Abstract
Di(2-ethylhexyl) phthalate (DEHP) is a commonly used plasticizer and known endocrine disrupting chemical, which causes transgenerational reproductive toxicity in female rodents. However, the mechanisms of action underlying the transgenerational toxicity of DEHP are not understood. Therefore, this study determined the effects of prenatal and ancestral DEHP exposure on various ovarian pathways in the F1, F2, and F3 generations of mice. Pregnant CD-1 dams were orally exposed to corn oil (vehicle control) or DEHP (20 μg/kg/day-750 mg/kg/day) from gestation day 10.5 until birth. At postnatal day 21 for all generations, ovaries were removed for gene expression analysis of various ovarian pathways and for 5-methyl cytosine (5-mC) quantification. In the F1 generation, prenatal DEHP exposure disrupted the expression of cell cycle regulators, the expression of peroxisome-proliferator activating receptors, and the percentage of 5-mC compared to control. In the F2 generation, exposure to DEHP decreased the expression of steroidogenic enzymes, apoptosis factors, and ten-eleven translocation compared to controls. It also dysregulated the expression of phosphoinositide 3-kinase (PI3K) factors. In the F3 generation, ancestral DEHP exposure decreased the expression of steroidogenic enzymes, PI3K factors, cell cycle regulators, apoptosis factors, Esr2, DNA methylation mediators, and the percentage of 5-mC compared to controls. Overall, the data show that prenatal and ancestral DEHP greatly suppresses gene expression of pathways required for folliculogenesis and steroidogenesis in the ovary in a transgenerational manner and that gene expression may be in influenced by DNA methylation. These results provide insight into some of the mechanisms of DEHP-mediated toxicity in the ovary across generations.
Keywords: Ovary, DEHP, endocrine disruptor, transgenerational, methylation
Introduction
Phthalates are a family of synthetic chemicals that act as plasticizers to confer flexibility and reduce breakage [1]. Phthalates are critical for the production of consumer goods. Many types of phthalates exist, but di(2-ethylhexyl) phthalate (DEHP) is a common plasticizer found in polyvinyl chloride products. DEHP is incorporated into a multitude of products including personal care products, medical equipment (i.e., blood and I.V. bags), car upholstery, food and beverage packaging, and building materials, particularly vinyl products [1–3]. DEHP is non-covalently bound to the polymer chains within these products; therefore, DEHP may leach from the products and into the environment after repeated use, heating, and cleaning [4]. Humans are exposed to DEHP by oral ingestion, inhalation, and dermal contact. However, the most common route of exposure to DEHP and phthalates in general is by ingestion. The estimated range of human exposure to DEHP is between 3 – 30 μg/kg/day [2, 5–7]. Human urine samples persistently test positive for DEHP and its metabolites, indicating that humans are repeatedly and continuously exposed to DEHP [8]. This is further supported by the detection of DEHP in human tissues such as blood, amniotic fluid, umbilical cord blood, breast milk, and ovarian follicular fluids in humans [2, 9–12].
DEHP is an endocrine disrupting chemical (EDC) [2, 13, 14], and reproductive tissues such as the gonads are particularly susceptible to EDCs. In humans, in utero exposure to DEHP is associated with decreased free testosterone and free testosterone:estradiol ratio the in cord blood of both male and female newborns [15, 16]. DEHP exposure also interferes with obstetrical outcomes, puberty, and gonadal function [17]. DEHP exposure is also associated with an early age of pubic hair development in young girls, an indicator of precocious puberty [18]. Further, in utero exposure to DEHP metabolites is associated with an earlier age of menarche in young girls [19]. Finally, urinary concentrations of DEHP metabolites are negatively associated with total oocytes, mature oocytes, fertilized oocytes, and top quality embryos, indicating that DEHP impairs oocyte parameters [20].
