Significance
Hepatitis B viruses (HBVs) have existed for millions of years. We describe divergent HBV species in shrews, which are ancient insectivorous mammals. The shrew viruses corroborated ancient origins of mammalian HBV and showed mutations in genomic domains required for production of the hepatitis B e antigen, crucial for vertical transmission and chronicity of HBV in humans. Shrew HBV infection patterns showed important similarities with human HBV, and shrews might be susceptible to human HBV. In contrast, shrew HBVs could neither infect human cells nor use the canonical HBV receptor molecule for cellular entry. The discovery and characterization of shrew HBVs allow conceptualizing urgently needed new animal models relying on engineered shrew and human HBV homologs to elucidate HBV pathogenesis.
Keywords: hepatitis B virus, viral evolution, zoonosis, shrew, E antigen
Abstract
Shrews, insectivorous small mammals, pertain to an ancient mammalian order. We screened 693 European and African shrews for hepatitis B virus (HBV) homologs to elucidate the enigmatic genealogy of HBV. Shrews host HBVs at low prevalence (2.5%) across a broad geographic and host range. The phylogenetically divergent shrew HBVs comprise separate species termed crowned shrew HBV (CSHBV) and musk shrew HBV (MSHBV), each containing distinct genotypes. Recombination events across host orders, evolutionary reconstructions, and antigenic divergence of shrew HBVs corroborated ancient origins of mammalian HBVs dating back about 80 million years. Resurrected CSHBV replicated in human hepatoma cells, but human- and tupaia-derived primary hepatocytes were resistant to hepatitis D viruses pseudotyped with CSHBV surface proteins. Functional characterization of the shrew sodium taurocholate cotransporting polypeptide (Ntcp), CSHBV/MSHBV surface peptide binding patterns, and infection experiments revealed lack of Ntcp-mediated entry of shrew HBV. Contrastingly, HBV entry was enabled by the shrew Ntcp. Shrew HBVs universally showed mutations in their genomic preCore domains impeding hepatitis B e antigen (HBeAg) production and resembling those observed in HBeAg-negative human HBV. Deep sequencing and in situ hybridization suggest that HBeAg-negative shrew HBVs cause intense hepatotropic monoinfections and low within-host genomic heterogeneity. Geographical clustering and low MSHBV/CSHBV-specific seroprevalence suggest focal transmission and high virulence of shrew HBVs. HBeAg negativity is thus an ancient HBV infection pattern, whereas Ntcp usage for entry is not evolutionarily conserved. Shrew infection models relying on CSHBV/MSHBV revertants and human HBV will allow comparative assessments of HBeAg-mediated HBV pathogenesis, entry, and species barriers.
The hepatitis B virus (HBV, genus Orthohepadnavirus) is a ubiquitous pathogen that causes 887,000 deaths annually, predominantly due to cirrhosis and hepatocellular carcinoma after chronic hepatitis B (CHB) (1). Distantly related hepadnaviruses were identified recently in animals other than humans and apes (1). The newly discovered animal viruses revealed that prototypic properties of HBV such as envelopment (2) and presence of an X gene (3) emerged de novo during orthohepadnavirus evolution.
Hepadnaviruses are ancient pathogens, likely infecting vertebrates for over 200 million years (3). Placental mammals evolved ∼99 million years ago (mya) and form 2 major clades termed Laurasiatheria and Euarchontoglires (4). The known laurasiatherian HBV hosts belong to several species within the orders Chiroptera (bats) and to one species each within the orders Carnivora (cat) and Artiodactyla (duiker). HBV hosts within the Euarchontoglires include humans and several nonhuman primate species within the order Primates and 3 closely related species within the order Rodentia (rodents) (1, 5). Bats may play an important role for Orthohepadnavirus evolution because they host a relatively higher genetic diversity of orthohepadnaviruses than other mammals (1, 6). However, bats are not the oldest laurasiatherian order. Predating the rise of bats, the order Eulipotyphla emerged ∼80 mya (4). This order comprises small insectivorous animals such as moles, hedgehogs, and shrews. After decades at the periphery of the life sciences, diverse homologs of major human viruses have been found in eulipotyphlan hosts, suggesting they may play an underestimated role in the evolution of mammalian viruses (7 –10). Very recently, a divergent hepadnavirus sequence was detected in Chinese shrews (11). Shrews represent about 90% of all eulipotyphlan species (12). Scarce virological data on shrews are partly due to difficulties in sampling these solitary animals that rapidly decay in traps due to their high metabolic rate (13).
Because hepadnaviruses may coevolve with their hosts (14) and because infection patterns in animal reservoirs can inform pathogenesis in humans (15), HBV homologs from shrews may provide important insights into the genealogy of HBV. Here, we conducted a systematic study into hepadnavirus genetic diversity and infection patterns in European and African shrews.
Results
Shrews Carry HBVs across a Broad Geographic and Host Range.
