Significance
Cancer genomes are characterized by focal increases in DNA methylation, co-occurring with widespread hypomethylation. We show that TET deficiency in diverse cell types (ESCs, NPCs, HSCs, pro-B cells, and T cells) results in a similar methylation landscape, with the expected localized increases in DNA methylation in active euchromatic regions, concurrently with unexpected losses of DNA methylation, reactivation of repeat elements, and enrichment for single-nucleotide alterations primarily in heterochromatic compartments. Thus, TET loss of function may be a primary mechanism underlying the characteristic pattern of global hypomethylation coupled to regional hypermethylation observed in diverse cancer genomes. Our data potentially explain the synergy between DNMT3A and TET2 mutations in hematopoietic malignancies, as well as the recurrent association of TET loss of function with cancer.
Keywords: epigenetics, heterochromatin, TET proteins, DNA hypomethylation, heterochromatin dysfunction
Abstract
Cancer genomes are characterized by focal increases in DNA methylation, co-occurring with widespread hypomethylation. Here, we show that TET loss of function results in a similar genomic footprint. Both 5hmC in wild-type (WT) genomes and DNA hypermethylation in TET-deficient genomes are largely confined to the active euchromatic compartment, consistent with the known functions of TET proteins in DNA demethylation and the known distribution of 5hmC at transcribed genes and active enhancers. In contrast, an unexpected DNA hypomethylation noted in multiple TET-deficient genomes is primarily observed in the heterochromatin compartment. In a mouse model of T cell lymphoma driven by TET deficiency (Tet2/3 DKO T cells), genomic analysis of malignant T cells revealed DNA hypomethylation in the heterochromatic genomic compartment, as well as reactivation of repeat elements and enrichment for single-nucleotide alterations, primarily in heterochromatic regions of the genome. Moreover, hematopoietic stem/precursor cells (HSPCs) doubly deficient for Tet2 and Dnmt3a displayed greater losses of DNA methylation than HSPCs singly deficient for Tet2 or Dnmt3a alone, potentially explaining the unexpected synergy between DNMT3A and TET2 mutations in myeloid and lymphoid malignancies. Tet1-deficient cells showed decreased localization of DNMT3A in the heterochromatin compartment compared with WT cells, pointing to a functional interaction between TET and DNMT proteins and providing a potential explanation for the hypomethylation observed in TET-deficient genomes. Our data suggest that TET loss of function may at least partially underlie the characteristic pattern of global hypomethylation coupled to regional hypermethylation observed in diverse cancer genomes, and highlight the potential contribution of heterochromatin hypomethylation to oncogenesis.
TET enzymes are Fe(II) and α-ketoglutarate–dependent dioxygenases that mediate DNA demethylation through sequential oxidation of the methyl group of 5-methylcytosine (5mC) to 5-hydroxymethyl, 5-formyl, and 5-carboxylcytosine (5hmC, 5fC, and 5caC) (1–3). The oxidized methylcytosines (oxi-mC) generated by TET proteins are intermediates in at least 2 pathways of DNA demethylation: 1) replication-dependent loss of methylation, reflecting inability of the DNMT1/UHRF1 complex to methylate unmodified CpGs on newly replicated DNA strands if an oxi-mC (rather than 5mC) is present on the template strand, and 2) a replication-independent process in which thymine DNA glycosylase (TDG) excises 5fC and 5caC, which are then replaced with unmodified cytosine through base excision repair (4).
Even in the absence of TET coding region mutations, TET loss of function and low 5hmC levels are strongly associated with cancer (5). TET2 mutations are frequent in diverse hematopoietic malignancies, including myelodysplastic syndromes, acute myeloid leukemias, and peripheral T cell lymphomas (6–8). However, both solid tumors and hematopoietic malignancies display TET loss of function without TET coding region mutations, as a result of TET promoter methylation, increased degradation of TET proteins, or aberrant microRNA expression (9–11). In addition, hypoxia and a variety of metabolic alterations impair the enzymatic activity of TET and other dioxygenases, by decreasing the levels of the substrates α-ketoglutarate and molecular oxygen or increasing the levels of the inhibitor 2-hydroxyglutarate (10, 11).
Based on the known biochemical activities of TET-family proteins in oxidizing 5mC (4), TET loss-of-function mutations are expected to result in gains of DNA methylation. In fact, increased methylation as a result of TET loss of function has been documented at many genomic regions including promoters, enhancers, and CTCF sites (9, 12–14). Unexpectedly, however, a small number of studies have noted widespread decreases of DNA methylation in the genomes of TET-deficient cells, by mapping differentially methylated regions or by comparing methylation values at random genomic regions after whole-genome bisulfite sequencing (WGBS) (12, 14–16). Because the hypomethylated regions observed in TET loss of function did not overlap with active or regulatory regions of the genome, these findings were largely ignored.
Principal component analysis of the interaction matrix obtained from Hi-C data has been used to compartmentalize the genome into an A compartment (positive PC1 values) and a B compartment (negative PC1 values) that exhibit the hallmark characteristics of euchromatin and heterochromatin, respectively (17, 18). The euchromatic A compartment is rich in expressed genes in the cell type under consideration, whereas the heterochromatic B compartment is gene-poor and bears epigenetically “repressive” chromatin marks, including H3K9me2/3 (17, 18). Moreover, the Hi-C B compartment overlaps with lamina-associated domains and corresponds to late-replicating regions of the genome, whereas the Hi-C A compartment corresponds to early replicating genomic regions and is not lamina associated (18, 19). Notably, the extended partially methylated domains (PMDs) observed in cancer genomes overlap with Hi-C B compartment, late-replicating, nuclear lamina-associated domains (20–22). In the remainder of this study, we will refer to the Hi-C A and B compartments as euchromatic and heterochromatic compartments, respectively.
Cancer genomes are characterized by 2 opposing patterns of aberrant DNA methylation: focal hypermethylation and widespread DNA hypomethylation (23). DNA hypermethylation at promoters and enhancers contributes to oncogenesis through transcriptional silencing of genes involved in DNA damage repair and tumor suppressors (23), and has been shown to reflect the impaired expression or activity of TET proteins. Despite our understanding of the biochemical and biological consequences of local hypermethylation, however, the causes and consequences of DNA hypomethylation in cancer genomes are less well understood.
Here, we use a combination of Hi-C and WGBS data to document the DNA methylation changes associated with TET loss of function in diverse TET-deficient cell types. We show that 5hmC in wild-type (WT) genomes, and DNA hypermethylation in TET-deficient genomes, are largely confined to the euchromatic Hi-C A compartment. This finding is consistent with the known functions of TET proteins in DNA demethylation and the known distribution of 5hmC at active enhancers and in the gene bodies of highly transcribed genes. In contrast, we show that the unexpected DNA hypomethylation noted in TET-deficient genomes is primarily present in the heterochromatic Hi-C B compartment. TET-deficient cells showed reactivation of repeat elements and pronounced enrichment for single-nucleotide variations (SNVs) in the heterochromatic Hi-C B compartment; this feature is characteristic of cancer genomes, in which mutation rates are elevated in genome compartments marked by H3K9me3 (24). We also show that DNMT3A relocalizes from the heterochromatic compartment to the euchromatic compartment in Tet1-deficient mouse embryonic stem cells (mESCs), providing a potential mechanism for the heterochromatin hypomethylation observed in TET-deficient genomes. Our results are consistent with the co-occurrence of DNMT3A and TET2 mutations in human cancers and the more pronounced leukemic phenotype observed in double Tet2/Dnmt3a-deficient mice compared with mice with individual disruption of Tet2 or Dnmt3a alone. Taken together, these data point to a functional interaction between TET proteins and DNMTs, and highlight the potential contribution of heterochromatic dysfunction to oncogenesis.
Results
Widespread DNA Hypomethylation in TET-Deficient mESCs.
To understand the impact of TET loss of function on genome-wide patterns of DNA modification, we reanalyzed data from several publicly available WGBS datasets across a diverse range of TET-deficient murine cell types: ESCs (12, 15, 25), neural precursor cells differentiated from ESCs (NPCs) (12); pro-B cells (26); hematopoietic stem/precursor cells (HSPCs) (27); and a mouse model of TET-deficient T cells (14) (see Figs. 1–3 and 5 and SI Appendix, Figs. S1 and S2; and see below). Bisulfite sequencing estimates the sum of 5mC and 5hmC (28), but 5hmC is always <10% of 5mC in the control cell types considered here, and lower or absent in their TET-deficient counterparts. Thus, throughout this manuscript, we refer to the values obtained from WGBS as “DNA methylation.”
Fig. 1.
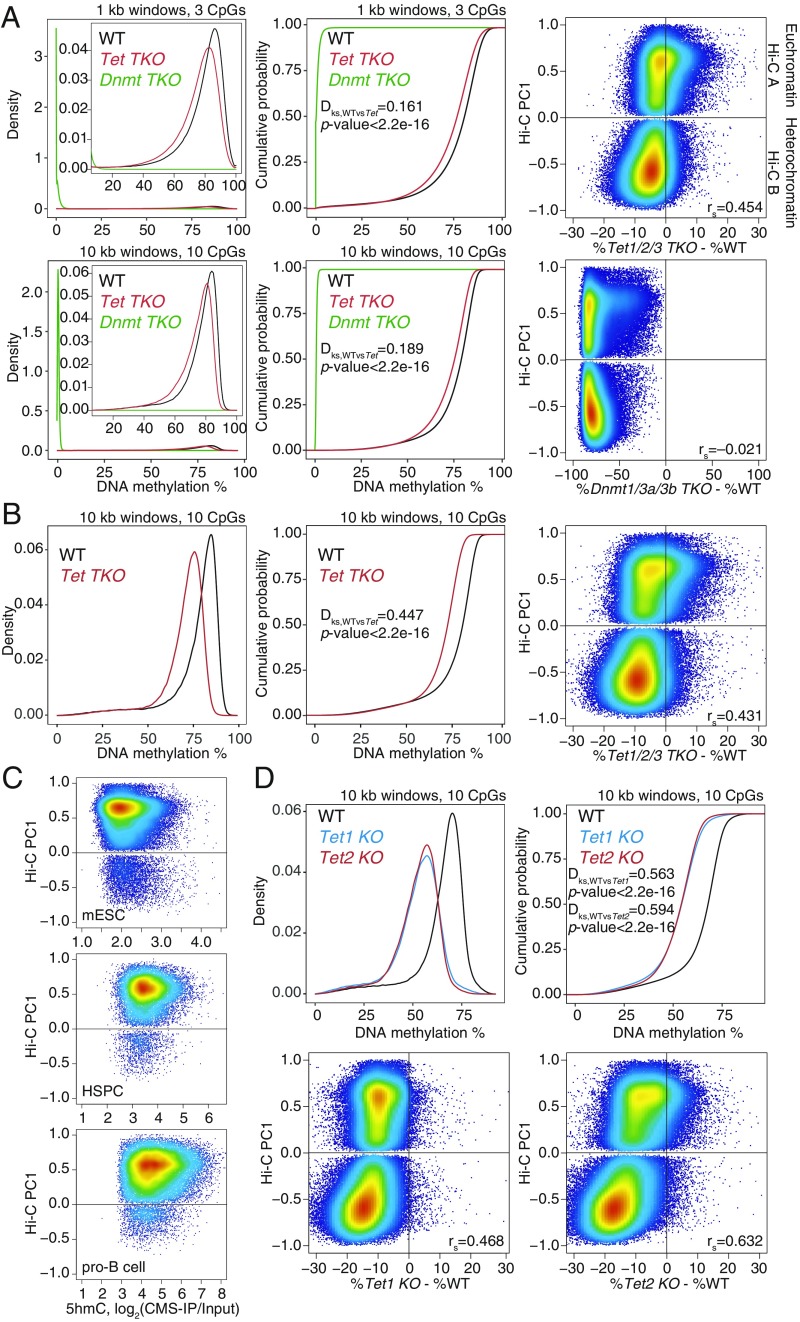
Widespread DNA hypomethylation in TET-deficient mouse embryonic stem cells (mESCs). (A) Density distribution (Left) and cumulative distribution (Middle) of average DNA methylation values within 1- and 10-kb windows across the genome in wild-type (WT), Dnmt TKO, and Tet TKO mESCs (data from ref. 25). Only windows with 3 or more CpGs per 1 kb, or 10 or more CpGs per 10 kb, covered by at least 5 WGBS reads per CpG, were considered for the analysis. Two-sample Kolmogorov–Smirnov test comparing the distributions of WT and Tet TKO was used to calculate the Dks statistic and P value. (Right) Correlation between DNA methylation changes (difference in cytosine modification percentage, mutant minus WT) and euchromatin/heterochromatin compartments (positive versus negative Hi-C PC1 values), in Tet TKO (Top) and Dnmt TKO (Bottom) mESCs. Spearman correlation coefficient is shown (rs value). (B) Density distribution (Left) and cumulative distribution (Middle) of average DNA methylation values within 10-kb windows across the genome in WT and Tet TKO mESCs (data from ref. 15). (Right) Correlation between DNA methylation changes and euchromatin/heterochromatin compartments. Spearman correlation coefficient is shown (rs value). (C) Relationship between 5hmC distribution (CMS-IP) and euchromatin/heterochromatin compartments (Hi-C PC1 values) in WT mESCs, HSCs, and pro-B cells. CMS-IP enrichment was calculated for 1-kb windows. (D) Density distribution (Left) and cumulative distribution (Right) of average DNA methylation values within 10-kb windows across the genome, in WT, Tet1 KO, and Tet2 KO mESCs (data from ref. 12). (Bottom) Correlation between DNA methylation changes and euchromatin/heterochromatin compartments. Spearman correlation coefficient is shown (rs value).
Fig. 3.
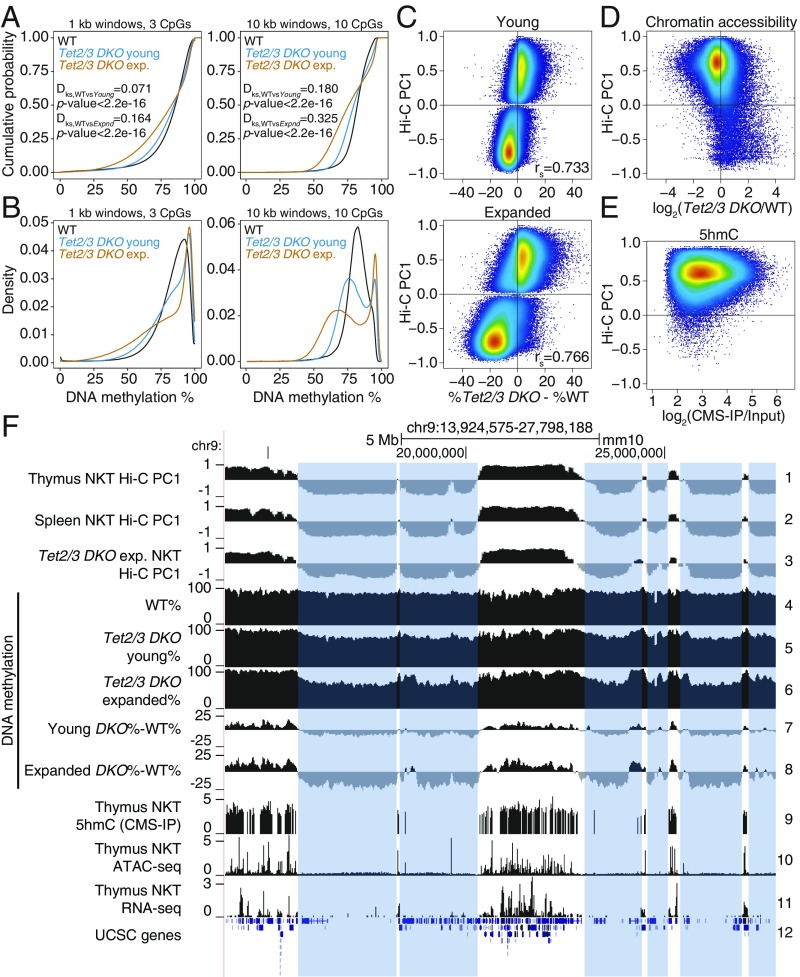
DNA hypermethylation in euchromatin and hypomethylation in heterochromatin in NKT-cell lymphoma from Tet2/3 DKO mice. (A and B) Cumulative (A) and density distribution (B) of average DNA methylation values within 1- and 10-kb windows across the genome, in wild-type (WT), young, and transferred and expanded Tet2/3 DKO NKT cells. In A, 2-sample Kolmogorov–Smirnov test comparing the distributions of WT and Tet2/3 DKO (young or expanded, as indicated) was used to calculate the Dks statistic and P value. (C) Correlation between DNA methylation changes (mutant minus WT) and euchromatin/heterochromatin compartments in Tet2/3 DKO NKT cells at 2 stages of expansion (young, and transferred and expanded). Spearman correlation coefficient is shown (rs value). (D) Correlation between changes in chromatin accessibility (ATAC-seq) in WT versus Tet2/3 DKO cells, and euchromatin/heterochromatin compartments, in thymic NKT cells from young mice. Accessibility differences were calculated at 1-kb resolution. (E) Correlation between 5hmC distribution (CMS-IP) and euchromatin/heterochromatin compartments in WT thymic NKT cells. CMS-IP enrichment was calculated for 1-kb windows. (F) Genome tracks showing a correspondence between Hi-C-defined B compartments in thymic and splenic WT NKT cells (tracks 1–2) and expanded splenic Tet2/3 DKO NKT cells (track 3), and large hypomethylated domains in WT, young and expanded Tet2/3 DKO NKT cells (tracks 4–8). Notice progressive hypomethylation of the heterochromatic compartment. 5hmC distribution (CMS-IP, track 9), chromatin accessibility (ATAC-seq, track 10), and gene expression (RNA-seq, track 11) in WT NKT cells are shown to contextualize methylation changes. The heterochromatic regions that lose DNA methylation in Tet2/3 DKO T cells are highlighted.
Fig. 5.
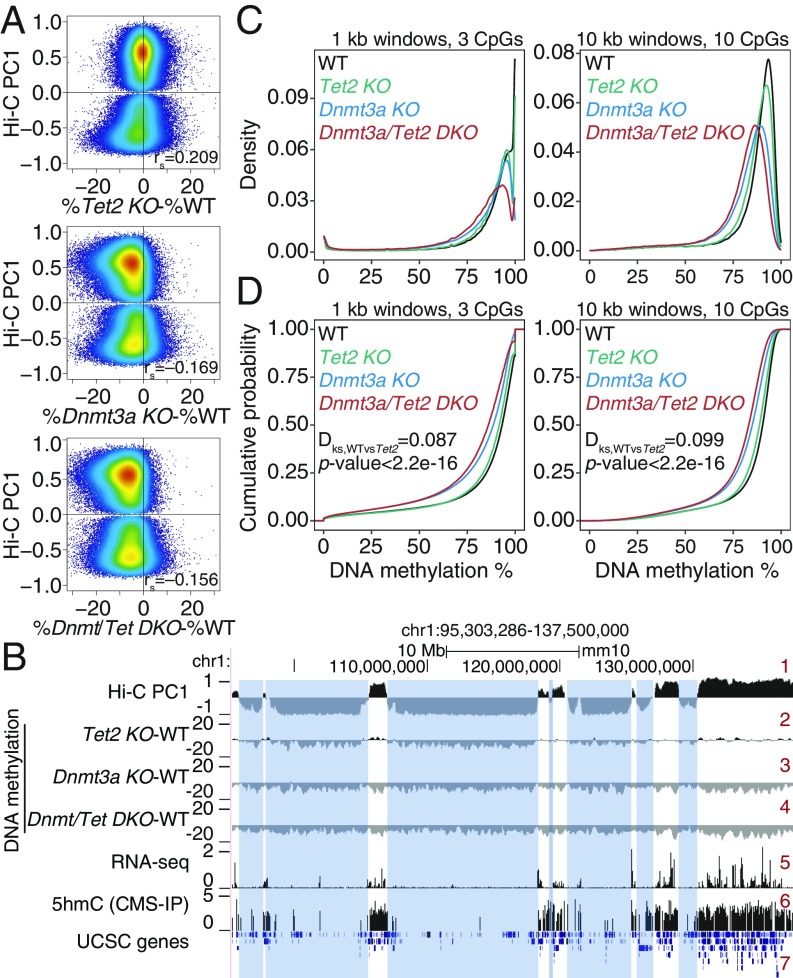
Increased DNA hypomethylation in double DNMT3A and TET2 mutant mice. (A) Correlation between DNA methylation changes (mutant minus wild type [WT]) and euchromatin/heterochromatin compartments, in Tet2 KO (Top), Dnmt3a KO (Middle), and Dnmt3a/Tet2 DKO (Bottom) hematopoietic stem cells (HSPCs). Spearman correlation coefficient is shown (rs value). DNA methylation data from refs. 27 and 46. (B) Genome tracks showing a correspondence between the Hi-C B compartment (negative Hi-C PC1 values, track 1) and large hypomethylated domains in Tet2 KO (track 2). In contrast, Dnmt3a KO and Dnmt3a/Tet2 DKO show a global, compartment-independent DNA hypomethylation (tracks 3 and 4). Gene expression (track 5) and 5hmC distribution (track 6) in WT HSPCs are shown for reference. (C and D) Density (C) and cumulative distribution (D) of average DNA methylation values within 1-kb (Left) and 10-kb (Right) windows across the genome, in WT, Dnmt3a KO, Tet2 KO, and Dnmt3a/Tet2 DKO HSPCs.
We compared mESCs triply deficient in all 3 TET proteins (Tet1, Tet2, and Tet3; Tet TKO mESCs) with mESCs triply deficient in all 3 DNMTs (Dnmt1, Dnmt3a, and Dnmt3b; Dnmt TKO mESCs) (25) (Fig. 1A). We plotted the distribution of average DNA methylation values in 1- and 10-kb windows containing at least 3 and at least 10 CpGs, respectively (Fig. 1A). As expected, Dnmt TKO mESCs lost all DNA methylation (5mC+5hmC; Fig. 1 A, Left and Middle). Unexpectedly, however, Tet TKO mESCs also showed pronounced and widespread loss of methylation compared with WT ESCs, with the shift in the distribution of window methylation percentages visible at both 1- and 10-kb resolution (Fig. 1A, compare red and black traces). Hypomethylation in Tet TKO mESCs could also be visualized in dot plots comparing DNA methylation levels in WT and mutant ESCs (SI Appendix, Fig. S1A). Each dot shows DNA methylation levels in 1-kb windows with at least 3 CpGs: 73.7% of the 1-kb windows showed decreased methylation in Tet TKO mESCs compared with WT, whereas only 26.1% showed increased methylation (SI Appendix, Fig. S1A). Analysis of WGBS data from an independent study of Tet TKO mESCs, generated using CRISPR/Cas9 technology (15), yielded very similar results (Fig. 1B and SI Appendix, Fig. S1B). The differences in global hypomethylation observed between WT and Tet TKO mESCs in the 2 studies may be due to different culture conditions or different lengths of time that the cells were maintained in culture (compare Fig. 1 A and B).
DNA Hypermethylation in Euchromatin and Hypomethylation in Heterochromatin in Diverse TET-Deficient Cells.
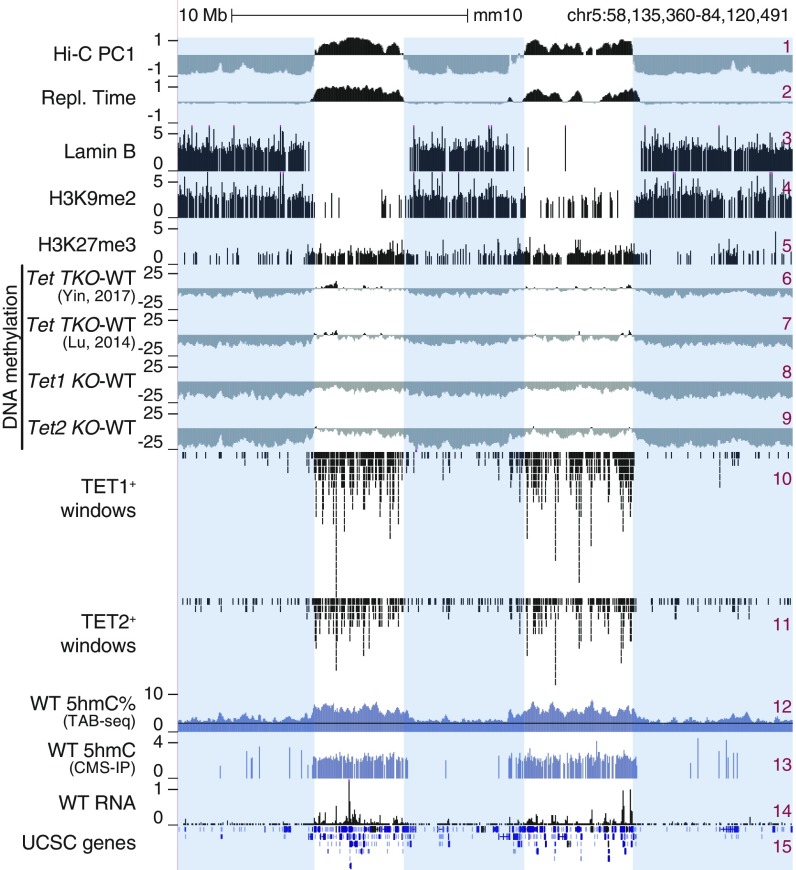
In genome browser views, DNA methylation changes were most striking when viewed at megabase-scale resolution (Fig. 2) and were reminiscent of the large PMDs noted in cancer genomes (20, 21). Drawing on publicly available Hi-C (29), replication timing (19), and Lamina B and H3K9me2 chromatin immunoprecipitation-sequencing (ChIP-seq) data (30) on mESCs, we found that regions that lost DNA methylation in all Tet-mutant mESCs (Fig. 2, tracks 6–9) mostly overlapped with the heterochromatic Hi-C B compartment (track 1), late replicating regions (track 2), lamina-associated domains (track 3), and regions marked by H3K9me2 (track 4). In contrast, H3K27me3 was present primarily in the euchromatic Hi-C A compartment (track 5). Likewise, TET1 and TET2 were primarily in the euchromatic compartment (tracks 10 and 11), consistent with the 5hmC distribution in WT mESCs, as shown by 2 independent methods of mapping 5hmC: TAB-seq (12) and CMS-IP (31) (Fig. 2, tracks 12 and 13; the background signal for TAB-seq is noisier than for CMS-IP). The preferential presence of 5hmC in the euchromatic Hi-C A compartment was observed in all mouse cell types investigated, including ESCs, HSPCs, and pro-B cells (Fig. 1C; for similar data on mouse T cells, see Fig. 3E). The majority of hypermethylated genomic regions in TET-deficient cells are located in the euchromatic compartment; this finding, coupled with the presence of TET1, TET2, and 5hmC in the same euchromatic compartment in WT cells, confirms the by now well-established connection between DNA demethylation, TET-generated 5hmC, and 5hmC localization in actively transcribed genes and enhancers (4).
Fig. 2.
DNA hypomethylation in heterochromatin compartment in TET-deficient mESCs. Genome tracks showing an overlap between Hi-C-defined A/B compartments (track 1), early/late replicating sites (early replicating: positive values; track 2), lamina-associated domains (track 3), regions marked by histone marks H3K9me2 (track 4) and H3K27me3 (track 5), and large hypomethylated domains in different Tet KO mESCs (shown as subtraction of DNA methylation percentage, mutant minus WT; tracks 6–9). TET1 and TET2 binding (ChIP-seq; tracks 10–11), 5hmC distribution (TAB-seq and CMS-IP; tracks 12–13), and gene expression (RNA-seq; tracks 14) in WT mESCs are shown for reference. Heterochromatic Hi-C B compartment regions are highlighted.
Dnmt TKO mESCs showed genome-wide hypomethylation as expected (Fig. 1 A, Right), but the gains and losses of DNA methylation in Tet TKO mESCs mostly occurred in the euchromatic and heterochromatic compartments, respectively (Figs. 1 A and B and 2, tracks 6–7; SI Appendix, Fig. S1 A and B). Even in singly deficient Tet1 KO and Tet2 KO mESCs (12), there is striking global DNA hypomethylation in both the euchromatic and heterochromatic compartments, most notably in the latter, which far exceeded the extent of the expected DNA hypermethylation (Figs. 1D and 2, tracks 8–9). Hypermethylated genomic regions were clearly apparent in Tet2 KO mESCs, but there was only marginal hypermethylation in Tet1 KO mESCs (12) (Fig. 1D and SI Appendix, Fig. S1C). Even when Tet2 KO mESCs were differentiated to NPCs for 3 d (12), the differentiated NPCs showed heterochromatin hypomethylation (SI Appendix, Fig. S2 A and B).
