Significance
Different mutations of the von Hippel–Lindau (VHL) tumor suppressor gene cause different subtypes of VHL hereditary cancer syndrome. Dysregulation of VHL’s canonical substrate, HIFα, cannot fully explain the complex genotype/phenotype manifestation within VHL disease. We describe an oxygen-sensitive function of VHL that regulates hydroxylated BIM-EL protein stability. Type 2C VHL mutants, which cause pheochromocytoma and paraganglioma (PCC/PGL) despite repressing HIFα, destabilize BIM-EL. Other genetic mutations linked to PCC/PGL, similar to loss of BIM-EL, protect from apoptosis upon nerve growth factor withdrawal. Dysregulation of BIM-EL might therefore contribute to the pathogenesis of VHL-related PCC/PGL. Loss of BIM-EL expression can lead to chemotherapy resistance in other VHL-related neoplasms and renders clear cell renal cell carcinoma cells insensitive to cisplatin.
Keywords: VHL, tumor suppression, hydroxylation, BIM-EL, pheochromocytoma
Abstract
Despite the discovery of the oxygen-sensitive regulation of HIFα by the von Hippel–Lindau (VHL) protein, the mechanisms underlying the complex genotype/phenotype correlations in VHL disease remain unknown. Some germline VHL mutations cause familial pheochromocytoma and encode proteins that preserve their ability to down-regulate HIFα. While type 1, 2A, and 2B VHL mutants are defective in regulating HIFα, type 2C mutants encode proteins that preserve their ability to down-regulate HIFα. Here, we identified an oxygen-sensitive function of VHL that is abolished by VHL type 2C mutations. We found that BIM-EL, a proapoptotic BH3-only protein, is hydroxylated by EglN3 and subsequently bound by VHL. VHL mutants fail to bind hydroxylated BIM-EL, regardless of whether they have the ability to bind hydroxylated HIFα or not. VHL binding inhibits BIM-EL phosphorylation by extracellular signal-related kinase (ERK) on serine 69. This causes BIM-EL to escape from proteasomal degradation, allowing it to enhance EglN3-induced apoptosis. BIM-EL was rapidly degraded in cells lacking wild-type VHL or in which EglN3 was inactivated genetically or by lack of oxygen, leading to enhanced cell survival and chemotherapy resistance. Combination therapy using ERK inhibitors, however, resensitizes VHL- and EglN3-deficient cells that are otherwise cisplatin-resistant.
Von Hippel–Lindau (VHL) disease is caused by germline mutations of the VHL gene and predisposes to a variety of tumors, including hemangioblastoma (HB) of the retina and nervous system, clear cell renal cell carcinoma (ccRCC; the most common form of kidney cancer), and tumors of sympathoadrenal origin such as pheochromocytoma (PCC)/paraganglioma (PGL) (1). HIFα activation is necessary and sufficient for many of the manifestations of VHL loss of function. For example, HIFα deregulation appears to play a causal role in VHL-defective ccRCC and in VHL-defective HB (1). Nonetheless, a number of other biochemical functions have been ascribed to VHL protein (pVHL), including binding to fibronectin, collagen, protein kinase C, and AKT (2–6). Different VHL germline mutations cause different organ-specific cancer risks in VHL disease (1). Type 1 VHL disease is defined as ccRCC and HB with low risk of PCC/PGL and is associated with disruptive mutations and gross deletions in VHL, leading to very high HIFα expression. Type 2 VHL disease, which is predominantly associated with VHL missense mutations, is defined by the occurrence of PCC/PGL, either alone (type 2C) or in combination with HBs (type 2A) or HBs and RCCs (type 2B). Some type 2C mutants retain the ability to suppress HIFα (7, 8). This suggests that PCC/PGL in type 2C VHL disease is not caused by HIFα activation; thus, VHL type 2C mutants would be a powerful tool for identifying HIFα-independent VHL functions.
We previously reported that genetic alterations, including VHL alterations, linked to familial PCC/PGL may act by decreasing the activity of a 2-oxoglutarate–dependent oxygenase, EglN3, thereby reducing apoptosis of neural crest cells during development (9). Furthermore, EglN3-induced apoptosis requires its hydroxylase activity, but is independent of its role in suppressing HIFα (9). Using an unbiased genome-wide short hairpin RNA (shRNA) screen, we identified the candidate 1p36 tumor suppressor KIF1Bβ, which is required for EglN3-dependent apoptosis, and we and others identified loss-of-function germline KIF1Bβ mutations in subsets of PCC/PGL and neuroblastomas (10–12). However, EglN3 does not appear to hydroxylate KIF1Bβ, which led us to search for EglN3 proapoptotic substrates. Here, we identified the proapoptotic BH3-only protein BIM-EL as a direct EglN3 hydroxylation substrate. BIM-EL represents an isoform of the BCL2L11 gene. The 3 major BIM isoforms are BIM-EL, BIM-L, and BIM-S, and all contain a BH3 domain but have different proapoptotic potencies. We found that hydroxylation of BIM-EL, but not BIM-L and BIM-S, enables direct VHL binding, which masks the phosphorylation site that is critical for BIM-EL protein degradation. Importantly, VHL recognition of hydroxylated BIM-EL is abolished by VHL type 2C mutations, including type 2C mutations that do not result in HIFα stabilization. Furthermore, reintroduction of wild-type (WT) VHL, but not type 2C VHL mutants, into VHL−/− cells restores the ability of EglN3 to induce apoptosis. These studies suggest that PCC/PGLs linked to VHL mutations are caused by defects in EglN3-induced apoptosis because of a failure to stabilize BIM-EL.
Results
EglN3-Induced Apoptosis and Regulation of BIM-EL Is Hydroxylation- and VHL-Dependent.
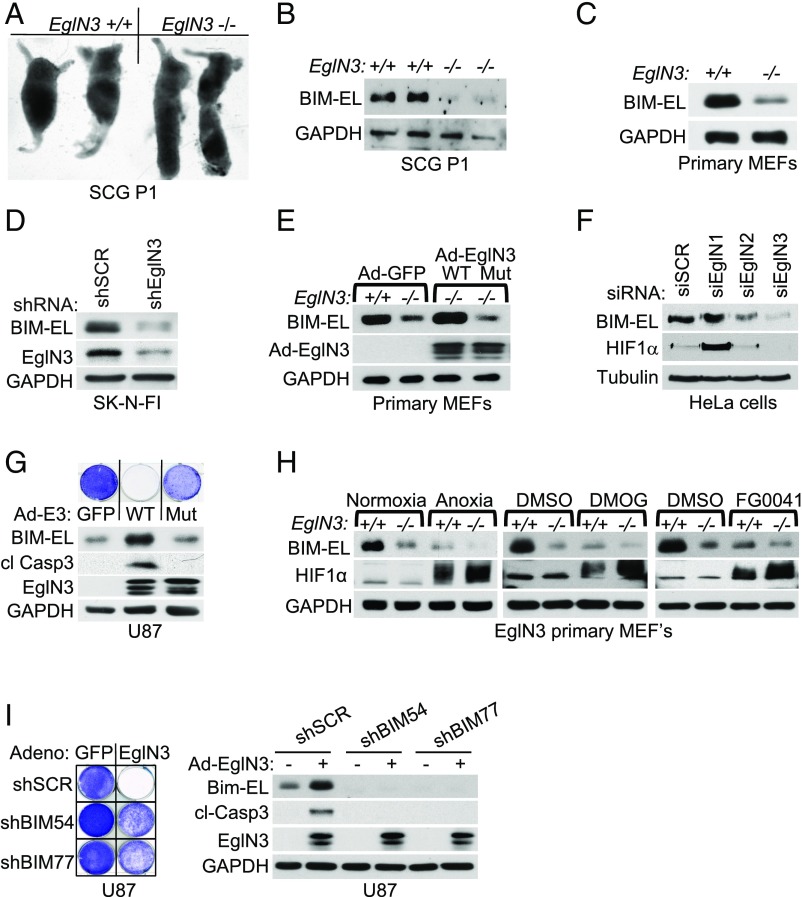
We previously demonstrated that EglN3 activity is required for apoptosis when sympathetic neurons are deprived of nerve growth factor (NGF) in vitro (9, 10). We also confirmed that the superior cervical ganglia (SCG), which are composed of such neurons, from postnatal day 1 (P1) mice lacking EglN3 are enlarged compared with their WT littermates (Fig. 1A), in keeping with a prior study (13). The proapoptotic BCL-2 family member BIM-EL is critical for neuronal apoptosis (14), and hypoxia’s ability to suppress apoptosis after NGF withdrawal involves suppression of BIM-EL (15). Thus, we investigated if EglN3 hydroxylation activity can induce BIM-EL.
Fig. 1.
