Significance
Lipids have essential roles in cellular energy homeostasis and are key structural components of membranes and thereby provide the basis of cellular compartmentalization. The maintenance of lipid homeostasis is of fundamental importance to cellular physiology. Lipid droplets (LDs) are central organelles orchestrating lipid fluxes inside cells. By examining pristinely preserved frozen-hydrated HeLa cells with cryoelectron microscopy, we show that LDs exhibit different internal organizations, as well as organelle associations, depending on cellular states. We demonstrate the presence of a liquid-crystalline phase under certain conditions, which are likely to impact the physiological functions of LDs. Furthermore, crystalline droplets are a major component of atherosclerotic lesions in human arteries. Crystalline LDs secreted by cells may therefore have a direct link to pathologies.
Keywords: cryoelectron tomography, cryofocused ion beam, correlative light and electron microscopy, membrane contact sites, cholesteryl ester
Abstract
Lipid droplets (LDs) are ubiquitous organelles comprising a central hub for cellular lipid metabolism and trafficking. This role is tightly associated with their interactions with several cellular organelles. Here, we provide a systematic and quantitative structural description of LDs in their native state in HeLa cells enabled by cellular cryoelectron microscopy. LDs consist of a hydrophobic neutral lipid mixture of triacylglycerols (TAG) and cholesteryl esters (CE), surrounded by a single monolayer of phospholipids. We show that under normal culture conditions, LDs are amorphous and that they transition into a smectic liquid-crystalline phase surrounding an amorphous core at physiological temperature under certain cell-cycle stages or metabolic scenarios. Following determination of the crystal lattice spacing of 3.5 nm and of a phase transition temperature below 43 °C, we attributed the liquid-crystalline phase to CE. We suggest that under mitotic arrest and starvation, relative CE levels increase, presumably due to the consumption of TAG metabolites for membrane synthesis and mitochondrial respiration, respectively, supported by direct visualization of LD–mitochondrial membrane contact sites. We hypothesize that the structural phase transition may have a major impact on the accessibility of lipids in LDs to enzymes or lipid transporters. These may become restricted in the smectic phase, affecting the exchange rate of lipids with surrounding membranes and lead to a different surface occupancy of LD-associated proteins. Therefore, the composition and the resulting internal structure of LDs is expected to play a key role in their function as hubs of cellular lipid flux.
Lipids have essential structural roles in the formation of physical barriers that delineate cells and allow for intracellular compartmentalization, as well as in energy homeostasis. Lipid droplets (LDs) are ubiquitous and highly dynamic cellular organelles present in many organisms, ranging from yeast to invertebrates, plants, and mammals. Neutral lipids, predominantly triacylglycerols (TAG) and cholesteryl esters (CE), are stored in LDs surrounded by a single monolayer of phospholipids to protect cells from the toxic effect of free lipid, but can also be mobilized for energy generation or membrane synthesis through interactions with other organelles (1). While LDs have long been regarded as a passive lipid storage organelle, it has been shown more recently that they act as central hubs for lipid metabolism and trafficking (2). This central role is further demonstrated by the interactions that LDs form with a variety of cellular organelles: for lipogenesis (endoplasmic reticulum, ER), fat accumulation (LD–LD interactions), and lipolysis (mitochondria, lysosomes, autophagosomes, ER) (3, 4).
Due to their central role in orchestrating lipid storage and use, LD biology has recently attracted much interest. However, our current knowledge of the structure, protein organization, biogenesis, and lipolysis of LDs relies heavily on microscopy techniques that either lack resolution ([live-cell] light microscopy) or the ability to preserve native cellular lipid and protein compositions (conventional transmission electron microscopy [TEM]) (5, 6). These technical limitations have restricted our understanding of LD native organization and their interaction mechanisms with cellular organelles at the molecular-structural level. To overcome this problem and achieve a more realistic view of these organelles, we obtained high-resolution cryoelectron microscopy (cryo-EM) images of LDs within cells unaltered by sample preparation for EM. Cryoelectron tomography (cryo-ET) is currently the only method providing in situ structural information at molecular resolution, covering the widest range of dimensions from whole cells to individual macromolecules (7, 8). Vitrification, the freezing of water in an amorphous state (9), arrests cells in a close-to-native state (i.e., without chemical fixation, dehydration, or staining), and tomograms under such conditions provide snapshots of molecular landscapes inside cells (10). Here, we combined LD vital staining, followed by cell vitrification, correlative light and electron microscopy, focused ion beam thinning at liquid nitrogen temperature (cryo-FIB), and cryoelectron microscopy and tomography (cryo-EM/ET), to provide a systematic and quantitative characterization of the structure of LDs within the context of HeLa cells under different cellular states.