The ability of DEHP to cause adverse reproductive outcomes is a major concern for the F1 and subsequent generations. DEHP-induced alterations in the ovary may be passed to the subsequent generations through the female germ cells [21]. Transmission of disease due to direct prenatal DEHP exposure may cause multigenerational effects and ancestral exposure to DEHP may cause transgenerational effects in the F3 generation. This is because during a developmental exposure window, the pregnant mouse (F0 generation) is exposed to DEHP via ingestion. Therefore, the F1 generation is directly exposed to DEHP as a fetus, and the F2 generation is directly exposed to DEHP as the developing germ cells in the F1 fetus, causing multigenerational effects in the F1 and F2 generations [22]. Effects observed in the F3 generation are not due to direct exposure, but instead are due to ancestral exposure, therefore demonstrating transgenerational inheritance [22].
Experimental studies show that DEHP exposure causes numerous multigenerational and transgenerational phenotypes in female reproduction. DEHP exposure during prenatal development dysregulates folliculogenesis, alters sex steroid hormone levels, and increases the presence of ovarian cysts in a multigenerational manner [23, 24]. Further, prenatal DEHP exposure decreases the percentage of dams that give birth in the F2 generation of mice [25]. Ancestral DEHP exposure accelerates ovarian follicle formation, onset of puberty, and reproductive senescence in the F3 generation of female mice [23, 25]. In addition, ancestral exposure to DEHP accelerates early folliculogenesis in a transgenerational manner [24]. Although studies demonstrate that phthalate exposure causes transgenerational effects on the ovary, the mechanisms underlying these effects are not well understood.
Epigenetic modification is thought to be the mechanism by which transgenerational effects are inherited [26]. Epigenetics are mitotically and meiotically heritable changes in gene function, without changing DNA sequences [27, 28]. These heritable changes in the epigenome define and control cell and tissue development by controlling gene expression [29]. Multiple molecular mechanisms alter the epigenome, however, DNA methylation is the most commonly studied epigenetic mechanism [30]. DNA methylation patterns are mediated by DNA methyltransferases (DNMTs) and ten-eleven translocation (TET) enzymes [31–33]. DNMTs are a family of enzymes that methylate CpG dinucleotides in DNA. DNMT1 is the maintenance DNMT that maintains original DNA methylation patterns in a cell lineage; it methylates CpG sites during DNA replication so that both daughter cells have the same DNA methylation patterns [34–36]. DNMT3A and DNMT3B methylate CpG sites on naked DNA outside of DNA replication and are required for genome-wide de novo methylation [31]. TETs are enzymes that oxidize 5-methyl cytosine (5-mC) as a demethylation mechanism [32, 37]. TET1 is primarily responsible for oxidizing 5-mC into 5-hydroxymethyl cytosine (5-hmC), whereas TET2 and TET3 primarily oxidize 5-hmC into further oxidized cytosines that are replaced with an unmethylated, unmodified cytosine [32, 38].
Studies have demonstrated that DEHP exposure modulates DNA methylation. Specifically, prenatal DEHP exposure induces long-lasting and robust promoter methylation-related silencing of fundamental genes in sperm physiology [39]. In utero DEHP exposure is associated with an enrichment of DNA methylation of genes involved in the androgen response, estrogen response, and spermatogenesis [40]. Prenatal exposure to DEHP differentially expressed 406 genes related to reproductive processes in rat ovaries [41]. Although these studies indicate that DEHP acts through methylation, studies do not indicate if DEHP modifies methylation statuses throughout generations. Therefore, the current study was designed to evaluate the pathways and mechanisms by which prenatal and ancestral exposure to DEHP influence key ovarian functions in the F1, F2, and F3 generations of mice. Specifically, this study tested the hypothesis that prenatal and ancestral DEHP exposure disrupt ovarian functions by altering gene expression of several ovarian pathways critical for cell growth, proliferation, and function (i.e., the sex steroid hormone synthesis pathway, phosphoinositide 3-kinase pathway (PI3K), cell cycle regulators, apoptosis and oxidative stress factors, steroid hormone receptors, and insulin-like growth factors) [24, 42–49], DNA methylation, and DNA methylation effectors such as DNMTs and TET enzymes.