EglN3 regulation of BIM-EL protein is hydroxylation-dependent. (A) Photographs of dissected mouse SCG of the indicated genotype and age. (B) Anti–BIM-EL immunoblot of dissected SCG from 1-wk-old mice of the indicated genotype. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. (C) Anti–BIM-EL immunoblot of EglN3 primary MEFs of the indicated genotype. (D) Anti–BIM-EL immunoblot of SK-N-FI cells that were stably transduced with lentivirus encoding shRNA targeting EglN3 (shEglN3) or scramble control (shSCR). (E) Immunoblot analysis of EglN3 MEFs transduced with adenovirus (Ad) encoding WT EglN3, catalytic-dead mutant (H196A; Mut), or GFP control (Ad-GFP). (F) Anti–BIM-EL immunoblot analysis of HeLa cells transfected with siRNA targeting EglN1, EglN2, EglN3, or nontargeting control (SCR) as indicated. si, small interfering. (G) Immunoblot and crystal violet staining of U87 cells transduced with adenovirus encoding WT EglN3, catalytic dead mutant (H196A), or GFP-control. Forty-eight hours after adenovirus transduction, cells were supplemented with medium and maintained in culture for 1 additional week, followed by crystal violet staining. cl, cleaved. (H) Immunoblot of EglN3 primary MEFs with the indicated genotype upon normoxic or anoxic conditions for 16 h or treated with 1 mM DMOG or 50 μM FG0041. (I, Left) Crystal violet staining of U87 cells stably transduced with lentiviral pLKO shRNA targeting BIM (shBIM) or nontargeting control (shSCR) and subsequently transiently transduced for 48 h with adenovirus encoding EglN3 or GFP control. (I, Right) Corresponding immunoblot.
We observed that BIM-EL protein was dramatically reduced in EglN3−/− SCG and primary mouse embryonic fibroblasts (MEFs) compared with littermate control EglN3+/+ SCG and MEFs, respectively (Fig. 1 B and C), and in neuroblastoma cells in which EglN3 was down-regulated with shRNA (Fig. 1D). To understand if BIM-EL protein levels are regulated by EglN3 hydroxylation activity, we made adenoviruses encoding WT EglN3 or a catalytic-dead EglN3 mutant (H196A). BIM-EL levels were restored in EglN3−/− MEFs infected with adenovirus (Ad)-EglN3-WT, but not Ad-EglN3-H196A (Fig. 1E). Thus, EglN3 hydroxylation activity regulates BIM-EL protein level.
To investigate if this effect on BIM-EL was specific to EglN3 among the 3 EglN paralogs, we specifically silenced EglN1, EglN2, or EglN3 in HeLa cervical carcinoma cells using small interfering RNAs (siRNAs) (Fig. 1F and SI Appendix, Fig. S2A). BIM-EL protein levels were reduced by silencing EglN3, and to a much lesser extent by silencing EglN2, but not by silencing EglN1. Conversely, silencing EglN1 caused the greatest effect on HIF1α, consistent with earlier studies (16), whereas silencing EglN3 had no measurable effect on HIF1α, suggesting specificity of function within the EglN family members.
We next overexpressed EglN3 in cell lines of sympathoadrenal (PC12, rat) (SI Appendix, Fig. S1A) or neuronal (U87, human) origin (Fig. 1G) using the adenovirus described above. In these cells, WT, but not catalytic-dead, EglN3 induced BIM-EL and apoptosis as measured by caspase 3 cleavage and crystal violet staining. Moreover, the effect of EglN3 on BIM-EL was blocked under anoxic conditions and by the hydroxylase inhibitors dimethyloxaloylglycine (DMOG) and FG0041 in EglN3 primary MEFs (Fig. 1H).
To address whether induction of BIM-EL is necessary for EglN3 to induce cell death, we silenced BIM-EL with 2 independent shRNAs in U87 cells (Fig. 1I) or using 2 independent siRNAs in PC12 cells (SI Appendix, Fig. S1B). Silencing BIM-EL protected against EglN3-induced apoptosis as measured by crystal violet staining, caspase 3 cleavage, and nuclear fragmentation.
Since we observed that BIM-EL protein abundance is dependent on EglN3 hydroxylase activity, we investigated if this observation is VHL-dependent. Cre-mediated deletion of VHL in MEFs homozygous for a floxed VHL allele robustly decreased BIM-EL protein levels (Fig. 2A), reminiscent of the loss of BIM-EL in EglN3−/− MEFs. Furthermore, reintroducing hemagglutinin (HA)-VHL (WT) into VHL-deficient RCC cells (786-O) restored BIM-EL protein expression (Fig. 2B) unless EglN3 was inactivated with an effective shRNA (Fig. 2C) or by anoxia (Fig. 2D).
Fig. 2.
BIM-EL protein regulation by EglN3 is dependent on pVHL. (A) Anti–BIM-EL immunoblot of VHL+/+ and VHLfl/fl MEFs (immortalized by K1 large T antigen) that were transiently transduced with adenovirus encoding Cre-recombinase (Ad-Cre) or GFP control. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. (B) Anti–BIM-EL immunoblot analysis of 786-O VHL-null cells (−/−) stably transfected to generate WT VHL. (C) Immunoblot of 786-O cells with the indicated VHL status transduced with lentiviral pLKO shRNA targeting EglN3 or nontargeting control (shSCR). (D) Immunoblot of 786-O WT-VHL and VHL-null cells (−/−) under normoxic or anoxic conditions for 16 h. (E) Immunoblot of 786-O cells stably transfected to generate the indicated VHL species. (F, Left) Crystal violet staining of 786-O cells with the indicated VHL status. Cells were transiently transduced for 48 h with adenovirus (Adeno) encoding either WT EglN3 or GFP control. Cells were maintained in culture for an additional week before being subjected to crystal violet staining. (F, Right) Corresponding immunoblot is shown 48 h after infection. cl, cleaved. (G) Crystal violet staining and immunoblot of 786-O cells expressing HA-VHL that were stably transduced with lentiviral-encoded shRNA targeting BIM (shBIM54) or nontargeting control (shSCR). Cells were transiently transduced with adenovirus encoding EglN3 (Ad-E3) or GFP control. (H) Immunoblot analysis of stable polyclonal PC12 cells expressing the indicated human VHL (huVHL) species. PC12 clones were transiently transduced for 72 h with lentivirus encoding shRNA targeting endogenous rat VHL (endg. sh-ratVHL) or scramble control (shSCR). (Note: VHL long-term knockdown was not tolerated in PC12 cells.) (I) Percentage of fragmented nuclei of stable PC12 clones 3 d after cotransfection to generate GFP-histone and Flag-EglN3 along with pLKO plasmid targeting rat VHL (sh-rVHL) or scramble control (sh-SCR). One-way ANOVA was used to determine the statistical significance using GraphPad Prism software (****P < 0.0001). (J) Anti–BIM-EL immunoblot analysis of human normal adrenal glands and human primary PCC and PGL tumors. VHL mutation and 1p36 status have been characterized as indicated.
Type 2C VHL mutants predispose to PCC/PGL without grossly deregulating HIFα (7). We therefore asked if VHL type 2C mutations are loss of function with respect to BIM-EL regulation and if this is reflected in VHL-defective human PCC/PGL. Compared with WT HA-VHL, type 2C HA-VHL (L188V and R64P) was clearly defective with respect to BIM-EL induction when reintroduced into 786-O cells despite their ability to repress HIF2α (Fig. 2E). Moreover, VHL−/− 786-O cells were resistant to EglN3-induced apoptosis (Fig. 2F). Importantly, reintroduction of WT VHL, but not the type 2C VHL mutant L188V, rescued the ability of EglN3 to induce apoptosis as measured by crystal violet staining and caspase 3 cleavage (Fig. 2F). Consistent with the data in Fig. 1I, silencing BIM-EL also prevented EglN3-induced apoptosis in the context of 786-O cells (Fig. 2G). EglN3 expression also caused some growth inhibition seen in crystal violet staining 2 wk postinfection in VHL−/− and mutant cells, consistent with previous reports that EgLN3 can suppress cell proliferation (17, 18). Similar to 786-O cells, VHL knockdown in rat PC12 PCC cells decreased BIM-EL levels (Fig. 2H) and protected against EglN3-induced apoptosis (Fig. 2I). Reintroduction of human HA-VHL WT, but not VHL 2C-L188V, restored BIM-EL expression and EglN3-induced apoptosis, suggesting that EglN3’s apoptotic function is linked to VHL type 2C PCC/PGL pathogenesis.
Thus, we assayed BIM-EL protein level in a panel of human primary PCC/PGL tumors that were sequenced for PCC/PGL susceptibility genes, including VHL, and analyzed for 1p36 deletion (12). Normal human adrenal glands were included as controls (Fig. 2J and SI Appendix, Fig. S2B). We observed low BIM-EL expression only in the PCC/PGL tumors carrying VHL mutations and not in other PCC/PGL tumors, supporting our in vitro findings that BIM-EL protein regulation is VHL-dependent.
EglN3 Hydroxylates BIM-EL at the Proline67/70 Residues, Enabling Interaction with WT VHL but Not VHL Type 2C Disease Mutants.
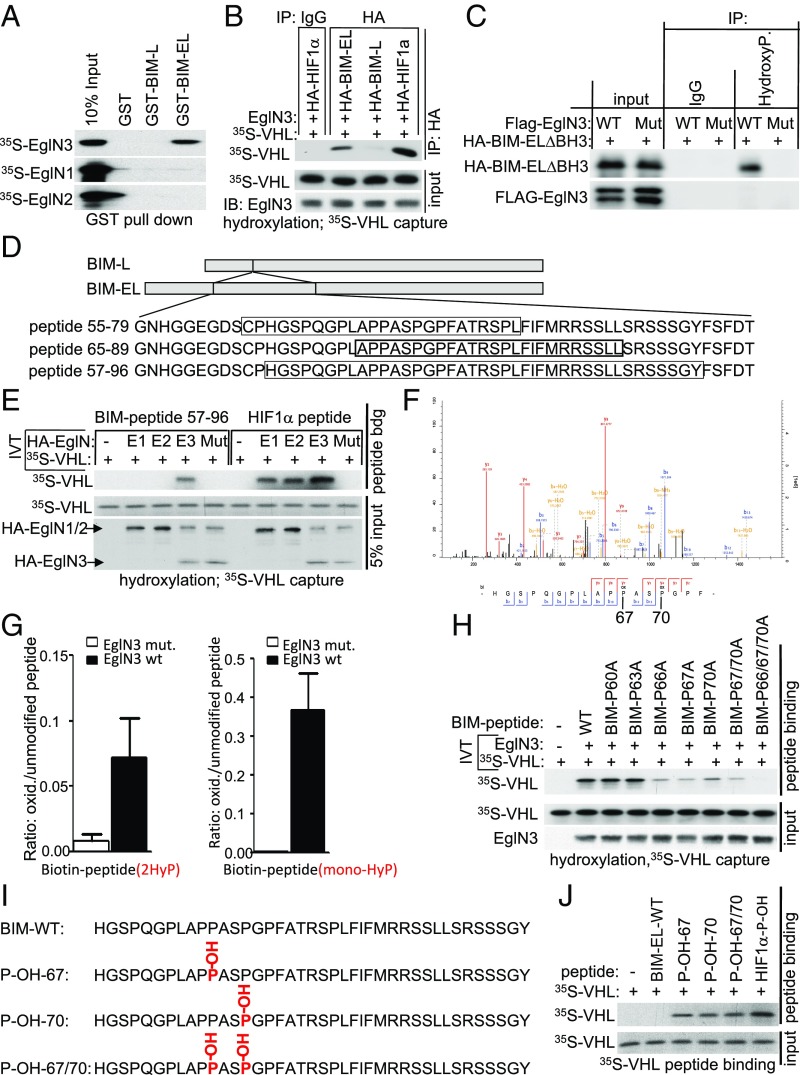
To test whether prolyl-hydroxylation of BIM-EL by EglN3 is responsible for the VHL-dependent regulation of BIM-EL abundance, we first performed EglN3-binding studies with BIM-EL using glutathione S-transferase (GST) pulldown assays. GST-BIM-EL bound to 35S-labeled EglN3 produced by in vitro translation (IVT), but not to 35S-EglN1 or 35S-EglN2 (Fig. 3A and SI Appendix, Fig. S3A). No interaction was observed between 35S-EglN3 and GST-BIM-L, a shorter BIM variant missing 116 amino acids generated by alternative splicing (Fig. 3A and SI Appendix, Fig. S3A). Thus, we investigated if BIM-EL could be hydroxylated by EglN3 and subsequently recognized by VHL. In vitro-translated HA-BIM-EL and HA-BIM-L were HA-immunoprecipitated and used in EglN3 hydroxylation assays. HA-HIF1α was tested in parallel as a positive control. HA-BIM-EL and HIF1α, but not HA-BIM-L, captured 35S-labeled VHL after incubation with EglN3 (Fig. 3B), suggesting that EglN3 hydroxylates proline(s) within the amino acid region that is missing in BIM-L. EglN3 enzymatic activity was required for VHL binding, because BIM-EL subjected to hydroxylation reactions using EglN3 catalytic-dead mutant (H196A) failed to capture 35S-VHL (SI Appendix, Fig. S3B). Furthermore, a pan-hydroxyproline antibody immunoprecipitated an apoptosis-defective HA-BIM-EL apoptotic variant (BIM-EL-∆BH3) from 293 cells (the apoptotic-defective variant was used to allow stable expression) that expressed exogenously EglN3 unless the EglN3 was catalytically inactive or the cells were treated with DMOG (Fig. 3C and SI Appendix, Fig. S3C).
Fig. 3.
BIM-EL is hydroxylated by EglN3 at proline 67/70, causing pVHL recognition. (A) Autoradiograms showing recovery of 35S-labeled EglN1, EglN2, EglN3, proteins bound to GST-BIM-EL, or GST-BIM-L fusion proteins (with GST protein as a negative control). 35S labeling was carried out by an IVT reaction with reticulocyte lysate. (B) Autoradiograms showing recovery of 35S-labeled VHL protein bound to HA-immunoprecipitated full-length BIM-EL, BIM-L, or HIF1α that was first subjected to hydroxylation by EglN3 WT. IgG served as a negative control. 35S labeling was carried out by IVT with retic lysate. EglN3, BIM-EL, BIM-L, or HIF1α was produced by cold IVT. Flag-EglN3 expression was verified by immunoblotting (IB; 5% input). IP, immunoprecipitation. (C) Immunoprecipitation using antihydroxyproline antibody (HydroxyP) from 293T cells that were transiently transfected with plasmids encoding HA-BIM-EL apoptotic mutant (∆BH3) and Flag-EglN3 WT or catalytic-dead mutant (Mut). Immunoblots show coimmunoprecipitation of HA-BIM-EL-∆BH3. (D) Schematic representation of the various biotinylated synthetic BIM-EL peptides used for the EglN3 binding and hydroxylation assays. (E) Autoradiograms showing recovery of 35S-labeled VHL protein bound to biotinylated BIM-EL-peptide 57 to 96 or HIF1α peptide 556 to 575. Before pulldown, peptides were incubated with EglN1, EglN2, EglN3, or EglN3 catalytic mutant generated by IVT or unprogrammed reticulocyte lysate (−). The expression of IVT-produced EglN proteins in each reaction was verified by immunoblotting. (F) MS of biotinylated BIM-EL-peptide subjected to an EglN3 hydroxylation assay. Representative fragmentation spectra of hydroxylated biotin-HGSPQGPLAPP(ox)ASP(ox)GP are shown. (G) Hydroxylation levels of proline residues 67 and 70 of BIM-EL peptide (monohydroxylation and double hydroxylation are indicated as mono-HyP and 2HyP, respectively) following hydroxylation with EglN3 WT (wt) or catalytic mutant (mut.) generated via IVT. The detected intensity of each hydroxylated peptide was normalized to the nonhydroxylated peptide. (H) Autoradiograms of EglN3 hydroxylation and 35S-VHL capture as shown in E using biotinylated BIM-EL peptides containing proline-to-alanine substitutions or no substitution (WT). (I) Schematic illustration of synthetic biotinylated BIM-EL peptides unmodified (WT) or hydroxylated at P-OH-67, P-OH-70, and P-OH-67/70. (J) Autoradiograms showing recovery of 35S-labeled VHL protein bound to biotinylated BIM-EL peptides with indicated hydroxyl-prolines as outlined in I. Synthetic biotinylated HIF1α peptide (residues 556 to 575) with hydroxylated proline 564 (HIF1α-P-OH) was included as a control. Unmodified peptide is indicated as BIM-EL-WT.
Next, we generated different peptides within the 116-amino acid region of BIM-EL to narrow the EglN3-binding domain and potential hydroxylation site (Fig. 3D). We observed 35S-EglN3 binding to biotinylated BIM-EL peptide corresponding to residues 65 to 89 (BIM-EL 65 to 89), but not to BIM-EL peptide corresponding to residues 55 to 79 (SI Appendix, Fig. S3D). However, despite EglN3 binding, we could not detect binding of VHL to BIM-EL 65 to 89 after incubation with EglN3 (SI Appendix, Fig. S3E). This prompted us to generate a longer peptide (40 amino acids), corresponding to BIM-EL residues 57 to 96. This longer biotinylated BIM-EL peptide captured 35S-VHL after the EglN3 hydroxylation reaction, similar to that of the biotinylated HIF1α-peptide residues 556 to 575 (SI Appendix, Fig. S3E). Next, we tested if BIM-EL hydroxylation is a specific function of EglN3 among the EglN paralogs. Incubation of the BIM-EL-peptide 57 to 96 with EglN3, but not with EglN1 or EglN2, promoted the capture of 35S-VHL (Fig. 3E), consistent with our earlier observations that regulation of BIM-EL abundance (Fig. 1F) and binding to BIM-EL (Fig. 3A) are distinguishing features of EglN3. In contrast, and as previously reported (19), the HIF1α peptide was hydroxylated in vitro by all 3 EglN family members (Fig. 3E). Hydroxylation of the BIM-EL-peptide 57 to 96 was confirmed by liquid chromatography-tandem mass spectrometry (MS/MS) analysis (Fig. 3F and SI Appendix, Fig. S3 F–I). MS confirmed that EglN3, but not the catalytic-dead mutant, hydroxylated prolines 67 and 70 (Fig. 3F and SI Appendix, Fig. S3F). We detected monohydroxylated peptide on either proline 67 (SI Appendix, Fig. S3G) or proline 70 (SI Appendix, Fig. S3H), as well as dihydroxylated peptides on prolines 67 and 70 (Fig. 3F). In addition to proline hydroxylation, we detected oxidation of the biotin residue, which is a common artifact, in hydroxylation reactions containing WT EglN3 or catalytic-dead EglN3 (20). The detected intensity of each proline hydroxylated peptide was quantified and normalized to the nonhydroxylated peptide (Fig. 3G). Peptides monohydroxylated on proline 67 or proline 70 coeluted in the chromatography, which prevented us from quantifying the hydroxylation of either site individually. To understand the importance of mono- or dihydroxylation of the respective proline residues for VHL binding, we synthesized BIM-EL 57 to 96 peptides with the proline-to-alanine substitutions (P60A, P63A, P66A, P67A, P70A, P67/70A, and P66/67/70A), and measured their hydroxylation by EglN3 using the 35S-VHL capture assay (Fig. 3H). The P60A and P63A proline substitutions did not alter hydroxylation relative to the WT peptide. In contrast, the P66A, P67A, and P70A substitutions significantly impaired 35S-VHL capture, and the triple substitution P66/67/70A completely abolished 35S-VHL recognition (Fig. 3H). It is possible that the mutated P67A peptide generates some substrate recognition for proline 66. Jumping to neighboring lower affinity sites has been previously observed in in vitro enzymatic assays of other posttranslational modifications (21). In a reciprocal experiment, we synthesized BIM-EL 57 to 96 peptides in which proline 67 (P-OH-67), proline 70 (P-OH-70), or both (P-OH-67/70) were hydroxylated (Fig. 3I). As expected, all 3 hydroxylated peptides could, similar to hydroxylated HIF1α peptide (556 to 575), capture 35S-VHL (Fig. 3J). In contrast, nonhydroxylated BIM-EL peptide (BIM-EL-WT) did not capture 35S-VHL.
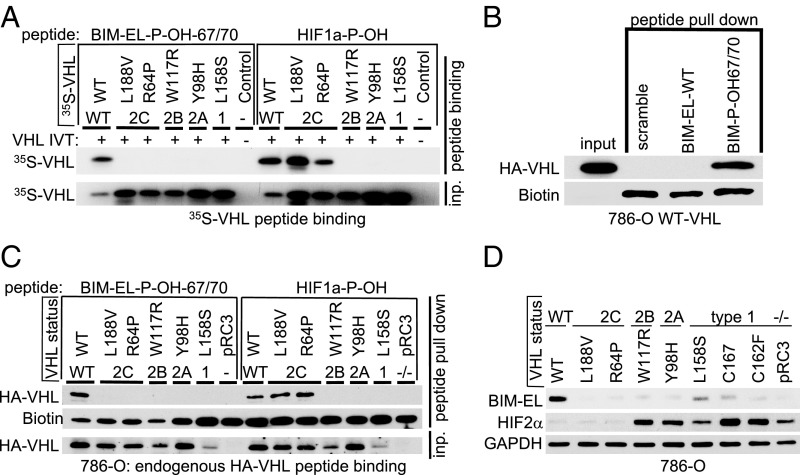
Our finding that BIM-EL expression can be restored by WT VHL, but not type 2C VHL mutants (Fig. 2E), suggested that the latter cannot recognize hydroxylated BIM-EL. Indeed, type 2C VHL mutants bound to a hydroxylated HIF1α peptide, but failed to bind hydroxylated BIM-EL peptide (P-OH-67/70) (Fig. 4A). Type 1 and type 2A/B VHL mutants also failed to recognize hydroxylated BIM-EL (Fig. 4A). We complimented these studies by expressing either WT HA-VHL or the corresponding type 1 or 2A/B/C VHL mutants in 786-O cells and performing pulldown assays with immobilized BIM-EL or HIF1α peptides. As expected, WT VHL bound to the dihydroxy-BIM-EL peptide (P-OH-67/70), but not to a nonhydroxylated BIM-EL peptide or scrambled peptide (Fig. 4B). Consistent with our findings in Fig. 4A, the type 2C VHL mutants bound to the hydroxylated HIF1α peptide, but not to the hydroxylated BIM-EL peptide, while the type 1 and type 2A/B VHL mutants were defective for both (Fig. 4C). Consistent with these findings, only WT VHL fully restored BIM-EL levels when introduced into 786-O cells (Fig. 4D). In contrast, but in keeping with prior studies, both type 2C VHL mutants suppressed HIF2α (Fig. 4D).
Fig. 4.
VHL disease mutations fail to recognize proline hydroxylated BIM-EL. (A) Autoradiograms showing recovery of 35S-labeled VHL protein (WT) or corresponding disease mutants (as indicated) bound to biotinylated BIM-EL-peptides synthesized with double hydroxyl-prolines on prolines 67 and 70 (BIM-EL-P-OH-67/70). Synthetic biotinylated HIF1α peptide (residues 556 to 575) with hydroxylated proline 564 (HIF1α-P-OH [HIF1a-P-OH]) was included as a control. An unprogrammed IVT was used as a negative control. (B) Immunoblot of HA-VHL pulldown using biotinylated BIM-EL-P-OH-67/70-peptide, unmodified BIM-EL peptide (WT), or control peptide (scramble) incubated with whole-cell lysate from 786-O HA-VHL–expressing cells. (C) Peptide pulldown as shown in B using either biotinylated BIM-EL-P-OH-67/70 peptide or biotinylated HIF1αP-OH peptide incubated with whole-cell lysates from 786-O cells expressing either HA-VHL WT or HA-VHL disease mutant. (D) Immunoblot analysis of endogenous BIM-EL and HIF2α protein expression in 786-O cells expressing either HA-VHL WT or HA-VHL disease mutant. GADPH, glyceraldehyde-3-phosphate dehydrogenase.
VHL-Mediated Regulation of BIM-EL Protein Stability Is Dependent on EglN3 Enzymatic Activity.
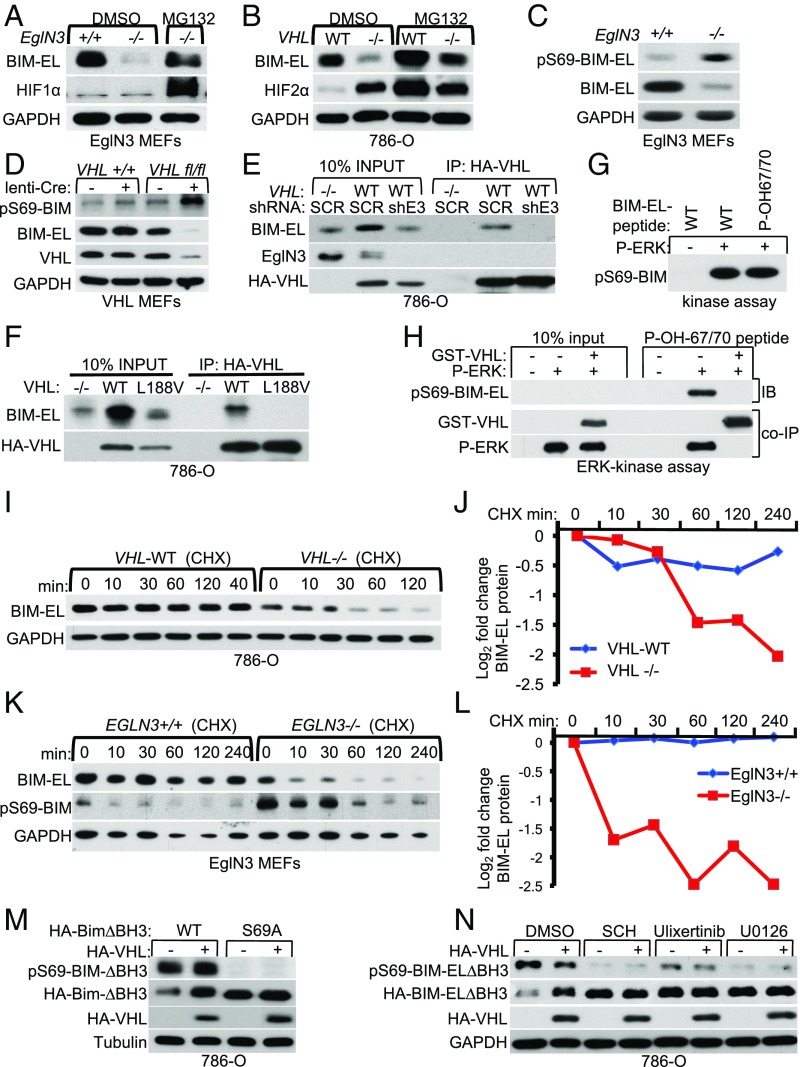
VHL stabilizes BIM-EL in RCC cells, but the mechanism is unknown (22). Since we observed that VHL binds to hydroxylated BIM-EL, we asked if this physical association stabilizes BIM-EL despite VHL’s well-established role as a component of an E3 ubiquitin ligase. The BIM-EL sites hydroxylated by EglN3 (P67 and P70) are near a critical serine residue (Ser69) that dictates BIM-EL proteasomal degradation upon extracellular signal-regulated kinase (ERK) phosphorylation (23). Thus, we hypothesized that binding of VHL to hydroxy-P67/P70 masks the S69 phosphorylation site, and thus prevents the proteasomal degradation of BIM-EL. First, we used the proteasome inhibitor MG132 to determine if the loss of BIM-EL in EglN3-knockout (KO) (−/−) primary MEFs was due to proteasomal degradation. MG132 increased BIM-EL protein abundance in EglN3-KO MEFs to the level of EglN3 WT (+/+) MEFs (Fig. 5A). An HIF1α immunoblot served as a positive control to ensure MG132 activity. Likewise, MG132 treatment of VHL−/− cells (786-O) restored BIM-EL levels (Fig. 5B), providing evidence that loss of either EglN3 or VHL accelerates BIM-EL proteasomal degradation. Importantly, phosphorylation of BIM-EL on the critical Ser69 residue required for protein turnover was markedly increased in both EglN3−/− MEFs (Fig. 5C and SI Appendix, Fig. S4A) and VHL−/− MEFs (Fig. 5D and SI Appendix, Fig. S4B), consistent with the idea that EglN3 and VHL block Ser69 phosphorylation.
Fig. 5.
VHL regulates BIM-EL protein stability depending on EglN3 nzymatic activity. (A) Immunoblot of EglN3-MEFs with the indicated genotype treated for 6 h with the proteasomal inhibitor MG132 (10 μM) or dimethyl sulfoxide (DMSO). GADPH, glyceraldehyde-3-phosphate dehydrogenase. (B) Immunoblot of 786-O VHL-null cells (−/−) or stably cells transfected to generate WT HA-VHL treated for 6 h with the proteasomal inhibitor MG132 (10 μM) or DMSO as a control. (C) Anti–phospho-BIM-EL(Ser69) immunoblot analysis of whole-cell lysates from EglN3 MEFs with the indicated genotype. (D) Anti–phospho-BIM-EL(Ser69) immunoblot of whole-cell lysates derived from VHL MEFs with the indicated genotype transduced with lentivirus encoding either Cre-recombinase or empty control lentivirus (lenti). (E) HA-VHL immunoprecipitation from 786-O cells transduced with lentivirus encoding shRNA targeting EglN3 (shE3) or scramble control (SCR). Immunoblots show coimmunoprecipitation of endogenous BIM-EL and HA-VHL expression. IP, immunoprecipitation. (F) HA-VHL immunoprecipitation from 786-O cells with stable expression of either HA-VHL WT or HA-VHL type 2C mutant (L188V). VHL-null cells (−/−) served as a negative control. Immunoblots show coimmunoprecipitation of endogenous BIM-EL and HA-VHL expression. (G) ERK activity measured by using WT BIM-EL-peptide or hydroxylated peptide BIM-EL-P-OH-67/70 as a substrate. Peptides were incubated with purified active ERK as indicated, and kinase activity was measured by phospho-BIM-EL(Ser69) immunoblot analysis. (H) ERK activity was measured as in G using hydroxylated BIM-EL-P-OH-67/70 peptide. Before the ERK reaction, the peptide was incubated with purified GST-VHL as indicated. Shown is an immunoblot (IB) of phospho-BIM-EL(Ser69) peptide indicating that the presence of VHL prevents substrate recognition and phosphorylation by ERK. (I) The 786-O VHL-null cells (−/−) or stable HA-VHL WT-expressing cells were treated with 10 μg/mL cycloheximide (CHX). At the indicated time points, whole-cell lysates were prepared for immunoblot analysis. (J) Quantification of the band intensities in I. (K) EglN3 MEFs with the indicated genotype were treated with cycloheximide at a concentration of 10 μg/mL, and whole-cell lysates were prepared for immunoblot analysis at the indicated time points. (L) Quantification of the band intensities in K. (M) Immunoblot of 786-O VHL-null (−/−) cells cotransfected with HA-VHL together with apoptotic defective HA-BIM-EL-∆BH3 (indicated as WT) or HA-BIM-EL-∆BH3-S69A mutant. (N) Immunoblot of 786-0 VHL−/− cells that were transiently transfected with plasmids encoding apoptotic defective HA-BIM-EL-∆BH3 and HA-VHL as indicated. Twenty-four hours after transfection, cells were treated for 24 h with the different ERK inhibitors SCH772984 (1 μM), Ulixertinib (1 μM), and U0126 (10 μM).
Next, we investigated if VHL interacts with endogenous BIM-EL in 786-O cells expressing HA-VHL (WT) and if this was dependent on EglN3. BIM-EL was readily detected in anti-HA immunoprecipitates of cells expressing HA-VHL unless EglN3 was down-regulated with an effective shRNA (Fig. 5E). Furthermore, BIM-EL coimmunoprecipitated only with WT VHL, but not with VHL type 2C mutant L188V (Fig. 5F). To test if VHL binding to hydroxylated BIM-EL might block Ser69 phosphorylation, we performed in vitro ERK assays. ERK activity was first measured using a WT BIM-EL peptide or a hydroxylated BIM-EL peptide (BIM-EL-P-OH-67/70) as an ERK substrate. Both peptides were similarly phosphorylated by ERK in vitro as measured by phospho-BIM-EL (Ser69) immunoblot analysis (Fig. 5G). However, preincubation with GST-VHL prevented the hydroxylated BIM-EL peptide (P-OH-67/70) from being phosphorylated by ERK (Fig. 5H).
To ask if EglN3 and VHL stabilize BIM-EL protein, we treated 786-O cells with the protein synthesis inhibitor cycloheximide. The half-life of BIM-EL was shorter in VHL−/− 786-O cells compared with WT VHL (Fig. 5 I and J). Similarly, the half-life of BIM-EL was shorter in EglN3-KO MEFs, accompanied by increased phosphorylation on Ser69, compared with WT MEFs (Fig. 5 K and L). These results demonstrate that VHL and EglN3 specifically stabilize BIM-EL in cells by preventing Ser69 phosphorylation. In accord with these data, we show that exogenous expression of the apoptosis-defective mutant of BIM-EL (HA-BIM-ELΔBH3) harboring the serine 69-to-alanine substitution (S69A) was stabilized in 786-O cells in the absence of VHL (Fig. 5M). Furthermore, exogenous BIM-EL WT (HA-BIM-ELΔBH3) protein was stabilized in VHL-null cells (786-O) that were treated with different ERK inhibitors (Fig. 5N).
Thus, we conclude that binding of VHL to hydroxylated BIM-EL prevents BIM-EL-Ser69 phosphorylation and degron recognition, and thus allows BIM-EL to escape from proteasomal degradation (SI Appendix, Fig. S4B).
Loss of EglN3 or VHL Causes Cisplatin Drug Resistance.
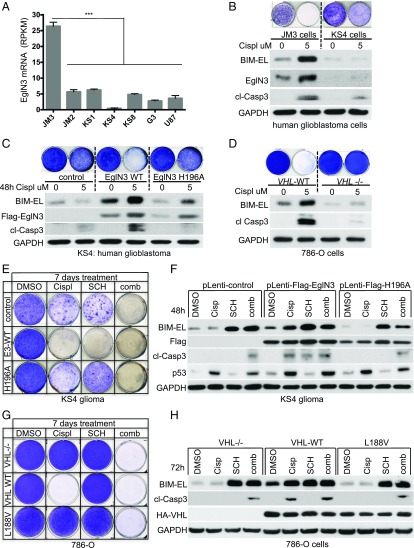
BIM protein degradation has been linked to cisplatin resistance in cancer therapy (24). The ccRCC cells were derived from renal epithelial cells, which are normally cisplatin-sensitive, and yet ccRCC and other VHL-associated neoplasms are generally resistant to chemotherapy, including cisplatin. Thus, we investigated if loss of EglN3 or VHL can likewise contribute to cisplatin resistance. Silencing of EglN3 has been recently described in high-grade glioma (17), and VHL loss is well characterized in ccRCC (1). Therefore, we explored cisplatin responsiveness in isogenic 786-O (VHL+/+ and VHL−/−) ccRCC cells, in neuroblastoma cells (SK-N-FI) that express EglN3 and VHL, and in primary (JM3, JM2, KS1, KS4, KS8, G3) and the established (U87) glioblastoma (GBM) cell lines in which EglN3 messenger RNA levels were determined by RNA sequencing (25) (Fig. 6A). We identified only 1 human primary GBM cell line (JM3) expressing EglN3. In addition, higher EglN3 expression was observed in human mature astrocytes compared with other normal human tissues (24) (SI Appendix, Fig. S5A), while EglN3 expression in the remaining primary GBM cultures was either significantly lower or undetectable (Fig. 6A). We assessed the response of JM3 (EglN3-positive) and KS4 (EglN3-negative) cells to address their response to Cisplatin. Cisplatin treatment caused a robust induction of endogenous EglN3 accompanied by the induction of BIM-EL and caspase 3 cleavage in JM3 cells, but not in KS4 cells (Fig. 6B). To understand if cisplatin resistance was caused by the lack of functional EglN3, we reintroduced EglN3 into KS4 cells using lentivirus encoding WT EglN3 or catalytic-dead mutant (H196A). Stable reintroduction of EglN3 WT, but not mutant, caused a marked increase of BIM-EL protein in KS4 cells that was further induced by cisplatin, leading to sensitization of these cells to cisplatin-induced apoptosis (Fig. 6C). Although catalytic-dead EglN3 failed to rescue BIM-EL protein expression in untreated cells, it did cause a modest induction of BIM-EL in the presence of cisplatin. The significance of this finding is not clear, but it could reflect a hydroxylase-independent function of EglN3 that is unmasked by cisplatin (Fig. 6C). Nonetheless, the cells expressing EglN3 catalytic-dead mutant remained resistant to cisplatin-induced apoptosis (Fig. 6C).
Fig. 6.
Loss of EglN3 or VHL contributes to cisplatin drug resistance. (A) Expression levels of EglN3, as represented by RPKM values, in human high-grade GBM cells (JM3, JM2, KS1, KS4, KS8, and G3) and the human U87 glioma cell line. Data are shown as mean ± SEM (***P < 0.001). mRNA, messenger RNA. RPKM, reads per kilobase million. (B) Crystal violet staining and immunoblot analysis of human GBM JM3 (EglN3-positive) and KS4 (EglN3-negative) cells treated with cisplatin (Cispl; 5 μM) for 48 h. For crystal violet staining, cells were maintained in culture for an additional 5 d. Immunoblot analysis was performed 48 h after cisplatin treatment. cl, cleaved; GADPH, glyceraldehyde-3-phosphate dehydrogenase. (C) Crystal violet staining and immunoblot analysis of KS4 cells stably transduced with lentivirus encoding Flag-EglN3 WT, catalytic dead mutant (H196A), or empty control. Stable clones were treated with cisplatin (5 μM) as indicated. Immunoblotting was performed 48 h after cisplatin treatment. For crystal violet staining, cells were maintained for an additional 5 d in culture. (D) Crystal violet staining and immunoblot analysis of 786-O cells with the indicated VHL status. Cells were treated with cisplatin once (5 μM). Immunoblotting was performed 72 h after cisplatin treatment. For crystal violet staining, cells were maintained in culture for an additional 4 d. (E) Crystal violet staining of KS4 stably transduced cells as in C and treated once with cisplatin (5 μM), Erk inhibitor SCH772984 (SCH; 5 μM), or a combination (comb) of these 2 drugs as indicated. For crystal violet staining, cells were maintained for 1 wk. DMSO, dimethyl sulfoxide. (F) Corresponding immunoblot shown in 48 h posttreatment. Cisp, cisplatin. Crystal violet staining (G) and a corresponding immunoblot (H) of stable 786-O cells treated once with cisplatin (5 μM), ERK inhibitor SCH772984 (SCH; 1 μM), or a combination of both as indicated for 72 h are shown. For crystal violet staining, cells were maintained for 7 d.
Similarly, lowering EglN3 levels in JM3 cells (SI Appendix, Fig. S5B) or SK-N-FI cells (SI Appendix, Fig. S5C) with an effective shRNA lowered BIM-EL levels and conferred cisplatin resistance (SI Appendix, Fig. S5C). Furthermore, reintroducing HA-VHL into VHL−/− 786-O cells restored the ability of cisplatin to induce BIM-EL and apoptosis in these cells (Fig. 6D).
Next, we tested if EglN3-null or VHL-null cells can be sensitized to cisplatin-induced apoptosis in combination with ERK inhibitors. BIM-EL expression was restored to that of EglN3 WT expressing KS4 glioma cells 24 h after treatment with various ERK inhibitors (SI Appendix, Fig. S5D). Despite BIM-EL induction in EglN3-null or EglN3-H196A mutant cells, ERK inhibitor alone caused significant apoptosis only in EglN3 WT-expressing cells (Fig. 6 E and F). Combined treatment of cisplatin and ERK inhibitor caused apoptosis, irrespective of EglN3 status (Fig. 6 E and F), without showing any further increase of BIM-EL protein compared with ERK inhibitor alone. Similarly, treatment of VHL-null cells with ERK inhibitor alone did not cause apoptosis, despite BIM-EL induction (Fig. 6 G and H), but did so when combined with cisplatin (Fig. 6G and SI Appendix, Fig. S5E). Thus, highly efficient killing by BIM-EL induced by ERK inhibitors in cells lacking EglN3 or VHL requires one or more collateral signals that can be provided by Cisplatin.
Discussion
The canonical function of VHL is to ubiquitinate hydroxylated HIFα for proteasomal degradation. HIFα deregulation can cause PCC and PGL, as evidenced by gain-of-function HIF2α and loss-of-function EglN1 mutations in rare familial cases. However, the mechanisms underlying the complex genotype/phenotype correlations in VHL disease cannot be explained by HIFα regulation. VHL type 2C mutations are not related to HIFα activation and preserve their ability to down-regulate HIFα (7, 8). Here, we identified an oxygen-sensitive function of pVHL that is abolished by VHL type 2C mutations. We identified BIM-EL as a direct EglN3 hydroxylation substrate and that hydroxylated BIM-EL is bound and stabilized by binding to VHL, but not by VHL type 2C mutants. BIM-EL hydroxylation on P67 and P70 is in near proximity to the critical serine 69, which dictates BIM-EL proteasomal degradation upon ERK phosphorylation (23). We observed that pVHL binds to hydroxylated BIM-EL at P67/P70 and prevents ERK phosphorylation on serine 69, leading to BIM-EL protein stabilization. Indeed, low BIM-EL expression in the PCC/PGL tumors carrying VHL mutations confirms that BIM-EL protein regulation is VHL-dependent.
We previously reported that EglN3 hydroxylase activity is linked to PCC pathogenesis through regulation of apoptosis during sympathoadrenal development, which is independent of HIFα hydroxylation (9). Here, we identified that VHL loss causes resistance to EglN3-induced apoptosis and that this can be restored by WT VHL, but not by VHL type 2C mutants. Furthermore, we found that EglN3-induced apoptosis requires hydroxylation of BIM-EL, which, in turn, allows for VHL recognition of the protein, thus leading to BIM-EL protein stabilization. Bim deletion has been reported to cause protection against developmental and induced neuronal apoptosis in both central and peripheral neuron populations (14). Furthermore, inhibition of NGF deprivation-induced death by low oxygen levels during sympathetic neuronal development involves suppression of BIM-EL (15). By identifying BIM-EL as a direct EglN3 hydroxylation substrate, we provide the missing mechanistic link between VHL type 2C PCC mutations, EglN3 hydroxylase activity, and escape from apoptosis during sympathoadrenal development.
Thus, our work provides insights as to why VHL type 2C mutations contribute to PCC/PGL pathogenesis. We identified a biochemical activity that is lost by type 2C pVHL mutants, leading to a loss of BIM-EL. Moreover, this biochemical activity affects neuronal apoptosis after NGF withdrawal, as do many of the other PCC/PGL-associated genes. Therefore, the law of parsimony suggests that dysregulation of BIM-EL contributes to the pathogenesis of PGLs caused by VHL mutations (including type 2C mutations). Nonetheless, type 2C mutants have also been shown to be defective with respect to other biochemical properties, including fibronectin and collagen binding (2–4). It is possible that these other biochemical properties are relevant to PGL as well.
Why a germline type 1 VHL mutation (deleterious mutants) seldomly causes PCC/PGL despite BIM-EL reduction has not been addressed in this work. However, we predict that complete loss of VHL during sympathoadrenal development is not tolerated, and thus does not result in PCC/PGL outcome. This argument is in line with observations in VHL conditional KO studies in mice (Tyrosine Hydroxylase [TH]-Cre), which show that loss of VHL during development results in the disappearance of catecholaminergic cells (26), in contrast to conditional TH-EglN1KO mice, which display hyperplasia in the carotid body (27). This suggests that unknown VHL functions are required for the formation of the sympathoadrenal cell lineage, and that complete loss of VHL (type 1) therefore might not be permissive in PCC/PGL pathogenesis. In contrast, in missense mutated VHL cells, reduction of BIM-EL protein might not impair the formation of the sympathoadrenal lineage. Instead it provides a mechanism to escape developmental apoptosis independent of HIFα regulation contributing to PCC/PGL. Our observation that all VHL-mutated PCC/PGL tumors show reduced BIM-EL expression in contrast to other non–VHL-mutated PCC/PGL tumors supports this. However, that would not exclude other VHL substrates contributing to VHL-related PCC.
Evading apoptosis is a hallmark of cancer. Every VHL mutation we have examined to date, whether type 1 or type 2, leads to loss of BIM-EL. It is therefore likely that loss of BIM-EL contributes to the pathogenesis of VHL-related neoplasms other than PGLs, including ccRCC and HB, as well as their characteristic insensitivity to chemotherapy. In this regard, cisplatin damages renal epithelial cells, leading to renal dysfunction, and yet is inactive against ccRCCs. We showed that loss of VHL, and subsequent down-regulation of BIM-EL, renders ccRCC cells insensitive to cisplatin. Thus, we further investigated the role of EglN3 during the cisplatin response. Cisplatin treatment induced EglN3 in VHL WT-expressing cells, accompanied by BIM-EL induction and apoptosis, whereas EglN3-silenced neuroblastoma cells or VHL-null ccRCC cells were resistant. Reintroduction of WT VHL, but not type 2C VHL mutants in ccRCC cells, restored sensitivity. We also studied resistance to cisplatin in high-grade glioma cells, since EglN3 has been reported to be silenced during glioma progression (17). We observed that EglN3-silenced GBM cells were cisplatin-resistant, but sensitized upon reintroduction of WT EglN3. In line with our observations, other studies also reported that EglN3 and BIM-EL are required for successful proapoptotic activity of various DNA damage-inducing chemotherapeutical agents (28, 29). By unraveling the precise molecular mechanism of BIM-EL stabilization by VHL and EglN3, we provide further insights into the chemoresistance that is typical of tumors lacking VHL or oxygen. Thus, restoration of BIM-EL protein by combination therapy using ERK inhibitors together with Cisplatin might provide a promising approach for cancer therapy (SI Appendix, Fig. S5F).
Methods
Cell Culture.
Human neuroblastoma cell line SK-N-FI and the rat PCC cell line PC12 were maintained as previously described (10, 30). Human GBM cell line U87 was maintained in Dulbecco’s modified Eagle’s medium (DMEM) (1 g/L glucose) containing 10% fetal bovine serum (FBS). MEFs and a human RCC cell line (786-O) were cultured in DMEM (4.5 g/L glucose) containing 10% FBS in 5% CO2 at 37 °C. The 786-O, SK-N-FI, and PC12 cell lines were purchased from the American Type Culture Collection. VHLfl/fl immortalized MEFs have been previously described (31), containing a blasticidin resistance cassette and directing the expression of large T antigen K1 (K1 T antigen inactivates p53, but not retinoblastoma protein, pRB). Human primary GBM cell cultures were established as previously described (25).
EglN3 KO Mice.
Generation of the EglN3 mouse strain is described by Schlisio et al. (10). P1 superior cervical ganglia dissection of EglN3 pups is described by Fell et al. (32).
Human Tissue Specimen.
PCC and abdominal PGL samples were collected from patients operated on and diagnosed at the Karolinska University Hospital (Stockholm, Sweden), and previously characterized for mutations in 14 proposed PCC/PGL susceptibility genes (12) (SI Appendix, Fig. S2). Four histologically confirmed normal adrenal glands from deidentified patients were included as controls. VHL mutations in samples 21, 25, 96, and 108 have been previously described (12):
Sample 21: Diagnosed with cerebellar HB and PCC the same year. This was a syndromic VHL patient with a germline VHL mutation: c.217C > T p.Gln73X (frameshift mutation resulting in truncated VHL, type 1). Please note that this sample has a truncating mutation. The risk of developing PCC in VHL type 1 patients is not 0, but lower than for type 2 patients.
Sample 25: Diagnosed with PCC at the age of 13 y with normal epinephrine and norepinephrine levels and no evidence of metastasis or relapse and no additional tumors. This was a syndromic VHL patient with a germline VHL mutation: c.193T > G p.Ser65Ala.
Sample 96: PCC, with trichoepithelioma 8 y earlier (benign skin lesion). This was an apparently sporadic patient (no VHL syndrome) with a somatic VHL mutation. Mutations identified in sample 96 are as follows: EglN1 c.799G > A p.Glu267Lys + SDHA c.223C > T p.Arg75X + VHL c.386T > C p.Leu129Pro.
Sample 108: PGL in 1998, basal cell carcinoma of the skin in 2013, and follicular lymphoma in 2018. This was an apparently sporadic patient (no VHL syndrome) with a somatic VHL mutation: VHL c.593T > G p.Leu198Arg.
Ethical Considerations.
Human samples (normal tissues, PCCs, and PGLs) are covered by an existing ethical approval (Drn 01-136, Karolinska Institutet [KI] forskningsetikkommitté Nord). Human GBM tissue specimens were collected via surgical resection under ethical permit KI 02-254 (22) or 2013/576-31, issued by the Ethical Review Board at the KI, in accordance with the Declaration of Helsinki. All samples were obtained following informed patient consent. All animal experiments were performed in accordance with Swedish animal welfare laws authorized by the Stockholm Animal Ethics Committee (Dnr 7694/17).
Peptide Pulldown Assay.
Streptavidin beads were incubated with biotinylated peptides (synthesized by peptides&elephants GmbH) and rotated for 1 h at room temperature. The 35S-VHL produced by IVT was captured as previously described (9). For pulldown assays using VHL-expressing 786-O cells, conjugated streptavidin beads with biotinylated peptides were incubated with 786-O cell lysate overnight at 4 °C with rotation. Samples were then washed 4 times with immunoprecipitation wash buffer (0.5% Nonidet P-40, 150 mM NaCl, 10 mM Tris⋅HCl) and eluted with 30 μL of Laemmli buffer, boiled for 5 min, and centrifuged at 8,000 × g for 30 s. Eluted supernatant was subjected to immunoblot analysis.
Peptide Hydroxylation and 35S-VHL–Binding Assay.
FLAG-EglN1/2/3 and VHL were produced by in vitro transcription/translation reactions (IVT) using TnT T7 Quick Master Mix. BIM-EL (1 μg) and HIF1α (1 μg) biotinylated peptides were conjugated with streptavidin agarose beads in 1 mL of phosphate-buffered saline (PBS) at room temperature with rotation for 1 h. The bead pellet was washed twice with PBS and once with hydroxylation buffer (40 mM Hepes [pH 7.4], 80 mM KCl). The pellet was resuspended with 300 μL of hydroxylation reaction buffer supplemented with 100 μM FeCl2, 2 mM ascorbate, and 5 mM 2-oxolgutarate. Fifteen microliters of IVT-synthesized EglN was added to start the hydroxylation reaction. The hydroxylation reaction was processed for 2 h at room temperature with rotation. Five hundred microliters of EBC lysis buffer (50 mM Tris [pH 8.0], 120 mM NaCl, 0.5% Nonidet P-40) was added to the reaction buffer to stop the hydroxylation; 15 μL of IVT-synthesized 35S radioactive-labeled VHL was subsequently added, and this mixture was incubated overnight. Samples were centrifuged at 8,000 × g for 30 s and washed 5 times with wash buffer (0.5% Nonidet P-40, 150 mM NaCl, 10 mM Tris⋅HCl). Bound protein complexes were eluted with 30 μL of Laemmli buffer, boiled for 5 min, and centrifuged at 8,000 × g for 30 s. Eluted supernatant was analyzed by immunoblotting.
Full-Length BIM-EL Hydroxylation and 35S-VHL Capture.
EglN3 WT, EglN3 mutant, 35S-VHL, and BIM-EL were produced by in vitro transcription/translation (IVT) reactions using TnT T7 Quick Master Mix. In vitro-translated BIM-EL was added to 300 μL of hydroxylation reaction buffer supplemented with 100 μM FeCl2, 2 mM ascorbate, and 5 mM 2-oxoglutarate. Fifteen microliters of unprogrammed reticulocyte lysate, IVT-synthesized WT EglN3, or EglN3 mutant was added to start the hydroxylation reaction. The hydroxylation reaction was processed for 2 h at room temperature with rotation. Five hundred microliters of EBC buffer was added to the reaction buffer to stop the hydroxylation reaction. Fifteen microliters of IVT-synthesized 35S-VHL was added, and this mixture was incubated for 2 h at room temperature with rotation. BIM-EL was immunoprecipitated with anti–BIM-EL antibody overnight at 4 °C with rotation and captured with protein G beads. The bead pellet was then washed 5 times with immunoprecipitation wash buffer (0.5% Nonidet P-40, 150 mM NaCl, 10 mM Tris⋅HCl). Bound protein complexes were eluted with 30 μL of Laemmli buffer, boiled for 5 min, and centrifuged at 8,000 × g for 30 s. Eluted supernatant was analyzed by immunoblot or 35S autoradiography.
Peptide Hydroxylation Assay and MS Analysis.
The hydroxylation assay with BIM-EL peptide was performed as described above. After hydroxylation, BIM-EL peptide-conjugated beads were washed 1 time with hydroxylation buffer and 3 times with immunoprecipitation buffer without detergent. Samples were processed as described by Rodriguez et al. (20). Biotinylated peptides were digested with chymotrypsin and directly analyzed by MS. Peptides were analyzed on a Q-Exactive mass spectrometer connected to an Ultimate Ultra3000 chromatography system incorporating an autosampler (both from Thermo Fisher Scientific). Proteolytic or synthesized peptides for each sample (5 μL) were applied to a home-made column (250-mm length, 75-μm inside diameter; packed with 1.8 μm of UChrom C18) and separated using a 40-min reverse-phase acetonitrile gradient (5 to 70% [vol/vol] acetonitrile) with a 250-nL⋅min−1 flow rate. The mass spectrometer was operated in positive ion mode with a capillary temperature of 220 °C and a 2,000-V potential applied to the column. Variable modifications were N-terminal biotin, N-terminal biotin hydroxylation, and proline hydroxylation.
Expression Plasmids, shRNA, and siRNAs.
pcDNA3 Flag-EglN3, Flag-H196A-mutant, and pcDNA3-VHL (including VHL-missense mutations) have been described previously (9). pcDNA3 HA-BIM-EL (mouse) and pcDNA3 HA-BIM-EL apoptotic mutant (rat BIM-EL∆BH3) were kind gifts from Stanley J. Korsmeyer, Dana-Farber Cancer Institute. The rat BIM-EL∆BH3 has a mutation in the BH3 domain, L150RRIGDEFN158, mutated to A150RRIAAEFN158. Amino acid numbering refers to the rat BIM-EL sequence. The rat BIM-EL∆BH3_S65A mutant was made using a site-directed mutagenesis kit (Agilent Technologies). The rat BIM-EL serine 65 residue corresponds to the human BIM-EL serine 69 residue. GST-BIM-EL and GST-BIM-L were kind gifts from the S. Cook laboratory, Babraham Institute, Cambridge, UK. Lentivirus encoding FLAG-EglN3 and FLAG-EglN3-H196A was generated in 293FT cells as previously described (10). siRNAs targeting EglN1, EglN2, or EglN3 have been previously described and validated (33). Lentivirus encoding shRNAs targeting BIM and EglN3 were generated using the pLKO.1 plasmid using the following sequences: human BIM-EL#54 5′-CCGGAGCCGAAGACCACCCACGAATCTCGAGATTCGTGGGTGGTCTTCGGCTTTTTT-3′; human BIM-EL#77 5′-CCGGGACCACCCAC GAATGGTTATCCTCGAGGATAACCATTCGTGGGTGGTCTTTTTG-3′; human EglN3 5′-CCGGGTTCTTCTGGTCAGATCGTAGCTCGAGCTACGATCTGAC CAGAAGAACTTTTTG-3′.
Phospho-ERK Activity Assay.
One microgram of biotinylated hydroxylated BIM-EL-peptides was conjugated with streptavidin agarose beads in 1 mL of PBS at room temperature with rotation for 1 h. The bead pellet was washed 3 times with PBS and resuspended with 500 μL of PBS supplemented with 1 μg of purified GST-VHL protein for 2 h at room temperature. Samples were centrifuged at 8,000 × g for 30 s, washed 3 times with PBS, and resuspended with 40 μL of assay dilution buffer I [ADBI; 20 mM 3-(N-morpholino)propanesulfonic acid (pH 7.2), 25 mM β-glycerol phosphate, 5 mM ethylene glycol bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid , 1 mM sodium orthovanadate, 1 mM dithiothreitol] supplemented with 10 μL of Mg2+/adenosine 5′-triphosphate (ATP) mixture (75 mM magnesium chloride, 500 μM ATP in ADBI) and 200 ng of purified phospho-ERK (14-173; Merck Millipore). The phosphorylation assay (MAP kinase/Erk assay, nonradioactive, 17-191; Merck Millipore) was processed for 30 min at room temperature with rotation at 30 °C. Samples were centrifuged at 8,000 × g for 30 s, washed 3 times with PBS, eluted with 30 μL of Laemmli buffer, boiled for 5 min, and centrifuged at 8,000 × g for 30 s. Eluted supernatant was analyzed by immunoblotting using phospho-specific BIM antibody (p-Ser69 [D7E11]).
BIM-EL Half-Life.
The 786-O cells were cultured in 6-well plates in 2 mL of DMEM to reach 50% confluency. Cells were treated with 10 μM cycloheximide at the indicated times, harvested, and lysed in 150 μL of EBC lysis buffer. Lysates were analyzed by immunoblotting with rabbit anti–BIM-EL antibody. ImageJ densitometry analysis was performed to quantify BIM-EL expression and determine protein half-life.
Statistics.
Statistical analyses were performed when appropriate, and P values are indicated by asterisks in the figure legends. Significant differences between means for multiple comparison analyses were conducted using ANOVA, followed by Bonferroni correction for post hoc analysis.
Supplementary Material
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. Q.Z. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1900748116/-/DCSupplemental.
References
- 1.Kaelin W. G., Jr, The von Hippel-Lindau tumour suppressor protein: O2 sensing and cancer. Nat. Rev. Cancer 8, 865–873 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Kurban G., et al. , Collagen matrix assembly is driven by the interaction of von Hippel-Lindau tumor suppressor protein with hydroxylated collagen IV alpha 2. Oncogene 27, 1004–1012 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Grosfeld A., et al. , Interaction of hydroxylated collagen IV with the von hippel-lindau tumor suppressor. J. Biol. Chem. 282, 13264–13269 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Ohh M., et al. , The von Hippel-Lindau tumor suppressor protein is required for proper assembly of an extracellular fibronectin matrix. Mol. Cell 1, 959–968 (1998). [DOI] [PubMed] [Google Scholar]
- 5.Okuda H., et al. , Direct interaction of the beta-domain of VHL tumor suppressor protein with the regulatory domain of atypical PKC isotypes. Biochem. Biophys. Res. Commun. 263, 491–497 (1999). [DOI] [PubMed] [Google Scholar]
- 6.Guo J., et al. , pVHL suppresses kinase activity of Akt in a proline-hydroxylation-dependent manner. Science 353, 929–932 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoffman M. A., et al. , von Hippel-Lindau protein mutants linked to type 2C VHL disease preserve the ability to downregulate HIF. Hum. Mol. Genet. 10, 1019–1027 (2001). [DOI] [PubMed] [Google Scholar]
- 8.Clifford S. C., et al. , Contrasting effects on HIF-1alpha regulation by disease-causing pVHL mutations correlate with patterns of tumourigenesis in von Hippel-Lindau disease. Hum. Mol. Genet. 10, 1029–1038 (2001). [DOI] [PubMed] [Google Scholar]
- 9.Lee S., et al. , Neuronal apoptosis linked to EglN3 prolyl hydroxylase and familial pheochromocytoma genes: Developmental culling and cancer. Cancer Cell 8, 155–167 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Schlisio S., et al. , The kinesin KIF1Bbeta acts downstream from EglN3 to induce apoptosis and is a potential 1p36 tumor suppressor. Genes Dev. 22, 884–893 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yeh I. T., et al. , A germline mutation of the KIF1B β gene on 1p36 in a family with neural and nonneural tumors. Hum. Genet. 124, 279–285 (2008). [DOI] [PubMed] [Google Scholar]
- 12.Welander J., et al. , Rare germline mutations identified by targeted next-generation sequencing of susceptibility genes in pheochromocytoma and paraganglioma. J. Clin. Endocrinol. Metab. 99, E1352–E1360 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bishop T., et al. , Abnormal sympathoadrenal development and systemic hypotension in PHD3-/- mice. Mol. Cell. Biol. 28, 3386–3400 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Putcha G. V., et al. , Induction of BIM, a proapoptotic BH3-only BCL-2 family member, is critical for neuronal apoptosis. Neuron 29, 615–628 (2001). [DOI] [PubMed] [Google Scholar]
- 15.Xie L., Johnson R. S., Freeman R. S., Inhibition of NGF deprivation-induced death by low oxygen involves suppression of BIMEL and activation of HIF-1. J. Cell Biol. 168, 911–920 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berra E., et al. , HIF prolyl-hydroxylase 2 is the key oxygen sensor setting low steady-state levels of HIF-1alpha in normoxia. EMBO J. 22, 4082–4090 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henze A.-T., et al. , Loss of PHD3 allows tumours to overcome hypoxic growth inhibition and sustain proliferation through EGFR. Nat. Commun. 5, 5582 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Högel H., Rantanen K., Jokilehto T., Grenman R., Jaakkola P. M., Prolyl hydroxylase PHD3 enhances the hypoxic survival and G1 to S transition of carcinoma cells. PLoS One 6, e27112 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaelin W. G. Jr, Ratcliffe P. J., Oxygen sensing by metazoans: The central role of the HIF hydroxylase pathway. Mol. Cell 30, 393–402 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Rodriguez J., et al. , Substrate-trapped interactors of PHD3 and FIH cluster in distinct signaling pathways. Cell Rep. 14, 2745–2760 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Treier M., Staszewski L. M., Bohmann D., Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell 78, 787–798 (1994). [DOI] [PubMed] [Google Scholar]
- 22.Guo Y., Schoell M. C., Freeman R. S., The von Hippel-Lindau protein sensitizes renal carcinoma cells to apoptotic stimuli through stabilization of BIM(EL). Oncogene 28, 1864–1874 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luciano F., et al. , Phosphorylation of Bim-EL by Erk1/2 on serine 69 promotes its degradation via the proteasome pathway and regulates its proapoptotic function. Oncogene 22, 6785–6793 (2003). [DOI] [PubMed] [Google Scholar]
- 24.Wang J., Zhou J. Y., Wu G. S., Bim protein degradation contributes to cisplatin resistance. J. Biol. Chem. 286, 22384–22392 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kurtsdotter I., et al. , SOX5/6/21 prevent oncogene-driven transformation of brain stem cells. Cancer Res. 77, 4985–4997 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Macías D., Fernández-Agüera M. C., Bonilla-Henao V., López-Barneo J., Deletion of the von Hippel-Lindau gene causes sympathoadrenal cell death and impairs chemoreceptor-mediated adaptation to hypoxia. EMBO Mol. Med. 6, 1577–1592 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fielding J. W., et al. , PHD2 inactivation in Type I cells drives HIF-2α-dependent multilineage hyperplasia and the formation of paraganglioma-like carotid bodies. J. Physiol. 10.1113/JP275996 (19 June 2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Delbridge A. R., et al. , RAG-induced DNA lesions activate proapoptotic BIM to suppress lymphomagenesis in p53-deficient mice. J. Exp. Med. 213, 2039–2048 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xie L., et al. , PHD3-dependent hydroxylation of HCLK2 promotes the DNA damage response. J. Clin. Invest. 122, 2827–2836 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li S., et al. , The 1p36 tumor suppressor KIF 1Bβ is required for calcineurin activation, controlling mitochondrial fission and apoptosis. Dev. Cell 36, 164–178 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Young A. P., et al. , VHL loss actuates a HIF-independent senescence programme mediated by Rb and p400. Nat. Cell Biol. 10, 361–369 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Fell S. M., et al. , Neuroblast differentiation during development and in neuroblastoma requires KIF1Bβ-mediated transport of TRKA. Genes Dev. 31, 1036–1053 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Q., et al. , Control of cyclin D1 and breast tumorigenesis by the EglN2 prolyl hydroxylase. Cancer Cell 16, 413–424 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.