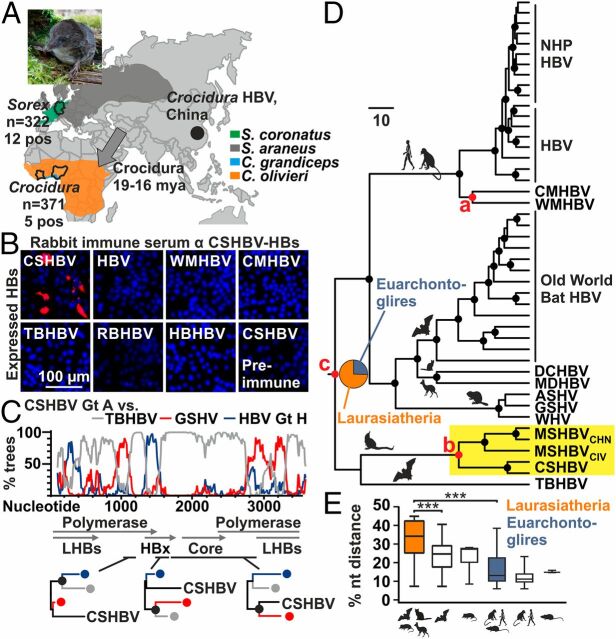
Shrews originated in Eurasia from where they populated Africa (Fig. 1A ) (16). To investigate shrews for HBV along their geographic dispersal history, we sampled 693 animals pertaining to the genera Sorex and Crocidura from Europe and Western Africa during 2004–2016 ( SI Appendix, Table S1). Liver specimens were tested for HBV DNA using a broadly reactive and highly sensitive nested PCR assay (17). In total, 17 animals were PCR-positive, including 1 common shrew (Sorex araneus) and 12 crowned shrews (Sorex coronatus) sampled in Germany during 2008–2009 and 4 African giant shrews (Crocidura olivieri) sampled in Ivory Coast in 2004 and 1 large-headed shrew (Crocidura grandiceps) sampled in Sierra Leone in 2014. The detection rate in European shrews (3.7%, 95% confidence interval [CI], 2.1–6.5) was not significantly higher than in African shrews (1.4%, 95% CI, 0.5–3.2; Fisher exact, P = 0.05). While the overall detection rate was low at 2.5% (95% CI, 1.5–3.9), it did not differ significantly from bat HBV detection rates in natural habitats (2.2%; Fisher exact, P = 1.0) (18) and may thus be representative of diverse HBV animal reservoirs. The discovery of shrew HBVs expands the mammalian orders known to be infected by orthohepadnaviruses to six ( SI Appendix, Fig. S1A ).
Fig. 1.
Evolutionary and antigenic characteristics of shrew HBVs. (A) HBV-positive shrew species, sample. Gray arrows, Soricidae dispersion routes (16). Sorex picture, Ulrike Rosenfeld. (B) Antigenic divergence of CSHBV. (C) Bootscan analysis with maximum likelihood (ML) phylogenies of highlighted genomic regions. Filled circles in A, B, and D, bootstrap support >90% (black) and >80% (gray). (D) Bayesian ancestral state reconstruction and hypothesis testing (42). Pie charts, posterior probabilities for ancestral traits. Filled circles at nodes, posterior probability >0.95. Red nodes, priors used for hypothesis testing (Scale bars, genetic distance). (E) Pairwise HBV nucleotide sequence distance comparisons per host order. Boxplots, interquartile range and median; whiskers, minimum to maximum. Gt, genotype; AA, amino acid; WMHBV, woolly monkey HBV; RBHBV, roundleaf bat HBV; HBHBV, horseshoe bat HBV; LHBs, large HBV surface protein; HBx, HBV X protein; NHP, nonhuman primates; DCHBV, domestic cat HBV; MDHBV, Maxwell’s duiker HBV; ASHV, arctic squirrel hepatitis virus; GSHV, ground squirrel hepatitis virus.
Shrew HBVs Form Separate Species and Genotypes.
To determine the genomic structure of shrew HBVs, we characterized 7 complete genome sequences by sets of nested PCR assays as described previously (17). Complete genomes were recovered from 5 HBVs hosted by S. coronatus, 1 HBV hosted by S. araneus, and 4 HBVs hosted by C. olivieri. An additional shrew HBV hosted by C. grandiceps was partially characterized in a genomic region encompassing 1,433 nucleotides within the surface/polymerase and X/preC/core open reading frames (ORFs) (GenBank accession nos. MK345460–MK345467, MK345470, MK345474, and MK345475; SI Appendix, Fig. S1B ). The genomes encompassed 3,089 nucleotides for the crowned shrew HBV from Germany (CSHBV) and 3,172 nucleotides for the musk shrew HBV from Ivory Coast (MSHBVCIV). All shrew viruses comprised 4 predicted ORFs encoding the polymerase, surface, X, and core proteins, comparable to other HBVs ( SI Appendix, Table S2). Averaged over the full genome, CSHBV, MSHBVCIV, and the recently described Chinese musk shrew HBV variant (MSHBVCHN) showed 22.9–28.9% mutual nucleotide sequence distances and 39.0–43.7% compared with other orthohepadnaviruses ( SI Appendix, Table S3). MSHBVCHN, MSHBVCIV, and CSHBV thus formed 3 Orthohepadnavirus species (19). The CSHBV comprised 2 divergent lineages that differed from one another by 11.6–12.0% genomic sequence distance. Because primate HBV genotypes differ by at least 8% over the full genomes, these 2 lineages represent distinct genotypes. CSHBV genotype A was recovered only from S. coronatus, whereas genotype B was recovered from both S. araneus and S. coronatus, likely representing spillover infections into S. araneus in sympatric animals, resembling those observed in ape-associated HBV infecting gorillas and chimpanzees (20) ( SI Appendix, Fig. S1B ). The partially characterized MSHBVCIV from Sierra Leone differed from the fully characterized MSHBVCIV from Ivory Coast by 8.4% within the characterized genomic region. The 2 mutually most closely related HBV genotypes from orangutans and gibbons differ by 6.0% in the homologous genomic region. The 2 MSHBVCIV lineages thus likely also correspond to distinct genotypes. Similarly, MSHBVCHN formed 2 genotypes with 8.5–8.7% genomic distance (11). In sum, shrews host at least 3 HBV species and 6 genotypes, equaling primates in HBV species richness.
Shrew HBVs Are Phylogenetically and Antigenically Divergent.
