for most xenobiotics.
Gastrointestinal pathology is common in people using pharmaceuticals, including nonsteroidal antiinflammatory drugs (NSAIDs) (11). While gastric ulceration has historically been a defining clinical presentation of NSAID-induced enteropathy, small intestinal pathology has also been observed, although the incidence may be underreported due to diagnostic limitations (e.g., the small bowel is less accessible to endoscopy than the stomach) (12). Both acute and chronic NSAID exposure increases intestinal inflammation, ulceration, and intestinal permeability in murine models (13). However, in vivo investigation of dynamic molecular and cellular responses to xenobiotic toxicity in the intestine has remained problematic, as experimental endpoints in rodent models are typically terminal. While a number of mechanisms of action have been ascribed for NSAID intestinal toxicity (including mitochondrial damage, intercalation into lipid bilayers, decreased mucus production, and increased intestinal permeability) (13, 14), NSAIDs are a diverse class of drugs and other mechanisms remain possible.
In the past decade, it has become increasingly apparent that intestinal microbiota serve important functions in drug pharmacology and metabolism, affecting both efficacy and tolerability (3, 15, 16). For example, microbial metabolism contributes to the retoxification of certain pharmaceuticals, including NSAIDs (e.g., diclofenac and ketoprofen) (13, 17) and chemotherapeutic agents (e.g., irinotecan) (18), leading to intestinal pathology. However, potential interactions between microbiota, pharmaceuticals, and host physiology remain unexplored for the vast majority of xenobiotics.
There is therefore a pressing need for improved animal models to elucidate the complex relationships between host, microbiota, and xenobiotic compounds. Here, we investigate mechanisms underlying xenobiotic-induced intestinal toxicity by leveraging the strengths of the zebrafish model system, including in vivo imaging, genetic tractability, and facile chemical and gnotobiotic manipulations. We previously demonstrated that acute (12 h) exposure of zebrafish larvae to the NSAID Glafenine elicited dramatic IEC sloughing (19). Our present study provides a substantial vertical advance in understanding intestinal toxicity, microbiota responses, as well as the physiological significance of IEC delamination following Glafenine exposure. Glafenine was used as an oral over-the-counter analgesic in Europe and the Middle East for more than 30 y before global withdrawal due to hepatic and renal toxicity, as well as anaphylaxis, yet the underlying mechanisms of toxicity were never defined (20–22). Intestinal pathology, however, was not reported in individuals taking Glafenine. Using a serial exposure regimen relevant to human chronic NSAID use, we demonstrate here that Glafenine-induced IEC loss is associated with a robust inflammatory response and UPR, as well as shifts in both host and environmental microbiotas. Our data indicate that IEC delamination depends on intact UPR signaling through the sensor Ire1α, and that delamination is a protective response that serves to limit inflammation and mortality. Finally, we provide evidence that Glafenine-induced pathology is caused not by its NSAID activity but instead by its off-target effects as an MDR efflux pump inhibitor.
Results
Serial Glafenine Exposure Results in IEC Delamination.
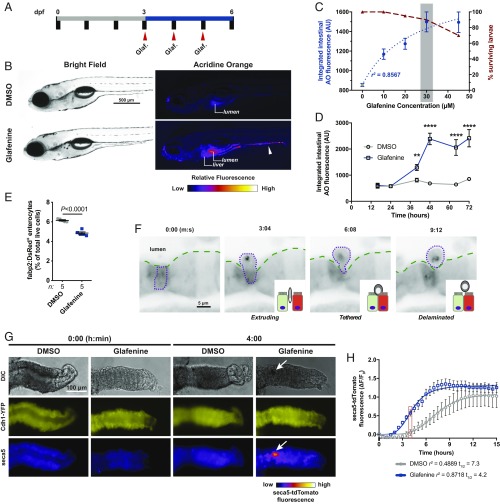
To establish a chronic intestinal injury model in larval zebrafish, we devised a serial exposure regimen in which animals are dosed with Glafenine at 24-h intervals from 3 to 6 days postfertilization (dpf) (Fig. 1A). Consistent with acute Glafenine exposure (19), we observed positive staining with the vital dye acridine orange (AO) in debris accumulating in the intestinal lumen as well as the liver (Fig. 1B and SI Appendix, Fig. S1A) (23) and quantitative analysis revealed a strong correlation between Glafenine concentration and AO+ intestinal material up to the 45-μM limit of solubility in zebrafish media (Fig. 1C and SI Appendix, Fig. S1 A–D). While survival was typically >90%, we noted minor alterations in standard developmental metrics, indicating serial Glafenine exposure causes slight developmental delay in zebrafish larvae (SI Appendix, Fig. S2).
Fig. 1.
Serial Glafenine exposure results in IEC delamination. (A) Schematic of dosing regimen used for serial Glafenine (Glaf.) exposure. (B) Representative brightfield and AO fluorescence images of 6 dpf DMSO- and Glafenine-treated larvae (arrowhead points to AO+ material in the intestinal lumen). (C) Glafenine dose–response for quantified intestinal AO fluorescence (left y axis, blue, 3-parameter least-squares fit) and survival (right y axis, maroon). (D) Kinetics of intestinal AO response with 30 μM Glafenine (n = 20 larvae per condition per time point; significance was determined between treatment groups within each time point by unpaired 2-sided Student’s t test; **P < 0.01, ****P < 0.0001). (E) Flow cytometry analysis of relative abundance of viable (7-AAD−) Tg(fabp2:DsRed)+ enterocytes from DMSO- and Glafenine-treated larvae (each point is a pool of 20 larvae; significance was determined in by unpaired 2-sided Student’s t test). (F) Sequential frames from live confocal imaging of Tg(fabp2:DsRed) larvae at 40 h into the treatment regimen. (G) Representative images of dissected larval zebrafish intestines exposed to DMSO or Glafenine ex vivo at 0 and 4 h (arrow points to mass of apoptotic cells in the intestinal lumen). (H) Quantification of seca5-tdTomato fluorescence in larval intestinal explants (n = 6 DMSO-treated and 5 Glafenine-treated intestines; 4-parameter least-squares fit; comparison of t1/2: P = 0.0002 [extra sum-of-squares F test]).
Based on survival and solubility data, we selected a Glafenine dose of 30 μM for all subsequent experiments (Fig. 1C). Kinetic analysis revealed a significant increase in intestinal AO fluorescence as early as 36 h into the exposure regimen, typically reaching peak intensity by 48 h, and persisting until the experimental endpoint at 72 h (Fig. 1D). Thus, serial Glafenine exposure results in time- and dose-dependent accumulation of AO+ material in the intestinal lumen. We hypothesized that the material accumulating in the intestinal lumen of Glafenine-treated larvae was composed of dead or dying IECs. Analysis of transgenic zebrafish ubiquitously expressing a secreted AnnexinV–tdTomato fusion protein [Tg(ubb:seca5-tdTomato)xt24], which labels apoptotic cells, revealed accumulation of seca5-tdTomato+ signal in the intestinal lumen. This indicates that the AO+ material indeed contains apoptotic cells (SI Appendix, Fig. S1 E and F and Movie S1).
Microscopic analysis revealed nucleated cells labeled with the absorptive cell marker 4E8 in the intestinal lumen of Glafenine-treated larvae (SI Appendix, Fig. S3B). On numerous occasions we observed the mass of apoptotic material passed out of the intestine (SI Appendix, Fig. S3A). We reasoned that if IECs are the predominant component of this mass, and enterocytes constitute a majority of IECs, we could detect enterocyte-specific epitopes in the surrounding media. Dot blot analysis of media samples with the enterocyte antibody 4E8 (24) demonstrated enrichment of reactive epitopes in media from Glafenine-treated animals at 24, 48, and 72 h after treatment (SI Appendix, Fig. S3 C and D), indicating that enterocytes are shed and passed out of the intestine following Glafenine treatment.