Results
Due to the high demand for lipids in proliferating, especially of cancer-derived, cells for growth and membrane expansion (11, 12), we initiated our work on cellular LDs in synchronized and mitotically arrested HeLa cell cultures (details are provided in SI Appendix, SI Materials and Methods). Mitotically arrested cells exhibited the typical internal organization of a monopolar spindle (SI Appendix, Fig. S1): the centrosome positioned roughly in the center of the cell nucleates a radial array of microtubules with condensed chromosomes organized at the periphery of this spindle. The cells were incubated with a cell-membrane–permeable fluorescent neutral lipid dye, BODIPY 493/503, and with a red BODIPY conjugated to fatty acids (13). Cells were deposited on EM grids, vitrified by plunge-freezing and imaged by confocal fluorescence light microscopy at liquid nitrogen temperature (cryo-FLM; SI Appendix, Fig. S2A). The grids were then transferred to the cryo-FIB dual-beam microscope. Coordinates of BODIPY-stained LDs in cryo-FLM stacks were identified in the cryo-FIB images following 3D correlation (14) and targeted to produce electron-transparent lamellae (SI Appendix, Fig. S2 B–F). The resulting vitreous lamellae are ∼200 nm in thickness, thus allowing direct observation in the TEM (SI Appendix, Fig. S2G).
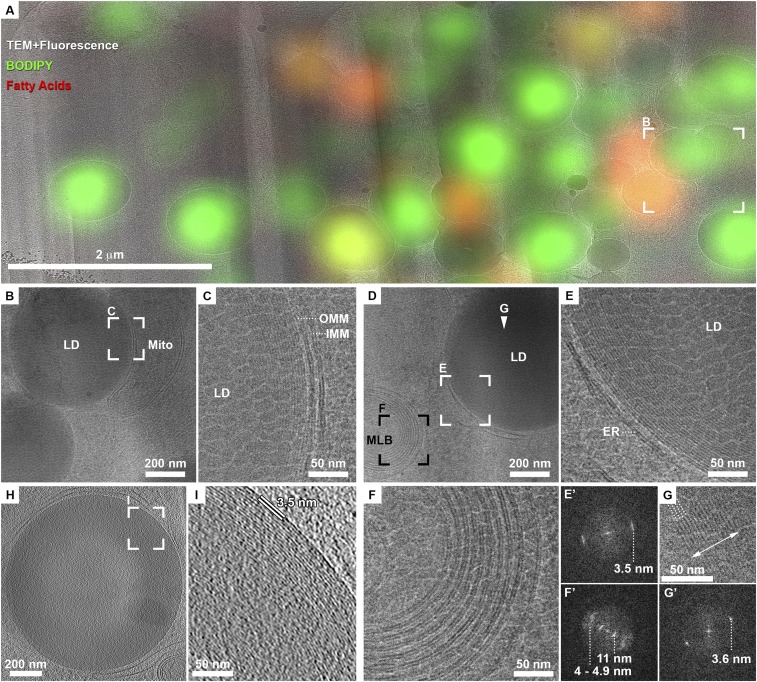
Cryo-TEM images of lamellae from synchronized cells show agglomerates of high density, almost perfectly round particles, predominantly concentrated at the cell center (SI Appendix, Fig. S2G). Calculation of 3D coordinate transformation allowed us to superimpose a computationally extracted slice from the original cryo-FLM data (SI Appendix, Fig. S2A) onto the cryo-TEM image of the lamella (SI Appendix, Fig. S2H and Fig. 1A) and confirmed that the high-density particles correspond to BODIPY-stained organelles: LDs. The observed diameter of the LDs in the ∼200-nm lamellae highly depends on the plane at which the LDs were sectioned. High-resolution 2D TEM images of LDs revealed an organization of concentric layers in all LDs at their periphery (Fig. 1 C and E) and occasionally in ordered nanodomains at the center of LDs (Fig. 1G). A quantitative measure of the layered organization was obtained by Fourier analysis restricted to the individual LDs (Fig. 1 E' and G'). The Fourier transforms showed sharp peaks at 3.5-nm spacing, indicating that LDs have a crystalline lattice. This organization and spacing are distinctly different from that of phospholipid bilayers (membranes in Fig. 1 C and E) or in multilamellar bodies (Fig. 1F) observed in the same lamella. Here, the polar heads of phospholipids appear as dense dark lines separated by a lower-density zone corresponding to the aliphatic chains. The spacing between the two membrane leaflets had an expectedly broad distribution of 4 to 4.9 nm, whereas the spacing between the individual membranes in multilamellar bodies was ∼11 nm (Fig. 1F'). There are multiple examples of stacked lipid bilayers in different cell types (15). Most of these systems are characterized by an asymmetric repeating unit of alternating dark bilayers and lighter spaces separating them. Although a few instances exist where the bilayer thickness is equal to the intermembrane space, the difference in density clearly defines these as two distinct milieus. To eliminate the possibility that the spacing measured in LDs here may be a misrepresentation of the 3D structure projected into a 2D image, we further acquired cryo-ET data and show that the spacing measured from 3D reconstructions is consistent with the 2D measurements (Fig. 1 H and I). Therefore, the internal organization observed in LDs is unique to these organelles and cannot be explained by a model of stacked lipid bilayers.