In an unrooted full genome phylogeny comprising the complete genetic diversity of the genus Orthohepadnavirus, shrew HBVs formed a distinct clade in distant sister relationship with the tent-making bat HBV (TBHBV) found in Central American bats (17, 18) ( SI Appendix, Fig. S1C ). The long branches segregating both bat-borne and shrew-borne HBVs in phylogenetic reconstructions relying on the full genomes and the translated ORFs suggest ancient evolutionary relationships for both host orders with HBVs. In the surface ORF, shrew HBVs clustered in basal relationship to all other orthohepadnaviruses at high statistical support ( SI Appendix, Fig. S1D ). To assess whether the evolutionary divergence was consistent with antigenic divergence, we expressed subviral shrew hepatitis B surface (HBs) particles in yeast and generated CSHBV-specific rabbit antisera ( SI Appendix, Fig. S1E ). Serological analyses confirmed complete lack of reactivity of CSHBV antisera with other orthohepadnavirus HBs (Fig. 1B ). This was consistent with only 33% sequence conservation within the HBs antigenic loop, a major immunogenic region of HBV ( SI Appendix, Fig. S1F ). These data together suggested ancient evolutionary origins of shrew HBVs.
Recombination Events Shaped the Genealogy of Shrew HBVs.
Shrew HBVs were about equidistant from other orthohepadnaviruses along the complete genome ( SI Appendix, Table S3), whereas relatively higher similarity with the TBHBV was observed in a genome region encoding the N-terminal part of the polymerase ORF (cyan line, SI Appendix, Fig. S1G ). The sequence distance comparisons and the divergent topology of shrew HBVs in phylogenetic reconstructions of individual ORFs ( SI Appendix, Fig. S1D ) jointly suggested recombination events during the genealogy of shrew HBVs, similar to other orthohepadnaviruses (21, 22). Formal bioinformatic analyses strongly supported ancient recombination events between shrew HBVs and bat-, rodent- and primate-associated HBV ancestors (Fig. 1C ) and within shrew viruses ( SI Appendix, Fig. S1H ).
Shrew HBVs Support Ancient Orthohepadnavirus Origins.
Next, we used the shrew HBVs to reconstruct the Orthohepadnavirus evolution. Bats are major reservoirs of mammalian viruses, likely including HBVs (6, 23). To investigate the origins of orthohepadnaviruses in light of the shrew HBVs, we compared different evolutionary hypotheses in a Bayesian framework. Under the assumption of a long-term association of shrew HBVs and their hosts resembling that of New World primate HBVs and their host (24), we calibrated the most recent common ancestors (MRCA) of the monkey-associated HBV species (24) (Fig. 1D , node a) and the MRCA of shrew-associated HBV species (node b) by the MRCAs of their hosts. We then compared whether calibration of the overall Orthohepadnavirus MRCA (node c) by a prior describing the rise of the order Chiroptera about 66 mya or by a prior describing the rise of the clade Laurasiatheria about 81 mya was most compatible with the data. Calibration of the root with a laurasiatherian prior yielded strong statistical support over a calibration with a chiropteran prior, expanding the Orthohepadnavirus origins beyond bats (Bayes factor [BF], 2.63; ref. 25). Control runs excluding the shrew-associated calibration also favored a laurasiatherian MRCA, but yielded lower statistical support (BF, 1.75), highlighting the importance of including shrew HBVs in reconstructions of the Orthohepadnavirus origins. Of note, the MRCA of MSHBVCIV and MSHBVCHN was projected to 18.4 mya (95% highest posterior density, 13.3–24.1), which was consistent with the time of dispersal of the genus Crocidura to the African continent (Fig. 1A ), suggesting robustness of our calibrations. Next, we conducted ancestral state reconstructions (ASR) in a Bayesian framework to confirm the relevance of laurasiatherian hosts for HBV evolution. ASRs yielded substantial evidence for a laurasiatherian host over a rodent or primate host (those HBV hosts belonging to the Euarchontoglires) at the Orthohepadnavirus root (BF, 2.91) (Fig. 1D ). Finally, recombination events within the dataset may influence our phylogenetic reconstructions. Therefore, we generated 3 different subgenomic datasets without reliably supported evidence for recombination ( SI Appendix, Fig. S1I ) and repeated all evolutionary reconstructions. A laurasiatherian host at the root of orthohepadnaviruses was confirmed independently in all 3 nonrecombinant datasets at similar or stronger statistical support ( SI Appendix, Fig. S1J ). This interpretation was consistent with significantly higher HBV genetic diversity in laurasiatherian hosts compared with hosts representing Euarchontoglires and Chiroptera (Fig. 1E ). In sum, our analyses suggest an ancient laurasiatherian origin of orthohepadnaviruses.
Laurasiatherian Hosts Share Selection Pressure Signatures.
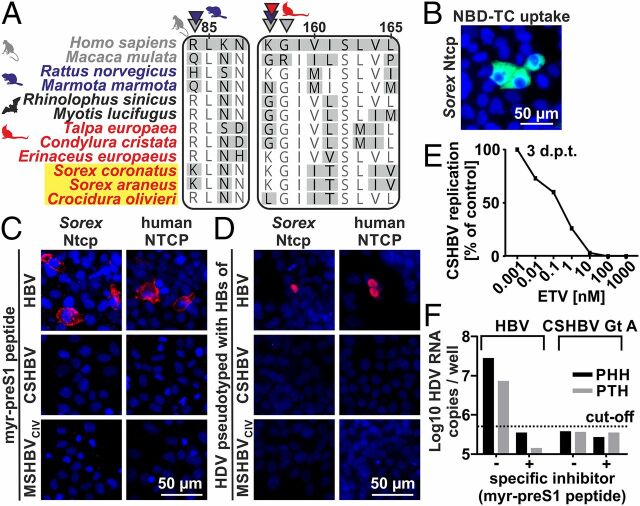
In human HBV, hepatotropism and host range are determined by specific binding of preS1 surface domains to the cellular receptor sodium taurocholate cotransporting polypeptide (NTCP in humans, Ntcp in animals) (26). Recent data suggest that Ntcp homologs from rodents and primates, but not those from bats, evolved under positive selection at HBV binding sites (27). This might hint at differences in receptor usage among HBVs infecting Euarchontoglires and Laurasiatheria. Only 3 eulipotyphlan Ntcp sequences were available in public databases, including the European hedgehog (Erinaceus europaeus), the star-nosed mole (Condylura cristata), and the common shrew (S. araneus). Therefore, we characterized 3 additional eulipotyphlan Ntcp sequences (24) from the European mole (Talpa europaea), the crowned shrew (S. coronatus), and the African giant shrew (C. olivieri) (GenBank accession nos. MN170823–MN170825). Only 1 of 9 sites under positive selection was predicted within the canonical HBV binding site in the eulipotyphlan Ntcp (K/L157; red arrows, Fig. 2A and SI Appendix, Table S4), suggesting similarities between the interactions of HBVs hosted by bats and shrews and their Ntcp orthologs. Notably, the prevalence of both shrew and bat HBV is low, and whether other factors than HBV infection may shape Ntcp evolution remains to be determined. Until now, NTCP usage is known only for primate HBVs and the zoonotic TBHBV (17, 24, 28). Because shrew HBVs strongly differed from primate HBVs in the NTCP-binding preS1 domains ( SI Appendix, Fig. S2A ), the potential interaction of shrew HBV surface proteins with Ntcp orthologs required experimental assessments.