Materials and Methods
Chemicals
DEHP (99% purity) was purchased from Sigma-Aldrich (St. Louis, MO). Stock solutions of DEHP (0.022, 0.224, 560, and 840 mg/mL) were prepared by diluting DEHP in tocopherol-stripped corn oil (MP Biomedicals, Solon, OH). These stock solutions were diluted to create doses of 20 μg/kg/day, 200 μg/kg/day, 500 mg/kg/day, and 750 mg/kg/day of DEHP. DEHP concentrations were chosen based on previous studies and their environmental relevance [45, 46, 50–53]. Specifically, the 20 μg/kg/day dose of DEHP was selected because the U.S. Environmental Protection Agency established the chronic oral reference dose as 20 μg/kg/day of DEHP. The reference dose is an estimate of the daily oral exposure of DEHP in the general population that has a low risk of adverse effects during the lifetime [54]. In addition, 20 μg/kg/day of DEHP falls within the estimated human exposure range based on urinary metabolite levels [5]. The 200 μg/kg/day dose of DEHP was used because it falls within the estimated occupational range of exposure [2]. In addition, adult exposure to 200 μg/kg/day of DEHP has been shown to cause abnormal estrous cyclicity and accelerate primordial follicle recruitment in female CD-1 mice [45]. The 500 mg/kg/day dose of DEHP was selected because it has been shown to cause abnormalities in spermatagonial stem cells across multiple generations in male CD-1 mice [52]. The 750 mg/kg/day dose of DEHP was selected because adult exposure has been shown to cause abnormal estrous cyclicity and accelerate primordial follicle recruitment in adult female CD-1 mice [45].
Animals and dosing paradigm
Adult female and male CD-1 mice (Charles River, USA) were housed at 25°C in conventional polysulfone, ventilated cages on 12L:12D cycles. The mice were fed Teklad Rodent Diet 8604 (Harlan) and provided highly purified water (reverse osmosis filtered water) in polysulfone water bottles ad libitum. All animal procedures were approved by the University of Illinois Institutional Animal Care and Use Committee and abide by the guidelines set forth by the National Institute of Health for the Care and Use of Laboratory Animals.
At 8 weeks of age, female mice (F0) were mated with control male mice of the same age. The female mice were monitored twice a day for the presence of a copulatory vaginal sperm plug to confirm mating. Once a copulatory vaginal sperm plug was confirmed, the presence of which was considered gestational day (GD) 0.5, the females were removed, weighed, and individually housed. Subsequently, the mice were weighed twice a week to confirm successful pregnancy. From GD 10.5 until birth of the pups, pregnant dams (F0) were orally dosed once a day with the vehicle control (tocopherol-stripped corn oil) or with DEHP (20 μg/kg/day, 200 μg/kg/day, 500 mg/kg/day, or 750 mg/kg/day) by placing a pipette tip with the dosing solution into the cheek pouch of the mouse. This dosing regimen was selected to mimic oral exposure to DEHP in humans [1, 45, 51]. The doses were calculated and adjusted based on daily body weights, and delivered in 25 – 33 μL of tocopherol-stripped corn oil. The treatment window was chosen because it is a critical time period of ovarian development. Specifically, this is when primordial germ cells arrive at the gonad [55, 56], sex determination occurs [57], and global demethylation and imprint erasure of primordial germ cells occur [58].
Pregnant mice were allowed to deliver naturally and the day of birth was considered postnatal day (PND) 0. Mice born from the F0 generation were labeled the F1 generation. Female mice from the F1 generation were mated with non-treated male CD-1 mice to produce the F2 generation. Females from the F2 generation were mated with non-treated male CD-1 mice to produce the F3 generation. No mice were mated with family members. At PND 21, mice (n = 3 – 15 dams/treatment group) were euthanized by CO2 affiliation followed by cervical dislocation. PND 21 was selected because mice are juvenile, not sexually mature, and no corpora lutea are present. Whole ovaries were collected from each mouse. One ovary was immediately frozen in liquid nitrogen and stored at −80°C for RNA and DNA extraction.