To ask whether our findings were generally applicable to other cell types, we integrated previously published Hi-C data from mouse pro-B cells (32) with WGBS data from WT or Tet2−/− Tet3fl/fl Mb1Cre pro-B cells (26) (SI Appendix, Fig. S2 C–E). Deficiency of Tet2 and Tet3 in Tet2−/− Tet3fl/fl Mb1Cre mice results in defective immunoglobulin light chain rearrangement and a consequent block of B cell development at the pro-B to pre-B transition (26). Again, we observed concurrent increases and decreases of DNA methylation in pro-B cells genome-wide, with hypermethylation again occurring in the euchromatic compartment and hypomethylation in the heterochromatic compartment (SI Appendix, Fig. S2 C–E). Very similar results were obtained for mouse T cells and HSPCs (see below; see Figs. 3 and 5).
Antigen-Driven Expansion, Increased Clonality, and DNA Damage in TET-Deficient T Cell Leukemia/Lymphoma.
To study hypomethylation induced by TET loss of function in the context of oncogenic transformation, we used a mouse model in which mice with profound TET loss of function (Tet2−/− Tet3fl/fl CD4Cre [Tet2/3 DKO] lacking Tet2 and Tet3 in T cells) rapidly developed an aggressive T cell leukemia/lymphoma with 100% penetrance (14). The disease involves a normally minor subset of T cells (iNKT cells, hereafter referred to simply as NKT cells), which recognize lipid antigens presented on a nonclassical major histocompatibility complex (MHC) protein known as CD1d (33). Tet2/3 DKO mice showed >10-fold expansion of NKT cells in the thymus as early as 20 d after birth and in the spleen by 3–4 wk, and succumbed to an NKT cell leukemia starting at 5 wk (14). Transfer of purified NKT cells from young mice, even into fully immunocompetent recipients, resulted in transfer of the leukemia, but transfer to recipient mice lacking CD1d, the MHC protein that presents lipid antigen to NKT cells (33), resulted in minimal expansion (14) (SI Appendix, Fig. S3A), indicating that the leukemia was driven by NKT-cell expansion arising from presentation of lipid antigens by CD1d. The leukemia was transmissible indefinitely, and secondary transfers could be performed with as few as 50 cells (SI Appendix, Fig. S3 A and B).
Sequencing of T cell receptor β-chain variable regions showed that Tet2/3 DKO NKT cells were oligoclonal even in young mice; after transfer to recipient mice, they displayed a remarkable increase in number (SI Appendix, Fig. S3B) and clonality (SI Appendix, Fig. S3C), likely due to due to strong selective expansion of cells with specific genomic characteristics (see below). Given that the expanded cells were effectively monoclonal and constituted >95% of lymphocytes in the peripheral lymphoid organs of recipient mice, we were able to perform whole-genome sequencing (WGS), together with WGBS and Hi-C, at relatively low cost.
TET-Deficient T Cell Lymphomas Show Euchromatic Compartment Hypermethylation and Heterochromatic Compartment Hypomethylation.
To define DNA modification patterns in the active and inactive genome compartments of TET-deficient NKT-cell lymphomas, we generated and analyzed WGBS and Hi-C data for WT, young, and transferred and expanded Tet2/3 DKO NKT cells. Like TET-deficient mESCs and pro-B cells (Figs. 1 and 2 and SI Appendix, Figs. S1 and S2), both young and expanded Tet2/3 DKO NKT cells showed increased methylation in the euchromatic compartment and decreased methylation in the heterochromatic compartment relative to WT NKT cells (Fig. 3 A–C and SI Appendix, Fig. S4 A and B). Moreover, as in the cell types considered above, the euchromatic compartment was gene-rich and contained the majority of expressed genes, accessible chromatin regions, and 5hmC (Fig. 3 D and E and SI Appendix, Fig. S4C; also see Fig. 3F, tracks 9–11). In genome browser views, there were only minor changes in euchromatic and heterochromatic compartments between WT thymus and spleen (Fig. 3F, tracks 1 and 2, and SI Appendix, Fig. S4D, Top Left), but somewhat greater differences between WT and expanded Tet2/3 DKO splenic NKT cells (Fig. 3F, tracks 2, 3; SI Appendix, Fig. S4D, Bottom). Windows that were less accessible in Tet2/3 DKO NKT cells compared with WT were primarily in the euchromatic compartment; the few windows that were more accessible in Tet2/3 DKO cells compared with WT were present in the heterochromatic compartment (Fig. 3D). Finally, both young and transferred/expanded Tet2/3 DKO NKT cells showed extended regions of increased and decreased methylation compared with WT NKT cells, and these largely coincided with the euchromatic and heterochromatic compartments, respectively (Fig. 3F, tracks 4–8).
Overall, the data on TET-deficient NKT-cell lymphomas were completely concordant with those for the other TET-deficient cell types considered above. Regardless of cell type, TET deficiency was broadly associated with DNA hypermethylation in the euchromatic compartment, concurrently with DNA hypomethylation in the heterochromatic compartment. In the remainder of this study, we examine other genomic features of Tet2/3 DKO NKT cells reported to be associated with hypomethylation, including reactivation of repeat elements and increased mutational load.
Mutational Signatures in TET-Deficient T Cell Lymphomas.
Hypomethylation has been previously associated with increased mutation rates (34) and genome instability (35, 36), and we and others have observed increased levels of DNA damage after TET deletion (9, 16). Expansion of Tet2/3 DKO NKT cells after transfer was accompanied by a striking increase in DNA double-strand breaks (DSBs): Expanded Tet2/3 DKO NKT cells showed increased staining for γH2AX, compared with WT NKT cells (SI Appendix, Fig. S3 D and E). In contrast, Tet2/3 DKO NKT cells transferred to CD1d KO recipient mice, which undergo only minimal expansion (14) (SI Appendix, Fig. S3A), displayed only a slight increase in DSBs compared with WT NKT cells (SI Appendix, Fig. S3 D and E). These results are consistent with the reports of increased DNA DSBs in Tet1-deficient B cells (9) and in acute myeloid leukemias resulting from inducible deletion of Tet2 and Tet3 (16).
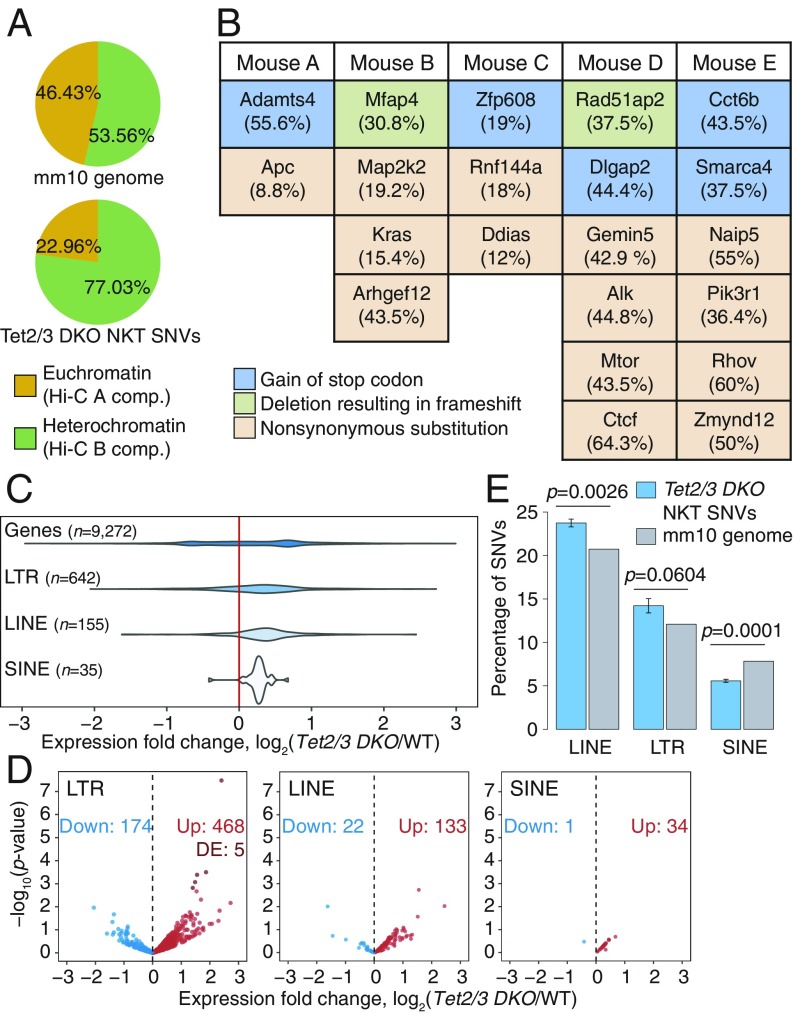
We performed WGS at >20× coverage. We examined our WGS data to identify SNVs in Tet2/3 DKO NKT cells. WGS on expanded NKT cells showed that most SNVs occurred in the heterochromatic compartment, which constitutes 54% of the genome but contains 77% of the SNVs (Fig. 4A). Furthermore, the SNVs identified in gene coding regions in the 5 different samples of transferred and expanded NKT cells from independent recipient mice (Fig. 4B) were not recurrently observed, suggesting that the selective advantage potentially conferred by any given SNV is limited to individual mice. Thus, most SNVs observed in Tet2/3 DKO NKT cells after transfer and expansion arise through a stochastic process occurring primarily in the heterochromatic compartment, as also observed for H3K9me3-marked heterochromatic genome regions in human cancers (24).
Fig. 4.
Transposable element (TE) reactivation and increased mutations predominate in heterochromatin of TET-deficient NKT-cell lymphoma. (A) Genome-wide distribution (Top), and percentage of SNVs (Bottom) located within euchromatin and heterochromatin compartments in Tet2/3 DKO expanded NKT cells. (B) Coding mutations resulting in change in the amino acid sequence in 5 independent WGS samples from Tet2/3 DKO expanded NKT cells. Mutant allele frequencies are shown in parentheses. (C) Distribution of the changes in expression (log2 fold change) of TEs belonging to the LTR, LINE, and SINE families in Tet2/3 DKO young NKT cells compared with WT, obtained from analysis of total RNA-seq data. Fold change differences for all genes in the genome are shown for reference. (D) Volcano plots of expression of TEs belonging to the LTR, LINE, and SINE families, in Tet2/3 DKO young NKT cells compared with WT. Differentially expressed (DE) TEs (adjusted P value < 0.1) are highlighted. (E) Percentage of SNVs in Tet2/3 DKO expanded NKT cells that overlap with LINEs, LTRs, and SINEs, compared with the mm10 genome distribution of each TE families. P values were calculated by 1-sample t test.
The mutational signature of the SNVs, based on nucleotide substitutions and sequence context at the 5′ and 3′ ends (37), clustered separately between the euchromatic and heterochromatic compartments (SI Appendix, Fig. S5 A and B). In both compartments, the signature was predominantly characterized by transitions (C>T, T>C, G>A, and A>G). Even though SNVs in general were more prevalent in the heterochromatic compartment, SNVs at cytosines in the CpG context were more prevalent in the hypermethylated euchromatic compartment (14%) compared with the hypomethylated heterochromatic compartment (8.6%) (SI Appendix, Fig. S5A, compare C>T red bars in Top and Bottom), as expected from the tendency of 5mC to undergo spontaneous deamination (37). Indeed, for CpGs for which DNA methylation data were available from WGBS analysis, we observed that C>T mutations in the CpG context occurred at CpG sites that were largely methylated (SI Appendix, Fig. S5C). Rainfall plots of intermutational distance against genomic location from 3 independent Tet2/3 DKO mice showed that mutations were often clustered in similar chromosomal locations but did not occur at the same nucleotides (SI Appendix, Fig. S5D).
Reactivation of Transposable Elements in TET-Deficient T Cell Lymphomas.
DNA hypomethylation has been widely associated with reactivation of transposable elements (TEs) (38). In light of the hypomethylation in heterochromatin of Tet2/3 DKO NKT cells, we analyzed the expression levels of distinct families of TEs in young Tet2/3 DKO NKT cells by RNA-seq (Fig. 4 C and D), keeping in mind that long interspersed elements (LINEs) are primarily located in heterochromatin while short interspersed elements (SINEs) are found in euchromatin (18). Indeed, using RNA-seq datasets from our previous study (14) as well as newly generated RNA-seq data from total ribodepleted RNA (Fig. 4 C and D and SI Appendix, Fig. S5E), we found that for those TEs for which we were able to detect transcripts reliably in at least 1 biological replicate, long terminal repeats (LTR) retrotransposons and LINEs were more highly expressed in Tet2/3 DKO NKT cells with respect to WT, whereas SINEs remained largely unchanged. Furthermore, a substantial fraction of the identified mutations fell within TEs, such as LINEs and LTRs, but appeared underrepresented in SINEs with respect to the genome average (Fig. 4E). These data support the hypothesis that the reactivation of LINEs and LTRs results from TET-associated hypomethylation occurring in heterochromatin, whereas the euchromatic compartment undergoes TET-associated hypermethylation and therefore most SINEs remain silent.
Reactivation and spurious transcription of repeat elements have been associated with formation of R-loops and genome instability (39, 40), linked to DNA damage and DNA DSBs (41). Indeed, we found an increase of R-loops in expanded Tet2/3 DKO compared with WT NKT cells, as detected by flow cytometry and DNA dot blots using the S9.6 antibody against RNA:DNA hybrids (42) (SI Appendix, Fig. S3 F and G). Further analysis will require methods for mapping DSBs and R-loops that can be applied to the very small numbers of NKT cells that can be isolated from normal WT mice, so as to determine whether these features show differential distribution in the euchromatic and heterochromatic compartments in WT versus TET-deficient cells.
Paradoxical Increase in Heterochromatic DNA Hypomethylation in HSPCs from Dnmt3a-Tet2 DKO Mice.
DNMT3A and TET2 mutations are frequently observed, both individually and together, in diverse myeloid and lymphoid malignancies (43, 44). Based on the biochemical activities of the encoded proteins, we originally expected opposing effects of DNMT3A and TET2 loss of function, both in terms of biological outcomes and in genome-wide analyses. Specifically, we expected first that the disease phenotypes of Dnmt3a and Tet2-deficient mice might be ameliorated in mice deficient for both enzymes, and second that there would be widespread losses of DNA methylation in the case of Dnmt3a mutations but widespread gains of DNA methylation in the case of Tet2 mutations. However, a previous collaborative study (27) unexpectedly showed that mice with dual Tet2 and Dnmt3a deficiency in HSPCs displayed more severe phenotypes than mice with individual Tet2 or Dnmt3a deletions alone.
Given these data, we postulated that one explanation for the paradoxical synergy between Dnmt3a and Tet2 mutations might be heterochromatic hypomethylation in both types of mutant cells. To test this hypothesis, we used previously published Hi-C data on WT HSPCs (45) and WGBS data for WT, Tet2 KO, Dnmt3a KO, and Dnmt3a/Tet2 DKO HSPCs (27, 46) to localize DNA methylation changes to the 2 genomic compartments defined by Hi-C (Fig. 5 A and B, top track). Indeed, both Tet2 and Dnmt3a deficiency were characterized by widespread losses of DNA methylation in HSPCs; moreover, HSPCs from doubly Tet2/Dnmt3a-deficient mice showed greater hypomethylation than HSPCs with either mutation alone (Fig. 5). The synergistic loss of DNA methylation was striking when 10-kb windows were considered (Fig. 5 C and D, Right), although the small fraction of fully methylated regions as well as completely or almost completely demethylated regions (e.g., CpG islands) were best observed in 1-kb windows (Fig. 5 C and D, Left). Specifically, Dnmt3a-deficient (Dnmt3a KO) HSPCs displayed greater loss of methylation compared with Tet2-deficient (Tet2 KO) HSPCs as expected, but HSPCs doubly deficient in Tet2 and Dnmt3a (Dnmt3a/Tet2 DKO) showed even greater loss of methylation compared with HSPCs singly deficient in either enzyme alone (Fig. 5 C and D).
In genome browser views, extended domains of hypomethylation were observed in Tet2 KO HSCs (Fig. 5B, track 2), as in all of the other cell types considered above (Figs. 1–3 and SI Appendix, Figs. S1 and S2). These PMDs mostly overlapped with the heterochromatic compartment, whereas regions in the euchromatic compartment showed a slight gain of methylation (Fig. 5A, Top, and Fig. 5B, track 2). In contrast, in both the Dnmt3a KO and Dnmt3a/Tet2 DKO HSPCs (Fig. 5A, Middle and Bottom, and Fig. 5B, tracks 3 and 4), we observed widespread DNA hypomethylation in both the euchromatic and heterochromatic compartments, as expected from the loss of Dnmt3a activity. Thus, both Tet2 and Dnmt3a mutations result in widespread hypomethylation in heterochromatic regions of the genome.
Decreased Localization of DNMT3A in the Heterochromatin Compartment of TET-Deficient ESCs.
We investigated the mechanisms underlying the loss of methylation in heterochromatin in TET-deficient cells. One potential mechanism stems from the observation that similar hypomethylated domains are observed in rapidly proliferating cells (22). This argument is plausible for the NKT-cell lymphoma in which TET deficiency accounts for rapid proliferation but cannot apply in the case of ESCs. PMDs are not observed in WT ESCs and induced pluripotent stem cells despite the high proliferation rates of these cells (47); moreover, Tet1/2/3 TKO mESCs do not proliferate faster than their WT counterparts (figure S6B from ref. 48 reproduced here in SI Appendix, Fig. S6A for the reader’s convenience) but clearly show decreased DNA methylation at late-replicating, lamina-associated domains (Fig. 2). Finally, senescent cells that have stopped proliferating also show partially methylated domains, with loss of methylation occurring predominantly in the heterochromatic compartment (49).
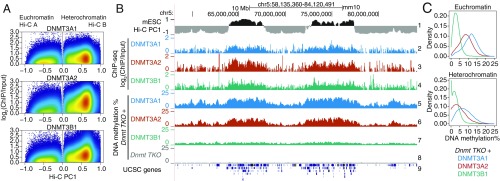
Since TET-deficient mESCs showed heterochromatin hypomethylation without increased proliferation, we asked whether hypomethylation in mESCs could be attributed to alterations of DNMT localization or function. We focused on the de novo DNMTs, DNMT3A, and DNMT3B, because of a previous report in which they were both shown by immunocytochemistry to colocalize with HP-1a in the heterochromatic compartment of mouse 3T3 cells and ESCs (50). To infer the contribution of each DNMT to methylation in the euchromatic and heterochromatic compartments, we reanalyzed a dataset in which DNMT3B1 and the 2 splice variants DNMT3A1 and DNMT3A2 were reconstituted in mESCs lacking all DNMTs (51, 52). Mapping of these 3 DNMT3 proteins in WT mESCs showed that all 3 were primarily present in the euchromatic compartment but were also significantly represented in the heterochromatic compartment (Fig. 6 A and B, tracks 1–4). WGBS performed on the DNMT-reconstituted cells showed that all 3 DNMT3 proteins contributed to methylation in both the euchromatic and heterochromatic compartments (Fig. 6B, tracks 5–8, and Fig. 6C). Notably, the major contribution to DNA methylation in the heterochromatic compartment was from DNMT3A1 (Fig. 6C).
Fig. 6.
Contributions of DNMT3 enzymes to DNA methylation in euchromatin and heterochromatin. (A) Genome-wide distribution of the de novo DNA methyltransferases DNMT3A1, DNMT3A2, and DNMT3B1 within euchromatin and heterochromatin compartments in WT mESCs. DNMT ChIP-seq enrichment (log2 fold change) was calculated at 1-kb resolution. Data from refs. 51 and 52. (B) Genome tracks showing the distribution of DNMT3A1, DNMT3A2, and DNMT3B1 (tracks 2–4) within Hi-C defined compartments (track 1), and the contribution to DNA methylation of the de novo DNMT proteins after reconstitution in a Dnmt TKO background in mESCs (tracks 5–7). Dnmt TKO methylation is shown for reference (track 8). (C) Density distribution of average DNA methylation values within 10-kb windows across the euchromatin and heterochromatin compartments, in Dnmt TKO mESCs reconstituted with DNMT3A1, DNMT3A2, and DNMT3B1.
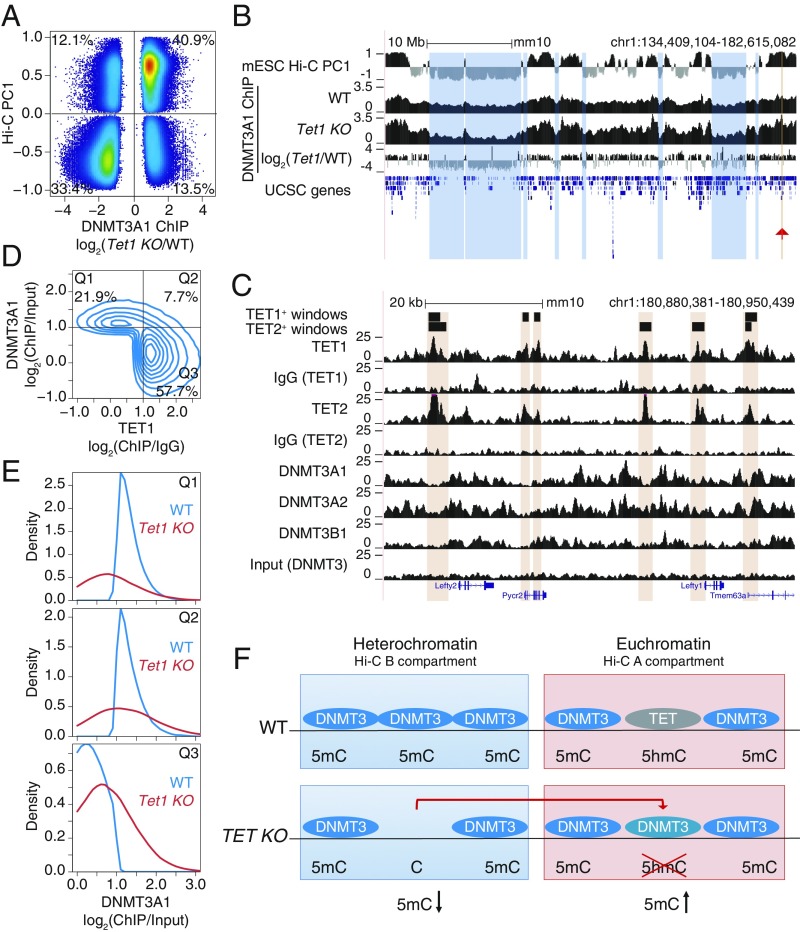
Heterochromatic hypomethylation in TET-deficient cells could reflect either altered distribution or function of DNMT3A1 (these 2 scenarios are not mutually exclusive). To examine alterations in DNMT3A1 localization, we used a dataset from a study in which DNMT3A1 tagged with the biotin acceptor peptide for Escherichia coli BirA was expressed in WT and Tet1-deficient mESCs (53). The data show unambiguously that, compared with WT mESCs, DNMT3A1 was enriched in the euchromatic compartment and depleted from the heterochromatic compartment in Tet1-deficient mESCs (Fig. 7A). A genome browser view illustrating the relocalization is shown in Fig. 7B; tracks 2 and 3 show the normalized ChIP-seq coverage, and track 4 shows the difference in DNMT3A1 binding in WT vs. Tet1-deficient mESCs. Together, these data indicate that DNMT3A1 relocalizes from the heterochromatic to the euchromatic compartment in Tet1-deficient mESCs.
Fig. 7.
Relocalization of DNMT3A away from the heterochromatin compartment of TET-deficient mESCs. (A) Comparison of changes in DNMT3A1 binding in Tet1 KO versus WT mESCs, and euchromatin versus heterochromatin compartments (Hi-C PC1 values) in WT mESCs. Differential binding was calculated at 1-kb windows and displayed are those with a value of P < 0.05. Percentage of windows within each quadrant is indicated. (B) Genome tracks showing a correspondence between the heterochromatic Hi-C B compartment (track 1) and decrease in DNMT3A1 occupancy in Tet1 KO with respect to WT (tracks 2–4). Tracks 2 and 3 show DNMT3A1 occupancy in WT and Tet1 KO mESCs, respectively (shown as reads per kilobase per million reads). Track 4 shows the difference in DNMT3A1 occupancy (visualized as log2 fold-change Tet1 KO/WT). A region containing the Lefty1/2 locus is highlighted by the red arrowhead and is shown in C. (C) Zoomed-in view of the Lefty1/2 locus within the euchromatic compartment. Genome tracks showing mutually exclusive localization of TET1 and TET2 (tracks 1–6) and de novo DNMT proteins (tracks 7–10). (D) Mutually exclusive localization between DNMT3A1 and TET1 binding in the euchromatic Hi-C A compartment in WT mESCs. Percentage of windows within each quadrant is indicated. ChIP-seq enrichment (log2 fold change) was calculated for 1-kb windows. DNMT3A1 data from ref. 53; TET1 data from ref. 54. (E) Distribution of DNMT3A1 occupancy in WT and Tet1 KO mESCs (displayed as enrichment over input) within each of the quadrants defined in D. (F) Schematic model illustrating that loss of TET1 in mESCs results in the relocalization of DNMT3A1, on the one hand from the heterochromatic to the euchromatic compartment as shown in B, and on the other hand to regions within euchromatin that were previously occupied by TET1 as shown in D and E.
We used the same datasets described above (51–54) to determine how DNMT3A1 relocalized within the euchromatin compartment in Tet1-deficient mESCs. A zoomed-in view of the Lefty1/2 locus within the euchromatic compartment revealed strong mutually exclusive localization of TET1/2 and DNMT3A (Fig. 7C). A contour plot illustrating this mutually exclusive localization is shown in Fig. 7D; Q1 contains the regions in the euchromatic Hi-C A compartment occupied by DNMT3A1 but not by TET1, whereas Q3 contains the regions occupied by TET1 but not by DNMT3A1. In Tet1-deficient mESCs, this distribution is altered: The density plots for Q1 show that binding of DNMT3A1 to its exclusive sites in WT mESCs is substantially decreased in Tet1 KO mESCs, whereas the density plots for Q3 show that DNMT3A1 shows increased binding in Tet1 KO compared with WT mESCs (Fig. 7E). This mutually exclusive localization seems to be a general feature of the relation between TET proteins and DNMT3s (SI Appendix, Fig. S6 B and C), as previously noted for TET1 and DNMT3A1 by ref. 53. Together, the data indicate that loss of TET1 (almost exclusively from the euchromatic compartment) in mESCs results in relocalization of DNMT3A1, on the one hand from the heterochromatic to the euchromatic compartment and on the other hand to regions within euchromatin that were previously occupied by TET1 (Fig. 7F). It is plausible that this relocalization contributes at least partly to the paradoxical loss of DNA methylation in the heterochromatic compartment that we observe in TET-deficient cells, as well as to the hypermethylation observed in the euchromatic compartment of the same cells.
Discussion
In this study, we document an unexpected similarity between the DNA methylation patterns of diverse TET-deficient cell types and those of cancer genomes. Cancer genomes show local hypermethylation combined with widespread hypomethylation (23), and we reproducibly observed both features in TET-deficient cells. As expected from the biochemical activities of TET enzymes in generating oxi-mC bases and their involvement in DNA demethylation (4), local DNA hypermethylation was consistently observed in the euchromatic Hi-C A compartment of TET-deficient cells; this compartment contains the vast majority of 5hmC, a stable modification that is most highly enriched in the gene bodies of the most highly expressed genes and at the most active enhancers (12, 14). Unexpectedly, however, we also observed large domains of DNA hypomethylation in the heterochromatic Hi-C B compartment of diverse TET-deficient cell types, including ESCs, NPCs, HSPCs, T cells, and pro-B cells. The existence of these hypomethylated domains cannot be explained by our current understanding of TET enzymatic activity, but their presence in TET-deficient cells suggests strongly that TET proteins are required, directly or indirectly, for optimal DNMT-mediated DNA methylation in heterochromatin.
To explore the biological consequences of TET loss of function in vivo, we used a mouse model of profound TET deficiency in T cells. Mice with deletion of Tet2 and Tet3 genes in T cells showed early signal-dependent expansion and increased clonality, which rapidly progressed to an aggressive NKT-cell lymphoma. The expanded Tet2/3 DKO NKT cells developed the same aberrations in DNA methylation—hypermethylation in the euchromatic compartment and hypomethylation in the heterochromatic compartment—that occur in cancer genomes and that we have noted above for multiple TET-deficient cell types. The cells accumulated SNVs, largely in the hypomethylated heterochromatic compartment through an apparently stochastic process that differed in each individual mouse. We also observed reactivation of TEs, particularly LTRs and LINEs that are primarily located in heterochromatin; these repetitive elements were also more prone to mutations compared with the genome in general, recalling the genome instability produced by spurious transcription of repeat elements (39, 40). As described in more detail below, DNA hypomethylation in heterochromatin may at least partly explain the oncogenic transformation, genome instability, and DNA damage observed in diverse mouse models of partial or profound TET deficiency (9, 16). The latency and penetrance of oncogenic transformation in these models likely depend on the extent of TET loss of function. Loss-of-function mutations in DNMT3A or TET2 are associated with clonal hematopoiesis in humans (55); similarly, TET deficiency in mouse models promotes the clonal expansion of TET-deficient cells. In both cases, full-blown oncogenesis requires the stochastic appearance of second hit mutations that vary from cell to cell but are subject to selection, driving clonal expansion and cancer evolution and explaining cancer heterogeneity.
The large hypomethylated domains we observe in the heterochromatic compartment of TET-deficient cells are very reminiscent of the extended PMDs observed in cancer genomes. Based on their overlap with nuclear lamina-associated, late-replicating domains, cancer-associated PMDs occur in the heterochromatic compartment (20, 21); their presence has been attributed to ineffective DNMT1-mediated remethylation of late-replicating genomic regions in rapidly proliferating cells (22). PMDs have also been documented in CD4+ T cells from a 103-y-old individual compared with those from a newborn human (22), suggesting that DNA methylation is also progressively lost in the heterochromatin of cells undergoing sustained long-term proliferation. While the presence of hypomethylated domains in heterochromatin of Tet2/3 DKO compared with WT NKT cells may indeed be a consequence of more rapid proliferation, especially since expanded Tet2/3 DKO NKT cells that have undergone many more cell divisions show more extensive hypomethylation than Tet2/3 DKO NKT cells from young mice (Fig. 3F), the PMDs observed in TET-deficient mESCs cannot be explained by increased proliferation (SI Appendix, Fig. S6A). Moreover, PMDs have also been observed in senescent IMR90 cells, which are no longer engaged in active proliferation (49). Thus, increased proliferation might contribute to, but is not the only mechanism underlying, the widespread losses of DNA methylation in heterochromatin of TET-deficient cells.
DNA hypomethylation has been associated with many biological consequences, including reactivation of TEs (38), sharply increased mutation rates (34), and genome instability with chromosome segregation defects and aneuploidies (35, 36). Mice with a hypomorphic mutation in Dnmt1 displayed genome-wide hypomethylation in all tissues and developed T cell lymphomas that occurred in 80% of mice and were characterized by recurrent aneuploidies (36). Reactivation of TEs is prevalent in cancer genomes, and is associated with the formation of RNA–DNA hybrids and R-loops (39, 40), which in turn have been linked to DNA damage and the appearance of DNA DSBs (41). Each of these features was observed, together with heterochromatin hypomethylation, in expanded Tet2/3 DKO NKT cells. Thus, in addition to their well-established role in promoting and maintaining DNA demethylation at promoters, gene bodies, and enhancers, TET proteins participate in maintaining physiological levels of DNA methylation in heterochromatic compartments of the genome.
Our findings may explain the unexpected synergy between TET2 and DNMT3A mutations in humans as well as mice. TET2 and DNMT3A are recurrently comutated in a diverse range of myeloid and lymphoid malignancies (43, 44). In a previous collaborative study, we found that the phenotypes of mice with dual Tet2 and Dnmt3a deficiency in HSPCs were considerably more severe than those of mice with individual Tet2 or Dnmt3a deletions alone (27). Dnmt3a and Tet2 deficiency would both result in loss of oxi-mC at specific genomic regions, through a direct decrease in DNA methylation in the case of Dnmt3a deficiency and through loss of the 5hmC substrate in the case of Tet2 deficiency. Thus, the stronger defects (e.g., in erythrocyte differentiation) in Tet2/Dnmt3a DKO mice compared with mice with Tet2 or Dnmt3a deficiency alone (27) could potentially arise from loss of “cooperation” between DNMT3A and TET2, leading to decreased 5hmC and increased 5mC at specific euchromatic locations (promoters, gene bodies, enhancers) in both humans and mice. Based on our data, however, we speculate that pronounced DNA hypomethylation in the heterochromatic compartment of Tet2/Dnmt3a DKO HSPCs (Fig. 5) could also be a major contributor to the observed synergy of oncogenic transformation resulting from loss-of-function mutations of both Dnmt3a and Tet2 (27).
Our reanalysis of published data suggests a potential mechanism for the synergistic actions of DNMT3A and TET proteins. TET1 and DNMT3A occupy mutually exclusive locations in the euchromatic compartment of mESCs, and loss of TET proteins from euchromatin results in relocalization of DNMT3A1 to regions previously occupied by TET1 (see model in Fig. 7F). Broadly, this observation suggests that the DNMT3 enzymes responsible for de novo methylation are recruited to euchromatin through a scaffold complex or other recruitment mechanism in common with TET enzymes, but for which the DNMTs have lower affinities than TETs under normal physiological conditions. Assuming that the DNMT3 enzymes are present at limiting concentrations, loss of TET proteins would cause them to relocalize away from heterochromatin and into euchromatic regions, resulting in the observed dual loss of DNA methylation in heterochromatin and increased DNA methylation in euchromatin. This observation is consistent with the notable finding that every animal genome that encodes a DNMT also harbors at least 1 functional TET/JBP protein (56). Further studies in specific cancer models will be required to advance our global understanding of the biochemistry underlying the functional interactions between TETs and DNMTs.
Our data suggest that loss of DNA methylation in heterochromatin results in “heterochromatin dysfunction” (57). This phenomenon has many manifestations, including aneuploidies resulting from chromosome instability related to centromere dysfunction, as observed in immunodeficiency/centromere instability/facial abnormalities (ICF) patients with germline DNMT3B mutations (58), as well as reactivation of TEs and increased R-loops. These features are all observed in Tet2/3 DKO NKT cells, as well as in cells with hypomorphic mutations in DNMT1 (36) (the increased copy number variations and aneuploidies observed in Tet2/3 DKO NKT cells will be described in a separate study). Based on these considerations, we speculate that cancers related to TET loss of function are initiated at least partly through defects in the maintenance of heterochromatin function. By inference, the functional interactions between DNMT and TET proteins that we document here are likely to be important for maintaining heterochromatin integrity.
In many hematopoietic and most solid cancers, TET loss of function is observed without coding region mutations in TET genes (5, 10). Early studies suggested that TET loss of function was secondary to TET promoter methylation, increased degradation of TET proteins, or aberrant microRNA expression (9–11). More recently, however, TET loss of function in solid cancers has been increasingly attributed to hypoxia (59), or to a variety of metabolic alterations that decrease α-ketoglutarate levels or increase the levels of 2-hydroxyglutarate (10, 11). Thus, loss of DNA methylation in the heterochromatic compartment, and the consequent development of heterochromatin dysfunction, could be the first steps in the development of many cancers characterized by TET loss of function. Moreover, mutations in proteins associated with the maintenance of heterochromatin integrity are frequent in cancer and many of them (e.g., NPM1) co-occur with TET2 mutations, leading to the postulate that heterochromatin dysfunction is not only a common feature of sporadic (nonhereditary) human cancers but also potentially an initiating event in oncogenic transformation (57).
The methylation losses that we observe are fractional, only around 25% in our T cell lymphoma model, meaning that only a quarter of the total alleles in the transformed Tet2/3 DKO T cell population have lost the methyl mark at any given CpG. This heterogeneity of DNA methylation could affect the reactivation of TEs, the binding of methyl-sensitive proteins and transcription factors (60), thus potentially contributing to the initiating events of transformation. An interesting question that cannot be addressed with current (short reads) sequencing methods is whether DNA demethylation occurs concordantly across long regions of the genome, and if so, whether cells with broadly demethylated alleles are more subject to oncogenic transformation. In either case, however, heterogeneity of DNA methylation could make a substantial contribution to genome diversity, population heterogeneity, and clonal evolution in cancer genomes.
Methods
WGBS, Hi-C, and RNA-seq library preparations were performed as previously described. Detailed experimental protocols can be found in SI Appendix, SI Materials and Methods. Methods applied to analysis of WGBS, Hi-C, ChIP-seq, and RNA-seq data are described in detail in SI Appendix, SI Materials and Methods.
Data Availability.
Sequencing data produced as part of this study are available in the Gene Expression Omnibus (GEO) database under accession no. GSE134396 and the NCBI BioProject database under accession no. PRJNA555063. GEO and European Nucleotide Archive accession codes from the previously published sequencing data used in this study are available in SI Appendix, Table S2.
Supplementary Material
Acknowledgments
We thank C. Kim, L. Nosworthy, D. Hinz, and R. Simmons (La Jolla Institute for Immunology [LJI] Flow Cytometry Core) for help with cell sorting; J. Day, N. Wlodychak, and C. Kim (LJI Functional Genomics Center) for assistance with next-generation sequencing; and Dr. David Adams (Sanger Institute) for advice and discussions about the WGS mutation calling. This work was supported by National Institutes of Health (NIH) Grant R35 CA210043 to A.R. I.F.L.-M. was supported by a University of California Institute for Mexico and the United States–Consejo Nacional de Ciencia y Tecnología Fellowship. A.T. was supported by a Cancer Research Institute (CRI) Irvington Postdoctoral Fellowship. H.S. was supported by American Association for Cancer Research Genentech Immuno-oncology Research Fellowship and the CRI Irvington Fellowship. B.D. was supported by a Jane Coffin Childs Memorial Fund Postdoctoral Fellowship. Funding for Illumina HiSeq 2500 and BD FACSAria II is supported by NIH (NIH S10OD016262 and NIH S10RR027366). Hi-C sequencing was supported by the Salk Institute’s NGS Core Facility with funding from NIH-NCI Cancer Center Support Grant P30 014195, the Chapman Foundation and the Helmsley Charitable Trust.
Footnotes
Conflict of interest statement: A.R. is on the scientific advisory board of Cambridge Epigenetix (Cambridge, UK). The other authors declare no competing interests.
Data deposition: The sequencing data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE134396), and the NCBI BioProject database, https://www.ncbi.nlm.nih.gov/bioproject (accession no. PRJNA555063).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1903059116/-/DCSupplemental.
References
- 1.Tahiliani M., et al. , Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ito S., et al. , Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333, 1300–1303 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.He Y. F., et al. , Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333, 1303–1307 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu X., Zhang Y., TET-mediated active DNA demethylation: Mechanism, function and beyond. Nat. Rev. Genet. 18, 517–534 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Ko M., et al. , Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature 468, 839–843 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delhommeau F., et al. , Mutation in TET2 in myeloid cancers. N. Engl. J. Med. 360, 2289–2301 (2009). [DOI] [PubMed] [Google Scholar]
- 7.Langemeijer S. M., et al. , Acquired mutations in TET2 are common in myelodysplastic syndromes. Nat. Genet. 41, 838–842 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Lemonnier F., et al. , Recurrent TET2 mutations in peripheral T-cell lymphomas correlate with TFH-like features and adverse clinical parameters. Blood 120, 1466–1469 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Cimmino L., et al. , TET1 is a tumor suppressor of hematopoietic malignancy. Nat. Immunol. 16, 653–662 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ko M., An J., Rao A., DNA methylation and hydroxymethylation in hematologic differentiation and transformation. Curr. Opin. Cell Biol. 37, 91–101 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raffel S., et al. , BCAT1 restricts αKG levels in AML stem cells leading to IDHmut-like DNA hypermethylation. Nature 551, 384–388 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Hon G. C., et al. , 5mC oxidation by Tet2 modulates enhancer activity and timing of transcriptome reprogramming during differentiation. Mol. Cell 56, 286–297 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flavahan W. A., et al. , Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature 529, 110–114 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsagaratou A., et al. , TET proteins regulate the lineage specification and TCR-mediated expansion of iNKT cells. Nat. Immunol. 18, 45–53 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu F., Liu Y., Jiang L., Yamaguchi S., Zhang Y., Role of Tet proteins in enhancer activity and telomere elongation. Genes Dev. 28, 2103–2119 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.An J., et al. , Acute loss of TET function results in aggressive myeloid cancer in mice. Nat. Commun. 6, 10071 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lieberman-Aiden E., et al. , Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Steensel B., Belmont A. S., Lamina-associated domains: Links with chromosome architecture, heterochromatin, and gene repression. Cell 169, 780–791 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hiratani I., et al. , Global reorganization of replication domains during embryonic stem cell differentiation. PLoS Biol. 6, e245 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berman B. P., et al. , Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina-associated domains. Nat. Genet. 44, 40–46 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hon G. C., et al. , Global DNA hypomethylation coupled to repressive chromatin domain formation and gene silencing in breast cancer. Genome Res. 22, 246–258 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou W., et al. , DNA methylation loss in late-replicating domains is linked to mitotic cell division. Nat. Genet. 50, 591–602 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baylin S. B., Jones P. A., Epigenetic determinants of cancer. Cold Spring Harb. Perspect. Biol. 8, a019505 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schuster-Böckler B., Lehner B., Chromatin organization is a major influence on regional mutation rates in human cancer cells. Nature 488, 504–507 (2012). [DOI] [PubMed] [Google Scholar]
- 25.Yin Y., et al. , Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science 356, eaaj2239 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lio C. W., et al. , Tet2 and Tet3 cooperate with B-lineage transcription factors to regulate DNA modification and chromatin accessibility. eLife 5, e18290 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang X., et al. , DNMT3A and TET2 compete and cooperate to repress lineage-specific transcription factors in hematopoietic stem cells. Nat. Genet. 48, 1014–1023 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang Y., et al. , The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS One 5, e8888 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bonev B., et al. , Multiscale 3D genome rewiring during mouse neural development. Cell 171, 557–572.e24 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poleshko A., et al. , Genome-nuclear lamina interactions regulate cardiac stem cell lineage restriction. Cell 171, 573–587.e14 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang Y., et al. , Distinct roles of the methylcytosine oxidases Tet1 and Tet2 in mouse embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 111, 1361–1366 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin Y. C., et al. , Global changes in the nuclear positioning of genes and intra- and interdomain genomic interactions that orchestrate B cell fate. Nat. Immunol. 13, 1196–1204 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bendelac A., Savage P. B., Teyton L., The biology of NKT cells. Annu. Rev. Immunol. 25, 297–336 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Chen R. Z., Pettersson U., Beard C., Jackson-Grusby L., Jaenisch R., DNA hypomethylation leads to elevated mutation rates. Nature 395, 89–93 (1998). [DOI] [PubMed] [Google Scholar]
- 35.Eden A., Gaudet F., Waghmare A., Jaenisch R., Chromosomal instability and tumors promoted by DNA hypomethylation. Science 300, 455 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Gaudet F., et al. , Induction of tumors in mice by genomic hypomethylation. Science 300, 489–492 (2003). [DOI] [PubMed] [Google Scholar]
- 37.Alexandrov L. B., et al. ; Australian Pancreatic Cancer Genome Initiative; ICGC Breast Cancer Consortium; ICGC MMML-Seq Consortium; ICGC PedBrain , Signatures of mutational processes in human cancer. Nature 500, 415–421 (2013). Erratum in: Nature502, 258 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walsh C. P., Chaillet J. R., Bestor T. H., Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat. Genet. 20, 116–117 (1998). [DOI] [PubMed] [Google Scholar]
- 39.Zeller P., et al. , Histone H3K9 methylation is dispensable for Caenorhabditis elegans development but suppresses RNA:DNA hybrid-associated repeat instability. Nat. Genet. 48, 1385–1395 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Zhu Q., et al. , Heterochromatin-encoded satellite RNAs induce breast cancer. Mol. Cell 70, 842–853.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Crossley M. P., Bocek M., Cimprich K. A., R-loops as cellular regulators and genomic threats. Mol. Cell 73, 398–411 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boguslawski S. J., et al. , Characterization of monoclonal antibody to DNA.RNA and its application to immunodetection of hybrids. J. Immunol. Methods 89, 123–130 (1986). [DOI] [PubMed] [Google Scholar]
- 43.Couronné L., Bastard C., Bernard O. A., TET2 and DNMT3A mutations in human T-cell lymphoma. N. Engl. J. Med. 366, 95–96 (2012). [DOI] [PubMed] [Google Scholar]
- 44.Odejide O., et al. , A targeted mutational landscape of angioimmunoblastic T-cell lymphoma. Blood 123, 1293–1296 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hu G., et al. , Transformation of accessible chromatin and 3D nucleome underlies lineage commitment of early T cells. Immunity 48, 227–242.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jeong M., et al. , Large conserved domains of low DNA methylation maintained by Dnmt3a. Nat. Genet. 46, 17–23 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lister R., et al. , Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature 471, 68–73 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li X., et al. , Tet proteins influence the balance between neuroectodermal and mesodermal fate choice by inhibiting Wnt signaling. Proc. Natl. Acad. Sci. U.S.A. 113, E8267–E8276 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cruickshanks H. A., et al. , Senescent cells harbour features of the cancer epigenome. Nat. Cell Biol. 15, 1495–1506 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bachman K. E., Rountree M. R., Baylin S. B., Dnmt3a and Dnmt3b are transcriptional repressors that exhibit unique localization properties to heterochromatin. J. Biol. Chem. 276, 32282–32287 (2001). [DOI] [PubMed] [Google Scholar]
- 51.Baubec T., et al. , Genomic profiling of DNA methyltransferases reveals a role for DNMT3B in genic methylation. Nature 520, 243–247 (2015). [DOI] [PubMed] [Google Scholar]
- 52.Manzo M., et al. , Isoform-specific localization of DNMT3A regulates DNA methylation fidelity at bivalent CpG islands. EMBO J. 36, 3421–3434 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gu T., et al. , DNMT3A and TET1 cooperate to regulate promoter epigenetic landscapes in mouse embryonic stem cells. Genome Biol. 19, 88 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Williams K., et al. , TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature 473, 343–348 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jaiswal S., et al. , Age-related clonal hematopoiesis associated with adverse outcomes. N. Engl. J. Med. 371, 2488–2498 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Iyer L. M., et al. , Lineage-specific expansions of TET/JBP genes and a new class of DNA transposons shape fungal genomic and epigenetic landscapes. Proc. Natl. Acad. Sci. U.S.A. 111, 1676–1683 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Janssen A., Colmenares S. U., Karpen G. H., Heterochromatin: Guardian of the genome. Annu. Rev. Cell Dev. Biol. 34, 265–288 (2018). [DOI] [PubMed] [Google Scholar]
- 58.Xu G. L., et al. , Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature 402, 187–191 (1999). [DOI] [PubMed] [Google Scholar]
- 59.Thienpont B., et al. , Tumour hypoxia causes DNA hypermethylation by reducing TET activity. Nature 537, 63–68 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mazor T., Pankov A., Song J. S., Costello J. F., Intratumoral heterogeneity of the epigenome. Cancer Cell 29, 440–451 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data produced as part of this study are available in the Gene Expression Omnibus (GEO) database under accession no. GSE134396 and the NCBI BioProject database under accession no. PRJNA555063. GEO and European Nucleotide Archive accession codes from the previously published sequencing data used in this study are available in SI Appendix, Table S2.