We next tested whether initiation of apoptosis or necroptosis in IECs precedes their exit from the epithelial layer. Coexposure with neither pan-caspase inhibitors nor the Ripk1 inhibitor Nec-1 altered the accumulation of AO+ material in the intestinal lumen (SI Appendix, Fig. S4 A and B), indicating that caspase- and Ripk1-dependent cell death pathways are dispensable for Glafenine-induced IEC loss. This also suggests IEC death occurs after shedding into the lumen.
Flow cytometry analysis of transgenic zebrafish with fluorescently labeled absorptive enterocytes [Tg(-4.5kb fabp2:DsRed) (25, 26); hereafter referred to as Tg(fabp2:DsRed)], revealed a 15 to 20% reduction in viable fabp2:DsRed+ cells after 72 h of serial Glafenine exposure, further validating enterocyte loss (Fig. 1E and SI Appendix, Fig. S5). Intestinal morphometry revealed increased lumen area and decreased epithelial thickness in Glafenine-treated larvae, hallmarks of intestinal injury (SI Appendix, Fig. S6 A and B). Consistent with our previous findings following acute Glafenine treatment (19), serial Glafenine exposure did not lead to increased intestinal permeability or profound rearrangements of epithelial junctions, although expression of genes encoding junctional components was slightly reduced in fabp2:DsRed+ enterocytes (SI Appendix, Fig. S6 C–I). These data demonstrate that absorptive enterocytes are sensitive to Glafenine treatment, but that enterocyte loss is achieved without major impairments to overall epithelial barrier function.
In vivo confocal microscopy imaging of Glafenine-treated Tg(fabp2:DsRed) larvae yielded further insights into Glafenine-induced IEC loss (Fig. 1F and Movies S2–S5). IEC shedding resembled epithelial cell delamination (27), with a stepwise progression of morphological events proceeding from rounding, extrusion, tethering, and finally detachment. Given that cell shedding was Caspase- and Ripk-independent (SI Appendix, Fig. S4 A and B), our results suggest Glafenine induces live-cell apical extrusion of IECs (27).
Considering that Glafenine induces profound enterocyte loss yet modestly effects animal mortality, we asked if IEC proliferation was altered in Glafenine-treated larvae. The proportion of EdU+ epithelial cells as well as cells in the underlying mesenchyme and muscle (subepithelium) (SI Appendix, Fig. S3 E and F) was markedly increased. These findings suggest that enhanced proliferation may be 1 mechanism by which zebrafish larvae tolerate substantial IEC loss.
Our previous study suggested that the IEC shedding phenomenon was uniquely elicited by the NSAID Glafenine (19). We explored this further in our serial exposure model, using a structurally diverse panel of selective and nonselective NSAIDs and COX inhibitors, and found that none of the other NSAIDs evaluated induced IEC shedding at any tested concentration (SI Appendix, Fig. S7 C and D and Dataset S1). Given this result, we asked if Glafenine possessed NSAID activity in zebrafish larvae. Considering that most NSAIDs function by inhibiting COX-dependent prostaglandin biosynthesis, we measured PGE metabolite levels in whole larvae and found significant and comparable reductions with both Glafenine and Indomethacin treatment relative to controls (SI Appendix, Fig. S7E). Supplementation with exogenous PGE2, however, failed to ameliorate Glafenine-induced IEC shedding (SI Appendix, Fig. S7F). These data suggest that while Glafenine does inhibit COX activity in zebrafish, the mechanism of IEC loss may be independent of decreased prostaglandin biosynthesis.
To investigate whether intestinal toxicity was a direct effect of Glafenine, we devised an explant culture assay using dissected larval zebrafish intestines cultured ex vivo and exposed to DMSO or Glafenine. Quantification from time-lapse imaging of intestines from cdh1-YFP;Tg(ubb:seca5-tdTomato) larvae revealed Glafenine accelerated apoptosis of IECs ex vivo, achieving half-maximal fluorescence at 4.2 h (vs. 7.3 h for DMSO-treated intestines) (Fig. 1 G and H and Movies S6 and S7). Although both historical human data and our previous findings suggested Glafenine induces hepatic damage, raising the possibility of enterohepatic recirculation mediating intestinal injury (19–22), these explant experiments demonstrate that Glafenine can directly induce IEC apoptosis.
Serial Glafenine Exposure Results in Intestinal Inflammation.
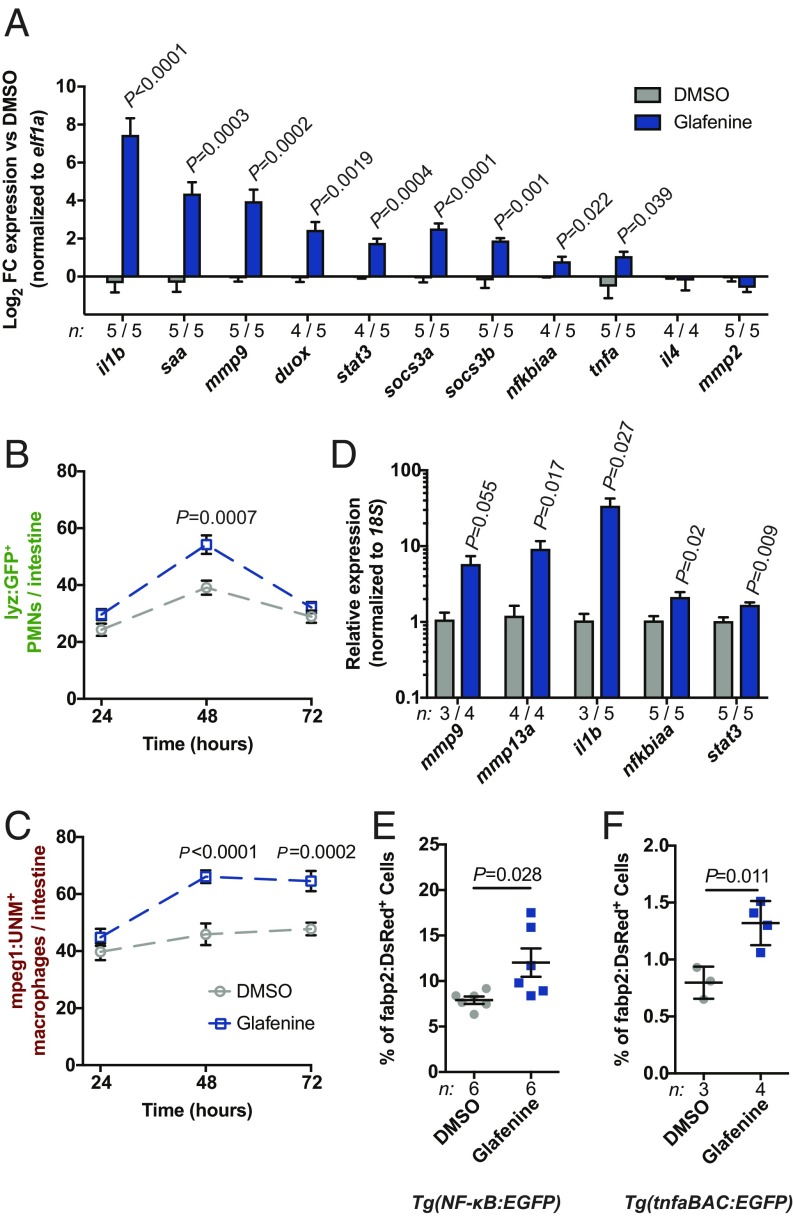
We next tested if serial Glafenine exposure resulted in intestinal inflammation. Gene-expression analysis of dissected digestive tracts revealed marked induction of mRNAs encoding proinflammatory effectors (il1b, saa, duox, mmp9, mmp13a, and tnfa) and regulators of innate immune signaling (stat3, socs3a, socs3b, and nfkbiaa) (Fig. 2A) in Glafenine-treated larvae. Moreover, we observed intestinal leukocyte infiltration, with increased numbers of intestine-associated lyz+ polymorphonuclear cells (PMNs) (28) and mpeg1+ macrophages (29) at 48 h into the treatment regimen. By 72 h, PMN numbers typically returned to control levels (although in some experiments we observed significant PMN infiltration at this time point) (Fig. 2B and SI Appendix, Fig. S8A), while macrophage numbers remained elevated (Fig. 2C and SI Appendix, Fig. S8B). Together, these findings demonstrate Glafenine induces intestinal inflammation concomitant with IEC loss.