Fig. 2.
NTCP usage and zoonotic potential of shrew HBVs. (A) NTCP/Ntcp-preS1 binding sites. Arrows, sites under positive selection for primates (gray), rodents (blue) (27), and Eulipotyphla (red). Yellow background, shrew HBV hosts. (B) Taurocholate (NBD-TC) uptake (green) by Sorex Ntcp. Blue, nuclei. (C) Myristoylated preS1 peptide binding (red) to human or Sorex NTCP/Ntcp. (D) Infection of HDV pseudotypes via human or Sorex NTCP/Ntcp. Red, newly produced Delta antigen. Blue, nuclei. (E) Inhibition of CSHBV Gt A replication. ETV, Entecavir. (F) Infection of primary human/tupaia hepatocytes by HDV pseudotypes. d.p.t, days posttransfection; GE, genome equivalents; Gt, genotype.
Shrew HBV Surface Peptides Do Not Interact with NTCP Orthologs.
In a first step, the functionality of the Sorex Ntcp was proven by taurocholate transport assay after transfection into human hepatoma cells (Fig. 2B ). Myristoylated (myr) peptides comprising the 48 N-terminal preS1 residues ( SI Appendix, Fig. S2A ) are sufficient for highly specific NTCP binding and interfering with HBV infection (29). While the human HBV myr-preS1 peptide could bind efficiently to the human NTCP and, to a lesser extent, to the Sorex Ntcp (Fig. 2C ), the homologous peptides of CSHBV and MSHBVCIV were unable to bind to either the human NTCP or the shrew Ntcp. Next, we successfully pseudotyped human hepatitis delta virus (HDV) with the surface proteins of HBV, MSHBVCIV, and CSHBV ( SI Appendix, Fig. S2B ). Infection experiments with pseudotyped HDV (psHDV) were performed to rule out that other domains of the surface proteins of shrew HBVs are important for Sorex Ntcp interaction and infection. These experiments confirmed that in contrast to primate HBV, shrew HBV does not use the Ntcp as a primary portal for infection. In contrast, human HBV constructs were able to enter and infect cells both via the human NTCP and via the Sorex Ntcp (Fig. 2D ). These results were consistent with the sequence analysis of NTCP orthologs, as the Sorex Ntcp has a conserved glycine at position 158 (Fig. 2A ), which is essential for preS1 binding of HBV (28). The Sorex Ntcp thus seems to be sufficient for HBV infection in the background of human hepatoma cells.
Zoonotic Potential of Shrew HBVs.
The bat-borne TBHBV can infect human hepatocytes (17). The genetic relationship of shrew HBVs with TBHBV in some genomic domains prompted us to investigate the zoonotic potential of shrew HBVs. First, we resurrected overlength CSHBV resembling the HBV pregenomic mRNA. The CSHBV was replication-competent in human hepatoma cells and could be blocked by the reverse transcriptase inhibitor Entecavir (Fig. 2E ). Reverse transcription of the HBV genome only takes place in a complex formed by the viral polymerase and the pregenomic mRNA encapsidated by the core protein. Therefore, proof of reverse transcription is also proof of successful pregenomic CSHBV mRNA transcription and of the translation of functional polymerase and core gene products. Next, we used pseudotyped viruses to infect primary hepatocytes from humans (PHH) and Tupaia belangeri (PTH), which are susceptible to human HBV. Notably, tupaias, commonly termed tree shrews, are not closely related to shrews (4). Unlike the TBHBV and HDVHBV, HDVCSHBV was unable to infect PHH and PTH (Fig. 2F ). Because entry is a major species barrier preventing HBV host switches, these data speak against a zoonotic potential of shrew HBVs despite the ability of CSHBV to replicate within human-derived cells.
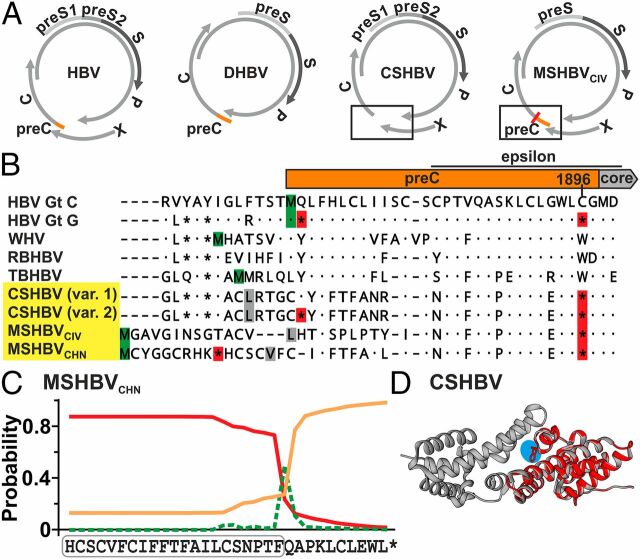
Unusual Genomic Signature of the Shrew HBV PreCore.
In HBV, the nonstructural hepatitis B e antigen (HBeAg) comprises an N-terminal portion translated from a 5′-extension of the core ORF termed preCore (preC) and a core ORF-derived C terminus that is posttranslationally cleaved. The existence of the preC-encoding region is conserved among distantly related hepadnaviruses, such as the duck HBV (DHBV) (Fig. 3A ). Strikingly, both CSHBV and MSHBVCHN lack a methionine to initiate preC translation. However, alternative start codons hypothetically allowing translation initiation by leaky ribosomal scanning occurred (Fig. 3B , gray). The MSHBVCIV may contain a methionine, but the resulting preC would be unusually long (Fig. 3B , green). We confirmed lack of a canonical preC start codon in all 6 additional CSHBV-positive animals for which full genomes had not been generated (GenBank accession nos. MK345468, MK345469, MK345471–MK345473, and MK345476), irrespective of host genus, sampling year, and site. Primary HBeAg negativity is mainly known from HBV genotype G, due to a conserved nonsense G to A mutation at genome position 1896 that creates a stop codon within the preC and an additional nonsense mutation creating a stop codon immediately after the preC start codon. The same nonsense mutations can occur at late stages of CHB caused by diverse HBV genotypes. Interestingly, all 17 shrew HBVs characterized in this study and the MSHBVCHN showed the G1896A nonsense mutation. One CSHBV genotype also showed the nonsense mutation following the preC start codon ( SI Appendix, Fig. S3A ). Additional similarities between between human and shrew HBV HBeAg-encoding regions included the prediction of a signal sequence in the preC region of MSHBVCHN (Fig. 3C and SI Appendix, Fig. S3B ) and the conservation of a C-terminal arginine-rich region, both of which are characteristic for posttranslational cleavage of the premature HBeAg ( SI Appendix, Fig. S3C ), and the conservation of the RNA encapsidation signal epsilon located at the 3′ end of the preC domain ( SI Appendix, Fig. S3D ). In addition, despite low amino acid identity of 54.1–55.3% between predicted HBeAg sequences of shrew and human HBVs, their 3D structures were closely related (Fig. 3D and SI Appendix, Fig. S3E ). Finally, ASR supported the existence of a conserved preC-encoding region at the Orthohepadnavirus MRCA (BF, 2.67), jointly suggesting loss of HBeAg coding capacity during the genealogy of extant shrew HBVs ( SI Appendix, Fig. S3F ).
Fig. 3.
Shrew HBV HBeAg. (A) Genome structures. (B) Translated preC and N-terminal core domains. Red, stop codons; green, methionine; gray, alternative start codons. Var. 1/2, HTS minority variants (25–30% occurrence). (C) Signal peptide prediction (red line) of MSHBVCHN. Green, cleavage site; orange line, no signal peptide; boxed, predicted signal sequence. (D) CSHBV HBeAg monomer (cyan, reverted G1896A) modeled on the HBV HBeAg dimer (43). Gt, genotype; RBHBV, roundleaf bat HBV.
HBeAg-Negative Shrew HBVs Cause Monoinfections.
In human HBV, genotype G infections usually occur as coinfections with another HBV genotype providing HBeAg in trans (30). To exclude that unrecognized coinfection with a putative HBeAg-positive shrew HBV variant occurred, high-throughput sequencing (HTS) was conducted on a broadly reactive and highly sensitive nested PCR amplicon (17). HTS of 10 CSHBV-positive and 5 MSHBVCIV-positive samples did not detect any additional divergent shrew HBV sequence in any sample.
Low Within-Host Genomic Variability of Shrew HBVs.
In CHB, HBeAg loss is associated with increased HBV genomic variability within infected individuals. To analyze within-host genomic variability in shrew HBV, single-nucleotide polymorphisms (SNP) in surface/polymerase and X/preC/core genomic domains were analyzed in liver tissue by HTS. The overall genomic variability was low with a mean of 0.5 SNPs (occurring in >2% of reads) per 100 nucleotides. For comparison, the HBeAg-positive capuchin monkey HBV (CMHBV) showed significantly more SNPs in serum (1.8 SNPs per 100 nucleotides; χ2, P < 0.0001) (24). This difference is noteworthy because liver contains the covalently closed circular DNA reservoir showing relatively higher virus diversity than the relaxed circular DNA found in circulating virions (31). Of note, significant differences in SNP occurrence rates were observed neither between coding nor noncoding shrew HBV genomic regions, nor between single or overlapping ORFs, nor between the different shrew genera ( SI Appendix, Table S5). Importantly, there was no detectable minority variant reverting the lack of methionine codons in both CSHBV genotypes and the presence of preC stop codons in both CSHBV and MSHBVCIV. Low within-host variability of HBeAg-negative shrew HBV thus surprisingly resembled the immunotolerant phase of HBeAg-positive CHB in humans (32).
Shrew HBVs Cause Intense Hepatotropic Infections.
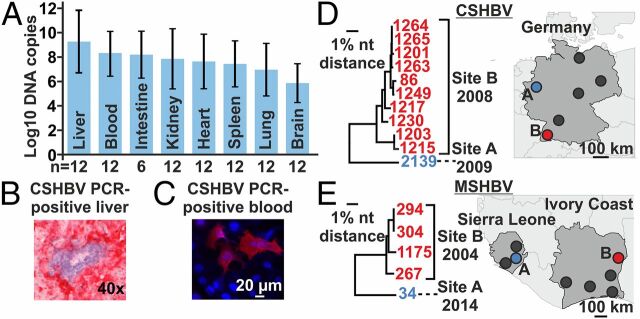
In humans, infections with HBeAg-negative HBV are associated with severe hepatitis (30). Infections with primarily HBeAg-negative HBV strains are associated with high viral loads, whereas HBeAg loss during CHB is commonly associated with low viral loads. To assess shrew HBV tropism and infection intensities, we used a strain-specific qPCR assay to quantify CSHBV concentrations in different tissues of 12 individual shrews. CSHBV concentrations were generally high. The highest mean concentrations occurred in the liver at 10e+9 CSHBV copies per gram, significantly exceeding those in other solid organs (t test, P = 0.043), whose similar virus concentrations likely originate from high viremia (Fig. 4A ). In situ hybridization with an RNA probe directed against CSHBV nucleic acid confirmed infection of hepatocytes by shrew HBVs (red; Fig. 4B ).
Fig. 4.
Shrew HBV infection patterns. (A) Mean CSHBV concentrations in solid organs (grams) and blood (milliliters), SDs. (B) In situ hybridization of CSHBV. (C) Shrew serum reacting with CSHBV proteins. Red, antibody binding; blue, nuclei. (D and E) Major sampling sites in Germany, Sierra Leone, and Ivory Coast; neighbor-joining phylogenies. Trees encompass 773 (CSHBV) and 1,432 (MSHBV) nucleotides. One CSHBV strain for which only a smaller fragment was characterized is not shown.
Low Shrew HBV Seroprevalence.
Next, we conducted serological analyses relying on resurrected full-length CSHBV and MSHBV to optimize test sensitivity and specificity (24). Using an immunofluorescence assay adapted for shrews, we tested 86 sera from Sorex and 57 sera from Crocidura. Strikingly, antibodies were only detected in 1 German and 1 African shrew (Germany, 1.2%, 95% CI 0.0–6.9; Sierra Leone, 1.8%, 95% CI 0.0–10.2, Fig. 4C ). Both antibody-positive animals were also PCR-positive. All other 11 PCR-positive shrews were seronegative, suggesting either sampling before seroconversion or chronic infections without detectable anti-HBc, similar to the monkey-associated CMHBV, but different from bat HBVs (17, 18, 24). Hypothetically, shrew hepatitis B may lead to death of the infected animals, thereby explaining the low seroprevalence. However, the sample amenable for serological investigation was small and it was not possible to conduct histopathological analyses due to advanced stages of decay after trapping.
Geographical Clustering of Shrew HBVs.
In Germany, 6 areas were sampled over 8 y. All 11 animals infected with CSHBV genotype B originated from one site only and were sampled in the same year (site B; Fig. 4D ). The detection rate in animals from that site was significantly higher compared with all other German sampling sites (Fisher exact, P < 0.0001). Similarly, all 4 Crocidura infected with MSHBVCIV from Ivory Coast originated from one site only, sampled in the same year (site B; Fig. 4E ). Again, the detection rate from this site was significantly higher compared with all other Ivorian sites (Fisher exact, P < 0.0001). Likewise, all 4 MSHBVCHN Gt A-positive samples originated from the same site (11). Notably, the Ivorian MSHBV strains sampled in 2004 differed by up to 1.9% of their complete genomes and the German CSHBV strains sampled in site B in 2008 differed by up to 4.1% of their partial genomes (Fig. 4 D and E ). High between-host genomic diversity is not necessarily at odds with focal transmission chains suggested by low molecular and serological detection rates, because a genomic diversity of up to 4.6% was observed in HBeAg-negative HBV strains during outbreaks in humans (33). Finally, information on sex was available for 320 shrews from Germany. No significant differences of shrew HBV detection rates were observed between male (4 of 161) and female (8 of 159) shrews (Fisher exact, P = 0.26), speaking against a sex bias potentially associated with shrew HBV transmission. In sum, regionally increased incidence and low seroprevalence jointly suggest focal transmission of shrew HBV and may hint at intense infections of HBeAg-negative shrew HBV.
Discussion
We identified previously undescribed shrew HBVs, revealed their distinct infection patterns, and reconstructed the genealogy of mammalian hepadnaviruses.
Hepadnaviruses as a family are phylogenetically much older than previously thought (1, 3), yet the origins of mammalian hepadnaviruses remain enigmatic. Our study projects the rise of mammalian hepadnaviruses toward the origins of the Laurasiatheria, 1 of the 2 large clades of placental mammals. Notably, many laurasiatherian species are insectivorous. Hypothetically, ancient insect-associated hepadnaviruses (34) may have been acquired by small insectivorous mammals like bats and shrews and further diversified within the mammalian lineage by complex macroevolutionary patterns likely including both long-term host–virus associations and host switches (1).
Ancient origins of shrew and bat HBVs are consistent with the evidence for recombination events between shrew HBVs and the TBHBV, a New World bat HBV. The ancestors of the Noctilionoidea, the bat superfamily to which the TBHBV host Uroderma bilobatum belongs, likely immigrated about 52 mya from the Old World into the New World (35), while shrews immigrated into the New World much later, about 13 mya (16). Targeted investigations of Old World bats for TBHBV-related hepadnaviruses and of New World shrews for CSHBV/MSHBV-related hepadnaviruses may allow reconstructions of host and hepadnavirus dispersal history at an unprecedented depth in time.
Given the ancient hepadnavirus origins, it may not be surprising that some hallmarks of human HBV are not evolutionarily conserved (1). Within the HBV species infecting primates, Ntcp usage seems to be conserved (24, 26). In contrast, our experimental and evolutionary data strongly support differences of receptor usage among distantly related mammalian hepadnaviruses. A different set of receptors or coreceptors for shrew HBV and potentially other HBV species may thus exist.