Fig. 1.
LDs exhibit an internal structure of concentric rings in mitotically arrested cells. (A) Overlay of cryo-TEM montage of FIB lamella with a computer-generated oblique slice through a confocal fluorescence microscopy volume (enlarged from yellow frame in SI Appendix, Fig. S2G.) Green, general neutral lipid dye. Red, fatty acids dye. LDs stained in green or red are distinguishable within the lamella. (B) Enlarged view of frame in A shows two dense LDs in close proximity to a mitochondrion (mito). (C) Enlarged view of frame in B shows on the right the two membranes of the mitochondrion (inner, IMM and outer, OMM), each consisting of a lipid bilayer. The lipid droplet on the left shows a layered structure at its periphery. Granulation observed in the image is due to surface sputter coating with platinum aimed at increasing the conductivity of lamellae for imaging with electrons. (D) Zoom-in of blue frame in SI Appendix, Fig. S2H shows an LD delineated by ER-membranes and a multilamellar body (MLB). (E) LD is enlarged: the periphery of the LD exhibits a layered organization. A Fourier transform of the image in E' exhibits a single peak of 3.5 nm. (F) Enlarged view of MLB in D shows membrane bilayers. A Fourier transform of the image in F' exhibits several peaks, prominently 4 to 4.9 nm of the lipid bilayer spacing and that of 11 nm corresponding to the space between the membranes. (G) Enlarged view of the center of LD in D (white arrowhead) shows a crystalline lattice (four lattice lines are marked with dotted lines) within a ∼50-nm domain (double-sided arrow delineates the width of the domain). A Fourier transform of the image in G' exhibits a single sharp peak of 3.6 nm. (H) Tomographic slice from a different FIB lamella showing a dense LD delineated by ER membranes. (I) Enlarged view of frame in H shows the layered structure at the periphery of the LD. Spacing between layers is directly measured to be 3.5 nm.
The reproducible and highly uniform cellular morphology of mitotically arrested cells and of the LDs under these conditions presented a unique opportunity for high-resolution cryo-EM imaging of a large number of LDs without further BODIPY staining to obtain a quantitative analysis of the native LD structure. LDs have so far been described to consist of an amorphous core of homogenously mixed TAG and CE (16). An organization of concentric layers in LDs similar to what we describe here was reported once in cryo-EM images of isolated LDs (16) and in freeze fracture replica experiments, where an onion-like structure was observed (17). However, these reports were interpreted as a preparation artifact. Cryo-ET of unperturbed cells, as employed here, has been repeatedly demonstrated to provide pristine structural preservation (8).
To characterize the LD structure in more detail and understand the molecular composition giving rise to it, we acquired high-dose cryo-EM 2D projections at 1.7 Å/pixel in the superresolution mode on a direct-electron detector (Fig. 2, Mitotic arrest). Defocus values of 0.5 to 2 μm were chosen such that the frequency-dependent signal loss (zero crossings of the contrast transfer function) does not fall in the frequency range expected for the spacing observed in the LDs. Furthermore, as LDs appeared to be not perfectly round, performing integrated analysis on whole LDs resulted in peak broadening. We therefore analyzed the layer spacing by plotting the density profile of individual LDs on polar coordinates, fitted a line to each layer, aligned the profiles for each layer, and averaged them to increase the signal-to-noise ratio. The layer spacing measured with higher accuracy ranged between 3.6 nm (typically at the LD periphery) and 3.45 nm (in the interior layers). Membrane bilayers in the vicinity of LDs were analyzed using the same methodology and consistently showed a spacing of 3.9 nm between the two leaflets, indicating that LDs are composed of molecules that pack more tightly and periodically than phospholipids.
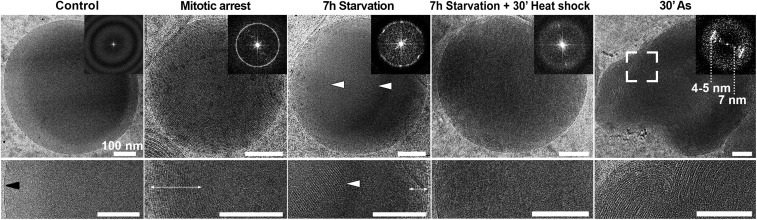
Fig. 2.