Fig. 2.
Serial Glafenine exposure results in intestinal inflammation. (A) qRT-PCR analysis of proinflammatory mRNAs from dissected digestive tissues of Glafenine- and DMSO-treated larvae (20 larval intestines per replicate; significance determined by unpaired 2-sided Student’s t test). (B and C) Quantification of intestine-associated PMNs (lyz:GFP+ cells, B) and macrophages (mpeg1:UNM+ cells, C)) (n = 20 larvae per condition per time point, statistical comparisons were performed between treatment conditions at individual time points; significance determined by unpaired 2-sided Student’s t test). (D–F) qRT-PCR analysis of FACS-isolated fabp2:DsRed+ enterocytes (D), and flow cytometry analysis of NFκB:EGFP (E) and tnfa:EGFP (F) reporter activity in fabp2:DsRed+ enterocytes (cells isolated from replicate pools of 20 larvae; significance determined by unpaired 2-sided Student’s t test).
We next investigated whether the inflammatory signatures we observed in dissected digestive tracts were induced in enterocytes. Isolated fabp2:DsRed+ cells from Glafenine-treated larvae exhibited significantly increased mRNA levels of inflammatory mediators (il1b, mmp9, mmp13a, and nfkbiaa) and regulators (stat3), mirroring our results from dissected digestive tissues (Fig. 2D). In corroboration, we also observed an ∼2-fold increase in the proportion of fabp2:DsRed+ enterocytes positive for either NF-κB (26) or tnfa (30) reporters (Fig. 2 E and F and SI Appendix, Fig. S5A). Serial Glafenine exposure therefore induces intestinal inflammation, with concomitant activation of innate immune signaling in enterocytes.
Glafenine Treatment Alters the Intestinal and Environmental Microbiotas.
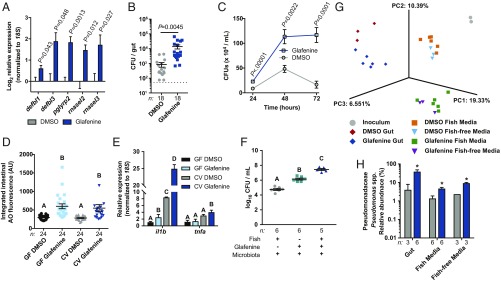
It is well established that changes in microbiota composition and activity can augment intestinal inflammation (31). Furthermore, recent studies in murine models have demonstrated that microbiota potentiate intestinal toxicity observed with certain pharmaceuticals, including NSAIDs (13, 17, 18). We first asked if Glafenine exposure was associated with perturbations in host–microbiota interactions. Expression of genes encoding antimicrobial proteins (AMPs) rnasel2, rnasel4, pglyrp2, and pglyrp5 (32) were significantly elevated in digestive tracts after Glafenine treatment (Fig. 3A). We next asked whether bacterial load was altered by Glafenine treatment by enumerating colony forming units from both dissected digestive tracts and media of DMSO- and Glafenine-treated larvae and observed increased abundance of culturable bacteria following Glafenine treatment (Fig. 3 B and C). Even with 50% (vol/vol) media changes at 24-h intervals, there was a substantial increase in the concentration of bacteria in media over time with Glafenine treatment (Fig. 3C). These results suggest that Glafenine treatment modify host–microbiota relationships.
Fig. 3.
Glafenine treatment alters the intestinal and environmental microbiotas. (A) qRT-PCR analysis of dissected digestive tracts from 6 dpf DMSO- and Glafenine-treated larvae for mRNAs encoding antimicrobial peptides (significance determined by unpaired 2-sided Student’s t test). (B and C) Quantification of culturable bacteria from dissected digestive tracts and media (in C, statistical comparisons performed between treatment groups at individual time points; significance determined by unpaired 2-sided Student’s t test). (D) Quantification of intestinal AO staining in GF and CV DMSO- and Glafenine-treated larvae (significance determined by 2-way ANOVA; letters indicate significantly different groups). (E) qRT-PCR analysis of dissected digestive tracts. (F) Media bacterial load at 72 h from the indicated groups. (G) Principal coordinates analysis of Jaccard β-diversity. (H) Relative abundance of Pseudomonas spp. from the indicated samples (significance was determined with LEfSe; asterisk indicates log10 LDA > 4.5). For E and F, significance was determined by 1-way ANOVA with Tukey’s multiple-comparison test; letters indicate groups determined to be statistically different. CFU, colony-forming unit.
Considering the up-regulated AMP gene expression and elevated bacterial load in digestive tissues and media, we hypothesized that microbiota may potentiate Glafenine-induced phenotypes. We first asked if microbiota are required for Glafenine-induced IEC loss by performing serial Glafenine exposure on gnotobiotic zebrafish larvae (33) (SI Appendix, Fig. S9A). IEC delamination was comparable between germ-free (GF) and conventionalized (CV) larvae, indicating that microbiota colonization was not required for Glafenine-induced IEC loss (Fig. 3D). Moreover, we observed no difference in mortality or IEC delamination between WT and myd88 mutant zebrafish, which have impaired detection of microbiota-derived signals (SI Appendix, Fig. S10) (34). However, we did observe augmented expression of proinflammatory mRNAs in CV animals treated with Glafenine (Fig. 3E), suggesting that the microbiota potentiate inflammation associated with Glafenine toxicity in zebrafish.
Previous studies have established that intestinal microbiota are able to metabolize and transform some xenobiotics, including pharmaceuticals (17, 18, 35, 36). Considering the relatively low nutrient availability in zebrafish media, we predicted that Glafenine may serve as a nutrient source that could affect microbial growth and ecology. Indeed, we found that Glafenine alone was sufficient to promote bacterial growth in media, and that the addition of zebrafish hosts and their associated products had an additive effect (Fig. 3F).
We hypothesized that Glafenine-induced changes in microbiota composition may underlie host inflammatory responses. We therefore serially exposed CV zebrafish larvae (inoculated with a conventional microbiota at 3 dpf) to DMSO or Glafenine, and used 16S rRNA gene sequencing to assess bacterial community composition in media and digestive tracts 72 h later (6 dpf) (SI Appendix, Fig. S9). To distinguish between fish-dependent and -independent effects of Glafenine on the media microbiota, we also used the same microbiota inoculum to colonize sterilized zebrafish media that lacked zebrafish (“fish-free”) (SI Appendix, Fig. S9B). Glafenine did not affect bacterial richness (α-diversity) in the gut but did significantly reduce richness in the media in both the presence and absence of zebrafish (Dataset S2). Principal coordinates (PC) analysis of community similarity (Jaccard index) revealed expected (37) compositional differences between gut and media samples, separating along the primary axis (PC1) (Fig. 3G and Dataset S3). PC2 separated DMSO- and Glafenine-treated samples, indicating Glafenine alters composition of the larval zebrafish microbiota. Surprisingly, this also altered community composition in the fish-free samples, demonstrating that the aquatic microbial community is directly responsive to Glafenine (Fig. 3 G and H, SI Appendix, Figs. S11 and S12, and Datasets S2–S5).
Examination of bacterial taxa that were affected by Glafenine administration uncovered a marked increase in the relative abundance of Pseudomonas spp. in all Glafenine-treated sample groups (Fig. 3H, SI Appendix, Fig. S11, and Datasets S4 and S5). We previously found that Pseudomonas spp. are sufficient to evoke robust proinflammatory responses in zebrafish larvae compared to other tested commensal bacteria (26, 38, 39). Since Glafenine exposure was associated with increased abundance of Pseudomonas spp. in the fish-free condition, we asked if other taxa were affected by Glafenine in a fish-independent manner. Indeed, we found that Magnetospirillum spp. increased with Glafenine treatment, while Peredibacter spp., and Aquicella spp. were depleted in Glafenine-treated media samples (SI Appendix, Figs. S11B and S12 A, B, and F). We also identified taxa that were affected by Glafenine only in the presence of fish: Shewanella spp. increased appreciably only in gut samples following Glafenine treatment, and Fluviicola spp. only increased in fish media samples (but not fish-free samples) with Glafenine treatment (SI Appendix, Figs. S11B and S12 E and G). These data demonstrate that microbiota alterations, including enrichment of potentially proinflammatory taxa, are associated with Glafenine-induced intestinal injury. Moreover, our fish-free experiments demonstrate that aquatic microbial community composition responds to the presence of Glafenine even in the absence of larval zebrafish hosts. Thus, Glafenine treatment of zebrafish leads to alterations of both host and environmental microbiotas, which may potentiate the host inflammatory response.
Ire1α Mediates Glafenine-Induced IEC Delamination to Restrict Inflammation and Mortality.
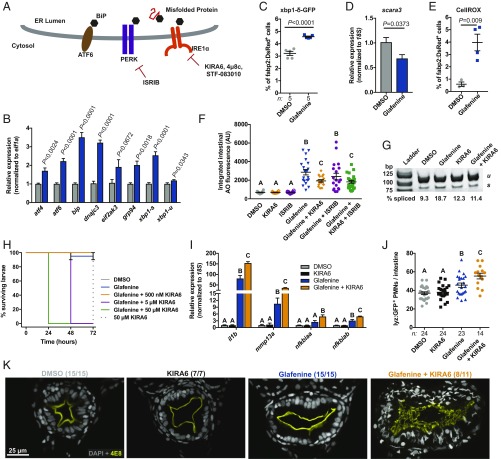
Because Glafenine-induced IEC shedding was microbiota-independent, we sought to identify other mechanisms by which Glafenine could regulate IEC loss. We previously observed organellar damage in IECs from Glafenine-treated larvae (19), and thus reasoned that Glafenine may induce endoplasmic reticulum (ER) stress. Three sensors (ATF6, PERK, and Ire1α) (Fig. 4A) span the ER membrane and respond to disrupted proteostasis in the ER lumen by initiating the UPR, which restores homeostasis by suppressing translation and up-regulating chaperone expression. Under conditions of unmitigated ER stress, however, the UPR sensors initiate apoptosis (40). We first asked if a UPR transcriptional signature was up-regulated in enterocytes following Glafenine treatment. qRT-PCR analysis of fabp2:DsRed+ enterocytes revealed a dramatic increase in UPR target gene mRNAs (41), including those encoding chaperones (bip and grp94), transcription factors (atf4, atf6, and xbp1), and translational regulators (dnajc3 and eif2ak3) (Fig. 4B), suggesting activation of all 3 UPR sensors.
Fig. 4.
Ire1α mediates Glafenine-induced IEC delamination to restrict inflammation and mortality. (A) Schematic of UPR sensors and small-molecule antagonists. (B) qRT-PCR analysis of UPR target gene expression in isolated fabp2:DsRed+ enterocytes (n = 5 replicates per group; significance determined by unpaired 2-sided Student’s t test). (C) Flow cytometry analysis for percent xbp1-δ-GFP+ of fabp2:DsRed+ cells (n = 4 replicates per condition, ≥5,000 cells per replicate; significance determined by unpaired 2-sided Student’s t test). (D) scara3 expression in isolated fabp2:DsRed+ enterocytes (n = 4 replicates per group; ≥5,000 cells per replicate; significance determined by unpaired 2-sided Student’s t test). (E) Proportion CellROX+ of fabp2:DsRed+ enterocytes (determined by flow cytometry; n = 4 replicates per group; ≥5,000 cells per replicate; significance determined by unpaired 2-sided Student’s t test). (F) Intestinal AO quantification from larvae treated with Glafenine or UPR inhibitors. (G) xbp1 splicing assay from fabp2:DsRed+ enterocytes isolated from larvae treated as indicated. (H) qRT-PCR analysis of inflammatory mRNAs in fabp2:DsRed+ enterocytes isolated from larvae treated as indicated (n = 3 replicates for DMSO and KIRA6, 4 replicates for Glafenine and Glafenine+KIRA6). (I) Quantification of intestine-associated lyz:GFP+ PMNs. (J) Survival of larvae treated as indicated. (K) Representative confocal micrographs of transverse sections from larvae of the specified treatment groups immunostained with the brush border reactive antibody 4E8. For F, H, and I, significance was determined by 1-way ANOVA with Tukey’s multiple comparison test; letters indicate groups determined to be statistically different.
When activated, the UPR sensor Ire1α functions both to splice xbp1 mRNA as well as to degrade a canonical set of cellular mRNAs through regulated Ire1α-dependent decay (RIDD) (42). We observe increased spliced xbp1 (xbp1-s) mRNA in enterocytes from Glafenine-treated larvae, which was confirmed by an increase in the proportion of enterocytes positive for an xbp1 splice reporter (43) (Fig. 4 B and C). Moreover, we detected an ∼40% decrease in expression of the RIDD target scara3 (42) (Fig. 4D). Collectively, these data demonstrate that Glafenine elicits UPR activation and Ire1α activity in enterocytes with distinct phenotypic outcomes compared to other ER stressors, which did not induce IEC delamination (SI Appendix, Fig. S13).
Since elevated intracellular reactive oxygen species levels are intimately related to ER protein folding and can lead to UPR activation (44), we asked if Glafenine perturbed enterocyte redox homeostasis. Indeed, flow cytometry analysis revealed a significant increase in the proportion of fabp2+ cells that were positive for the total intracellular reactive oxygen species probe CellROX (Fig. 4E).
We next assessed the role of distinct UPR sensors in Glafenine-induced enteropathy using pharmacological inhibitors of PERK and Ire1α. While the small-molecule PERK inhibitor ISRIB (45) had no effect on IEC delamination, coexposure with the Ire1α inhibitor KIRA6 significantly attenuated IEC loss (Fig. 4F). The zebrafish Ire1α amino acid sequence is highly conserved, with predicted cytosolic kinase and endonuclease domains, and conserved residues required for inhibitor activity (K599/600 and K907/909 [human/zebrafish]) (SI Appendix, Fig. S14 A, C, and D). RT-PCR analysis of isolated fabp2:DsRed+ enterocytes confirmed that 500 nM KIRA6 reduced Glafenine-induced xbp1 splicing (Fig. 4G).
Genetic and biochemical studies in mice and humans have linked Ire1α signaling and autophagy in the intestine (46, 47). We asked whether Glafenine elicited an autophagic response. We utilized double-transgenic Tg(CMV:GFP-LC3) (48) Tg(fabp2:DsRed) zebrafish to visualize autophagic structures in enterocytes (SI Appendix, Fig. S15A). We observed an ∼2-fold increase in autophagic punctae in enterocytes with Glafenine treatment, which was suppressed in larvae cotreated with KIRA6 (SI Appendix, Fig. S15B). Thus, serial Glafenine induced autophagy in IECs is downstream of Ire1α activity.
While optimizing inhibitor dosage, we observed a concentration-dependent increase in mortality and concomitant developmental delay in larvae cotreated with Glafenine and each of 3 different Ire1α inhibitors: KIRA6, 4μ8c, and STF-083010 (42, 49–52) (Fig. 4H and SI Appendix, Fig. S14 E and F and H–M). Surprisingly, gene-expression analysis of isolated enterocytes revealed that cotreatment with Glafenine and KIRA6 exacerbated Glafenine-induced induction of proinflammatory genes, and was associated with increased intestinal PMN infiltration (Fig. 4 I and J). Whereas Glafenine treatment alone did not markedly affect intestinal architecture, cotreatment with Glafenine and the Ire1α inhibitor KIRA6 led to a strikingly aberrant mesh-like patterning of the intestinal brush border (Fig. 4K). Moreover, we observed reduced expression of genes encoding the brush border components sucrase isomaltase (si), solute carrier family 5 member 1 (slc5a1), and intestinal alkaline phosphatase (alpi.1) in digestive tracts from larvae cotreated with Glafenine and KIRA6 (SI Appendix, Fig. S14G). Remarkably, we noted no alteration in intestinal permeability in larvae treated with both Glafenine and KIRA6 (SI Appendix, Fig. S6I).
Taken together, these data suggested 2 possible scenarios: 1) failed IEC delamination associated with Ire1α inhibition drives an augmented inflammatory response, or 2) elevated inflammation induced in larvae by treatment with both Glafenine and Ire1α inhibitors suppressed IEC delamination. In order test the possibility that inflammation suppresses cell shedding, we coexposed zebrafish larvae to combinations of Glafenine, KIRA6, and the antiinflammatory glucocorticoid Dexamethasone and quantified intestinal AO fluorescence (SI Appendix, Fig. S16A). Since therapeutic concentrations of Dexamethasone (53) did not restore AO signal in larvae treated with Glafenine and KIRA6 to levels observed in larvae treated Glafenine alone, we concluded that elevated inflammation was secondary to reduced IEC delamination. Together, these data demonstrate that Ire1α signaling is essential for proper IEC delamination and maintenance of intestinal architecture during Glafenine challenge, limiting inflammation and promoting animal survival.
In Vivo Screening Identifies MDR Efflux Pump Inhibitors Which Phenocopy the Effects of Glafenine.
Since Glafenine was the only NSAID we tested that induced profound IEC loss (SI Appendix, Fig. S7 B–D), we sought to define the essential chemical features required for this effect. We initially noted that the chemical structure of Glafenine was distinct from the other NSAIDs we had tested, belonging to the class of anthranilic acid-derived NSAIDs (SI Appendix, Fig. S7A). We undertook in vivo structure–activity relationship studies with the aim of identifying essential molecular features required for Glafenine-induced intestinal toxicity and applied our AO-based IEC delamination assay to larvae treated with compounds structurally similar to Glafenine (see Dataset S1 for Tanimoto coefficients and maximum common substructure scores). Floctafenine and 7-chloro-(4-hydroxyanilino)quinolone exhibited poor solubility in zebrafish media, restricting maximum doses to 10 and 20 μM, respectively (SI Appendix, Fig. S17). However, none of these Glafenine-related compounds were able to induce IEC delamination to the extent of Glafenine (SI Appendix, Fig. S17).
We next wondered whether Glafenine metabolic products could elicit IEC delamination. We predicted that the ester linkage in the 2,3-dihydroxypropyl acetate side chain of Glafenine could be cleaved by carboxylesterases (CES), yielding Glafenic acid and propylene glycol (PG) (SI Appendix, Fig. S18A). However, neither Glafenic acid nor PG could elicit IEC loss to the same extent as Glafenine (SI Appendix, Fig. S17B). Moreover, neither of the CES inhibitors, BNPP (54) or Paraoxon (55), were sufficient to alter Glafenine-induced IEC loss at tolerated doses (SI Appendix, Fig. S18 B and C), indicating that CES metabolism of Glafenine is not required for IEC delamination.
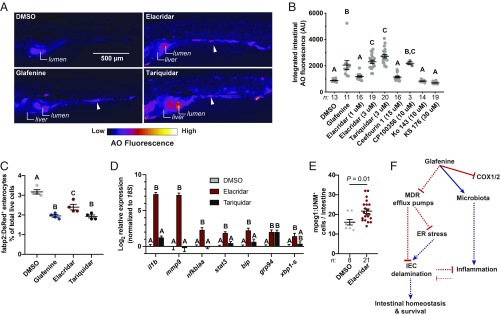
Although Glafenine was developed as an NSAID, 2 previous cell-culture screens identified Glafenine having an off-target effect as an inhibitor of the MDR efflux pumps MRP4 and ABCG2 (56, 57). MDRs belong to the family of ATP binding cassette (ABC) transporters, and are expressed in privileged tissues (e.g., the blood brain barrier) as well as mucosal surfaces (e.g., the intestine), where they function to eliminate xenobiotics (58, 59). Considering that other NSAIDs did not induce IEC delamination, and PGE2 supplementation failed to rescue Glafenine-induced IEC loss, we asked if MDR efflux pump inhibition could be an alternate mechanism underlying IEC delamination. We screened 9 structurally diverse MDR efflux pump inhibitors with various target specificities and identified 3 compounds that induced IEC delamination responses similar to Glafenine: Elacridar, Tariquidar, and CP100356 (Fig. 5 A and B, SI Appendix, Fig. S19 A–C, and Movie S8). While Glafenine induced AO staining in the intestine and liver (Fig. 1B and SI Appendix, Fig. S1A), we also observed AO fluorescence in the skin and other tissues of larvae treated with Elacridar and Tariquidar, suggesting that these drugs may have broader effects than Glafenine (Fig. 5A). Flow cytometry analysis of fabp2:DsRed+ larvae revealed that both Elacridar and Tariquidar treatment led to reduced enterocyte abundance (although the Elacridar’s effect was not as strong as Glafenine or Tariquidar) (Fig. 5C). Similar to the effects of Glafenine, we observed significantly elevated proinflammatory and UPR mRNA levels (Fig. 5D). There was also a significant increase in the abundance of intestine-associated macrophages in Elacridar-treated larvae compared to controls, which positively correlates with the increased proinflammatory mRNA levels (Fig. 5E). Thus, our findings indicate that MDR efflux pump inhibitors can drive IEC delamination, activation of the UPR, and intestinal inflammation. Collectively, these data support a model wherein Glafenine-dependent IEC delamination is NSAID-independent and instead driven by off-target activity of Glafenine against MDR efflux pumps.
Fig. 5.
A subset of MDR efflux pump inhibitors phenocopy the effects of Glafenine. (A) Representative fluorescence images of AO-stained larvae from the indicated treatment groups (arrowheads indicate AO+ material in the intestinal lumen). (B) Intestinal AO quantification for larvae treated with DMSO, Glafenine, or the indicated MDR inhibitors (each dot corresponds to an individual larva; significance determined by 1-way ANOVA with Tukey’s multiple comparison test). (C) Relative abundance of fabp2:DsRed+ enterocytes from (n = 4 replicates group, 20 larvae per replicate). (D) qRT-PCR analysis of inflammatory and UPR mRNAs (n = 4 replicates per condition, ≥5,000 cells per replicate). (E) Quantification of intestine associated macrophages (mpeg1+ cells) from DMSO- and Elacridar-treated larvae (each point corresponds to an individual larva; significance determined by unpaired 2-sided Student’s t test). For B, C, and D, significance was determined by 1-way ANOVA with Tukey’s multiple comparisons test; letters indicate groups determined to be statistically different. (F) Proposed model of Glafenine-induced intestinal toxicity (solid lines indicated experimentally confirmed direct interactions, while dashed lines indicate relationships that may be indirect).
Discussion
Applying pharmacological, molecular, and microbiological approaches in zebrafish larvae, this work yielded in vivo insights into the mechanisms underlying a model of pharmaceutical-induced enteropathy. Before its global withdrawal in the early 1990s, Glafenine was used as an over-the-counter oral analgesic in Europe and the Middle East for nearly 3 decades until global withdrawal due to hepatic and renal toxicity (20). We leveraged the strengths of the larval zebrafish model in combination with a robust and stereotyped phenotypic response to Glafenine to investigate underlying molecular and cellular mechanisms of pharmaceutical-induced intestinal injury.
We designed a serial Glafenine exposure protocol to model human drug usage patterns in zebrafish larvae. Notably, our exposure regimen complements previous reports which adapted murine colitogenic agents (trinitrobenzenesulfonic acid [TNBS] and dextran sodium sulfate [DSS]) to study chemically induced enterocolitis in zebrafish (60, 61). While these TNBS and DSS studies demonstrated promise, the broad range of injury to the host limits the utility of these agents for specific study of intestinal damage. Indeed, immersion exposure of zebrafish to TNBS (an acid) or DSS (a detergent) elicits inflammatory responses in the skin and other tissues. In contrast, Glafenine exposure is a more specific zebrafish intestinal injury model, as we find it induces injury restricted to digestive organs and is sufficient to induce IEC apoptosis ex vivo.
Our work establishes an experimental platform that recapitulates a subset of the clinical manifestations associated with pharmaceutical-induced enteropathy (epithelial cell loss, disruption of epithelial redox homeostasis, and inflammation), and offers improved tissue specificity for investigation of intestinal injury in zebrafish. While gastric and duodenal ulceration is a common clinical manifestation of NSAID-induced enteropathy, we observed no evidence of bleeding following Glafenine treatment. Glafenine-induced IEC loss was NSAID-independent and refractory to supplementation with exogenous PGE2.
We were surprised that Glafenine had direct effects on microbial community composition, an observation with 3 important implications. First, Glafenine treatment altered microbiota composition in the zebrafish gut and media, including enrichment of Pseudomonas spp., which have been shown to be proinflammatory (38). While Glafenine-induced IEC delamination was microbiota-independent, microbial colonization enhanced the inflammation driven by Glafenine. Second, a subset of Glafenine-driven alterations in the environmental microbiota were zebrafish-independent, suggesting that Glafenine can act directly on bacteria. Although xenobiotic exposure has been associated with altered microbiota composition in mammals (16), to our knowledge this has not been demonstrated in zebrafish. Our findings demonstrate the ability of xenobiotics to alter host-associated and environmental microbiotas is a conserved feature. Third, zebrafish have been used as a pharmacologically tractable model system for many years, for both directed pharmacological interventions and small-molecule screens (62, 63). Our results underscore that studies using pharmaceuticals or other chemical compounds in zebrafish and other animals should be interpreted carefully with attention to the potential reciprocal relationships between chemicals, microbiota, and host (64). For example, evaluation of chemical impacts on gnotobiotic or germ-free zebrafish and on the microbial community composition alone can be used to determine if the microbiota respond to the presence of study compounds and/or mediate their effects on the host.
While our gene-expression data indicate that all 3 arms of UPR signaling are activated in Glafenine-treated larvae, we found that Ire1α signaling is required for epithelial cell loss following Glafenine exposure. Our findings are congruent with reports from mice demonstrating that Ire1α/Xbp1 signaling in the intestine is protective both at baseline and following DSS-induced colitis (46, 47). The requirement for Ire1α could be attributed to the fact that, unlike the other canonical UPR sensors, Ire1α also signals to a number of other cellular pathways: Activating JNK to potentiate apoptotic programs, binding filamin A to regulate cytoskeletal dynamics and cell migration (65), inducing autophagy, promoting inflammasome formation (66), and activation of cellular innate immune pathways (9). Despite its pleiotropic functions, our pharmacological interventions offer some insight into how it may affect delamination: While all 3 of the Ire1α inhibitors we tested suppress Glafenine-induced IEC delamination, 2 inhibit both kinase and nuclease functions (KIRA6 and 4μ8c) (42, 50) and 1 (STF-083010) only inhibits endonuclease activity (51), suggesting that RNA processing by Ire1α is essential for Glafenine-induced IEC loss. Importantly however, induction of ER stress alone is insufficient to promote delamination (SI Appendix, Fig. S13). While the molecular events leading to IEC delamination downstream of Ire1α activation in this model have yet to be defined, our results show that this phenomenon depends on the effects Glafenine likely through inhibition of MDR efflux pumps.
It is curious that Ire1α inhibition in the context of Glafenine exposure is associated with increased proinflammatory gene expression, including regulators of NF-κB (nfkbiaa and nfkbiab) (Fig. 4H), since Ire1α signaling has been demonstrated to promote NF-κB activity through TRAF2 (67). We propose that failed IEC delamination in larvae cotreated with Glafenine and Ire1α inhibitors leads to a local increase in the concentration of damage-associated molecular pattern molecules, which may augment the inflammatory response (SI Appendix, Fig. S16). Thus, our findings demonstrate a protective function of Ire1α signaling in the intestine.
Our results establish that Glafenine-induced intestinal injury in zebrafish is not mediated by its NSAID activity but may instead be caused by off-target inhibition of MDR efflux pumps. The MDR efflux transporters are a large gene family in vertebrates, and act on a diverse set of substrates (5). Eukaryotic MDR efflux pumps belong to the ABC gene superfamily, which can be subdivided into as many as 8 subfamilies (depending on the species); 3 subfamilies include genes known to encode active drug transporters as well as proteins whose substrates have yet to be identified (68). Comparative genomic analyses identified 52 ABC genes in zebrafish (69), although the majority have not been functionally characterized. The Mdr1a−/− (Abcb1a) mouse develops spontaneous colitis (70), and the development of pathology in this model can be accelerated by colonization with pathogenic bacteria (71) or chemical insult (72). Glafenine was previously identified as an off-target inhibitor of MRP4 (ABCC4) and BCRP (ABCG2) in mammalian cell-culture screens using heterologous expression constructs. From the 27 structurally and functionally related chemicals that we tested in this study, the only 3 that induced IEC delamination similar to Glafenine were MDR inhibitors. The 2 MDR inhibitors that most robustly induced IEC delamination in zebrafish larvae, Elacridar and Tariquidar, have submicromolar BCRP inhibitory activity (127 nM and 100 nM IC50, respectively). Additionally, a selective MRP4 inhibitor (Ceefourin 1) failed to induce IEC loss, suggesting that Glafenine-induced intestinal injury may be due to inhibition of BCRP (ABCG2) homologs (Fig. 5 A and B and SI Appendix, Fig. S19D). The abcg2 family members a and b (as well as c, although at lower levels) are expressed in zebrafish IECs (73), as is abcc4 (73, 74). Our results predict 1 or more of these proteins have critically important roles during intestinal homeostasis, perhaps including efflux of endogenous molecular substrates that are toxic to IECs at high concentrations.
These data also suggest that Glafenine, Tariquidar, Elacridar, and perhaps other MDR efflux pump inhibitors share a pharmacophore, which selectively inhibits zebrafish MDR pumps. It is noteworthy that exposure to Elacridar and Tariquidar led to variable induction of proinflammatory mRNA in enterocytes (Fig. 5D), and that while Elacridar had no profound effects on intestinal architecture, Tariquidar exposure was associated with aberrant localization of brush border components to basolateral membranes (SI Appendix, Fig. S19). These phenotypic variations in zebrafish larvae treated with different MDR efflux pump inhibitors remain unclear but could be due to differences in target affinity or specificity. We note that Elacridar most accurately phenocopied Glafenine treatment overall, but both Tariquidar and Elacridar could now be pursued as new enteropathy models in zebrafish. Collectively, our data demonstrate that IEC delamination following pharmaceutical-induced enteropathy is a host-protective response and functions to limit inflammation and death, as experimental manipulations that block epithelial cell delamination are associated with increased inflammation and mortality (Fig. 5F). Our results also suggest that export of unknown molecular substrates by members of the ABCG2/BCRP family is required for maintenance of intestinal homeostasis.