Another hallmark of human HBV is the HBeAg, which is essential for the development of CHB. HBeAg functions as an immunomodulatory tolerogen, which down-regulates the immune response to HBcAg and, thereby, promotes chronicity (30). Additionally, antigen-presenting cells are conditioned in utero by the HBeAg to suppress the cytotoxic T lymphocyte response in vertically infected offspring by up-regulation of the inhibitory ligand PD-L1 (36). In humans, HBeAg negativity represents the wild type in HBV genotype G only, whereas HBeAg negativity can develop during the late phase of CHB in many genotypes. Our discovery of diverse HBV species completely lacking HBeAg in wild shrews is therefore surprising. In contrast, some characteristics of HBeAg-negative HBVs seem to be conserved between humans and shrews, including (i) the position and type of nonsense mutations precluding HBeAg production (30), (ii) intense infections with high virus concentrations such as in patients acutely infected with primary HBeAg-negative HBV strains or occasionally during reactivation in CHB (33), and (iii) potentially high virulence.
At first glance, it seems odd that shrew HBVs may persist on the population level without relying on a viral protein supporting chronically infected offspring as continuous sources of secondary infections. However, differences in viral infection patterns between humans and animal reservoirs are well documented, including persistent infection of rodents with arenaviruses and hantaviruses, and long-term population-level shedding of bat morbillivirus-related pathogens compared with predominantly acute human infections (37, 38). In shrews, territorial behavior leading to blood contact during fights may contribute to efficient horizontal transmission and population-level maintenance of HBeAg-negative HBVs (12). Hypothetically, the long-term evolutionary association between shrews and HBeAg-negative HBVs suggested by our data may have allowed for relatively decreased virulence despite an HBeAg-negative phenotype, reminiscent of the decreased virulence of rabbit myxoma virus over time (39), thereby supporting shrew HBV population-level survival.
Finally, lack of an effective therapy for CHB demands a suitable animal model. Experimental HBV infection of chimpanzees is restricted due to ethical concerns. HBV infection efficacy in tupaias is low, and autochthonous tupaia hepadnaviruses are unknown. Woodchuck hepatitis virus (WHV) models are generally suitable to study CHB (1), but keeping woodchucks is costly and naturally occurring HBeAg-negative WHV variants are unknown (40). Shrews are small and tractable animals that can be housed and experimentally infected, e.g., with bornaviruses (41). Our data suggest that a shrew HBV model to investigate HBeAg-mediated chronicity and HBV species barriers relying on shrew and human HBV mutants and revertants may be attainable.
Materials and Methods
National authorities approved sampling. Hepadnavirus screening, genomic characterization, evolutionary analysis, generation of pseudotypes and infectious clones, receptor characterizations, high-throughput sequencing, and in situ hybridization were done as described previously (17, 24) and detailed in SI Appendix .
Supplementary Material
Acknowledgments
This study was supported by funding from German Research Foundation Grants B08/SFB 1021/2, BO 3790/1-1, DR 810/1-1, FI 1781/1-1, GL 595/4-1; European INCO-DEV program Grant ICA4-CT2002-10050; European Foundation Initiative for African Research into Neglected Tropical Diseases Grant 1/85/022; and German Federal Ministry of Education and Research through the German Research Platform for Zoonoses Grants 01KI1018 and 01KI1303. The National Reference Center for Hepatitis B and D Viruses is supported by the German Ministry of Health via the Robert Koch Institute. We thank M. Eschbach-Bludau, S. Brünink, T. Bleicker, J. Schneider, S. Broehl, B. Allali, S. Kouassi Kan, R. Ansumana, D. Sondufu, J. Buanie, J. Lamin, M. Dawson, A. Oyeyiola, A. Obadare, J. Igbokwe, and numerous persons of the network “Rodent-borne pathogens” for technical assistance.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Sequence data have been submitted to GenBank, https://www.ncbi.nlm.nih.gov/genbank (accession nos. MK345460–MK345476 and MN170823–MN170825).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1908072116/-/DCSupplemental.
Change History
April 29, 2022: The affiliation of author Andris Kazaks has been updated.
References
- 1. Rasche A., Sander A. L., Corman V. M., Drexler J. F., Evolutionary biology of human hepatitis viruses. J. Hepatol. 70, 501–520 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lauber C., et al., Deciphering the origin and evolution of hepatitis B viruses by means of a family of non-enveloped fish viruses. Cell Host Microbe 22, 387–399.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Suh A., et al., Early mesozoic coexistence of amniotes and hepadnaviridae. PLoS Genet. 10, e1004559 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Foley N. M., Springer M. S., Teeling E. C., Mammal madness: Is the mammal tree of life not yet resolved? Philos. Trans. R. Soc. Lond. B Biol. Sci. 371, 20150140 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gogarten J. F., et al., A novel orthohepadnavirus identified in a dead Maxwell’s Duiker (Philantomba maxwellii) in Taï National Park, Côte d’Ivoire. Viruses 11, E279 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rasche A., Souza B. F. C. D., Drexler J. F., Bat hepadnaviruses and the origins of primate hepatitis B viruses. Curr. Opin. Virol. 16, 86–94 (2016). [DOI] [PubMed] [Google Scholar]
- 7. Drexler J. F., et al.; Hepatovirus Ecology Consortium , Evolutionary origins of hepatitis A virus in small mammals. Proc. Natl. Acad. Sci. U.S.A. 112, 15190–15195 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Corman V. M., et al., Characterization of a novel betacoronavirus related to middle East respiratory syndrome coronavirus in European hedgehogs. J. Virol. 88, 717–724 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hilbe M., et al., Shrews as reservoir hosts of borna disease virus. Emerg. Infect. Dis. 12, 675–677 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Guo W. P., et al., Phylogeny and origins of hantaviruses harbored by bats, insectivores, and rodents. PLoS Pathog. 9, e1003159 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nie F. Y., et al., Discovery of a highly divergent hepadnavirus in shrews from China. Virology 531, 162–170 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Szalay F. S., Walker’s mammals of the World. Ronald M. Nowak. Q. Rev. Biol. 75, 71–72 (2000). [Google Scholar]
- 13. Ochocińska D., Taylor J. R., Living at the physiological limits: Field and maximum metabolic rates of the common shrew (Sorex araneus). Physiol. Biochem. Zool. 78, 808–818 (2005). [DOI] [PubMed] [Google Scholar]
- 14. Geoghegan J. L., Duchêne S., Holmes E. C., Comparative analysis estimates the relative frequencies of co-divergence and cross-species transmission within viral families. PLoS Pathog. 13, e1006215 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Geoghegan J. L., Senior A. M., Di Giallonardo F., Holmes E. C., Virological factors that increase the transmissibility of emerging human viruses. Proc. Natl. Acad. Sci. U.S.A. 113, 4170–4175 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dubey S., Salamin N., Ohdachi S. D., Barrière P., Vogel P., Molecular phylogenetics of shrews (Mammalia: Soricidae) reveal timing of transcontinental colonizations. Mol. Phylogenet. Evol. 44, 126–137 (2007). [DOI] [PubMed] [Google Scholar]
- 17. Drexler J. F., et al., Bats carry pathogenic hepadnaviruses antigenically related to hepatitis B virus and capable of infecting human hepatocytes. Proc. Natl. Acad. Sci. U.S.A. 110, 16151–16156 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hiller T., et al., Host biology and anthropogenic factors affect hepadnavirus infection in a neotropical bat. EcoHealth 16, 82–94 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schaefer S., Hepatitis B virus taxonomy and hepatitis B virus genotypes. World J. Gastroenterol. 13, 14–21 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Starkman S. E., MacDonald D. M., Lewis J. C., Holmes E. C., Simmonds P., Geographic and species association of hepatitis B virus genotypes in non-human primates. Virology 314, 381–393 (2003). [DOI] [PubMed] [Google Scholar]
- 21. Locarnini S., Littlejohn M., Aziz M. N., Yuen L., Possible origins and evolution of the hepatitis B virus (HBV). Semin. Cancer Biol. 23, 561–575 (2013). [DOI] [PubMed] [Google Scholar]
- 22. Mühlemann B., et al., Ancient hepatitis B viruses from the bronze age to the medieval period. Nature 557, 418–423 (2018). [DOI] [PubMed] [Google Scholar]
- 23. Olival K. J., et al., Host and viral traits predict zoonotic spillover from mammals. Nature 546, 646–650 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. de Carvalho Dominguez Souza B. F., et al., A novel hepatitis B virus species discovered in capuchin monkeys sheds new light on the evolution of primate hepadnaviruses. J. Hepatol. 68, 1114–1122 (2018). [DOI] [PubMed] [Google Scholar]
- 25. Jeffreys H., The Theory of Probability (Clarendon Press, Oxford, 1961). [Google Scholar]
- 26. Yan H., et al., Sodium taurocholate co-transporting polypeptide is a functional receptor for human hepatitis B and D virus. eLife 1, e00049 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jacquet S., et al., Evolution of hepatitis B virus receptor NTCP reveals differential pathogenicities and species specificities of hepadnaviruses in primates, rodents, and bats. J. Virol. 93, e01738-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Müller S. F., König A., Döring B., Glebe D., Geyer J., Characterisation of the hepatitis B virus cross-species transmission pattern via Na+/taurocholate co-transporting polypeptides from 11 New World and Old World primate species. PLoS One 13, e0199200 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Glebe D., et al., Mapping of the hepatitis B virus attachment site by use of infection-inhibiting preS1 lipopeptides and tupaia hepatocytes. Gastroenterology 129, 234–245 (2005). [DOI] [PubMed] [Google Scholar]
- 30. Kramvis A., Kostaki E. G., Hatzakis A., Paraskevis D., Immunomodulatory function of HBeAg related to short-sighted evolution, transmissibility, and clinical manifestation of hepatitis B virus. Front. Microbiol. 9, 2521 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rybicka M., et al., Differences in sequences between HBV-relaxed circular DNA and covalently closed circular DNA. Emerg. Microbes Infect. 6, e55 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang H. Y., et al., Distinct hepatitis B virus dynamics in the immunotolerant and early immunoclearance phases. J. Virol. 84, 3454–3463 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Seiz P. L., et al., Studies of nosocomial outbreaks of hepatitis B in nursing homes in Germany suggest a major role of hepatitis B e antigen expression in disease severity and progression. Int. J. Med. Microbiol. 305, 663–672 (2015). [DOI] [PubMed] [Google Scholar]
- 34. Gong Z., Han G. Z., Insect retroelements provide novel insights into the origin of hepatitis B viruses. Mol. Biol. Evol. 35, 2254–2259 (2018). [DOI] [PubMed] [Google Scholar]
- 35. Lim B. K., Review of the origins and biogeography of bats in South America. Chiropt. Neotrop. 15, 391–410 (2009). [Google Scholar]
- 36. Tian Y., Kuo C. F., Akbari O., Ou J. H., Maternal-derived hepatitis B virus e antigen alters macrophage function in offspring to drive viral persistence after vertical transmission. Immunity 44, 1204–1214 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Charrel R. N., et al., Arenaviruses and hantaviruses: From epidemiology and genomics to antivirals. Antiviral Res. 90, 102–114 (2011). [DOI] [PubMed] [Google Scholar]
- 38. Drexler J. F., et al., Bats host major mammalian paramyxoviruses. Nat. Commun. 3, 796 (2012). Erratum in: Nat. Commun. 5, 3032 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kerr P. J., et al., Next step in the ongoing arms race between myxoma virus and wild rabbits in Australia is a novel disease phenotype. Proc. Natl. Acad. Sci. U.S.A. 114, 9397–9402 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chen H. S., et al., The precore gene of the woodchuck hepatitis virus genome is not essential for viral replication in the natural host. J. Virol. 66, 5682–5684 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nobach D., et al., Shedding of infectious borna disease virus-1 in living bicolored white-toothed shrews. PLoS One 10, e0137018 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Drummond A. J., Suchard M. A., Xie D., Rambaut A., Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29, 1969–1973 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Eren E., et al., Structures of hepatitis B virus core- and e-antigen immune complexes suggest multi-point inhibition. Structure 26, 1314–1326.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.