LDs exhibit varying internal structure under different cellular states. Top row shows entire LD with Insets representing the fast Fourier transforms (FFT). Bottom rows provide a zoomed-in view at a central slice through the LDs (periphery-center). (Scale bars, 100 nm.) LDs in normally cycling cultures (control) are amorphous, and the FFT only exhibits modulations associated with EM images acquired at defocus. Mitotically arrested cells show a uniform concentric layered structure at their periphery (double-sided arrow), represented by a ring in the FFT. Starved cells also exhibit a layered structure (double-sided arrow), as well as crystalline nanodomains at the center (arrowheads), represented by bright peaks superimposed on a ring of the same spacing in the FFT. Starved cells exposed to heat shock of 43 °C lose the crystalline structure and are amorphous. LDs in Arsenite-treated cells (30′ As) are distorted and exhibit bilayer-like structure at the periphery. The FFT was generated from the framed area.
Our data show that LDs have a core-shell organization of an amorphous interior and a liquid-crystalline shell with lattice spacing of 3.4 to 3.6 nm in mitotically arrested cells. Importantly, this organization exists at body temperature. It is known that pure CE arranges into different structural phases depending on temperature (18). Specifically, at 37 °C, pure cholesteryl oleate exists as a smectic phase, in which the extended, rod-like molecules are arranged in layers spaced regularly at 3.4 nm. In the plane of each layer, the molecules are still liquid-like. The lattice spacing depends on the particular cholesteryl species and ranges between 3.4 and 3.6 nm for the prominent acyl chains (oleate [18:1], linoleate [18:2], and palmitate [16:0]) in cells. Thus, we attribute the liquid-crystalline shell observed in LDs to CE molecules with variable acyl chains.
Studies in a system of similar composition to LDs, namely, human low-density lipoparticles (LDL), demonstrated that a mixture of TAG and CE results in an unstructured emulsion at ambient temperature. The transition temperature from an ordered to disordered state for cholesteryl oleate was determined to be 41 °C in the presence of 5% TAG (19) and is highly affected by the amount of TAG (20). We therefore suggest that following mitotic arrest, the relative concentration of the two lipid components in LDs is perturbed, either by accumulation of CE or by depletion of TAG. This results in an increased local concentration and a consequent phase separation of CE molecules within the droplets, eventually leading to a structural transition into a liquid-crystalline phase. These findings are in line with in vivo studies in yeast triple mutants for the synthesis of neutral lipids, which maintain the single ARE2+ contributing exclusively to CE production (21). LDs in these cells showed a phase transition into a crystalline arrangement due to CE. Mutants in which TAG was exclusively produced and incorporated into LDs did not exhibit order–disorder phase transitions over a wide range of temperatures.
The number of layers quantified in individual LDs ranged between 19 and 29. An average volume ratio occupied by ordered layers to the disordered core, approximated from the boundaries measured for each phase in the 2D projection, was 1.9 ± 1.2 (n = 14 LDs). The wide variation in the number of lattice layers and volume ratio is most likely attributed to the random section plane obtained through LDs, i.e., central vs. peripheral, in cryo-FIB lamellae. Nevertheless, this provides an indication of the relative amounts of the two major constituents, namely, TAG and CE, in LDs exhibiting two structural phases.
Images obtained from HeLa cells of normally cycling cultures following the same sample preparation and imaging showed that LDs are typically amorphous and are delineated by a single high-density layer (Fig. 2, Control) (16). Analysis of LD internal organization from a larger number of cells showed that while over 90% of mitotically arrested cells exhibited concentric rings, only about 20% of control cells showed layering to some degree (Table 1). We therefore followed up to characterize the difference in LDs between these two cellular states by light microscopy (SI Appendix, Fig. S3) (13, 22). LDs stained with BODIPY increased in number (141 vs. 78 LDs per cell) and size (0.36 ± 0.45 μm3 vs. 0.18 ± 0.08 μm3) in mitotically arrested cells in comparison with the control cultures. As LDs were larger in mitotically arrested cells and exceeded the diffraction limit, it appeared that the BODIPY staining recapitulates the core-shell structure observed by cryo-EM, presumably due to pi-stacking interactions between BODIPY and CE leading to preferential incorporation of the dye into the shell CE layers (SI Appendix, Fig. S3A, Inset and Movie S1). The light-microscopy data quantification indicated an increase in the total amount of neutral lipids in mitotically arrested cells.
Table 1.
Analysis of cellular LDs structure and abundance under different cellular states
| Treatment | # cells for EM analysis | % cells with crystalline LDs* | BODIPY/cell† |
| Control | 14 | 21.43 | 1.00 |
| Mitotically arrested | 34 | 91.18 | 1.27 |
| 7 h starvation | 11 | 72.73 | 1.39 |
| 7 h starvation + 30′ heat shock | 5 | 0.00 | — |
| Heat shock | 4 | 0.00 | 2.15 |
Percentage of cells with crystalline LDs quantified from 2D cryo-EM projections of FIB lamellae. Lamellae typically contained 5 to 20 LDs, with all LDs analyzed, and frequently exhibited the same structure within the same cell.
LD volume per cell was quantified to account for the observed increase in both LD number and size by light microscopy. BODIPY per cell is represented as a fold increase relative to the control cells.
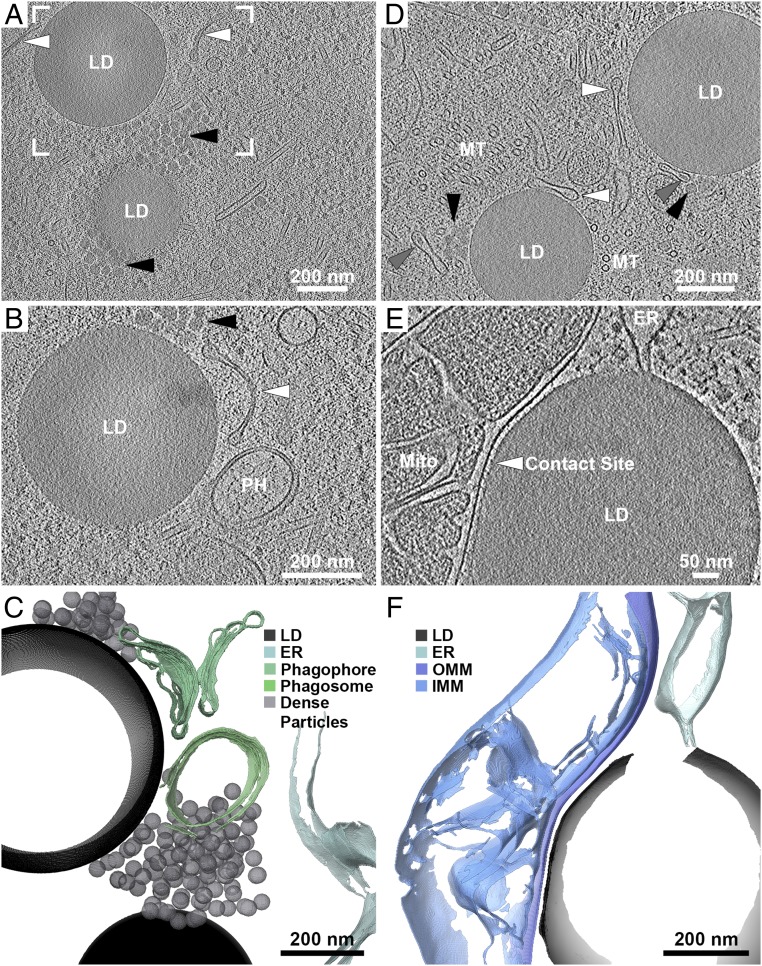
Examination of the intracellular ultrastructure under our experimental conditions may provide a hint as to the source of lipid accumulation in mitotic arrest. Cryo-ET data acquired from lamellae of mitotically arrested cells showed that LDs are commonly associated with elongated membranes and with ∼20-nm-diameter high-density particles (Fig. 3 A–D, white and black arrowheads, respectively, and Movies S2 and S3). The elongated membranes resembled the classical morphology of phagophores. Autophagosomes were also observed in close proximity (Fig. 3 B and C) (23). Many of the phagophore-resembling structures, and not the autophagosomes, were observed to contain dense material (Fig. 3D, gray arrowheads). Based on the observations of autophagy-related intracellular morphology, we extended our analysis to a variety of cellular stress conditions (e.g., starvation, heat shock, arsenite stress). Upon starvation, cells transition from glucose-based metabolism to mitochondrial fatty acid oxidation for energy production. Under these conditions, fatty acids are mobilized to mitochondria and are subsequently depleted from LDs (24, 25). Indeed, under starvation conditions, 70% of the cells analyzed exhibited crystalline LDs (Fig. 2, 7h Starvation, and Table 1), consistent with local increase of CE relative concentration following TAG consumption. Autophagy appeared to be an important mechanism for lipid trafficking into LDs (25). Inhibition of autophagosome fusion to lysosomes by Bafilomycin under starvation resulted in reduction of total BODIPY signal (1.39 vs. 0.59 in starvation without and with Bafilomycin) (26). Complementary to these results, cryo-ET data on starved cells showed tight membrane contact sites (16-nm spacing) between LDs and mitochondria, spanning ∼400 nm of the LD edge and containing densities of molecular tethers (Fig. 3 E and F and Movie S4). Direct ER-membrane–LD interactions were also observed (27). To further demonstrate that the crystalline structure is attributed specifically to crystallization of the CE, we subjected the starved cells to heat shock (43 °C). Heating beyond the transition temperature of CE resulted in “melting” of the liquid-crystalline smectic phase into a less ordered phase (Fig. 2, 7h Starvation + 30’ Heat shock, and Table 1). At the other extreme of metabolic stress, arsenite treatment of cells, which blocks mitochondrial respiration, resulted in either amorphous LDs or the appearance of stacked bilayers in LDs (Fig. 2, 30′ As). This is presumably due to the decreased mobilization of fatty acids to mitochondria and their subsequent accumulation in LDs. LDs in heat-shocked cells remained unstructured despite an increase in total LD lipids (Table 1). In conclusion, even though a number of conditions were found to increase lipid accumulation in LDs (Table 1), only some also result in the appearance of liquid-crystalline transition in LDs. Therefore, the internal organization of LDs is directly related to cellular metabolic state.
Fig. 3.
Cellular tomography of LDs reveals spatial association with organelles. (A) LDs in a mitotically arrested cell, spatially associated with elongated membranes (white arrowheads) and with 20- to 30-nm-diameter high-density particles (black arrowheads). (B) Enlarged view of frame in A at a different tomographic slice showing that the elongated membranes resemble the classical morphology of phagophores. A closed autophagosome is in close proximity (PH). (C) Annotated 3D segmentation of LDs, putative phagophores/autophagosomes, and high-density particles. (D) Two LDs in a different mitotically arrested cell, associated with the appearance of elongated membranes (white arrowheads) and high-density particles (black arrowheads). Elongated membrane structures with density present within their lumen are annotated by gray arrowheads. Microtubules (MT) are present at high abundance due to the mitotic cell-cycle stage. (E) LD in a starved cell associated with a mitochondrion (Mito) through a tight membrane contact site (arrowhead). An ER membrane is in direct physical contact with the LD. (F) Annotated 3D segmentation of LD contacts with mitochondrion and ER. IMM, inner mitochondrial membrane; OMM, outer mitochondrial membrane.
Discussion
LDs are organelles of great metabolic importance in most cell types. The neutral lipids (TGA and CE) stored in their cores provide a buffer for energy fluctuations and a reservoir for membrane lipid precursors. In this study, we employed cryoelectron microscopy and tomography to characterize LDs directly inside HeLa cells. We show that under normal culture conditions, LDs exhibit the expected morphology of an amorphous emulsion of neutral lipids surrounded by a phospholipid monolayer. However, under conditions of mitotic arrest and nutrient depravation, LDs structurally reorganize into a liquid-crystalline shell surrounding an amorphous core. By accurate quantification of the shell lattice spacing of 3.4 to 3.6 nm, we could attribute the crystalline phase to CE. These observations are consistent with previous reports in yeast (21). We further show that the volume ratio of the amorphous core to the crystalline shell falls in the range of 1:2. To further validate this hypothesis, we showed that heat shock treatment of cells with crystalline LDs beyond the phase transition temperature of predominant CE species in cells results in melting of the lattice. Our experimental conditions, consisting of hours-long mitotic arrest and starvation, led to a homogenously crystalline population of LDs throughout the cell. It remains to be explored whether this end-state is preceded by a stepwise transition in subpopulations of preexisting or newly forming LDs in the cell.
We hypothesize that the formation of the liquid-crystalline CE phase is induced by a relative increase in CE, concomitant with a decrease of TAG levels in LDs under these conditions. We show that HeLa cells accumulate more total lipid in LDs, an observation in agreement with a previous study on starvation in mouse embryonic fibroblasts (25). We further show that LDs in mitotic arrest are commonly found in close proximity to phagophore-resembling or autophagosome structures. It is possible that autophagy is the main mechanism contributing to the trafficking of lipids into these LDs (25). However, numerous 20-nm-diameter dense particles were also found in the proximity of LDs and the phagophore/autophagosome membranes. These could represent an alternative mechanism for cellular lipid trafficking into LDs in discrete packages, analogous to lipoprotein particles, rather than by membrane contact site-mediated lipid transfer (3). Alternatively, the particles may represent the documented Arf1/COPI machinery-mediated budding of nano-LDs from LD surfaces, leading to a phospholipid deprivation and, consequently, to increased surface tension (27, 28). We, however, could not discern the presence of COPI densities on the surface of the particles in our tomography data as suggested by Bykov et al. (29).
On the other side of the lipid flux pathways, namely, lipolysis, we show that under starvation known to increase the flux of TAG degradation products—predominantly fatty acids (FA) —from LDs to mitochondria for oxidative respiration, LD–mitochondrial membrane contact sites exist, which may facilitate FA–lipid transfer (3, 4, 24, 25). Lipolysis also generates TAG metabolites that serve as membrane lipid precursors or signaling lipids important for growth and division. Consistent with increased membrane requirements in proliferating cells (11), synthesis of phospholipids in mammalian cells is in part regulated in a cell-cycle dependent manner (30), explaining the liquid-crystalline phase of LDs in mitotically arrested cells. At this point, we could not exclude the possibility that the mitotic arrest may lead to a stress response contributing to crystalline LDs (31).
Altogether, these data demonstrate that LDs can exhibit different structural phases that are directly related to cellular conditions and metabolic scenarios and present different modes of interactions with various cellular organelles. There is a general consensus that cancer cells and immortalized cell cultures display metabolic reprogramming compared with healthy cells (reviewed in refs. 11 and 12). It remains to be validated whether the phase transitions we describe in HeLa cells exist in nontransformed cells. LDs also serve as localization sites for some proteins (17, 32), such as enzymes related to LD metabolism including neutral lipid synthesis or lipolysis, and have a fundamentally different architecture compared with bilayer-bound organelles, resulting in unique physical properties of the LD surface consisting of a phospholipid monolayer. It is not unlikely that the structural phase transitions described here could have a major impact on the organelle contact site with LDs and the accessibility of lipids in LDs to specific binding to enzymes or lipid transporters. These may become restricted when LDs exhibit the smectic-crystalline state, slow the exchange of lipids with surrounding membranes, and lead to a different surface occupancy of LD-associated proteins (33). Therefore, the composition, and, in turn, the resulting internal structure of LDs is expected to play a key role in their function as a central hub of cellular lipid metabolism.
CE droplets are also a major component of atherosclerotic lesions in human arteries (18, 34). Intriguingly, Rambold et al. (25) established that LDs in starved fibroblasts can be secreted to the extracellular milieu due to the limited LD storage capacity in cells of nonadipose origin. Our data raise the possibility that secreted crystalline LDs may directly contribute to the formation of atherosclerotic lesions, in analogy to LDL particles, and may therefore have a direct link to human pathologies.
Materials and Methods
Cell Culture for Cryo-ET.
HeLa Cells were cultured on gold Quantifoil TEM grids overnight, subjected to starvation, heat shock, and arsenite stress where specified, and vitrified by plunge-freezing using a Vitrobot Mark 4. For synchronization, cells were treated by Thymidine and released into fresh medium followed by an STLC block. Mitotic cells were detached from the culture flask by mechanical shake-off, applied to copper grids, and plunge-frozen. Details are reported in SI Appendix, SI Materials and Methods.
Cryo-3D Correlation, Cryo-FIB, and Cryo-TEM/ET.
All procedures for 3D cryocorrelative targeted FIB preparations and cryo-ET data acquisition on lamellae were reported in refs. 14 and 10 and are further described in SI Appendix, SI Materials and Methods.
Fluorescent Light Microscopy and Quantification of LDs.
Cells were cultured on glass coverslips, subjected to perturbations as detailed in SI Appendix, SI Materials and Methods, fixed with 2% formaldehyde, and stained with BODIPY. Stacks were acquired on a wide-field microscope. Images were processed in ImageJ using the 3D object counter plugin.
Supplementary Material
Acknowledgments
We are grateful to Ina Poser and Tony Hyman for providing the HeLa cell lines and to Friedrich Förster for valuable input on data processing. J.M. was supported by postdoctoral fellowships from the European Molecular Biology Organization and the Human Frontier Science Program, Weizmann Institute Women in Science Program, and acknowledges support from the European Molecular Biology Laboratory. W.B. and H.L. acknowledge the Center for Integrated Protein Science Munich.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1903642116/-/DCSupplemental.
References
- 1.Walther T. C., Farese R. V. Jr, The life of lipid droplets. Biochim. Biophys. Acta 1791, 459–466 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fujimoto T., Parton R. G., Not just fat: The structure and function of the lipid droplet. Cold Spring Harb. Perspect. Biol. 3, a004838 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schuldiner M., Bohnert M., A different kind of love—Lipid droplet contact sites. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1862, 1188–1196 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Valm A. M., et al. , Applying systems-level spectral imaging and analysis to reveal the organelle interactome. Nature 546, 162–167 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barbosa A. D., Savage D. B., Siniossoglou S., Lipid droplet-organelle interactions: Emerging roles in lipid metabolism. Curr. Opin. Cell Biol. 35, 91–97 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Fujimoto T., Ohsaki Y., Suzuki M., Cheng J., Imaging lipid droplets by electron microscopy. Methods Cell Biol. 116, 227–251 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Beck M., Baumeister W., Cryo-electron tomography: Can it reveal the molecular sociology of cells in atomic detail? Trends Cell Biol. 26, 825–837 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Pfeffer S., Mahamid J., Unravelling molecular complexity in structural cell biology. Curr. Opin. Struct. Biol. 52, 111–118 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Dubochet J., Mcdowall A. W., Vitrification of pure water for electron-microscopy. J. Microsc. 124, Rp3–Rp4 (1981). [Google Scholar]
- 10.Mahamid J., et al. , Visualizing the molecular sociology at the HeLa cell nuclear periphery. Science 351, 969–972 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Natter K., Kohlwein S. D., Yeast and cancer cells—Common principles in lipid metabolism. Biochim. Biophys. Acta 1831, 314–326 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tirinato L., et al. , An overview of lipid droplets in cancer and cancer stem cells. Stem Cells Int. 2017, 1656053 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Listenberger L. L., Studer A. M., Brown D. A., Wolins N. E., Fluorescent detection of lipid droplets and associated proteins. Curr. Protoc. Cell Biol. 71, 4.31.1–4.31.14 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Arnold J., et al. , Site-specific cryo-focused ion beam sample preparation guided by 3D correlative microscopy. Biophys. J. 110, 860–869 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leforestier A., Lemercier N., Livolant F., Contribution of cryoelectron microscopy of vitreous sections to the understanding of biological membrane structure. Proc. Natl. Acad. Sci. U.S.A. 109, 8959–8964 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tauchi-Sato K., Ozeki S., Houjou T., Taguchi R., Fujimoto T., The surface of lipid droplets is a phospholipid monolayer with a unique Fatty Acid composition. J. Biol. Chem. 277, 44507–44512 (2002). [DOI] [PubMed] [Google Scholar]
- 17.Bersuker K., Olzmann J. A., Establishing the lipid droplet proteome: Mechanisms of lipid droplet protein targeting and degradation. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1862, 1166–1177 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Engelman D. M., Hillman G. M., Molecular organization of the cholesteryl ester droplets in the fatty streaks of human aorta. J. Clin. Invest. 58, 997–1007 (1976). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kroon P. A., The order-disorder transition of the core cholesteryl esters of human plasma low density lipoprotein. A proton nuclear magnetic resonance study. J. Biol. Chem. 256, 5332–5339 (1981). [PubMed] [Google Scholar]
- 20.Mateu L., Kirchhausen T., Camejo G., A low temperature structural transition in human serum low density lipoproteins. Biochim. Biophys. Acta 487, 243–245 (1977). [DOI] [PubMed] [Google Scholar]
- 21.Czabany T., et al. , Structural and biochemical properties of lipid particles from the yeast Saccharomyces cerevisiae. J. Biol. Chem. 283, 17065–17074 (2008). [DOI] [PubMed] [Google Scholar]
- 22.DiDonato D., Brasaemle D. L., Fixation methods for the study of lipid droplets by immunofluorescence microscopy. J. Histochem. Cytochem. 51, 773–780 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Yang Z., Klionsky D. J., Eaten alive: A history of macroautophagy. Nat. Cell Biol. 12, 814–822 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herms A., et al. , AMPK activation promotes lipid droplet dispersion on detyrosinated microtubules to increase mitochondrial fatty acid oxidation. Nat. Commun. 6, 7176–7190 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rambold A. S., Cohen S., Lippincott-Schwartz J., Fatty acid trafficking in starved cells: Regulation by lipid droplet lipolysis, autophagy, and mitochondrial fusion dynamics. Dev. Cell 32, 678–692 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamamoto A., et al. , Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct. Funct. 23, 33–42 (1998). [DOI] [PubMed] [Google Scholar]
- 27.Wilfling F., et al. , Arf1/COPI machinery acts directly on lipid droplets and enables their connection to the ER for protein targeting. eLife 3, e01607 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thiam A. R., et al. , COPI buds 60-nm lipid droplets from reconstituted water-phospholipid-triacylglyceride interfaces, suggesting a tension clamp function. Proc. Natl. Acad. Sci. U.S.A. 110, 13244–13249 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bykov Y. S., et al. , The structure of the COPI coat determined within the cell. eLife 6, e32493 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jackowski S., Cell cycle regulation of membrane phospholipid metabolism. J. Biol. Chem. 271, 20219–20222 (1996). [DOI] [PubMed] [Google Scholar]
- 31.Bischof J., et al. , Clearing the outer mitochondrial membrane from harmful proteins via lipid droplets. Cell Death Discov. 3, 17016 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kory N., Farese R. V. Jr, Walther T. C., Targeting fat: Mechanisms of protein localization to lipid droplets. Trends Cell Biol. 26, 535–546 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hsieh K., et al. , Perilipin family members preferentially sequester to either triacylglycerol-specific or cholesteryl-ester-specific intracellular lipid storage droplets. J. Cell Sci. 125, 4067–4076 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Small D. M., Shipley G. G., Physical-chemical basis of lipid deposition in atherosclerosis. Science 185, 222–229 (1974). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.