RNA sequencing analysis
Frozen whole ovaries collected at PND 21 from control and 20 μg/kg/day (n = 3 ovaries/treatment group) from the F3 generation were used for RNA sequencing. Raw reads were checked for quality using FASTQC (v 0.11.5) then trimmed and filtered using Trimmomatic (v 0.36) to remove residual adapter content, low quality bases (Phred quality score < 28), and resulting reads shorter than 30 nt. Trimmed/filtered reads were aligned to NCBI’s Mus musculus GRCm38.p6 genome and gene model annotation release 106 using STAR (v 2.5.3a). Post-alignment gene counts were then determined for each NCBI EntrezGene ID using featureCounts from Subread (v 1.5.2-pl) with multi-mapping reads excluded.
The raw read counts were input into R [59] (v 3.4.3) for pre-processing and analysis together using Bioconductor [60] packages as listed below. Approximately ~23 million reads aligned uniquely within the 41,595 M. musculus genes. We used TMM method [61] in the edgeR package [62] (v 3.20.5) to normalize the counts to log2-transformed counts per million (logCPM), using the cpm() function with prior.count = 3. Specifically, 25,141 genes did not have logCPM > log2(0.5) in at least three samples and were filtered out, leaving 16,454 genes to be analyzed for differential expression. TMM-values and logCPM normalized values were re-calculated with prior.count = 3 after gene filtering. Principle components analysis clustering of the samples (data not shown) indicated one of the treatment replicates was more variable than the other two. Rather than remove this sample completely, we did a surrogate variables analysis [63, 64] on the logCPM values, which estimated one surrogate variable that corrected for the difference in this replicate (data not shown). This surrogate variable was added to edgeR’s quasi-likelihood negative binomial generalized log-linear model [65], which was fit on the original read counts + TMM values to find differential expression between the treated and control groups. Multiple hypothesis test correction was done using the False Discovery Rate method [66].
Data obtained from RNA sequencing were functionally analyzed using The Database of Annotation, Visualization, and Integrated Discovery Bioinformatics (DAVID) 6.8 following the previously published protocol [67, 68]. Genes with a false discovery rate < 0.62 and p < 0.007 were entered into DAVID for functional annotation analysis for a total of 177 genes. “Gene_Ontology” and “Pathways” and the denoted DAVID defined defaults were selected for functional annotation clustering. To determine if functional gene groups were valuable, annotation clusters with a significant enrichment score ≥ 1 were further explored [67].
Gene expression analysis
Frozen whole ovaries collected at PND 21 were used for quantitative real-time polymerase chain reaction (qPCR) analysis (n = 3 – 6 ovaries/treatment group). Total RNA (>100 ng) was extracted from the whole ovaries using the AllPrep DNA/RNA Mini Kit (Qiagen, Austin, TX, USA) according to the manufacturer’s protocol, including DNase digestion. Total RNA (100 ng) was reverse transcribed to complementary DNA (cDNA) using the iScript RT Kit (Bio-Rad Laboratories, Inc., Hercules, CA, USA) according to the manufacturer’s protocol. Each cDNA sample was diluted 1:8 using nuclease-free water prior to qPCR analysis. Analysis of qPCR was performed using the CFX96 C1000 Real-Time PCR Detection System and CFX Manager Software (Bio-Rad Laboratories, Inc., Hercules, CA, USA) according to the manufacturer’s protocol. Each qPCR reaction was done in duplicate using 2 μL of cDNA, forward and reverse primers (5 pmol) for select genes, nuclease-free water, and SsoFastEvaGreen Supermix for a final reaction volume of 10 μL. