Significance
Lateral branch suppression has been selected during crop domestication, and axillary branches need to be manually removed during fresh cucumber production. Auxin is a key repressor for shoot branching underlying apical dominance. The TEOSINTE BRANCHED1 (TB1)/BRANCHED1 (BRC1) gene acts as the focal point for multiple signals to inhibit branching. However, the relationship between auxin and BRC1 remains elusive. Here cucumber BRANCHED1 (CsBRC1) is found to inhibit shoot branching by directly repressing the auxin efflux carrier CsPIN3. Compared to its wild ancestor, cultivated cucumber displays reduced branches, higher CsBRC1 expression, reduced CsPIN3 expression, and higher auxin accumulation in buds. In this study, we find a regulatory pathway of CsBRC1–CsPIN3–auxin transport in suppressing shoot branching in cucumber.
Keywords: auxin transport, axillary bud outgrowth, CsBRC1, CsPIN3, cucumber
Abstract
Shoot branching is an important agronomic trait that directly determines plant architecture and affects crop productivity. To promote crop yield and quality, axillary branches need to be manually removed during cucumber production for fresh market and thus are undesirable. Auxin is well known as the primary signal imposing for apical dominance and acts as a repressor for lateral bud outgrowth indirectly. The TEOSINTE BRANCHED1/CYCLOIDEA/PCF (TCP) family gene BRANCHED1 (BRC1) has been shown to be the central integrator for multiple environmental and developmental factors that functions locally to inhibit shoot branching. However, the direct molecular link between auxin and BRC1 remains elusive. Here we find that cucumber BRANCHED1 (CsBRC1) is expressed in axillary buds and displays a higher expression level in cultivated cucumber than in its wild ancestor. Knockdown of CsBRC1 by RNAi leads to increased bud outgrowth and reduced auxin accumulation in buds. We further show that CsBRC1 directly binds to the auxin efflux carrier PIN-FORMED (CsPIN3) and negatively regulates its expression in vitro and in vivo. Elevated expression of CsPIN3 driven by the CsBRC1 promoter results in highly branched cucumber with decreased auxin levels in lateral buds. Therefore, our data suggest that CsBRC1 inhibits lateral bud outgrowth by direct suppression of CsPIN3 functioning and thus auxin accumulation in axillary buds in cucumber, providing a strategy to breed for cultivars with varying degrees of shoot branching grown in different cucumber production systems.
Shoot branching is an important agronomic trait that directly determines plant architecture and affects crop productivity. In flowering plants, the development of lateral shoots starts from axillary meristem (AM) initiation at the leaf axils. The AMs then develop into axillary buds comprising a few leaf primordia and a meristem, which either remain dormant at this stage or grow out to form a branch (1, 2). Excess of branches often compete for nutrient allocation and light harvesting and generally have inverse effects on crop yield. In fact, suppression of shoot branching is an important selection trait during crop domestication (3). Cucumber (Cucumis sativus L.), a worldwide cultivated vegetable crop that is consumed freshly or processed into pickles, is domesticated from its highly branched wild ancestor Cucumis sativus var. hardwickii (4). Despite the fact that lateral branches are greatly suppressed in cultivated cucumber as compared to hardwickii, the degree of branch suppression varies depending on cultivars, geographic regions, and market types. To promote crop yield and quality, extra branches need to be manually removed during cucumber production, especially for fresh market, which is labor intensive and time-consuming. Therefore, it is of great importance for cucumber production and breeding to understand the regulatory mechanisms of lateral branch arrest.
Axillary bud arrest or outgrowth is a plastic developmental process regulated by diverse environmental and developmental factors (5). Auxin is synthesized in the shoot apex and acts as a repressor for branching and serves as the primary signal imposing for apical dominance (6). The main shoot tip produces a massive amount of auxin, which moves downward within the stem and represses axillary bud outgrowth. Removal of the main shoot tip by decapitation leads to the outgrowth of axillary buds into branches (7, 8). Two nonexclusive hypotheses explain indirect regulation of shoot branching by auxin. According to the second messenger hypothesis, auxin acts outside the axillary buds and regulates the production of second messengers, cytokinins (CK) and strigolactones (SL), to control shoot branching (9, 10). A more recently proposed model is the auxin transport canalization-based hypothesis (11, 12), in which auxin flows between an auxin source and an auxin sink leading to up-regulation and polarization of auxin transporters to canalize auxin transport into cell files (11, 13–15). The shoot apex and axillary buds are auxin sources that compete for sink strength of the stem to transport auxin rootward (11, 16). The shoot apex continuously produces auxin and transports down the stem, which reduces stem sink strength and prevents auxin export from lateral buds, hence resulting in bud arrest (15).
The founder member of the class II TCP (TEOSINTE BRANCHED1/CYCLOIDEA/PCF) family of transcription factors, known as TEOSINTE BRANCHED1 (TB1)-like in monocots and BRANCHED1 (BRC1)-like in eudicots, has been shown to be an integrator of multiple internal and external signals that act inside the axillary buds to suppress shoot branching (17–20). The biosynthesis of CK can be repressed by auxin, resulting in increased expression of TB1/BRC1 to inhibit shoot branching in rice and pea (21, 22). On the contrary, auxin stimulates SL biosynthesis to promote transcription of TB1/BRC1 and repress shoot branching in Arabidopsis and pea (18, 22). BRC1 transcription is auxin responsive in Arabidopsis; BRC1 determines bud activation potential but is dispensable for bud growth inhibition (11). Sugar, an important nutrient substance, plays a pivotal role in axillary bud outgrowth through repressing TB1/BRC1 expression in wheat (23–25). Shade, generated by increased planting density or by decreased ratio of red to far-red light, induces the expression of TB1/BRC1 and leads to axillary bud dormancy (26–28). Therefore, TB1/BRC1 serves as the focal point for hormonal, nutritional, and environmental signals to inhibit lateral bud outgrowth.
Moreover, multiple regulatory pathways have been dissected for the TB1/BRC1-mediated suppression of shoot branching (5). The transcriptional repressor TIE1 (TCP interactor containing EAR motif protein 1) positively regulates shoot branching by directly repressing BRC1 activity in Arabidopsis, while the transcription factor IDEAL PLANT ARCHITECTURE1 (IPA1) promotes the expression of OsTB1 by directly binding to its promoter region in rice (29, 30). BRC1 can directly bind to 3 homeodomain leucine zipper protein (HD-ZIP)-encoding genes, HOMEOBOX PROTEIN 21, HOMEOBOX PROTEIN 40, and HOMEOBOX PROTEIN 53, and positively regulates their expression in Arabidopsis. BRC1, together with these 3 downstream genes, promotes the transcription of 9-CIS-EPOXICAROTENOID DIOXIGENASE 3, which encodes a key enzyme in abscisic acid (ABA) biosynthesis, and leads to ABA accumulation and branching suppression (9). Besides, maize TB1 was found to be a key domestication gene that directly activates the expression of TASSELS REPLACE UPPER EARS1 (TRU1) to mediate axillary bud dormancy (31). Modern maize with suppressed lateral branches largely resulted from overexpression of TB1 by retrotransposon insertion in the promoter during domestication from its wild ancestor teosinte (32).
Although the function of BRC1 in lateral bud arrest is widely conserved in the plant kingdom, the direct molecular link between BRC1 and auxin remains unknown. Here we show that the cucumber BRC1 (CsBRC1) directly binds to the auxin efflux carrier PIN-FORMED3 (CsPIN3) and negatively regulates its expression, resulting in auxin accumulation in axillary buds and arrest of bud outgrowth.
Results
Expression of CsBRC1 Is Negatively Correlated with Lateral Branch Outgrowth in Cucumber.
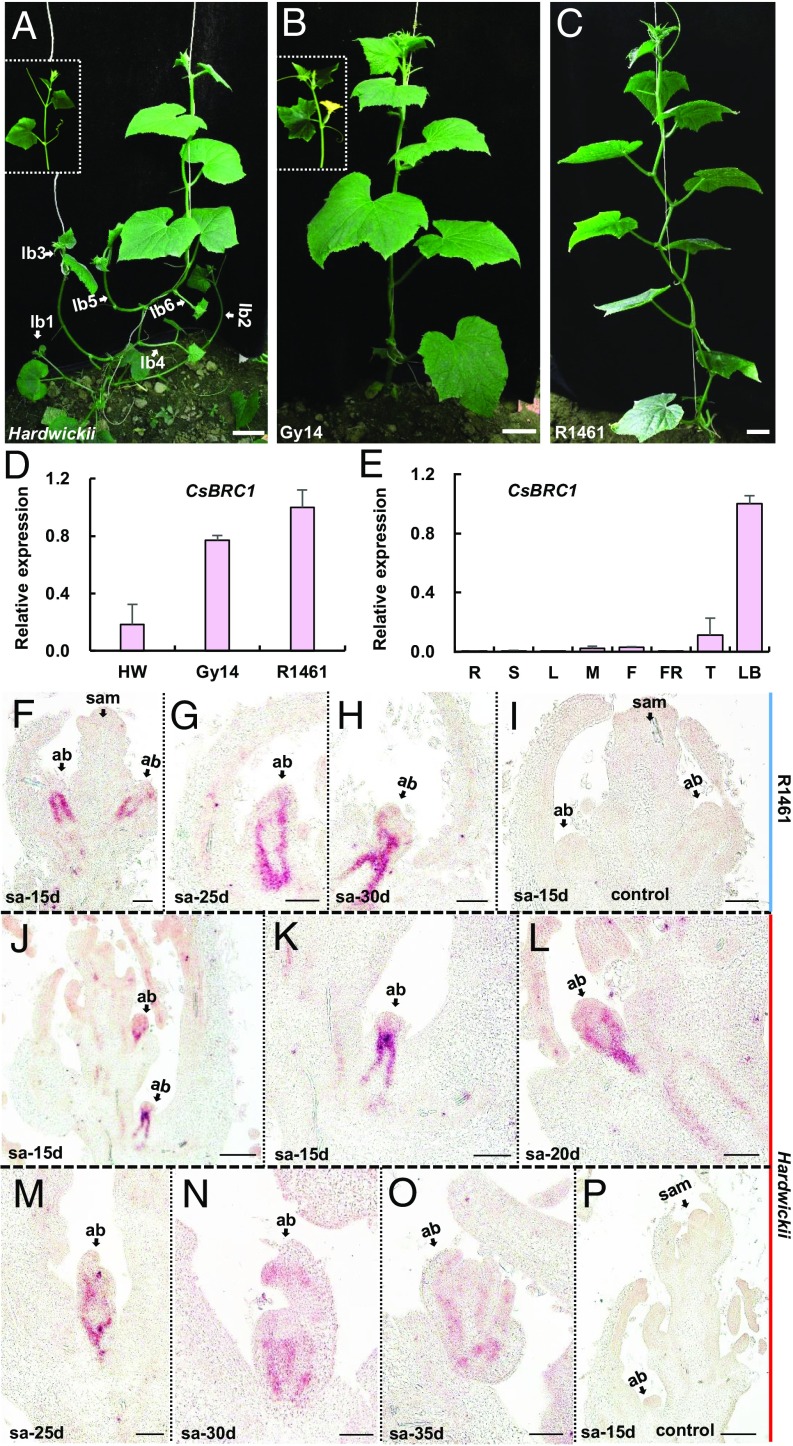
Compared to the highly branched wild ancestor Cucumis sativus var. hardwickii (Fig. 1A), the lateral branch is suppressed at different levels in cultivated cucumber varieties. The sequenced pickling cucumber Gy14 line displays moderate branching with lateral branch suppression mainly occurring in the early growth period (Fig. 1B), whereas East Asian line R1461 shows very few branches with bud outgrowth arrested throughout the life cycle (Fig. 1C). To identify which genes account for lateral branch suppression during cucumber domestication, we performed expression analysis with cucumber homologous genes to BRANCHED 1 (CsBRC1), LATERAL SUPPRESSOR (CsLS), BLIND (CsBLIND), and MORE AXILLARY BRANCHES 1, 2, 3, and 4 (CsMAX1 to CsMAX4) in hardwickii, Gy14, and R1461 (Fig. 1D and SI Appendix, Fig. S1). The expression of CsBRC1 (Csa1G020890) was negatively correlated with branching and displayed the greatest change between hardwickii and R1461 (Fig. 1D) and thus was chosen for further characterization in this study.
Fig. 1.
Expression of CsBRC1 in cultivated cucumbers and its wild ancestor Cucumis sativus var. hardwickii. (A–C) Morphology of highly branched cucumber hardwickii (A), moderately branched cucumber line Gy14 (B), and rarely branched line R1461 (C). lb, lateral branch. (D and E) Expression analyses of CsBRC1 by qRT-PCR in 3 cucumber lines (D) and in different organs of cucumber line R1461 (E). Values are means of 3 independent biological replicates from different plants. Error bars represent ±SD. HW, hardwickii; R, roots; S, stems; L, leaves; M, male buds; F, female buds; FR, fruits; T, shoot apex; LB, lateral buds. (F–P) In situ hybridization of CsBRC1 in cultivated cucumber R1461 (F–I) and wild ancestor hardwickii (J–P) during seedling development from 15 to 35 d after planting. K is the magnified image of the bottom axillary bud in J. sa, shoot apex; sam, shoot apical meristem; ab, axillary bud. (Scale bars represent 5 cm in A–C, 100 μm in F–I and K–O, and 200 μm in J and P.)
CsBRC1 contains 2 exons and encodes a 342-amino-acid-long putative TCP transcription factor with a typical bHLH motif (SI Appendix, Fig. S2). There is no difference in the coding sequence of CsBRC1 among the 3 cucumber varieties hardwickii, GY14, and R1461. Subcellular localization analysis indicated that CsBRC1 was localized in the nucleus (SI Appendix, Fig. S3). Quantitative real-time PCR indicated that CsBRC1 was highly enriched in axillary buds (Fig. 1E). In situ hybridization further showed that the transcripts of CsBRC1 were specifically accumulated in vascular tissues of axillary buds (Fig. 1 F–P). In the cultivated cucumber R1461, the expression of CsBRC1 remained at high levels at axillary buds during the development from 15- to 30-d seedlings, and no outgrowth of axillary buds was detected (Fig. 1 F–H). However, in the wild ancestor hardwickii, the CsBRC1 signals became weaker as the axillary buds grew up over time (Fig. 1 J–O). No signal was detected upon hybridization with the sense probe of CsBRC1 (Fig. 1 I and P).
Knockdown of CsBRC1 Resulted in Increased Outgrowth of Axillary Bud in Cucumber.
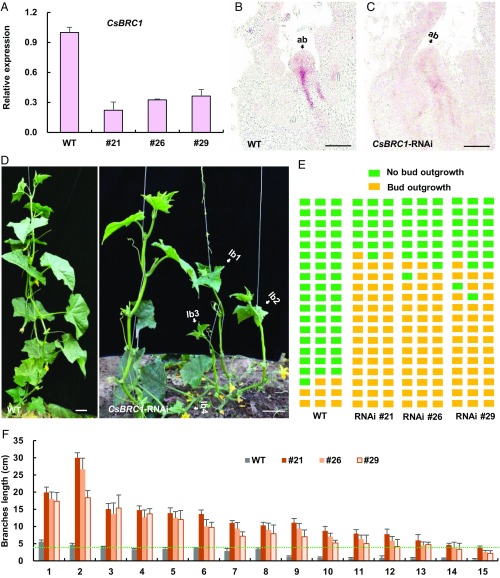
To gain insight into CsBRC1 function in lateral branch outgrowth in cucumber, we created a double-stranded RNAi construct containing the 585-bp coding sequence of CsBRC1 and transformed into the cucumber inbred line R1461 via Agrobacterium-mediated cotyledon transformation (33, 34). Twelve CsBRC1-RNAi lines were obtained, and 3 representative lines (21, 26, and 29) with different severities were chosen for further characterization (Fig. 2). Quantitative real-time PCR showed that the expression of CsBRC1 was reduced by 64% ∼78% in the transgenic CsBRC1-RNAi lines as compared to the empty vector control (WT) (Fig. 2A). In situ hybridization confirmed that the transcripts of CsBRC1 were dramatically decreased in lateral buds of CsBRC1-RNAi lines (Fig. 2 B and C). Consistent with its negative correlation with shoot branching (Fig. 1), CsBRC1-RNAi plants produced many more lateral branches with great increase in length (Fig. 2 D–F). In cucumber production, lateral branches longer than 4 cm are considered as manageable branches to be manually removed by farmers or technicians; we thus set 4 cm as the cutoff value of branch outgrowth. Using this standard, only 2 to 3 branches were formed out of the 20 examined nodes in WT plants, whereas 12 to 15 branches were produced in CsBRC1-RNAi transgenic lines (Fig. 2E). Similarly, the branch length was greatly increased, with a maximum of 5.5 cm in WT but of 18.3 to 30 cm in the CsBRC1-RNAi transgenic lines (Fig. 2F). These data suggested that CsBRC1 acts as an important repressor for axillary bud outgrowth in cucumber.
Fig. 2.
Knockdown of CsBRC1 by RNAi led to increased lateral bud outgrowth in cucumber. (A) Expression analysis of CsBRC1 in empty vector (WT) and CsBRC1-RNAi lines (21, 26, and 29). Values are means of 3 independent biological replicates of lateral buds from different plants. Error bars represent ±SD. (B and C) In situ hybridization of CsBRC1 in WT and CsBRC1-RNAi line 21. ab, axillary bud. (D) Representative images of WT and CsBRC1-RNAi transgenic plants. White arrows indicate lateral branches in CsBRC1-RNAi plants. lb, lateral branch. (E) Diagrammatic data showed the position of axillary bud outgrowth in WT and CsBRC1-RNAi lines. Each layer represents a node in cucumber. Green squares represent no bud outgrowth, and yellow squares represent bud outgrowth (longer than 4 cm). Each column represents an individual plant of WT or CsBRC1-RNAi lines. (F) The length of each lateral bud in WT and CsBRC1-RNAi lines from 1st to 15th nodes. Bud length higher than the green dotted line indicates bud outgrowth. Values are means of lateral branches from the same node of 3 independent plants. Error bars represent ±SD. (Scale bars represent 100 μm in B and C and 5 cm in D.)
CsBRC1 Inhibits Branching through Regulating Auxin Pathway in Cucumber.
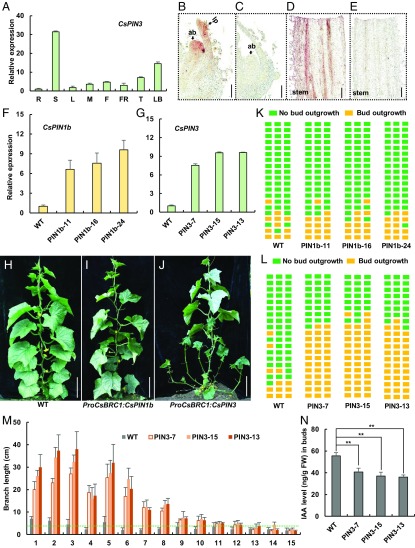
To identify the downstream targets and regulatory network of CsBRC1 in cucumber, transcriptome profiling by RNA-Seq was performed with the lateral buds from WT and CsBRC1-RNAi line 21. Using a false discovery rate (FDR) less than 0.05 and the fold change larger than 2 as thresholds, 1,680 up-regulated and 1,171 down-regulated genes were identified as differentially expressed genes (DEGs) (SI Appendix, Table S1 and Dataset S1) (35). Gene Ontology (GO) term enrichment analysis of the up-regulated genes in the CsBRC1-RNAi line indicated that the term “auxin efflux transmembrane transporter activity” was significantly enriched (SI Appendix, Fig. S4). Heat map analysis indicated that 23 genes related to auxin response, and 2 auxin transporters and 1 auxin signaling gene were differentially expressed in CsBRC1-RNAi vs. WT plants (Fig. 3A). Quantitative real-time PCR confirmed that 2 auxin efflux carrier genes, CsPIN1b (Csa1G025070) and CsPIN3 (Csa5G576590), were significantly up-regulated in CsBRC1-RNAi lines (Fig. 3B), with 7.4 fold and 4.4 fold higher in the CsBRC1-RNAi lines as compared to that in WT, respectively. Phylogenetic analysis indicated that there are 3 CsPIN1 genes (CsPIN1a, CsPIN1b, and CsPIN1c) in cucumber, and CsPIN3 is closely related to CsPIN4 (SI Appendix, Fig. S5).
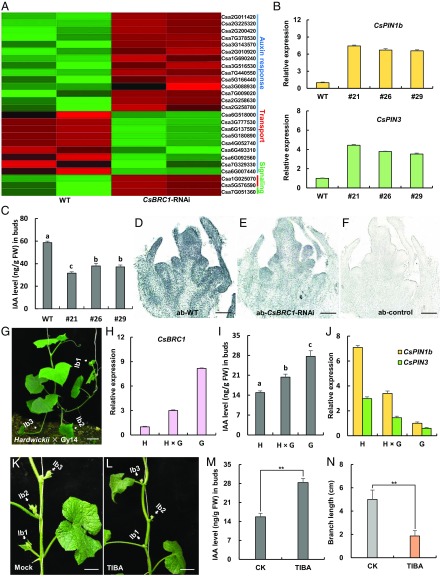
Fig. 3.
CsBRC1 regulates auxin transport in cucumber. (A) Heat map showing the expression levels of cucumber homologs of auxin pathway genes in WT and CsBRC1-RNAi lines. Red boxes mean up-regulation, and green boxes indicate down-regulation. Blue, red, and green lines represent genes involved in auxin response, transport, and signaling, respectively. (B) Expression verification by qRT-PCR of the 2 auxin efflux carrier genes CsPIN1b and CsPIN3 identified by RNA-seq analysis. (C) The content of endogenous auxin was greatly reduced in the axillary buds of CsBRC1-RNAi lines. (D–F) Immuno-gold localization of IAA in elongated axillary buds at the same node of WT and CsBRC1-RNAi plants. ab, axillary bud. (G) A representative image of an F1 plant resulted from a cross between hardwickii and Gy14. lb, lateral branch. (H–J) The expression of CsBRC1 (H), auxin content (I), and the expression of CsPIN1b/3 (J) in F1 plant as well as in the 2 parents (hardwickii and Gy14). (K and L) Images showing the axillary bud outgrowth after auxin inhibitor TIBA treatment of hardwickii lateral buds. (M) The IAA level was significantly increased upon TIBA treatment. (N) The branch length was significantly shorter in TIBA-treated plants than mock. Values are means of 3 biological replicates of lateral buds (B, C, H–J, and M) or 6 repeats of lateral branches (N) from different plants. Error bars represent ±SD. Significance analysis was conducted with the 1-way ANOVA in C and I (P < 0.01) and the 2-tailed Student’s t test in M and N (**P < 0.01). (Scale bars represent 100 μm in D–F and 5 cm in G, K, and L.)
Next, we measured the auxin level in lateral buds of WT and CsBRC1-RNAi lines. As expected, auxin concentration was reduced in CsBRC1-RNAi lines (Fig. 3C), consistent with the repressing role of auxin in lateral branch outgrowth (5). Immune colloidal gold assay of 3-Indole acetic acid (IAA) further showed the reduced auxin accumulation in lateral buds of CsBRC1-RNAi plants (Fig. 3 D–F), suggesting that decrease of auxin level may be responsible for the increased axillary branch outgrowth in CsBRC1-RNAi lines.
We next performed a cross experiment using hardwickii (more branches) and Gy14 (less branches) as parents. The resultant F1 plants displayed an intermediate phenotype (more branches than Gy14 but less than hardwickii) (Fig. 3G). Consistently, the expression of CsBRC1 and the auxin level in F1 plants were between the values of 2 parents (Fig. 3 H and I). Further, the expression of CsPIN1b and CsPIN3 displayed the same trend (Fig. 3J), suggesting that there may be a dosage effect of auxin on lateral branch suppression. To explore whether auxin efflux leads to increased axillary bud outgrowth, 0.5 mM 2,3,5-Triiodobenzoic acid (TIBA), a well-known auxin transport inhibitor (36), was applied to the axillary buds of hardwickii (Fig. 3 K and L). As compared to the mock treatment, the auxin content of the TIBA-treated axillary buds was significantly increased, and consequently, the outgrowth of axillary bud was dramatically inhibited (Fig. 3 M and N).
CsBRC1 Directly Binds to the Auxin Efflux Carrier CsPIN3 In Vitro and In Vivo.
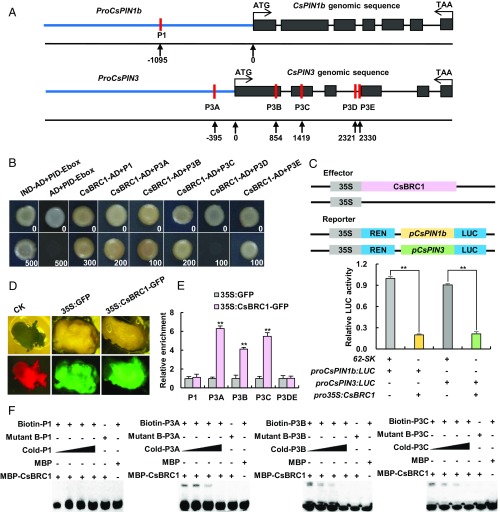
Previous studies showed that transcription factors of the TCP family directly bind to GGNCCCAC elements (9, 37). To further dissect the regulatory role of CsBRC1 on the auxin pathway, cis-acting elements on the genome sequences of PIN genes were analyzed. One putative element (P1) in the CsPIN1b promoter and 5 putative elements (P3A-P3E) in the CsPIN3 genome sequence were identified (Fig. 4A). No GGNCCCAC element was found on other PIN genes in cucumber. To explore whether CsBRC1 could bind to CsPIN1b and CsPIN3 directly, we carried out yeast 1-hybrid assay. The results showed that CsBRC1 bound to the P1 element in CsPIN1b and 4 (P3A, P3B, P3C, and P3E) out of the 5 elements of CsPIN3 (Fig. 4B). A dual-luciferase (LUC) transaction assay in tobacco leaves was then performed. The relative intensity of LUC signals significantly decreased upon cotransformation of 35S:CsBRC1 with ProCsPIN1b:LUC or ProCsPIN1b3:LUC, as compared to the vector control (Fig. 4C), suggesting that CsBRC1 may repress expression of CsPIN1b and CsPIN3. To verify such interactions in vivo, 35S:CsBRC1-GFP and 35S:GFP were separately transformed into cucumber calli from hypocotyl explants for chromatin immunoprecipitation (ChIP) assay (Fig. 4D). ChIP-qPCR results showed that the P3A, P3B, and P3C elements in CsPIN3 were significantly enriched by antibodies recognizing CsBRC1-GFP protein, while P3DE (containing P3D and P3E) in CsPIN3 and the P1 element in CsPIN1b were not (Fig. 4E). As expected, electrophoretic mobility shift assay (EMSA) indicated binding of P3A, P3B, and P3C elements in CsPIN3 by CsBRC1; trinucleotide mutations could abolish such binding activities (Fig. 4F). Together, CsBRC1 directly binds to CsPIN3 to repress its expression in vitro and in vivo.
Fig. 4.
CsBRC1 directly binds to the auxin efflux carrier gene CsPIN3 to repress its expression. (A) Schematic diagram of the putative cis-elements in CsPIN1b (P1) and CsPIN3 (P3A–P3E) used for CsBRC1 binding assay. (B) Yeast 1-hybrid assay showed binding of CsBRC1 to P1, P3A, P3B, P3C, and P3E. The numbers in the right corner indicate the basal concentration of Aureobasidin A (AbA; ng/mL). (C) Luciferase activity measurement in tobacco leaves after coexpression of Pro35S:CsBRC1 and ProCsPIN1b:LUC or ProCsPIN3:LUC. The empty vector 62-SK was used as the control. Four independent transfection experiments were performed. (D) Callus images from cucumber hypocotyl explants transfected with empty control, Pro35S:GFP, and Pro35S:CsBRC1-GFP used in ChIP assay. (E) ChIP-qPCR assay showed that CsBRC1 binds to P3A, P3B, and P3C regions of CsPIN3. Three technical replicates and 3 biological replicates from different calli were performed. Error bars represent ±SD. (F) EMSA assay showed that CsBRC1 binds to P3A, P3B, and P3C regions of CsPIN3. Significance analysis was conducted with the 2-tailed Student’s t tests (**P < 0.01).
Enhanced Expression of CsPIN3 Driven by the CsBRC1 Promoter Led to Excessive Shoot Branching in Cucumber.
The expression patterns of CsPIN3 were analyzed by qRT-PCR and in situ hybridization (Fig. 5 A–E). CsPIN3 was highly expressed in the young stem and lateral buds (Fig. 5A). In situ hybridization further showed that CsPIN3 transcripts specifically accumulated in lateral buds, leaf primordia, and stems in cucumber (Fig. 5 B–E). We next transformed CsPIN1b and CsPIN3, driven by the CsBRC1 promoter, into cucumber inbred line R1461. Three representative ProCsBRC1:CsPIN1b transgenic lines (PIN1b-11, PIN1b-16, and PIN1b-24) and 3 ProCsBRC1:CsPIN3 transgenic lines (PIN3-7, PIN3-13, and PIN3-15) were chosen for further characterization (Fig. 5 F–N). qRT-PCR showed that the expression of CsPIN1b and CsPIN3 were significantly increased in the transgenic lines as compared to that in lines harboring the empty vector (WT) (Fig. 5 F and G). Importantly, enhanced expression of CsPIN3 resulted in more lateral branches with a great increase in length, while elevated expression of CsPIN1b displayed no such significant difference as compared to WT (Fig. 5 H–J). Quantification data showed that only 3 to 5 branches were formed out of the 20 examined nodes in WT plants, while 11 to 14 branches were produced in CsPIN3 transgenic lines (PIN3-7, PIN3-13, and PIN3-15) (Fig. 5 K and L). Similarly, the maximum of lateral branch length in WT was 7.0 cm, while those of CsPIN3 transgenic lines were 27 to 38 cm (Fig. 5M). Moreover, the auxin level was significantly reduced in lateral buds of CsPIN3 transgenic lines (Fig. 5N). On the basis of the canalization hypothesis (12), we found that CsPIN3, but not CsPIN1b, plays an essential role in auxin export during bud outgrowth in cucumber.
Fig. 5.
Phenotype analysis of overexpression of CsPIN1b and CsPIN3 driven by the promoter of CsBRC1. (A) Expression analysis of CsPIN3 by qRT-PCR in different organs of cucumber inbred line R1461. Values are means of 3 biological replicates from different plants. Error bars represent ±SD. R, roots; S, stems; L, leaves; M, male buds; F, female buds; FR, fruits; T, shoot apex; LB, lateral buds. (B–E) In situ hybridization of CsPIN3 in axillary buds (B) and the stem (D). The sense CsPIN3 probe was hybridized as the negative control (C and E). ab, axillary bud; lp, leaf primordium. (F and G) Expression analysis of CsPIN1b (F) and CsPIN3 (G) by qRT-PCR in control (WT), ProCsBRC1:CsPIN1b (PIN1b-11, PIN1b-16, and PIN1b-24), and ProCsBRC1:CsPIN3 transgenic plants (PIN3-7, PIN3-15, and PIN3-13). Values are means of 3 biological replicates of lateral buds from different plants. (H–J) Representative images of WT (H), ProCsBRC1:CsPIN1b (I), and ProCsBRC1:CsPIN3 transgenic plants (J) 60 d after planting grown under the same greenhouse condition. (K and L) Diagrammatic data showed the position of axillary bud outgrowth in CsPIN1b (K) and CsPIN3 (L) transgenic cucumber lines. Each layer represents a node in cucumber. Green squares represent no bud outgrowth, and yellow squares represent bud outgrowth (longer than 4 cm). (M) The length of each lateral bud in WT and ProCsBRC1:CsPIN3 transgenic lines from the 1st to 15th nodes. Bud length higher than the green dotted line indicates bud outgrowth. Values are means of lateral branch length from the same node of 3 independent plants. (N) The content of endogenous IAA was greatly reduced in the axillary buds of ProCsBRC1:CsPIN3 transgenic lines. Values are means of 3 biological replicates of lateral buds from independent plants. Error bars represent ±SD. Significance analysis was conducted with the 2-tailed Student’s t tests (**P < 0.01). (Scale bars represent 100 μm in B and C, 200 μm in D and E, and 20 cm in H–J.)
CsBRC1 Shows Different Responses to Light Intensity in Wild and Cultivated Cucumbers.
Increase in planting density is an important trait that has been selected for high yield during crop domestication (26), which results in some degree of shading from neighboring plants. Recent studies showed that BRC1 promotes branch suppression in response to shade in Arabidopsis, sorghum, and potato (26–28). To investigate whether BRC1 responds to variations in light intensity related to shading in wild and cultivated cucumbers, we performed light intensity treatment (by blocking 90% light) to wild cucumber hardwickii and inbred line R1461. Transcript levels of CsBRC1 were increased by 2 to 3 fold in axillary buds of line R1461 upon 10 to 20 h shading (SI Appendix, Fig. S6A). In contrast, CsBRC1 expression was not induced by shading in wild cucumber hardwickii (SI Appendix, Fig. S6A). Promoter analysis indicated that R1461 possesses 2 additional light responsive elements (an AE box and a GATA motif) as compared to that in hardwickii (SI Appendix, Fig. S6B), which may be the reason for different responses to reduced light intensity between hardwickii and R1461.
Discussion
Apical dominance is a widely found phenomenon in plants where a main shoot dominates and inhibits the outgrowth of lateral branches, and auxin has been shown to be the key inhibitory signal produced by shoot tip responsible for apical dominance (7, 38). Auxin transport canalization hypothesis is one of the leading theories explaining apical dominance (11, 39). According to the theory, polar auxin transport from shoot apex to root leads to the auxin concentration in the main stem and thus reduces its sink strength, which will inhibit auxin flow from axillary buds to main stem, resulting in auxin accumulation in lateral buds and subsequent suppression of lateral branches (11, 16). On the other side, TB1/BRC1 has been well characterized to be the central integrator for multiple environmental and developmental factors inhibiting shoot branching (18, 22). BRC1 transcription in lateral buds is responsive to auxin application in Arabidopsis (11). However, the direct molecular link between BRC1 and auxin remains elusive.
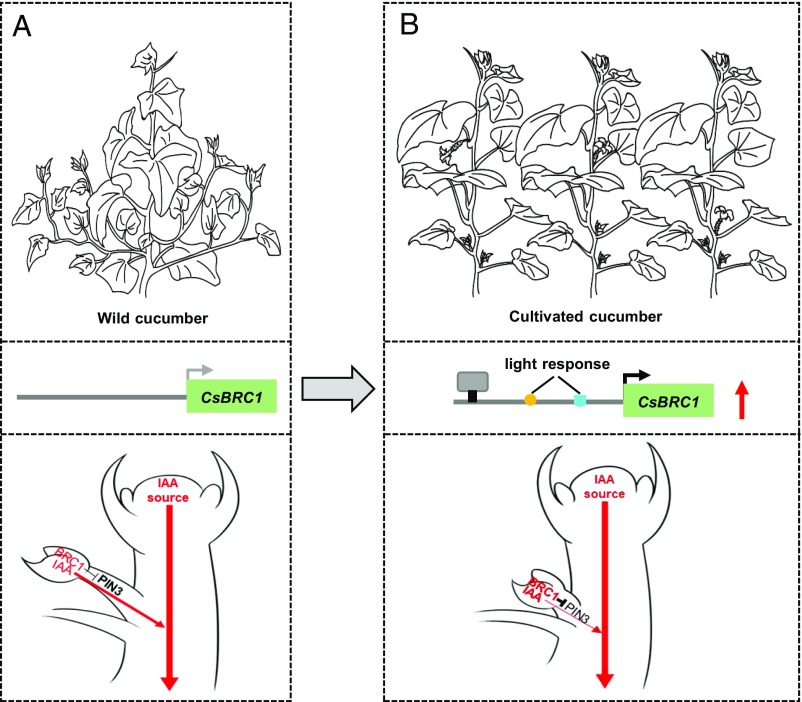
Here we found that CsBRC1 was specifically expressed in axillary buds to inhibit bud outgrowth in cucumber (Figs. 1–2). Further, CsPIN3 contains 5 CsBRC1-binding elements in its genome sequence (Fig. 4), and its expression was significantly up-regulated in CsBRC1-RNAi lines (Fig. 3). More importantly, both in vitro and in vivo biochemical assays suggested that CsBRC1 directly binds to the auxin transporter CsPIN3 and down-regulates its expression (Fig. 4). Elevated expression of CsPIN3 driven by the CsBRC1 promoter results in excessive shoot branching in cucumber (Fig. 5). We propose that CsBRC1 functions as a key repressor for shoot branching by directly inhibiting CsPIN3 activity, which down-regulates auxin efflux from buds, hence bud outgrowth arrests in cucumber (Fig. 6 A and B).
Fig. 6.
A working model of CsBRC1 in repressing axillary bud outgrowth in cucumber. The main shoot tip produces massive auxin, which moves downward in the stem. In wild cucumber, low BRC1 expression is unable to inhibit the PIN3-mediated auxin efflux from lateral buds, which leads to bud outgrowth (A). During domestication, insertion of 2 light response elements in CsBRC1 promoter may favor elevation of CsBRC1 expression and adaption to shade response from increased planting density. Alternatively, an unknown regulator (indicated as a gray box) promotes the transcription of CsBRC1. The elevated CsBRC1 expression inhibits PIN3-mediated auxin efflux from lateral buds. Consequently, auxin accumulates in axillary buds and no bud outgrowth in cultivated cucumber (B).
Considerable work has showed that PIN proteins are responsible for polar auxin transport underlying multiple aspects of plant development (40). The PIN1 protein is polarized on cell membrane and involved in plant organogenesis and leaf shape formation, as well as root development (41–44). PIN3 protein is expressed in gravity-sensing tissues and involved in differential growth in root (45), acts as a target for the CK signaling pathway to promote shoot branching (46), and mediates connective auxin transport facilitating communication between shoot apices to control shoot branching (40). Triple mutants of PIN3, PIN4, and PIN7 (pin3 pin4 pin7) show no marked difference in shoot branching, although triple mutation (pin3 pin4 pin7) partially suppresses excessive branching of SL-deficient mutants. Further, involvement of PIN3, PIN4, and PIN7 in regulation of shoot branching is independent of the BRC1 pathway in Arabidopsis (47). However, CsPIN3 is featured by its multiple cis elements for CsBRC1 binding in cucumber, and CsBRC1 indeed binds to CsPIN3 for transcriptional suppression, as indicated by a whole set of biochemical, transcriptomic, and transgenic data (Figs. 3–5). Enhanced expression of CsPIN3 dramatically stimulated bud outgrowth (Fig. 5). Thus, there may exist divergent regulatory mechanisms shaping shoot architecture in 2 different dicots Arabidopsis and cucumber. Such regulatory divergence may be attributed to following facts. First, the promoter and genic region of CsPIN3 in cucumber harbors cis elements for CsBRC1 binding (Fig. 4), while PIN3 does not contain BRC1 binding elements in Arabidopsis. Second, CsPIN3 transcripts were enriched in lateral buds of cucumber (Fig. 5) in contrast to no PIN3 transcription reported in lateral buds in Arabidopsis (48). Third, 2 dicots have dramatically different shoot branching patterns. In Arabidopsis, the shoot apical meristem produces rosette leaves with compact internodes during vegetative growth. The inflorescence meristem gives rise to a few cauline leaves and then flowers with elongated internodes during the reproductive stage (49). Between 2 types of branching (rosette and cauline) in Arabidopsis, BRC1 and PIN3 play a negative and positive role, respectively, in regulating rosette branching (18, 40, 47). In cucumber, however, the shoot apical meristem produces leaves with elongated internodes throughout the life cycle, and only 1 type of shoot branches (similar to cauline branches in Arabidopsis) emerges from the leaf axil of each node. Thus, genetic and developmental divergence enables CsBRC1 to inhibit cucumber branching by repressing CsPIN3 transcription and diminishing auxin export from lateral buds to the main stem (Figs. 2–5). Our finding paves a path to dissect BRC1-auxin interplay in shoot branching and yielding in closely related horticultural crops.
During crop domestication, a set of common traits have been selected and fixed to meet human needs and modern agronomic practices, such as reduced lateral branching, lack of shattering, greater fruit and seed size, and more determinate growth (50). TB1 is one of the few domestication genes that regulates many of the domestication traits including shoot branching, prolificacy, inflorescence sex, and glume hardness that distinguish maize from its wild ancestor teosinte (51). In cucumber, cultivated cucumber displays reduced branching, larger fruit size, nonbitterness, and increased planting density as compared to wild cucumber hardwickii (4). What genes or regulatory networks underlying above domestication traits are largely unknown. Here we found that the expression of CsBRC1, a repressor for shoot branching, was higher in cultivated cucumber lines R1461 and Gy14 than that in wild cucumber hardwickii (Fig. 1). Further, our data showed that there were 2 additional light-responsive elements in the promoter of CsBRC1 in cultivated cucumber other than in hardwickii, which might be potentially related to enhanced CsBRC1 expression when light intensity was dramatically reduced (SI Appendix, Fig. S6). Increase in planting density is a common feature during crop domestication (52). To maximize light harvesting and avoid shade from neighboring vegetation, plants have evolved the shade avoidance syndrome, among which suppression of shoot branching is one of the key developmental responses. In wild cucumber, low CsBRC1 expression allows higher levels of PIN3-mediated auxin efflux from buds to the stem, which leads to axillary bud outgrowth (Fig. 6A). In cultivated cucumber, 2 inserted light response cis elements in the CsBRC1 promoter during domestication possibly favor elevation of CsBRC1 expression and plant competition for light interception in the densely planting system. Another possibility is that yet-to-be-identified upstream regulators promote CsBRC1 transcription in cultivated cucumber. Considering that CsBRC1 expression varies among cultivated cucumber cultivars (Fig. 1), high CsBRC1 expression reduces CsPIN3-mediated auxin efflux from lateral buds and thus suppresses axillary bud outgrowth (Fig. 6B); our work suggests a strategy, via modulating CsBRC1 expression, to breed for cultivars with varying degrees of shoot branching grown in different cucumber production systems.
Materials and Methods
Plant Materials and Growth Conditions.
Cucumber (Cucumis sativus L.) inbred lines R1461 and Gy14, as well as wild ancestor hardwickii, were used in this study. The cucumber seeds were germinated at 28 °C in dark and then grown in a growth chamber at 16 h day (25 °C) and 8 h night (18 °C) until 2 true-leaf stage. Seedlings were then transferred to the greenhouse of China Agricultural University in Beijing. Water and pest control were performed according to standard protocol.
Gene Cloning and Phylogenetic Analysis.
The complete coding sequence of CsBRC1, CsPIN1b, and CsPIN3 was amplified from the axillary buds using gene-specific primers. The full-length amino acid sequence of cucumber CsBRC1, CsPIN1b, and CsPIN3 homologs were performed by BLASTP at Cucurbit Genomics Database (http://cucurbitgenomics.org/). The phylogenetic tree was generated with MEGA5 software, using the neighbor-joining method with 1,000 bootstrap replicates (53). Gene information for protein alignment and phylogenetic analysis is listed in SI Appendix, Table S2.
Expression Analysis by Quantitative Real-Time PCR.
The cucumber root, stem, leaf, flower bud, fruit, shoot tip, and axillary bud were used for total RNA extraction and cDNA synthesis. The qRT-PCR analysis was performed using ABI PRISM 7500 Real-Time PCR System (Applied Biosystems). The cucumber UBIQUITIN (Csa000874) was used as internal controls. Three biological and 3 technical replicates (3 × 3) were performed for each gene. The primer information is listed in SI Appendix, Table S3.
Subcellular Localization.
The full-length coding sequence of CsBRC1 without the termination codon was cloned into the plasmid pCAMBIA1300 to fuse with GFP. Subcellular localization was performed in the abaxial sides of tobacco leaves (4 to 6 wk old) as described previously (54). Fluorescence images were taken with a confocal laser-scanning microscope (Leica SP8) excited at a 488-nm wavelength. The primers are listed in SI Appendix, Table S3.
In Situ Hybridization.
Cucumber shoot apexes of 15-, 20-, 25-, 30-, and 35-d-old seedlings were fixed in 3.7% formol-acetic-alcohol. Sample fixation, sectioning, and hybridization were performed as described (55). The CsBRC1 probe was 501-bp and the CsPIN3 probe was 1,020-bp linearized fragments from the unique region of the corresponding coding sequence. The primers are listed in SI Appendix, Table S3.
Cucumber Transformation.
A 585-bp specific fragment of the CsBRC1 coding sequence was used for generating the CsBRC1-RNAi construct, and the empty PFGC-1008 vector was used as transformation control. The recombinant construct was introduced into Agrobacterium strain GV3101 and transformed into cucumber inbred line R1461 using a cotyledon transformation method as described (33, 34). To generate ProCsBRC1:CsPIN1b and ProCsBRC1:CsPIN3, the full-length coding sequence of CsPIN1b and CsPIN3 was driven by the 2-kb promoter sequence of CsBRC1, and the recombinant construct was transformed into cucumber line R1461 as described above. The primers are listed in SI Appendix, Table S3.
RNA-Seq Sample Preparation and Sequencing.
The lateral branches from the same node of WT and transgenic cucumber plants were collected for RNA-seq analysis. Two biological replicates were performed for each sample. Illumina HiSeq 2000 platform was used to generate 100-bp paired-end reads. Bioinformatic analyses of RNA-Seq data were performed as previously described (56).
Extraction and Quantification of Endogenous Auxins.
About 0.1 g samples were harvested from axillary buds of WT and CsBRC1-RNAi transgenic plants and used for measurement of auxin content. The extraction and quantification of endogenous auxin were performed using ELISAs according to methods previously described (57). Three biological repeats were performed for each sample.
Immuno-Gold Localization of IAA.
Cucumber axillary buds were fixed and incubated with anti-IAA monoclonal antibody as previously described (57). The primary antibody (IAA | indole-3-acetic acid [N1]) and secondary antibody (Anti-Rabbit IgG–Gold antibody produced in goat) were used in this experiment. Images were obtained using a light microscope (D72; Olympus).
Auxin Transport Inhibitor Treatment.
Axillary buds (about 2 mm) at the same nodes of wild cucumber hardwickii were treated with 0.5 mM 2, 3, 5-Triiodobenzoic acid (TIBA) as auxin transport inhibitor. The treatment was repeated every 72 h until 15 d. Three biological repeats were performed for each treatment.
Yeast 1-Hybrid Assay.
The coding region sequence of CsBRC1 was cloned into prey vector (pGADT7), and the oligonucleotides of CsBRC1-binding sites (P1 in CsPIN1 and P3A–P3E in CsPIN3) were synthesized and ligated into the pAbAi vectors (Clontech). The linearized pAbAi vectors containing the binding sites were transformed into the yeast strain Y1H Gold according to the manufacturer’s manual. Then pGADT7-CsBRC1 was transformed into the Y1H Gold strain with CsBRC1 binding sites and selected by AbA (Aureobasidin A) on SD/-LEU (Synthetic Dropout Medium/-Leucine) medium (Clontech). Interaction of IND-AD with the PID-E box was used as a positive control (57), and the empty AD and PID-E box served as a negative control. The primers are listed in SI Appendix, Table S3.
Dual Luciferase (LUC) Assay for Protein–DNA Interactions in Tobacco Leaves.
Promoters of CsPIN1b and CsPIN3 (2,000 bp) were cloned into the transient expression vector pGreenII 0800-Luc. The CsBRC1 coding sequence was cloned into pGreenII 62-SK as the effector. Tobacco (Nicotiana benthamiana) leaves were used for coexpression studies as previously described (57). The empty vector pGreenII 62-SK and the proCsPIN1b-LUC or proCsPIN3-LUC vector were used as negative controls. The dual luciferase assay reagents (Promega) were used for examining the firefly luciferase and renilla luciferase. The primers for all constructs are listed in SI Appendix, Table S3.
Electrophoretic Mobility Shift Assay.
The full-length CsBRC1 protein sequence was fused with MBP tag and introduced into Escherichia coli BL21 (DE3). Hot probes harboring elements (P1 in CsPIN1 and P3A-P3C in CsPIN3) were synthesized and labeled with biotin. The recombinant protein was purified by using maltose. An electrophoretic mobility shift assay (EMSA) was performed using the Light Shift Chemiluminescent EMSA kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. The primers are listed in SI Appendix, Table S3.
ChIP-qPCR Assay.
The recombinant pCAMBIA1300-CsBRC1-GFP construct and pCAMBIA1300-GFP constructs were transformed into cucumber calli of hypocotyl explants, and the ChIP assays were performed as described in the EpiQuik Plant ChIP Kit (Epigentek) with anti-GFP antibodies (Sigma-Aldrich). Probes of P1 and P3A–P3DE were analyzed to assess their enrichment. For each ChIP-qPCR assay, 3 biological repeats and 3 technical replicates (3 × 3) were included. The primers are listed in SI Appendix, Table S3.
Shade Treatment.
The hardwickii and R1461 plants with 15 nodes were selected for shade treatment. Half of the plants were shaded for 48 h (frame covered with a thick black fabric shading cloth which could block 90% of light). The other half of the plants placed under normal light conditions (10 h day/14 h night) were used as control. Axillary buds at the same node in plants were harvested to detect the expression level of CsBRC1 after 10 or 20 h of shading.
Supplementary Material
Acknowledgments
This study was supported by the National Key Research and Development Program (Grant 2018YFD1000800), National Natural Science Foundation of China (Grants 31572132 and 31772315), 111 Project (Grant B17043), and the Construction of Beijing Science and Technology Innovation and Service Capacity in Top Subjects (Grant CEFF-PXM2019_014207_000032). The authors are grateful to Dr. Yiqun Weng and Dr. Zhonghua Zhang for sharing the wild cucumber hardwickii seeds.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: RNA sequence data were deposited to the Gene Expression Omnibus database at the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/geo/) with accession number GSE123949.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1907968116/-/DCSupplemental.
References
- 1.Greb T., et al. , Molecular analysis of the LATERAL SUPPRESSOR gene in Arabidopsis reveals a conserved control mechanism for axillary meristem formation. Genes Dev. 17, 1175–1187 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schmitz G., Theres K., Shoot and inflorescence branching. Curr. Opin. Plant Biol. 8, 506–511 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Ross-Ibarra J., Morrell P. L., Gaut B. S., Plant domestication, a unique opportunity to identify the genetic basis of adaptation. Proc. Natl. Acad. Sci. U.S.A. 104 (suppl. 1), 8641–8648 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qi J., et al. , A genomic variation map provides insights into the genetic basis of cucumber domestication and diversity. Nat. Genet. 45, 1510–1515 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Wang B., Smith S. M., Li J., Genetic regulation of shoot architecture. Annu. Rev. Plant Biol. 69, 437–468 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Thimann K. V., Skoog F., Studies on the growth hormone of plants: III. The inhibiting action of the growth substance on bud development. Proc. Natl. Acad. Sci. U.S.A. 19, 714–716 (1933). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cline M. G., Apical dominance. Bot. Rev. 57, 318–358 (1991). [Google Scholar]
- 8.Leyser O., Regulation of shoot branching by auxin. Trends Plant Sci. 8, 541–545 (2003). [DOI] [PubMed] [Google Scholar]
- 9.González-Grandío E., et al. , Abscisic acid signaling is controlled by a BRANCHED1/HD-ZIP I cascade in Arabidopsis axillary buds. Proc. Natl. Acad. Sci. U.S.A. 114, E245–E254 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brewer P. B., Dun E. A., Ferguson B. J., Rameau C., Beveridge C. A., Strigolactone acts downstream of auxin to regulate bud outgrowth in pea and Arabidopsis. Plant Physiol. 150, 482–493 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seale M., Bennett T., Leyser O., BRC1 expression regulates bud activation potential but is not necessary or sufficient for bud growth inhibition in Arabidopsis. Development 144, 1661–1673 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balla J., Kalousek P., Reinöhl V., Friml J., Procházka S., Competitive canalization of PIN-dependent auxin flow from axillary buds controls pea bud outgrowth. Plant J. 65, 571–577 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Sachs T., Polarity and induction of organized vascular tissues. Ann. Bot. 33, 263–275 (1969). [Google Scholar]
- 14.Sachs T., The control of the patterned differentiation of vascular tissues. Adv. Bot. Res. 9, 151–262 (1981). [Google Scholar]
- 15.Bennett T., Hines G., Leyser O., Canalization: What the flux? Trends Genet. 30, 41–48 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Prusinkiewicz P., et al. , Control of bud activation by an auxin transport switch. Proc. Natl. Acad. Sci. U.S.A. 106, 17431–17436 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doebley J., Stec A., Hubbard L., The evolution of apical dominance in maize. Nature 386, 485–488 (1997). [DOI] [PubMed] [Google Scholar]
- 18.Aguilar-Martínez J. A., Poza-Carrión C., Cubas P., Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell 19, 458–472 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martín-Trillo M., et al. , Role of tomato BRANCHED1-like genes in the control of shoot branching. Plant J. 67, 701–714 (2011). [DOI] [PubMed] [Google Scholar]
- 20.Finlayson S. A., Arabidopsis Teosinte Branched1-like 1 regulates axillary bud outgrowth and is homologous to monocot Teosinte Branched1. Plant Cell Physiol. 48, 667–677 (2007). [DOI] [PubMed] [Google Scholar]
- 21.Minakuchi K., et al. , FINE CULM1 (FC1) works downstream of strigolactones to inhibit the outgrowth of axillary buds in rice. Plant Cell Physiol. 51, 1127–1135 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Braun N., et al. , The pea TCP transcription factor PsBRC1 acts downstream of strigolactones to control shoot branching. Plant Physiol. 158, 225–238 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mason M. G., Ross J. J., Babst B. A., Wienclaw B. N., Beveridge C. A., Sugar demand, not auxin, is the initial regulator of apical dominance. Proc. Natl. Acad. Sci. U.S.A. 111, 6092–6097 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barbier F., et al. , Sucrose is an early modulator of the key hormonal mechanisms controlling bud outgrowth in Rosa hybrida. J. Exp. Bot. 66, 2569–2582 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kebrom T. H., et al. , Inhibition of tiller bud outgrowth in the tin mutant of wheat is associated with precocious internode development. Plant Physiol. 160, 308–318 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.González-Grandío E., Poza-Carrión C., Sorzano C. O., Cubas P., BRANCHED1 promotes axillary bud dormancy in response to shade in Arabidopsis. Plant Cell 25, 834–850 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kebrom T. H., Brutnell T. P., Finlayson S. A., Suppression of sorghum axillary bud outgrowth by shade, phyB and defoliation signalling pathways. Plant Cell Environ. 33, 48–58 (2010). [DOI] [PubMed] [Google Scholar]
- 28.Nicolas M., Rodríguez-Buey M. L., Franco-Zorrilla J. M., Cubas P., A recently evolved alternative splice site in the BRANCHED1a gene controls potato plant architecture. Curr. Biol. 25, 1799–1809 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Yang Y., et al. , The TIE1 transcriptional repressor controls shoot branching by directly repressing BRANCHED1 in Arabidopsis. PLoS Genet. 14, e1007296 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu Z., et al. , Genome-wide binding analysis of the transcription activator ideal plant architecture1 reveals a complex network regulating rice plant architecture. Plant Cell 25, 3743–3759 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dong Z., et al. , Ideal crop plant architecture is mediated by tassels replace upper ears1, a BTB/POZ ankyrin repeat gene directly targeted by TEOSINTE BRANCHED1. Proc. Natl. Acad. Sci. U.S.A. 114, E8656–E8664 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hubbard L., McSteen P., Doebley J., Hake S., Expression patterns and mutant phenotype of teosinte branched1 correlate with growth suppression in maize and teosinte. Genetics 162, 1927–1935 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ding L., et al. , HANABA TARANU regulates the shoot apical meristem and leaf development in cucumber (Cucumis sativus L.). J. Exp. Bot. 66, 7075–7087 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao W., et al. , CsLFY is required for shoot meristem maintenance via interaction with WUSCHEL in cucumber (Cucumis sativus). New Phytol. 218, 344–356 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Robinson M. D., McCarthy D. J., Smyth G. K., edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jásik J., et al. , Effects of auxins on PIN-FORMED2 (PIN2) dynamics are not mediated by inhibiting PIN2 endocytosis. Plant Physiol. 172, 1019–1031 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martín-Trillo M., Cubas P., TCP genes: A family snapshot ten years later. Trends Plant Sci. 15, 31–39 (2010). [DOI] [PubMed] [Google Scholar]
- 38.Barbier F. F., Dun E. A., Beveridge C. A., Apical dominance. Curr. Biol. 27, R864–R865 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Domagalska M. A., Leyser O., Signal integration in the control of shoot branching. Nat. Rev. Mol. Cell Biol. 12, 211–221 (2011). [DOI] [PubMed] [Google Scholar]
- 40.Bennett T., et al. , Connective auxin transport in the shoot facilitates communication between shoot apices. PLoS Biol. 14, e1002446 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Crawford S., et al. , Strigolactones enhance competition between shoot branches by dampening auxin transport. Development 137, 2905–2913 (2010). [DOI] [PubMed] [Google Scholar]
- 42.Shinohara N., Taylor C., Leyser O., Strigolactone can promote or inhibit shoot branching by triggering rapid depletion of the auxin efflux protein PIN1 from the plasma membrane. PLoS Biol. 11, e1001474 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Geldner N., Friml J., Stierhof Y. D., Jürgens G., Palme K., Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature 413, 425–428 (2001). [DOI] [PubMed] [Google Scholar]
- 44.Band L. R., et al. , Systems analysis of auxin transport in the Arabidopsis root apex. Plant Cell 26, 862–875 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pernisova M., et al. , Cytokinins influence root gravitropism via differential regulation of auxin transporter expression and localization in Arabidopsis. New Phytol. 212, 497–509 (2016). [DOI] [PubMed] [Google Scholar]
- 46.Waldie T., Leyser O., Cytokinin targets auxin transport to promote shoot branching. Plant Physiol. 177, 803–818 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Rongen M., Bennett T., Ticchiarelli F., Leyser O., Connective auxin transport contributes to strigolactone-mediated shoot branching control independent of the transcription factor BRC1. PLoS Genet. 15, e1008023 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Friml J., Wiśniewska J., Benková E., Mendgen K., Palme K., Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 415, 806–809 (2002). [DOI] [PubMed] [Google Scholar]
- 49.Fletcher J. C., Shoot and floral meristem maintenance in Arabidopsis. Annu. Rev. Plant Biol. 53, 45–66 (2002). [DOI] [PubMed] [Google Scholar]
- 50.Doebley J. F., Gaut B. S., Smith B. D., The molecular genetics of crop domestication. Cell 127, 1309–1321 (2006). [DOI] [PubMed] [Google Scholar]
- 51.Dong Z., Alexander M., Chuck G., Understanding grass domestication through maize mutants. Trends Genet. 35, 118–128 (2019). [DOI] [PubMed] [Google Scholar]
- 52.Smith J. E., Jordan P. W., Stand density effects on branching in an annual legume (Senna obtusifolia). Ann. Bot. 74, 17–25 (1994). [DOI] [PubMed] [Google Scholar]
- 53.Tamura K., et al. , MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao J., et al. , A functional allele of CsFUL1 regulates fruit length through repressing CsSUP and inhibiting auxin transport in Cucumber. Plant Cell 31, 1289–1307 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang X., et al. , Transcription repressor HANABA TARANU controls flower development by integrating the actions of multiple hormones, floral organ specification genes, and GATA3 family genes in Arabidopsis. Plant Cell 25, 83–101 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sun C., et al. , Integration of hormonal and nutritional cues orchestrates progressive corolla opening. Plant Physiol. 171, 1209–1229 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu X., et al. , CsSPL functions as an adaptor between HD-ZIP III and CsWUS transcription factors regulating anther and ovule development in Cucumis sativus (cucumber). Plant J. 94, 535–547 (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.