Materials and Methods
Zebrafish Experiments.
All zebrafish experiments were performed in accordance with NIH guidelines and protocols approved by the Duke University Medical Center Institutional Animal Care and Use Committee (protocol numbers A115-16-05 and A096-19-04). Detailed methods are available in SI Appendix.
Statistical Analysis.
Statistical analyses were performed with GraphPad Prism v.7. All data are presented as mean ± SEM. For comparison between 2 groups, an unpaired 2-tailed Student’s t test was performed. For comparison between ≥3 groups, a 1-way ANOVA with Tukey’s multiple comparison test was applied. P < 0.05 was considered significant. Outcomes of statistical tests are defined in the figure legends. For 1-way ANOVA, groups with the same letter are not significantly different from each other. Where indicated, a 3- or 4-parameter least-squares regression was calculated. Data shown are representative of at least 3 independent experiments. Sample sizes are reported in figures and legends.
Supplementary Material
Acknowledgments
We thank Aadra Bhatt, Jennifer Heppert, Daniel Levic, Colin Lickwar, Caitlin Murdoch, and Jia Wen for critical reading of the manuscript. We acknowledge Holly Dressman and Zhengzheng Wei in the Duke Microbiome Shared Resource for microbial community DNA isolation and 16S rRNA gene sequencing; Nancy Martin, Bin Li, and Mike Cook at the Duke Cancer Institute Flow Cytometry Shared Resource for assistance with fluorescence activated cell sorting (FACS); Benjamin Carlson, Yasheng Gao, and Lisa Cameron of the Duke University Light Microscopy Core Facility for assistance with instrumentation; and Rebecca Graham, Eileen Gu, Sheila Janardhan, Justice Lu, and Patrick Williams for zebrafish husbandry and care. This work was supported by NIH Grants P01-DK094779 (to J.F.R.), R24-OD016761 (to J.F.R.), AI130236 (to D.M.T.), AI125517 (to D.M.T.), CA098468 (to M.R.R.), and CA207416 (to M.R.R.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the NCBI Sequence Read Archive (accession no. PRJNA512913).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1902596116/-/DCSupplemental.
References
- 1.Delgado M. E., Grabinger T., Brunner T., Cell death at the intestinal epithelial front line. FEBS J. 283, 2701–2719 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Bisanz J. E., Spanogiannopoulos P., Pieper L. M., Bustion A. E., Turnbaugh P. J., How to determine the role of the microbiome in drug disposition. Drug Metab. Dispos. 46, 1588–1595 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spanogiannopoulos P., Bess E. N., Carmody R. N., Turnbaugh P. J., The microbial pharmacists within us: A metagenomic view of xenobiotic metabolism. Nat. Rev. Microbiol. 14, 273–287 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou F., et al. , Toward a new age of cellular pharmacokinetics in drug discovery. Drug Metab. Rev. 43, 335–345 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Vasiliou V., Vasiliou K., Nebert D. W., Human ATP-binding cassette (ABC) transporter family. Hum. Genomics 3, 281–290 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ernst R., Kueppers P., Stindt J., Kuchler K., Schmitt L., Multidrug efflux pumps: Substrate selection in ATP-binding cassette multidrug efflux pumps—First come, first served? FEBS J. 277, 540–549 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Glavinas H., Krajcsi P., Cserepes J., Sarkadi B., The role of ABC transporters in drug resistance, metabolism and toxicity. Curr. Drug Deliv. 1, 27–42 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Patel H. R., et al. , Metabolic competence and susceptibility of intestinal epithelium to genotoxic injury during regeneration. Carcinogenesis 18, 2171–2177 (1997). [DOI] [PubMed] [Google Scholar]
- 9.Adolph T. E., Niederreiter L., Blumberg R. S., Kaser A., Endoplasmic reticulum stress and inflammation. Dig. Dis. 30, 341–346 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abraham C., Medzhitov R., Interactions between the host innate immune system and microbes in inflammatory bowel disease. Gastroenterology 140, 1729–1737 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Philpott H. L., Nandurkar S., Lubel J., Gibson P. R., Drug-induced gastrointestinal disorders. Frontline Gastroenterol. 5, 49–57 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marlicz W., Loniewski I., Grimes D. S., Quigley E. M., Nonsteroidal anti-inflammatory drugs, proton pump inhibitors, and gastrointestinal injury: Contrasting interactions in the stomach and small intestine. Mayo Clin. Proc. 89, 1699–1709 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Boelsterli U. A., Redinbo M. R., Saitta K. S., Multiple NSAID-induced hits injure the small intestine: Underlying mechanisms and novel strategies. Toxicol. Sci. 131, 654–667 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bjarnason I., et al. , Mechanisms of damage to the gastrointestinal tract from nonsteroidal anti-inflammatory drugs. Gastroenterology 154, 500–514 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Koppel N., Maini Rekdal V., Balskus E. P., Chemical transformation of xenobiotics by the human gut microbiota. Science 356, eaag2770 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patterson A. D., Turnbaugh P. J., Microbial determinants of biochemical individuality and their impact on toxicology and pharmacology. Cell Metab. 20, 761–768 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saitta K. S., et al. , Bacterial β-glucuronidase inhibition protects mice against enteropathy induced by indomethacin, ketoprofen or diclofenac: Mode of action and pharmacokinetics. Xenobiotica 44, 28–35 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wallace B. D., et al. , Alleviating cancer drug toxicity by inhibiting a bacterial enzyme. Science 330, 831–835 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldsmith J. R., Cocchiaro J. L., Rawls J. F., Jobin C., Glafenine-induced intestinal injury in zebrafish is ameliorated by μ-opioid signaling via enhancement of Atf6-dependent cellular stress responses. Dis. Model. Mech. 6, 146–159 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stricker B. H., Blok A. P., Bronkhorst F. B., Glafenine-associated hepatic injury. Analysis of 38 cases and review of the literature. Liver 6, 63–72 (1986). [PubMed] [Google Scholar]
- 21.Stricker B. H., de Groot R. R., Wilson J. H., Anaphylaxis to glafenine. Lancet 336, 943–944 (1990). [DOI] [PubMed] [Google Scholar]
- 22.Ypma R. T., Festen J. J., De Bruin C. D., Hepatotoxicity of glafenine. Lancet 2, 480–481 (1978). [DOI] [PubMed] [Google Scholar]
- 23.van Ham T. J., Mapes J., Kokel D., Peterson R. T., Live imaging of apoptotic cells in zebrafish. FASEB J. 24, 4336–4342 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crosnier C., et al. , Delta-Notch signalling controls commitment to a secretory fate in the zebrafish intestine. Development 132, 1093–1104 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Her G. M., Chiang C. C., Wu J. L., Zebrafish intestinal fatty acid binding protein (I-FABP) gene promoter drives gut-specific expression in stable transgenic fish. Genesis 38, 26–31 (2004). [DOI] [PubMed] [Google Scholar]
- 26.Kanther M., et al. , Microbial colonization induces dynamic temporal and spatial patterns of NF-κB activation in the zebrafish digestive tract. Gastroenterology 141, 197–207 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gudipaty S. A., Rosenblatt J., Epithelial cell extrusion: Pathways and pathologies. Semin. Cell Dev. Biol. 67, 132–140 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hall C., Flores M. V., Storm T., Crosier K., Crosier P., The zebrafish lysozyme C promoter drives myeloid-specific expression in transgenic fish. BMC Dev. Biol. 7, 42 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ellett F., Pase L., Hayman J. W., Andrianopoulos A., Lieschke G. J., mpeg1 promoter transgenes direct macrophage-lineage expression in zebrafish. Blood 117, e49–e56 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marjoram L., et al. , Epigenetic control of intestinal barrier function and inflammation in zebrafish. Proc. Natl. Acad. Sci. U.S.A. 112, 2770–2775 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Belkaid Y., Hand T. W., Role of the microbiota in immunity and inflammation. Cell 157, 121–141 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oehlers S. H., et al. , Topographical distribution of antimicrobial genes in the zebrafish intestine. Dev. Comp. Immunol. 35, 385–391 (2011). [DOI] [PubMed] [Google Scholar]
