Abstract
During the last decade, the endosymbiont bacterium Wolbachia has emerged as a biological tool for vector disease control. However, for long time, it was believed that Wolbachia was absent in natural populations of Anopheles. The recent discovery that species within the Anopheles gambiae complex host Wolbachia in natural conditions has opened new opportunities for malaria control research in Africa. Here, we investigated the prevalence and diversity of Wolbachia infection in 25 African Anopheles species in Gabon (Central Africa). Our results revealed the presence of Wolbachia in 16 of these species, including the major malaria vectors in this area. The infection prevalence varied greatly among species, confirming that sample size is a key factor to detect the infection. Moreover, our sequencing and phylogenetic analyses showed the important diversity of Wolbachia strains that infect Anopheles. Co‐evolutionary analysis unveiled patterns of Wolbachia transmission within some Anopheles species, suggesting that past independent acquisition events were followed by co‐cladogenesis. The large diversity of Wolbachia strains that infect natural populations of Anopheles offers a promising opportunity to select suitable phenotypes for suppressing Plasmodium transmission and/or manipulating Anopheles reproduction, which in turn could be used to reduce the malaria burden in Africa.
Keywords: Anopheles, co‐evolution, disease control, diversity, Wolbachia
1. INTRODUCTION
Malaria still affects millions of people and is the cause of thousands of deaths worldwide, although sub‐Saharan Africa pays the highest tribute (WHO, 2018). Currently, vector control measures (e.g., insecticide‐treated bed nets or indoor residual sprays) are the largest contributors to malaria eradication (Bhatt et al., 2015). If these interventions are maintained or increased, malaria burden should be drastically reduced in Africa before 2030 (Griffi et al., 2016). These predictions are based on the constant effectiveness of these methods. However, the spread of insecticide resistance (Ranson & Lissenden, 2016) and vector behavioural changes related to the massive use of bed nets (Pates & Curtis, 2005) might challenge malaria eradication in the coming decades. Therefore, it is vital to develop alternative and non‐insecticide‐based control strategies for malaria control, at it has been promoted by the Global Technical Strategy form Malaria 2016–2030, which look for “reducing global malaria incidence and mortality rates by at least 90% by 2030” (Newby et al., 2016; WHO, 2015).
Several methods have been proposed to accompany or replace the use of synthetic insecticides (McGraw & O'Neill, 2013). Among them, the use of the maternally inherited Wolbachia bacteria (α‐proteobacteria, Anaplasmataceae family) has emerged as a promising alternative biological tool for fighting malaria and other vector‐borne diseases (Bourtzis et al., 2014; Hoffmann, Ross, & Rasic, 2015; Iturbe‐Ormaetxe, Walker, & Neill, 2011; Kambris, Cook, Phuc, & Sinkins, 2009; McGraw & O'Neill, 2013). This bacterium exhibits a large spectrum of interactions with its hosts: from mutualism and commensalism to parasitism (Werren, Baldo, & Clark, 2008). Moreover, Wolbachia can invade mosquito populations and/or prevent vector‐borne infections in some of the most important mosquito vectors (Dodson et al., 2014; Hoffmann et al., 2015; Iturbe‐Ormaetxe et al., 2011). Indeed, Aedes aegypti populations that were artificially infected with Wolbachia have been successfully used to suppress dengue transmission in laboratory conditions and have been released in natural populations of this mosquito (Hoffmann et al., 2011; Schmidt et al., 2017). Similarly, laboratory studies showed that infection of Anopheles (the vector of human malaria) with Wolbachia strains has a negative impact on the transmission of Plasmodium parasites (Bian et al., 2013; Hughes, Koga, Xue, Fukatsu, & Rasgon, 2011; Kambris et al., 2010), providing a relevant alternative for malaria control. Unfortunately, only one stable transfected Wolbachia colony has been described in Anopheles stephensi (Bian et al., 2013). Therefore, data on the use Wolbachia for Anopheles control remain scarce and mainly concern experimental studies in laboratory conditions (Bian et al., 2013; Hughes, Vega‐Rodriguez, Xue, & Rasgon, 2012), due to technical (i.e., egg microinjection) and biological (i.e., competitive exclusion with the bacterium Asaia) difficulties in carrying out transinfections in Anopheles, despite multiple assays (Hughes, Dodson, et al., 2014; Jeffries, Golovko, et al., 2018; Jeffries, Lawrence, et al., 2018; Rossi et al., 2015). For a long time, it was assumed that Wolbachia was absent in natural populations of Anopheles (Hughes, Dodson, et al., 2014). However, in the last few years, three studies reported that Anopheles gambiae, Anopheles coluzzii and Anopheles arabiensis (three major malaria vectors) populations from Burkina Faso and Mali (West Africa) are naturally infected by Wolbachia (Baldini et al., 2014; Gomes et al., 2017; Shaw et al., 2016). Notably, they showed a negative correlation between Wolbachia infection and Plasmodium development (Gomes et al., 2017; Shaw et al., 2016). Moreover, a very recent report suggests that other Anopheles species also are infected with Wolbachia (Jeffries, Golovko, et al., 2018; Jeffries, Lawrence, et al., 2018). These findings support the development of novel vector control strategies based on Wolbachia–Anopheles interactions. However, although Wolbachia naturally infects 40%–60% of arthropods (Duron et al., 2008; Zug & Hammerstein, 2012), infection of Anopheles species is still not well documented. Moreover, during the last decade, screens in many other malaria mosquito species worldwide (n = 38) did not bring any evidence of Wolbachia infection (Bourtzis et al., 2014; Hughes, Dodson, et al., 2014; Osei‐Poku, Han, Mbogo, & Jiggins, 2012).
In this study, we investigated the presence of Wolbachia in 25 Anopheles species in Gabon, Central Africa. We sampled mosquitoes across the country and in a variety of ecological settings, from deep rainforest to urban habitats. By using a molecular approach, we confirmed Wolbachia presence in 16 species, including all the major malaria vectors in Central Africa (An. gambiae, An. coluzzii, Anopheles funestus, Anopheles nili and Anopheles moucheti). The prevalence of Wolbachia infection was particularly high in An. nili and An. moucheti. Phylogenetic analysis revealed that all the infected mosquito species hosted Wolbachia bacteria belonging to the supergroup A or B (both exhibit high genetic diversity). Finally, we explored the co‐evolution between Wolbachia and Anopheles. The results have direct implications for the development of new and non‐insecticide‐based vector control strategies and open new directions for research on pathogen transmission and reproductive manipulation.
2. MATERIAL AND METHODS
2.1. Research and ethics statements
Mosquitoes were collected in Gabon under the research authorization AR0013/16/MESRS/CENAREST/CG/CST/CSAR and the national park entry authorization AE16008/PR/ANPN/SE/CS/AEPN. Mosquito sampling using the human‐landing catch (HLC) method was performed under the protocol 0031/2014/SG/CNE approved by the National Research Ethics Committee of Gabon.
2.2. Mosquito sampling and DNA extraction
Mosquitoes were collected in eight sites across Gabon, Central Africa, from 2012 to 2016 (Figure 1, Table 1, Appendix S1). These sites included sylvatic (national parks) and domestic habitats (villages and cities). Adult females were collected using Center for Disease Control () light traps, BioGents (BG) traps and HLC. Overall, CDC and BG were used in sylvatic and HLC in domestic sites (see Figure 1, Table S1). Collected specimens were taxonomically identified according to standard morphological features (Gillies & Coetzee, 1987; Gillies & de Meillon, 1968). Then, they were individually stored in 1.5 ml tubes at −20°C and sent to Centre International de Recherches Scientifiques de Franceville for molecular analysis. When possible, at least 30 mosquitoes (from 1 to 58) for each Anopheles species from different sites were selected for genomic analysis. Total genomic DNA was extracted from the whole body using the DNeasy Blood and Tissue Kit (Qiagen), according to the manufacturer's instructions. Genomic DNA was eluted in 100 μl of TE buffer. Specimens belonging to the An. gambiae complex, An. funestus group, An. moucheti complex and An. nili complex were molecularly identified using PCR‐based diagnostic protocols (Cohuet et al., 2003; Fanello, Santolamazza, & della Torre, 2002; Kengne et al., 2007; Kengne, Awono‐Ambene, Nkondjio, Simard, & Fontenille, 2003; Santolamazza et al., 2008).
Figure 1.
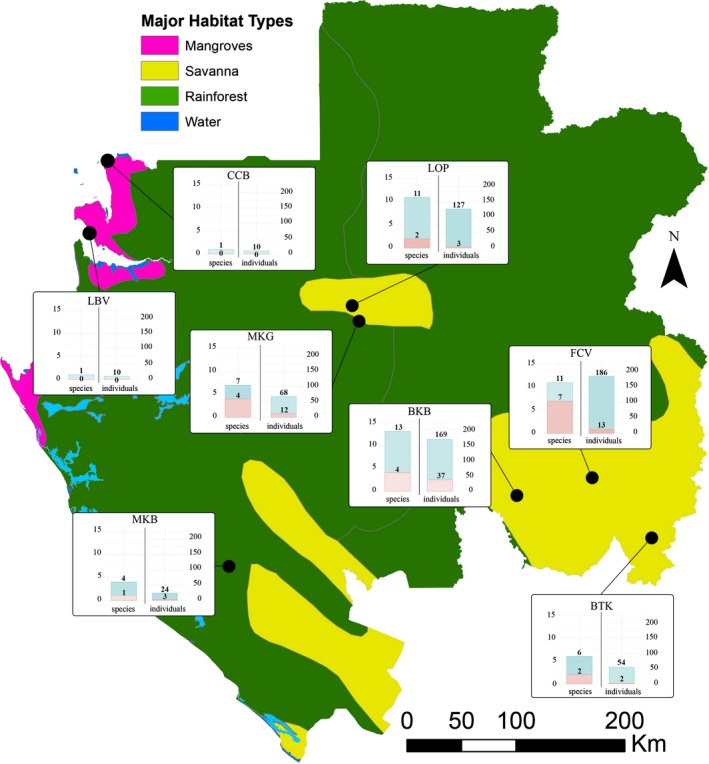
Sampling sites and Wolbachia infection prevalence. Map of Gabon showing the main African habitat types ((Olson et al., 2001), free ly available at http://maps.tnc.org/gis_data.html) and the villages where sampling took place (black dots). The map was drawn using ArcGIS Basic v.10. The prevalence of Wolbachia infection (number of infected Anopheles species and individuals) per site is presented in bar charts. The pink colour indicates positive species/individuals and blue the total number of species/individuals screened for Wolbachia infection at that site. BKB: Bakoumba; BTK: National Park of Plateaux Batékés; CCB: Cocobeach; FCV: Franceville; LBV: Libreville; LOP: Lopé; MKB: National Park of Moukalaba‐Doudou; MKG: Mikongo
Table 1.
Summary of the Anopheles species screened in this study
| Group/complex | Species | Malaria role | Infected | Tested | Infection (%) |
|---|---|---|---|---|---|
| gambiae | An. gambiae | H | 5 | 44 | 11 |
| An. coluzzii | H | 2 | 58 | 3 | |
| An. brunnipes | 0 | 1 | 0 | ||
| An. cinctus | 0 | 2 | 0 | ||
| moucheti | An. moucheti | H, P, A | 30 | 42 | 71 |
| An. nigeriensis | h | 1 | 27 | 4 | |
| An. "GAB−2" | 5 | 8 | 63 | ||
| An. "GAB−3" | 1 | 1 | 100 | ||
| An. gabonensis | A | 0 | 29 | 0 | |
| funestus | An. funestus | H | 2 | 37 | 5 |
| An. implexus | 1 | 26 | 4 | ||
| An. jebudensis | 1 | 2 | 50 | ||
| An. maculipalpis | 0 | 29 | 0 | ||
| nili | An. nili | H, A | 11 | 19 | 58 |
| An. carnevalei | h, A | 2 | 29 | 7 | |
| An. "GAB−1" | 0 | 19 | 0 | ||
| An. hancocki | h | 1 | 41 | 2 | |
| An. theileri | h | 0 | 24 | 0 | |
| An. rodhesiensis | 0 | 4 | 0 | ||
| coustani | An. coustani | h, A | 2 | 35 | 6 |
| An. paludis | h, A | 1 | 16 | 6 | |
| An. gr coustani | h | 0 | 51 | 0 | |
| An. squamosus | 0 | 32 | 0 | ||
| An. marshallii | h, P, A | 2 | 42 | 5 | |
| An. vinckei | P, A | 3 | 30 | 10 | |
| Total | 70 | 648 |
2.3. Wolbachia screening and multilocus sequence typing analysis
Wolbachia infection in adult females was detected by nested PCR amplification of a Wolbachia‐specific 16S rDNA fragment (~400 bp) using 2 μl of host genomic DNA, according to the protocol developed in Catteruccia's laboratory (Shaw et al., 2016). Amplification of this 16S rDNA fragment in infected Aedes albopictus and Culex pipiens genomic DNA (data not shown) confirmed the performance of this nested PCR protocol to detect Wolbachia in many different mosquito species (Shaw et al., 2016). To detect potential contaminations, Ae. albopictus and Culex quinquefasciatus from Gabon were used as positive controls, and water and Ae. aegypti as negative controls. Moreover, PCR amplifications for each species were carried out independently and on different days. The amplicon size was checked on 1.5% agarose gels, and amplified 16S rDNA fragments were sent to Genewiz (UK) for sequencing (forward and reverse) to confirm the presence of Wolbachia‐specific sequences. The DNA quality of all samples was confirmed by the successful amplification of a fragment (~800 bp) of the mitochondrial gene COII in all the Anopheles species under study (Ndo et al., 2010; Rahola et al., 2014). PCR products were run on 1.5% agarose gels, and COII fragments from 176 mosquito specimens of the 25 species were sequenced (forward and reverse) by Genewiz (UK) for the Anopheles phylogenetic studies. Wolbachia‐positive genomic DNA samples (2 μl/sample) were then genotyped by multilocus sequence typing (MLST) using three loci, coxA (~450 bp) ftsZ (~500 bp) and fbpA (~460 bp) (Baldo et al., 2006), and according to standard conditions (Baldo et al., 2006). If the three fragments could not be amplified, a newly developed nested PCR protocol was used. Specifically, after the first run with the standard primers, 2 μl of the obtained product was amplified again using internal primers specific for each gene: coxA (coxA_NF‐2: 5′‐TTTAACATGCGCGCAAAAGG‐3′; coxA_NR‐2: 5′‐TAAGCCCAACAGTGAACATATG‐3′), ftsZ (ftsZ_NF‐2: 5′‐ATGGGCGGTGGTACTGGAAC‐3′; ftsZ_NR‐2: 5′‐AGCACTAATTGCCCTATCTTCT‐3′) and fbpA (fbpA_NF‐1: 5′‐AGCTTAACTTCTGATCAAGCA‐3′; fbpA_NR‐1: 5′‐TTCTTTTTCCTGCAAAGCAAG‐3′). Cycling conditions for coxA and ftsZ were as follows: 94°C for 5 min, followed by 36 cycles at 94°C for 15 s, 55°C for 15 s and 72°C for 30 s, and a final extension step at 72°C for 10 min. For fbpA, they were: 94°C for 5 min followed by 36 cycles at 94°C for 30 s, 59°C for 45 s and 72°C for 90 s, and a final extension step at 72°C for 10 min. The resulting fragments (coxA, 357 bp; fbpA, 358 bp; and ftsZ, 424 bp) were sequenced bidirectionally by Genewiz. The new sequences obtained in this study were submitted to GenBank (Table S1). Unfortunately, the other three MLST genes (gatB, wsp and hcpA) could not be amplified, due to technical problems (i.e., multiple bands).
2.4. Phylogenetic and statistical analysis
All Wolbachia sequences for the 16S, coxA, fbpA and ftsZ gene fragments and for Anopheles COII were manually corrected using Geneious R10 (Kearse et al., 2012). The resulting consensus sequences for each gene were aligned with sequences that represent the main known Wolbachia supergroups obtained from GenBank (see Table S1). Only unique haplotypes for each species were included in the analysis (haplotype was defined as a unique allelic profile for each examined locus). Inference of phylogenetic trees was performed using the maximum likelihood (ML) method and RAxML (Stamatakis, 2014) with a substitution model GTR + CAT (Stamatakis, 2006) and 1,000 bootstrapping replicates. Finally, all MLST Wolbachia sequences were used to build phylogenetic trees using RAxML (GTR + CAT model, 1,000 bootstrapping replicates). Trees were visualized with iTOL v.3.4.3 (Letunic & Bork, 2007).
To quantify the accuracy of the observed Wolbachia infection prevalence, the influence of sample size on its estimation was assessed. For this, it was assumed that Wolbachia prevalence within a host species followed a beta binomial distribution (Zug & Hammerstein, 2012) yielding many species with a low or a high Wolbachia prevalence but few with an intermediate one. This allowed quantifying, for each sample size, the proportion of samples (over 1,000 realizations) that could yield an estimate that was not significantly different from the prevalence over the whole population with a z test and a significance threshold at 95%. As expected, sample size could be small for very low (<15%) or very high prevalence (>60%; 60 individuals are enough in 95% of cases for these extreme prevalence values), while it was much higher for intermediate prevalence values (up to 150 individuals for a prevalence value close to 50%).
All statistical analyses were performed using “R” v3.2.5 (R Development Core Team, http://cran.r-project.org/), with the addition of the “ggplot2” library (Wickham, 2009).
3. RESULTS
3.1. Wolbachia naturally infects a large number of Anopheles species from Gabon
In this study, we screened 648 mosquitoes from eight sites in Gabon (Figure 1, Table 1, Table S1). On the basis of their morphological traits (Gillies & Coetzee, 1987) and molecular analysis results (Cohuet et al., 2003; Kengne et al., 2007, 2003; Rahola et al., 2014; Santolamazza et al., 2008), we identified 25 Anopheles species (Appendix S1). Our sampling included all the species in which the presence of Wolbachia was previously investigated in Africa (An. gambiae, An. coluzzii, An. funestus and Anopheles coustani), with the exception of An. arabiensis that is absent in Gabon (Table 1) (Makanga et al., 2016). By PCR amplification of a 16S rRNA fragment (Shaw et al., 2016), we found 70 Wolbachia‐positive specimens that belonged to 16 different Anopheles species, distributed throughout the country (Figure 1, Table S1). When considering only species with more than 10 screened individuals, we observed that Wolbachia infection was commonly lower than 15% (11/13), as observed in other arthropods (Duron et al., 2008; Zug & Hammerstein, 2012). On the other hand, two species, and moreover major malaria vectors, An. moucheti and An. nili, exhibited more than 50% of Wolbachia infection (Table 1), as previously reported in other mosquito species where prevalence can be very high (Dumas et al., 2013; Duron et al., 2005).
3.2. Wolbachia is maternally inherited in An. moucheti
Although Wolbachia is mainly maternally transmitted (Werren et al., 2008), horizontal transmission may occasionally occur in natural conditions (Ahmed, De Barro, Ren, Greeff, & Qiu, 2013; Li et al., 2017; Werren, Zhang, & Guo, 1995). To confirm the maternal transmission in the infected mosquito species, we focused on An. moucheti for logistic reasons (i.e., highest Wolbachia prevalence and ease of sampling). Although no laboratory An. moucheti strain is currently available, we obtained eggs from six Wolbachia‐infected females. In total, we analysed the infectious status of 79 progeny by PCR amplification of the same 16S rRNA fragment (Shaw et al., 2016) (Table S3) and found that 70 were infected, with an average maternal transmission frequency of 97.54% (range: 90%–100%).
3.3. Naturally occurring Wolbachia strains in Anopheles reveal high genetic diversity
By sequence analysis of the 16S rRNA fragment PCR amplified from each Anopheles sample (Table 1), we could assign the Wolbachia strains to three pre‐existing supergroups: A (n = 5), B (n = 64) and C (n = 1; Figure 2). Specifically, we detected supergroup B Wolbachia in 64 mosquitoes belonging to all 16 infected Anopheles species. We found supergroup A Wolbachia in five individuals from four species (An. funestus, An. coluzzii, Anopheles vinckei and Anopheles carnevalei), thus providing examples of multiple infections, as previously observed in Ae. albopictus (Sinkins, Braig, & Oneill, 1995) (Figure 2). None of the mosquitoes examined was co‐infected by Wolbachia strains belonging, for instance, to the supergroups A and B. Moreover, we confirmed that the Wolbachia strains previously identified in An. gambiae s.l. from Burkina Faso and Mali are included in the supergroups A and B (Baldini et al., 2014; Gomes et al., 2017). Finally, we found that one An. coustani individual was infected by a Wolbachia strain from supergroup C that is known to infect only filarial worms. Therefore, we investigated the presence of filarial nematode DNA in the mosquito by PCR amplification and sequencing of a fragment of the COI filarial gene (Casiraghi, Anderson, Bandi, Bazzocchi, & Genchi, 2001), followed by phylogenetic analysis with RAxML. Our results confirmed the presence of Dirofilaria immitis in this specimen (Figure S1). This canine filarial parasite hosts Wolbachia and is transmitted by many mosquitoes, including Anopheles (Simon et al., 2012). Therefore, it is not surprising to find an An. coustani specimen infected by this filarial nematode.
Figure 2.
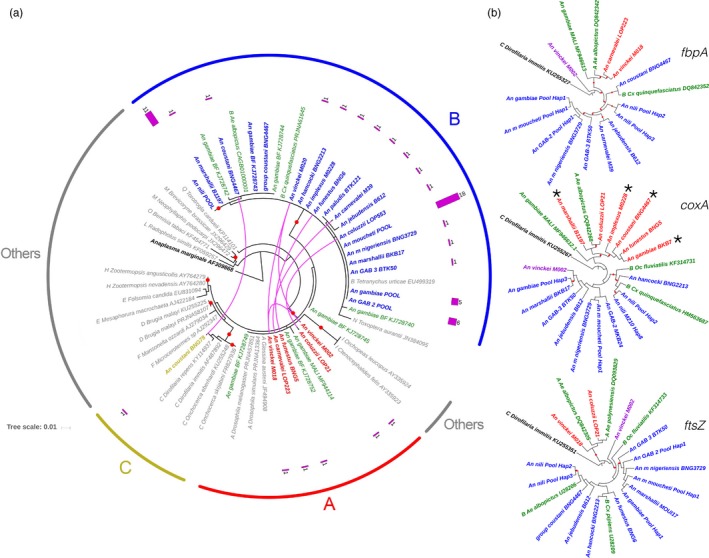
Circular phylograms of the Wolbachia strains isolated in the 16 Anopheles species. The phylogenetic trees were built with RAxML (Stamatakis, 2014). The names of the Anopheles species from which the Wolbachia‐specific sequences were isolated in this study are shown in blue (positive for Wolbachia supergroup B), red (positive for supergroup A) and brown (positive for supergroup C), while the names of mosquitoes species (Diptera) from which the previously published Wolbachia sequences were isolated are in green. Other Wolbachia strains sequences (“others,” in grey) were obtained directly from gene sequence repository ncbi (https://www.ncbi.nlm.nih.gov/). Red dots show branches supporting a bootstrap >70% from 1,000 replicates. (a) Circular phylogenetic tree using the Wolbachia‐specific 16S rRNA fragment and Anaplasma marginale as outgroup. Different Wolbachia strains found in the same Anopheles species are connected by pink lines. The pink bar charts indicate the number of identical Wolbachia haplotypes found in each species. Scale bar corresponds to nucleotide substitutions per site. (b) Circular phylogenetic trees based on the coxA, fbpA and ftsZ fragment sequences using Dirofilaria immitis (supergroup C) as outgroup. Specimens with a different supergroup assignation than 16S are marked with asterisks. Only, Anopheles vinckei M002 (purple) oscillated between groups B and A across the four genes
To expand our knowledge on the Wolbachia strains that infect natural Anopheles populations, we PCR amplified, sequenced and analysed fragments from three conserved Wolbachia genes (coxA, fbpA and ftsZ) that are commonly used for strain typing and evolutionary studies (Baldo et al., 2006) (Figure 2). We used a new nested PCR protocol (see section 22) for samples that could not be genotyped using the classical MLST primers (Table S1). Our phylogenetic analyses confirmed the 16S results, assigning most of the species to supergroups A and B. Few samples (asterisks in Figure 2, gene coxA) showed some incongruence relative to the 16S results. They suggest signals of recent recombination between the supergroups A and B, as previously demonstrated (Baldo et al., 2006). Detailed sequence analysis revealed that mosquito species belonging to the same group or complex (i.e., An. moucheti and An. gambiae) displayed a common Wolbachia haplotype (defined here as a unique allelic profile; Figures 2 and 3). Conversely, some species with lower prevalence (i.e., An. coluzzii, An. marshallii, An. vinckei or An. funestus) displayed a variety of haplotypes. The case of An. vinckei was particularly interesting because the three infected specimens displayed different haplotypes for the analysed Wolbachia genes. Moreover, one specimen (An. vinckei M002, Figure 2) was infected by a completely different Wolbachia strain. Overall, the Wolbachia haplotypes identified in this study were different from the allelic profiles of the previously annotated Wolbachia strains or of the strain that infects An. gambiae in Burkina Faso and Mali (Baldini et al., 2014; Gomes et al., 2017) (Figures 2 and 3). Within supergroup B, we could easily distinguish at least two strains. The strain infecting An. moucheti (wAnmo), which showed no variation across localities, was similar to the one identified in An. gambiae (in our study) or Anopheles marshallii, while the strain infecting An. nili (wAnni), which evidenced strains variation even in the same locality, was more closely related to those found in other mosquito species, such as Ae. albopictus or Cx. quinquefasciatus (Figures 2 and 3). Conversely, the other haplotypes were associated with one specific host.
Figure 3.
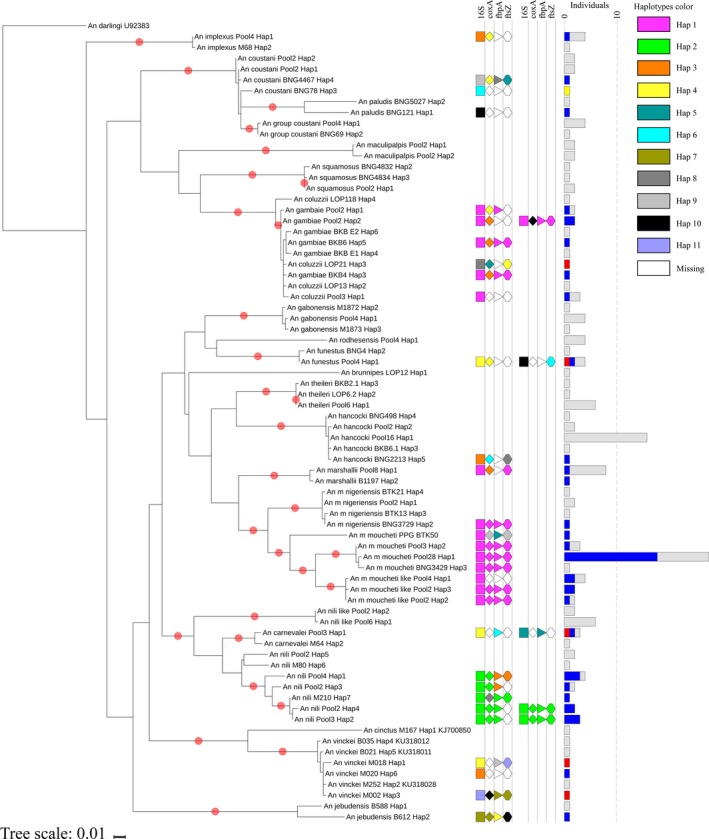
Maximum likelihood phylogeny of the 25 Anopheles species under study and Wolbachia haplotypes. The tree was inferred with RAxML (Stamatakis, 2014) using the sequences of the COII fragment from 176 Anopheles specimens belonging to the 25 species under study and rooted with Anopheles darlingi as outgroup (New World mosquito, diverged 100 Myr ago (Neafsey et al., 2015)). Red dots in branches represent bootstrap values >70% from 1,000 replicates. The shape of each field column represents the 16S (rectangle), coxA (rhombus), fbpA (triangle) and ftsZ (hexagon) genes. The different Wolbachia gene haplotypes (i.e., unique allelic profiles) are indicated with colour codes (all pink = the newly identified wAnmo strain). The bar chart size indicates the number of individuals of the same species with the same haplotype, and the colour represents their infection status: grey, noninfected; blue, infected by the Wolbachia supergroup B; red, infected by supergroup A; brown, infected by supergroup C
3.4. Wolbachia independently evolves in malaria‐transmitting mosquitoes
As Wolbachia is mainly a maternally inherited bacterium, the host mitochondrial DNA (mtDNA) is a suitable marker to study its evolutionary history in Anopheles (Richardson et al., 2012). Analysis of COII sequences from 176 specimens belonging to the 25 Anopheles species collected in Gabon provided the most exhaustive phylogenetic tree of Anopheles in Central Africa (Figure 3). This analysis highlighted the independent acquisition and apparent loss of Wolbachia across the different Anopheles species clades. Moreover, the genetic distances of Wolbachia strains and their Anopheles host were not correlated (Mantel test, p > 0.05; Figure S2). Nevertheless, mosquitoes from the An. moucheti complex, and therefore genetically very close, shared the same Wolbachia supergroup and haplotypes (Figure 3 and Figure S2). Finally, we investigated how Wolbachia evolved within each Anopheles species (Charlat et al., 2009). Our results revealed that Wolbachia‐infected and noninfected mosquitoes shared the same mtDNA haplotype (Figure 3), indicating that infection status and host haplotypes are not associated.
4. DISCUSSION
The present study provides three key findings. First, the genus Anopheles includes a large number of species that are naturally infected by Wolbachia (16/25), with high infection prevalence among major malaria vectors. Second, Anopheles‐infecting Wolbachia bacteria show high genetic diversity, with similar haplotypes detected in different Anopheles species. Third, the independent evolution of Wolbachia and Anopheles might be interpreted as multiple acquisition events with horizontal transmission. The large diversity of Wolbachia strains that infect many natural Anopheles populations could represent a major opportunity for reducing pathogen transmission and/or for reproductive manipulation in Anopheles with the aim of decreasing malaria burden in Africa.
During the last decades, the scientific community has evidenced an interest to find new ways to use Wolbachia for fighting vector‐borne diseases (Bourtzis et al., 2014; Hoffmann et al., 2015; Iturbe‐Ormaetxe et al., 2011; McGraw & O'Neill, 2013). In arthropods, Wolbachia infection is very common, including among Culex and Aedes mosquitoes. Conversely, the genus Anopheles revealed no infection to the bacteria. Until recently, Wolbachia infections were mainly limited to species within the gambiae complex (Baldini et al., 2014; Gomes et al., 2017) and few other species (Baldini et al., 2018; Jeffries, Golovko, et al., 2018; Jeffries, Lawrence, et al., 2018; Niang et al., 2018). Several hypotheses can be put forward to explain this. First, low infection prevalence or local variations could have hindered the discovery of Wolbachia infections, independently of the sampling effort. In our study, most Anopheles species exhibited a prevalence lower than 15% (Table 1). This pattern is common in many other arthropods (Duron et al., 2008; Zug & Hammerstein, 2012), and it is usually associated with a weak manipulation of the host reproduction and/or imperfect maternal transmission (Engelstadter & Hurst, 2009). In general, our sampling effort was higher than in previous studies (n < 30) (Bourtzis et al., 2014; Osei‐Poku et al., 2012), and this could explain why we found more infected species. Our statistical analysis showed that a sample size of 60 individuals per species is needed to quantify correct prevalence rates lower than 15%, with a probability of 95% (Figure S3). Moreover, local frequency variations among populations could also hinder the detection of Wolbachia infections (Dumas et al., 2013). For instance, we sampled An. coluzzii in three different sites, but we only found Wolbachia‐infected mosquitoes at La Lopé (Figure 1, Table S1). Therefore, sampling in different localities and in different seasons might improve detection rates. Second, it could be difficult to detect low‐density Wolbachia infections in Anopheles with the routinely used molecular tools, as previously reported for other arthropods (Arthofer, Riegler, Avtzis, & Stauffer, 2009; Augustinos et al., 2011) and recently in An. gambiae (Gomes et al., 2017). Our results indicate that conventional PCR amplification (wsp‐targeting primers (Baldo et al., 2006)) analysis allowed the detection of Wolbachia infection only in 6 of the 16 species (An. moucheti, Anopheles m. nigeriensis, An. “GAB‐3,” An. nili, Anopheles jebudensis and An. vinckei) under study, presumably because of the high Wolbachia density. Moreover, some Anopheles species with high Wolbachia infection rates, such as An. moucheti or An. nili, were never screened before.
Our work revealed that Anopheles species are infected by different Wolbachia strains. Although t previous studies reported Wolbachia infection in Anopheles (Baldini et al., 2014; Gomes et al., 2017; Jeffries, Golovko, et al., 2018; Jeffries, Lawrence, et al., 2018; Niang et al., 2018; Shaw et al., 2016), there exist the doubt if they are real infections (Chrostek & Gerth, 2018). The Wolbachia sequences found in our specimens were genetically close to those found in other Diptera, and no signal of extensive divergence was detected (Figures 2 and 3). Therefore, there is no risk that horizontal gene transfer (resulting in the insertion of Wolbachia genes within the mosquito genome) or parasitism (e.g., by filarial nematodes) could explain the detection of Wolbachia genes in our infected mosquitoes without maternal transmission. Moreover, the analysis of An. moucheti F1 progeny confirms, at least in this species, that no other biological Wolbachia contamination was present in our analysis. In conclusion, our data suggest that Wolbachia is naturally present in the Anopheles species of Central Africa analysed in our study, and that it is maternally inherited in An. moucheti (Table S2). In this sense and besides the challenge to rear An. moucheti under insectary conditions, this mosquito should be considered as potential model species to study the reproductive phenotypes of Wolbachia and its effect in Plasmodium infections.
In Central African Anopheles, Wolbachia acquisition seems to be independent of the host phylogeny (Figures 2 and 3). Our results revealed that the genetic distances between Wolbachia and Anopheles are not positively correlated (Mantel test, p > 0.05; Figure S2). The lack of correlation could lead to think that Wolbachia and the host lineage evolved independently. The different larval ecology of these species suggests other ways of lateral transfer (e.g., during nectar feeding (Li et al., 2017)). On the other hand, we found that species belonging to An. moucheti complex shared related Wolbachia strains (Figure 3). Permeable reproductive barriers among members of the same complex could facilitate the intermittent movement of the bacterium (Pombi et al., 2017). Interestingly, although they share similar Wolbachia strains, sibling species showed different infection prevalence. Indeed, An. carnevalei and An. m. nigeriensis exhibited frequencies lower than 15%, whereas An. nili and An. moucheti, their respective counterparts and the most important malaria vectors in their complex, displayed frequencies higher than 50% (Table 1). Moreover, our An. gambiae and An. coluzzii populations were infected by different Wolbachia strains than those detected in Burkina Faso and Mali. Similarly, in mosquitoes (Dumas et al., 2013) and ants (Tsutsui, Kauppinen, Oyafuso, & Grosberg, 2003), the same species is infected by different Wolbachia strains according to the region. The availability of whole‐genome sequences for Wolbachia strains (Gerth, Gansauge, Weigert, & Bleidorn, 2014) will enlighten the intricate phylogenetic relationships among the different strains in Anopheles.
5. CONCLUSIONS
Wolbachia has emerged as a biological tool for controlling vector‐borne diseases (Hoffmann et al., 2011; Schmidt et al., 2017). In this study, we demonstrated the natural presence of this endosymbiont bacterium in a large number of Anopheles species, including the five major malaria vectors in Central Africa. Previously, it has been shown that Wolbachia ability to interfere with pathogen transmission depends on the bacterium strain (Blagrove, Arias‐Goeta, Failloux, & Sinkins, 2012; Kambris et al., 2010; Walker et al., 2011). Therefore, our results offer the opportunity to determine whether the different Anopheles‐infecting Wolbachia strains affect Plasmodium transmission and/or Anopheles reproduction. Indeed, three major vectors of human and nonhuman malaria (An. moucheti, An. nili and An. vinckei) were infected by Wolbachia (Makanga et al., 2016; Paupy et al., 2013). Therefore, we could investigate both Wolbachia‐mediated decreases (Hughes, Rivero, & Rasgon, 2014; Zele et al., 2014) and increases (Shaw et al., 2016) in susceptibility of these natural vectors to Plasmodium. Moreover, the strongest effect on suppression of pathogen transmission or reproductive manipulation has been observed in Wolbachia transinfections (Bian et al., 2013; Bian, Xu, Lu, Xie, & Xi, 2010; Blagrove et al., 2012; Hughes et al., 2011; Joubert et al., 2016; Moreira et al., 2009; Walker et al., 2011). Therefore, the availability of Wolbachia strains that infect natural Anopheles populations offers promising opportunities for experimental and theoretical studies in Anopheles, and also in other mosquito families that are vectors of other diseases, including Ae. aegypti and Ae. albopictus. In conclusion, our findings are merely the “tip of the iceberg” of Wolbachia research in Anopheles. The selection of suitable phenotypes for suppressing Plasmodium transmission and/or manipulating Anopheles reproduction could greatly participate to reduce the malaria burden across the world.
CONFLICT OF INTEREST
None declared.
Supporting information
ACKNOWLEDGEMENTS
We thank the “Agence Nationale de la Preservation de la Nature” (ANPN) and the “Centre National de la Recherche Scientifique et Technologique of Gabon” (CENAREST) that authorized this study and facilitated the access to the national parks of La Lopé, Moukalaba‐Doudou and Plateaux Batékés.
Ayala D, Akone‐Ella O, Rahola N, et al. Natural Wolbachia infections are common in the major malaria vectors in Central Africa. Evol Appl. 2019;12:1583–1594. 10.1111/eva.12804
DATA AVAILABILITY
Data for this study are available at the Dryad digital Repository: https://doi.org/10.5061/dryad.sn81548 (Ayala et al., 2019). DNA sequences of Wolbachia and Anopheles recovered in this study and of those used as references for phylogenetic analyses are submitted at Genbank (MK755460–MK755837).
REFERENCES
- Ahmed, M. Z. , De Barro, P. J. , Ren, S. X. , Greeff, J. M. , & Qiu, B. L. (2013). Evidence for horizontal transmission of secondary endosymbionts in the Bemisia tabaci cryptic species complex. PLoS ONE, 8(1), e53084 10.1371/journal.pone.0053084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthofer, W. , Riegler, M. , Avtzis, D. N. , & Stauffer, C. (2009). Evidence for low‐titre infections in insect symbiosis: Wolbachia in the bark beetle Pityogenes chalcographus (Coleoptera, Scolytinae). Environmental Microbiology, 11(8), 1923–1933. 10.1111/j.1462-2920.2009.01914.x [DOI] [PubMed] [Google Scholar]
- Augustinos, A. A. , Santos‐Garcia, D. , Dionyssopoulou, E. , Moreira, M. , Papapanagiotou, A. , Scarvelakis, M. , … Bourtzis, K. (2011). Detection and characterization of Wolbachia Infections in Natural Populations of Aphids: Is the hidden diversity fully unraveled? PLoS ONE, 6(12), e28695 10.1371/journal.pone.0028695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala, D. , Akone‐Ella, O. , Rahola, N. , Kengne, P. , Ngangue, M. F. , Mezeme, F. , … Paupy, C. (2019). Natural Wolbachia infections are common in the major malaria vectors in Central Africa. Evolutionary Applications. Dryad digital Repository; 10.5061/dryad.sn81548 [DOI] [PMC free article] [PubMed]
- Baldini, F. , Rouge, J. , Kreppel, K. , Mkandawile, G. , Mapua, S. A. , Sikulu‐Lord, M. , … Okumu, F. O. (2018). First report of natural Wolbachia infection in the malaria mosquito Anopheles arabiensis in Tanzania. Parasit Vectors, 11(1), 635 10.1186/s13071-018-3249-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldini, F. , Segata, N. , Pompon, J. , Marcenac, P. , Shaw, W. R. , Dabire, R. K. , … Catteruccia, F. (2014). Evidence of natural Wolbachia infections in field populations of Anopheles gambiae. Nature Communications, 5, 3985 10.1038/ncomms4985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldo, L. , Dunning Hotopp, J. C. , Jolley, K. A. , Bordenstein, S. R. , Biber, S. A. , Choudhury, R. R. , … Werren, J. H. (2006). Multilocus sequence typing system for the endosymbiont Wolbachia pipientis. Applied and Environmental Microbiology, 72(11), 7098–7110. 10.1128/aem.00731-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt, S. , Weiss, D. J. , Cameron, E. , Bisanzio, D. , Mappin, B. , Dalrymple, U. , & Gething, P. W. (2015). The effect of malaria control on Plasmodium falciparum in Africa between 2000 and 2015. Nature, 526(7572), 207–211. 10.1038/nature15535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian, G. , Joshi, D. , Dong, Y. , Lu, P. , Zhou, G. , Pan, X. , … Xi, Z. (2013). Wolbachia invades Anopheles stephensi populations and induces refractoriness to plasmodium infection. Science, 340(6133), 748–751. 10.1126/science.1236192 [DOI] [PubMed] [Google Scholar]
- Bian, G. , Xu, Y. , Lu, P. , Xie, Y. , & Xi, Z. (2010). The endosymbiotic bacterium Wolbachia induces resistance to dengue virus in Aedes aegypti . PLoS Path, 6(4), e1000833 10.1371/journal.ppat.1000833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagrove, M. S. C. , Arias‐Goeta, C. , Failloux, A. B. , & Sinkins, S. P. (2012). Wolbachia strain wMel induces cytoplasmic incompatibility and blocks dengue transmission in Aedes albopictus . Proceedings of the National Academy of Sciences of the United States of America, 109(1), 255–260. 10.1073/pnas.1112021108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boundenga, L. , Makanga, B. , Ollomo, B. , Gilabert, A. , Rougeron, V. , Mve‐Ondo, B. , … Paupy, C. (2016). Haemosporidian parasites of antelopes and other vertebrates from Gabon, Central Africa. PLoS ONE, 11(2), e0148958 10.1371/journal.pone.0148958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourtzis, K. , Dobson, S. L. , Xi, Z. Y. , Rasgon, J. L. , Calvitti, M. , Moreira, L. A. , … Gilles, J. R. L. (2014). Harnessing mosquito‐Wolbachia symbiosis for vector and disease control. Acta Tropica, 132, S150–S163. 10.1016/j.actatropica.2013.11.004 [DOI] [PubMed] [Google Scholar]
- Casiraghi, M. , Anderson, T. J. C. , Bandi, C. , Bazzocchi, C. , & Genchi, C. (2001). A phylogenetic analysis of filarial nematodes: Comparison with the phylogeny of Wolbachia endosymbionts. Parasitology, 122, 93–103. 10.1017/s0031182000007149 [DOI] [PubMed] [Google Scholar]
- Charlat, S. , Duplouy, A. , Hornett, E. A. , Dyson, E. A. , Davies, N. , Roderick, G. K. , … Hurst, G. D. (2009). The joint evolutionary histories of Wolbachia and mitochondria in Hypolimnas bolina. BMC Evolutionary Biology, 9(1), 64 10.1186/1471-2148-9-64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrostek, E. , & Gerth, M. (2018). Is anopheles gambiae a natural host of Wolbachia? bioRxiv. 10.1101/491449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohuet, A. , Simard, F. , Toto, J. C. , Kengne, P. , Coetzee, M. , & Fontenille, D. (2003). Species identification within the Anopheles funestus group of malaria vectors in Cameroon and evidence for a new species. American Journal of Tropical Medicine and Hygiene, 69(2), 200–205. [PubMed] [Google Scholar]
- Dodson, B. L. , Hughes, G. L. , Paul, O. , Matacchiero, A. C. , Kramer, L. D. , & Rasgon, J. L. (2014). Wolbachia enhances West Nile Virus (WNV) infection in the mosquito Culex tarsalis. Plos Neglected Tropical Diseases, 8(7), e2965 10.1371/journal.pntd.0002965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumas, E. , Atyame, C. M. , Milesi, P. , Fonseca, D. M. , Shaikevich, E. V. , Unal, S. , … Duron, O. (2013). Population structure of Wolbachia and cytoplasmic introgression in a complex of mosquito species. BMC Evolutionary Biology, 13, 181 10.1186/1471-2148-13-181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duron, O. , Bouchon, D. , Boutin, S. , Bellamy, L. , Zhou, L. , Engelstaedter, J. , & Hurst, G. D. (2008). The diversity of reproductive parasites among arthropods: Wolbachia do not walk alone. BMC Biology, 6, 1–12. 10.1186/1741-7007-6-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duron, O. , Lagnel, J. , Raymond, M. , Bourtzis, K. , Fort, P. , & Weill, M. (2005). Transposable element polymorphism of Wolbachia in the mosquito Culex pipiens: Evidence of genetic diversity, superinfection and recombination. Molecular Ecology, 14, 1561–1573. 10.1111/j.1365-294X.2005.02495.x [DOI] [PubMed] [Google Scholar]
- Engelstadter, J. , & Hurst, G. D. D. (2009). The ecology and evolution of microbes that manipulate host reproduction. Annual Review of Ecology Evolution and Systematics, 40, 127–149. 10.1146/annurev.ecolsys.110308.120206 [DOI] [Google Scholar]
- Fanello, C. , Santolamazza, F. , & della Torre, A. (2002). Simultaneous identification of species and molecular forms of the Anopheles gambiae complex by PCR‐RFLP. Medical and Veterinary Entomology, 16(4), 461–464. [DOI] [PubMed] [Google Scholar]
- Gerth, M. , Gansauge, M. T. , Weigert, A. , & Bleidorn, C. (2014). Phylogenomic analyses uncover origin and spread of the Wolbachia pandemic. Nature Communications, 5, 1–7. 10.1038/ncomms6117 [DOI] [PubMed] [Google Scholar]
- Gillies, M. T. , & Coetzee, M. C. (1987). A supplement to the anophelinae of Africa south of the Sahara (Afrotropical region). Johannesburg, South Africa: The South African Institute for Medical Research. [Google Scholar]
- Gillies, M. T. , & de Meillon, B. (1968). The anophelinae of Africa, south of the Sahara, Vol. 54 Johannesburg, South Africa: The South African Institute for Medical Research. [Google Scholar]
- Gomes, F. M. , Hixson, B. L. , Tyner, M. D. W. , Ramirez, J. L. , Canepa, G. E. , Silva, T. , … Barillas‐Mury, C. (2017). Effect of naturally occurring Wolbachia in Anopheles gambiae s.l. mosquitoes from Mali on Plasmodium falciparum malaria transmission. PNAS, 114(47), 12566–12571. 10.1073/pnas.1716181114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffi, J. T. , Bhatt, S. , Sinka, M. E. , Gething, P. W. , Lynch, M. , Patouillard, E. , … Ghani, A. C. (2016). Potential for reduction of burden and local elimination of malaria by reducing Plasmodium falciparum malaria transmission: A mathematical modelling study. Lancet Infectious Diseases, 16(4), 465–472. 10.1016/s1473-3099(15)00423-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamon, J. , & Mouchet, J. (1961). Secondary vectors of human malaria in Africa. Medecine Tropicale, 21, 643–660. [PubMed] [Google Scholar]
- Hoffmann, A. A. , Montgomery, B. L. , Popovici, J. , Iturbe‐Ormaetxe, I. , Johnson, P. H. , Muzzi, F. , … O'Neill, S. L. (2011). Successful establishment of Wolbachia in Aedes populations to suppress dengue transmission. Nature, 476(7361), 454–U107. 10.1038/nature10356 [DOI] [PubMed] [Google Scholar]
- Hoffmann, A. A. , Ross, P. A. , & Rasic, G. (2015). Wolbachia strains for disease control: Ecological and evolutionary considerations. Evolutionary Applications, 8(8), 751–768. 10.1111/eva.12286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, G. L. , Dodson, B. L. , Johnson, R. M. , Murdock, C. C. , Tsujimoto, H. , Suzuki, Y. , … Rasgon, J. L. (2014). Native microbiome impedes vertical transmission of Wolbachia in Anopheles mosquitoes. Proceedings of the National Academy of Sciences, 111(34), 12498–12503. 10.1073/pnas.1408888111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, G. L. , Koga, R. , Xue, P. , Fukatsu, T. , & Rasgon, J. L. (2011). Wolbachia infections are virulent and inhibit the human malaria parasite Plasmodium falciparum in Anopheles gambiae . PLoS Path, 7(5), e1002043 10.1371/journal.ppat.1002043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, G. L. , Rivero, A. , & Rasgon, J. L. (2014). Wolbachia can enhance plasmodium infection in mosquitoes: Implications for malaria control? PLoS Path, 10(9), e1004182 10.1371/journal.ppat.1004182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, G. L. , Vega‐Rodriguez, J. , Xue, P. , & Rasgon, J. L. (2012). Wolbachia strain wAlbB enhances infection by the rodent malaria parasite Plasmodium berghei in Anopheles gambiae mosquitoes. Applied and Environmental Microbiology, 78(5), 1491–1495. 10.1128/aem.06751-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iturbe‐Ormaetxe, I. , Walker, T. , & Neill, S. L. O. (2011). Wolbachia and the biological control of mosquito‐borne disease. EMBO Reports, 12(6), 508–518. 10.1038/embor.2011.84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffries, C. L. , Golovko, G. , Kristan, M. , Orsborne, J. , Spence, K. , Hurn, E. , & Walker, T. (2018). Novel Wolbachia strains in Anopheles malaria vectors from Sub‐Saharan Africa. bioRxiv 10.1101/338434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffries, C. L. , Lawrence, G. G. , Golovko, G. , Kristan, M. , Orsborne, J. , Spence, K. , … Walker, T. (2018). Novel Wolbachia strains in Anopheles malaria vectors from Sub‐Saharan Africa. Wellcome Open Res, 3, 113 10.12688/wellcomeopenres.14765.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joubert, D. A. , Walker, T. , Carrington, L. B. , De Bruyne, J. T. , Kien, D. H. , Hoang Nle, T. , … O'Neill, S. L. (2016). Establishment of a wolbachia superinfection in Aedes aegypti mosquitoes as a potential approach for future resistance management. PLoS Path, 12(2), e1005434 10.1371/journal.ppat.1005434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambris, Z. , Blagborough, A. M. , Pinto, S. B. , Blagrove, M. S. C. , Godfray, H. C. J. , Sinden, R. E. , & Sinkins, S. P. (2010). Wolbachia stimulates immune gene expression and inhibits plasmodium development in Anopheles gambiae . PLoS Path, 6(10), e1001143 10.1371/journal.ppat.1001143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambris, Z. , Cook, P. E. , Phuc, H. K. , & Sinkins, S. P. (2009). Immune activation by life‐shortening wolbachia and reduced filarial competence in mosquitoes. Science, 326(5949), 134–136. 10.1126/science.1177531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse, M. , Moir, R. , Wilson, A. , Stones‐Havas, S. , Cheung, M. , Sturrock, S. , … Drummond, A. (2012). Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics, 28(12), 1647–1649. 10.1093/bioinformatics/bts199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kengne, P. , Antonio‐Nkondjio, C. , Awono‐Ambene, H. P. , Simard, F. , Awolola, T. S. , & Fontenille, D. (2007). Molecular differentiation of three closely related members of the mosquito species complex, Anopheles moucheti, by mitochondrial and ribosomal DNA polymorphism. Medical and Veterinary Entomology, 21(2), 177–182. [DOI] [PubMed] [Google Scholar]
- Kengne, P. , Awono‐Ambene, P. , Nkondjio, C. A. , Simard, F. , & Fontenille, D. (2003). Molecular identification of the Anopheles nili group of African malaria vectors. Medical and Veterinary Entomology, 17(1), 67–74. 10.1046/j.1365-2915.2003.00411.x [DOI] [PubMed] [Google Scholar]
- Letunic, I. , & Bork, P. (2007). Interactive Tree Of Life (iTOL): An online tool for phylogenetic tree display and annotation. Bioinformatics, 23(1), 127–128. 10.1093/bioinformatics/btl529 [DOI] [PubMed] [Google Scholar]
- Li, S. J. , Ahmed, M. Z. , Lv, N. , Shi, P. Q. , Wang, X. M. , Huang, J. L. , & Qiu, B. L. (2017). Plant‐mediated horizontal transmission of Wolbachia between whiteflies. ISME Journal, 11(4), 1019–1028. 10.1038/ismej.2016.164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makanga, B. , Yangari, P. , Rahola, N. , Rougeron, V. , Elguero, E. , Boundenga, L. , … Paupy, C. (2016). Ape malaria transmission and potential for ape‐to‐human transfers in Africa. PNAS, 113(19), 5329–5334. 10.1073/pnas.1603008113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGraw, E. A. , & O'Neill, S. L. (2013). Beyond insecticides: New thinking on an ancient problem. Nature Reviews Microbiology, 11(3), 181–193. [DOI] [PubMed] [Google Scholar]
- Moreira, L. A. , Iturbe‐Ormaetxe, I. , Jeffery, J. A. , Lu, G. , Pyke, A. T. , Hedges, L. M. , … O'Neill, S. L. (2009). A Wolbachia symbiont in Aedes aegypti limits infection with dengue, chikungunya, and plasmodium. Cell, 139(7), 1268–1278. 10.1016/j.cell.2009.11.042 [DOI] [PubMed] [Google Scholar]
- Ndo, C. , Antonio‐Nkondjio, C. , Cohuet, A. , Ayala, D. , Kengne, P. , Morlais, I. , … Simard, F. (2010). Population genetic structure of the malaria vector Anopheles nili in sub‐Saharan Africa. Malaria Journal, 9, 161 10.1186/1475-2875-9-161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neafsey, D. E. , Waterhouse, R. M. , Abai, M. R. , Aganezov, S. S. , Alekseyev, M. A. , Allen, J. E. , … Besansky, N. J. (2015). Highly evolvable malaria vectors: The genomes of 16 Anopheles mosquitoes. Science, 347(6217), 43 10.1126/science.1258522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newby, G. , Bennett, A. , Larson, E. , Cotter, C. , Shretta, R. , Phillips, A. A. , & Feachem, R. G. A. (2016). The path to eradication: A progress report on the malaria‐eliminating countries. Lancet, 387(10029), 1775–1784. [DOI] [PubMed] [Google Scholar]
- Niang, E. H. A. , Bassene, H. , Makoundou, P. , Fenollar, F. , Weill, M. , & Mediannikov, O. (2018). First report of natural Wolbachia infection in wild Anopheles funestus population in Senegal. Malaria Journal, 17(1), 408 10.1186/s12936-018-2559-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson, D. M. , Dinerstein, E. , Wikramanayake, E. D. , Burgess, N. D. , Powell, G. V. N. , Underwood, E. C. , … Kassem, K. R. (2001). Terrestrial ecoregions of the worlds: A new map of life on Earth. BioScience, 51(11), 933–938. 10.1641/0006-3568(2001)051[0933:teotwa]2.0.co;2 [DOI] [Google Scholar]
- Osei‐Poku, J. , Han, C. , Mbogo, C. M. , & Jiggins, F. M. (2012). Identification of wolbachia strains in mosquito disease vectors. PLoS ONE, 7(11), e49922 10.1371/journal.pone.0049922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pates, H. , & Curtis, C. (2005). Mosquito behavior and vector control. Annual Review of Entomology, 50, 53–70. 10.1146/annurev.ento.50.071803.130439 [DOI] [PubMed] [Google Scholar]
- Paupy, C. , Makanga, B. , Ollomo, B. , Rahola, N. , Durand, P. , Magnus, J. , … Prugnolle, F. (2013). Anopheles moucheti and Anopheles vinckei are candidate vectors of ape plasmodium parasites, including Plasmodium praefalciparum in Gabon. PLoS ONE, 8(2), e57294 10.1371/journal.pone.0057294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pombi, M. , Kengne, P. , Gimonneau, G. , Tene‐Fossog, B. , Ayala, D. , Kamdem, C. , … Costantini, C. (2017). Dissecting functional components of reproductive isolation among closely related sympatric species of the Anopheles gambiae complex. Evolutionary Applications, 10(10), 1102–1120. 10.1111/eva.12517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahola, N. , Makanga, B. , Yangari, P. , Jiolle, D. , Fontenille, D. , Renaud, F. , … Paupy, C. (2014). Description of Anopheles gabonensis, a new species potentially involved in rodent malaria transmission in Gabon, Central Africa. Infection Genetics and Evolution, 28, 628–634. 10.1016/j.meegid.2014.05.012 [DOI] [PubMed] [Google Scholar]
- Ranson, H. , & Lissenden, N. (2016). Insecticide resistance in African anopheles mosquitoes: A worsening situation that needs urgent action to maintain malaria control. Trends in Parasitology, 32(3), 187–196. 10.1016/j.pt.2015.11.010 [DOI] [PubMed] [Google Scholar]
- Richardson, M. F. , Weinert, L. A. , Welch, J. J. , Linheiro, R. S. , Magwire, M. M. , Jiggins, F. M. , & Bergman, C. M. (2012). Population Genomics of the Wolbachia Endosymbiont in Drosophila melanogaster. PLOS Genetics, 8(12), e1003129 10.1371/journal.pgen.1003129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert, V. , Ayala, D. , & Simard, F. (2017). Les anopheles In Duvallet G., Fontenille D. & Robert V. (Eds.), Entomologie médicale et vétérinaire (p. 687). Paris, France: IRD Editions. [Google Scholar]
- Rossi, P. , Ricci, I. , Cappelli, A. , Damiani, C. , Ulissi, U. , Mancini, M. V. , … Favia, G. (2015). Mutual exclusion of Asaia and Wolbachia in the reproductive organs of mosquito vectors. Parasit Vectors, 8, 1–10. 10.1186/s13071-015-0888-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santolamazza, F. , Mancini, E. , Simard, F. , Qi, Y. , Tu, Z. , & della Torre, A. (2008). Insertion polymorphisms of SINE200 retrotransposons within speciation islands of Anopheles gambiae molecular forms. Malaria Journal, 7, 163 10.1186/1475-2875-7-163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, T. L. , Barton, N. H. , Rašić, G. , Turley, A. P. , Montgomery, B. L. , Iturbe‐Ormaetxe, I. , … Turelli, M. (2017). Local introduction and heterogeneous spatial spread of dengue‐suppressing Wolbachia through an urban population of Aedes aegypti. PLOS Biology, 15(5), e2001894 10.1371/journal.pbio.2001894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw, W. R. , Marcenac, P. , Childs, L. M. , Buckee, C. O. , Baldini, F. , Sawadogo, S. P. , … Catteruccia, F. (2016). Wolbachia infections in natural Anopheles populations affect egg laying and negatively correlate with Plasmodium development. Nature Communications, 7, 11772 10.1038/ncomms11772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon, F. , Siles‐Lucas, M. , Morchon, R. , Gonzalez‐Miguel, J. , Mellado, I. , Carreton, E. , & Montoya‐Alonso, J. A. (2012). Human and animal dirofilariasis: The emergence of a zoonotic. Mosaic. Clinical Microbiology Reviews, 25(3), 507–544. 10.1128/cmr.00012-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinkins, S. P. , Braig, H. R. , & Oneill, S. L. (1995). Wolbachia superinfections and the expression of cytoplasmic incompatibility. Proceedings of the Royal Society B‐Biological Sciences, 261(1362), 325–330. 10.1098/rspb.1995.0154 [DOI] [PubMed] [Google Scholar]
- Stamatakis, A. (2006). Phylogenetic models of rate heterogeneity: A high performance computing perspective . Paper presented at the International Parallel and Distributed Processing Symposium, Rhodos, Greece: [Google Scholar]
- Stamatakis, A. (2014). RAxML version 8: A tool for phylogenetic analysis and post‐analysis of large phylogenies. Bioinformatics, 30(9), 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsui, N. D. , Kauppinen, S. N. , Oyafuso, A. F. , & Grosberg, R. K. (2003). The distribution and evolutionary history of Wolbachia infection in native and introduced populations of the invasive argentine ant (Linepithema humile). Molecular Ecology, 12(11), 3057–3068. 10.1046/j.1365-294X.2003.01979.x [DOI] [PubMed] [Google Scholar]
- Walker, T. , Johnson, P. H. , Moreira, L. A. , Iturbe‐Ormaetxe, I. , Frentiu, F. D. , McMeniman, C. J. , … Hoffmann, A. A. (2011). The wMel Wolbachia strain blocks dengue and invades caged Aedes aegypti populations. Nature, 476(7361), 450–U101. 10.1038/nature10355 [DOI] [PubMed] [Google Scholar]
- Werren, J. H. , Baldo, L. , & Clark, M. E. (2008). Wolbachia: Master manipulators of invertebrate biology. Nature Reviews Microbiology, 6(10), 741–751. 10.1038/nrmicro1969 [DOI] [PubMed] [Google Scholar]
- Werren, J. H. , Zhang, W. , & Guo, L. R. (1995). Evolution and phylogeny of Wolbachia: Reproductive parasites of arthropods. Proceedings of the Royal Society B: Biological Sciences, 261(1360), 55–63. 10.1098/rspb.1995.0117 [DOI] [PubMed] [Google Scholar]
- WHO (2015). Global technical strategy for malaria 2016–2030. Geneva, Switzerland: WHO. [Google Scholar]
- WHO (2018). World malaria report 2018 . Geneva, Switzerland: WHO; [Google Scholar]
- Wickham, H. (2009). ggplot2: Elegant graphics for data analysis. New York, NY: Springer Publishing Company, Incorporated. [Google Scholar]
- Zele, F. , Nicot, A. , Berthomieu, A. , Weill, M. , Duron, O. , & Rivero, A. (2014). Wolbachia increases susceptibility to Plasmodium infection in a natural system. Proceedings of the Royal Society B‐Biological Sciences, 281(1779), 20132837 10.1098/rspb.2013.2837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zug, R. , & Hammerstein, P. (2012). Still a host of hosts for wolbachia: analysis of recent data suggests that 40% of terrestrial arthropod species are infected. PLoS ONE, 7(6), e38544 10.1371/journal.pone.0038544 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Ayala, D. , Akone‐Ella, O. , Rahola, N. , Kengne, P. , Ngangue, M. F. , Mezeme, F. , … Paupy, C. (2019). Natural Wolbachia infections are common in the major malaria vectors in Central Africa. Evolutionary Applications. Dryad digital Repository; 10.5061/dryad.sn81548 [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
Data for this study are available at the Dryad digital Repository: https://doi.org/10.5061/dryad.sn81548 (Ayala et al., 2019). DNA sequences of Wolbachia and Anopheles recovered in this study and of those used as references for phylogenetic analyses are submitted at Genbank (MK755460–MK755837).