- 33.Pham L. N., Kanther M., Semova I., Rawls J. F., Methods for generating and colonizing gnotobiotic zebrafish. Nat. Protoc. 3, 1862–1875 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burns A. R., et al. , Interhost dispersal alters microbiome assembly and can overwhelm host innate immunity in an experimental zebrafish model. Proc. Natl. Acad. Sci. U.S.A. 114, 11181–11186 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haiser H. J., et al. , Predicting and manipulating cardiac drug inactivation by the human gut bacterium Eggerthella lenta. Science 341, 295–298 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maurice C. F., Haiser H. J., Turnbaugh P. J., Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell 152, 39–50 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wong S., et al. , Ontogenetic differences in dietary fat influence microbiota assembly in the zebrafish gut. MBio 6, e00687–e15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rawls J. F., Mahowald M. A., Ley R. E., Gordon J. I., Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell 127, 423–433 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rawls J. F., Samuel B. S., Gordon J. I., Gnotobiotic zebrafish reveal evolutionarily conserved responses to the gut microbiota. Proc. Natl. Acad. Sci. U.S.A. 101, 4596–4601 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walter P., Ron D., The unfolded protein response: From stress pathway to homeostatic regulation. Science 334, 1081–1086 (2011). [DOI] [PubMed] [Google Scholar]
- 41.Vacaru A. M., et al. , Molecularly defined unfolded protein response subclasses have distinct correlations with fatty liver disease in zebrafish. Dis. Model. Mech. 7, 823–835 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cross B. C., et al. , The molecular basis for selective inhibition of unconventional mRNA splicing by an IRE1-binding small molecule. Proc. Natl. Acad. Sci. U.S.A. 109, E869–E878 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li J., et al. , A transgenic zebrafish model for monitoring xbp1 splicing and endoplasmic reticulum stress in vivo. Mech. Dev. 137, 33–44 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Malhotra J. D., et al. , Antioxidants reduce endoplasmic reticulum stress and improve protein secretion. Proc. Natl. Acad. Sci. U.S.A. 105, 18525–18530 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sidrauski C., et al. , Pharmacological brake-release of mRNA translation enhances cognitive memory. eLife 2, e00498 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adolph T. E., et al. , Paneth cells as a site of origin for intestinal inflammation. Nature 503, 272–276 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tschurtschenthaler M., et al. , Defective ATG16L1-mediated removal of IRE1α drives Crohn’s disease-like ileitis. J. Exp. Med. 214, 401–422 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.He C., Bartholomew C. R., Zhou W., Klionsky D. J., Assaying autophagic activity in transgenic GFP-Lc3 and GFP-Gabarap zebrafish embryos. Autophagy 5, 520–526 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Feldman H. C., et al. , Structural and functional analysis of the allosteric inhibition of IRE1α with ATP-competitive ligands. ACS Chem. Biol. 11, 2195–2205 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ghosh R., et al. , Allosteric inhibition of the IRE1α RNase preserves cell viability and function during endoplasmic reticulum stress. Cell 158, 534–548 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Papandreou I., et al. , Identification of an Ire1alpha endonuclease specific inhibitor with cytotoxic activity against human multiple myeloma. Blood 117, 1311–1314 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang L., et al. , Divergent allosteric control of the IRE1α endoribonuclease using kinase inhibitors. Nat. Chem. Biol. 8, 982–989 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tobin D. M., et al. , Host genotype-specific therapies can optimize the inflammatory response to mycobacterial infections. Cell 148, 434–446 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nagashima A., Touhara K., Enzymatic conversion of odorants in nasal mucus affects olfactory glomerular activation patterns and odor perception. J. Neurosci. 30, 16391–16398 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hatfield M. J., Potter P. M., Carboxylesterase inhibitors. Expert Opin. Ther. Pat. 21, 1159–1171 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang Y., et al. , Identification of inhibitors of ABCG2 by a bioluminescence imaging-based high-throughput assay. Cancer Res. 69, 5867–5875 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cheung L., et al. , Identification of new MRP4 inhibitors from a library of FDA approved drugs using a high-throughput bioluminescence screen. Biochem. Pharmacol. 93, 380–388 (2015). [DOI] [PubMed] [Google Scholar]
- 58.Callaghan R., Luk F., Bebawy M., Inhibition of the multidrug resistance P-glycoprotein: Time for a change of strategy? Drug Metab. Dispos. 42, 623–631 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mao Q., Unadkat J. D., Role of the breast cancer resistance protein (BCRP/ABCG2) in drug transport—An update. AAPS J. 17, 65–82 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Oehlers S. H., et al. , Chemically induced intestinal damage models in zebrafish larvae. Zebrafish 10, 184–193 (2013). [DOI] [PubMed] [Google Scholar]
- 61.Oehlers S. H., et al. , A chemical enterocolitis model in zebrafish larvae that is dependent on microbiota and responsive to pharmacological agents. Dev. Dyn. 240, 288–298 (2011). [DOI] [PubMed] [Google Scholar]
- 62.Peal D. S., Peterson R. T., Milan D., Small molecule screening in zebrafish. J. Cardiovasc. Transl. Res. 3, 454–460 (2010). [DOI] [PubMed] [Google Scholar]
- 63.Wiley D. S., Redfield S. E., Zon L. I., Chemical screening in zebrafish for novel biological and therapeutic discovery. Methods Cell Biol. 138, 651–679 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.National Academies of Sciences, Engineering, and Medicine , Environmental Chemicals, the Human Microbiome, and Health Risk: A Research Strategy (The National Academies Press, Washington, DC, 2017). [PubMed] [Google Scholar]
- 65.Urra H., et al. , IRE1α governs cytoskeleton remodelling and cell migration through a direct interaction with filamin A. Nat. Cell Biol. 20, 942–953 (2018). [DOI] [PubMed] [Google Scholar]
- 66.Bronner D. N., et al. , Endoplasmic reticulum stress activates the inflammasome via NLRP3- and caspase-2-driven mitochondrial damage. Immunity 43, 451–462 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Keestra-Gounder A. M., et al. , NOD1 and NOD2 signalling links ER stress with inflammation. Nature 532, 394–397 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Luckenbach T., Fischer S., Sturm A., Current advances on ABC drug transporters in fish. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 165, 28–52 (2014). [DOI] [PubMed] [Google Scholar]
- 69.Annilo T., et al. , Evolution of the vertebrate ABC gene family: Analysis of gene birth and death. Genomics 88, 1–11 (2006). [DOI] [PubMed] [Google Scholar]
- 70.Panwala C. M., Jones J. C., Viney J. L., A novel model of inflammatory bowel disease: Mice deficient for the multiple drug resistance gene, mdr1a, spontaneously develop colitis. J. Immunol. 161, 5733–5744 (1998). [PubMed] [Google Scholar]
- 71.Maggio-Price L., et al. , Helicobacter bilis infection accelerates and H. hepaticus infection delays the development of colitis in multiple drug resistance-deficient (mdr1a-/-) mice. Am. J. Pathol. 160, 739–751 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Staley E. M., Schoeb T. R., Lorenz R. G., Differential susceptibility of P-glycoprotein deficient mice to colitis induction by environmental insults. Inflamm. Bowel Dis. 15, 684–696 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lickwar C. R., et al. , Genomic dissection of conserved transcriptional regulation in intestinal epithelial cells. PLoS Biol. 15, e2002054 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lu X., et al. , Characterization of zebrafish Abcc4 as an efflux transporter of organochlorine pesticides. PLoS One 9, e111664 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.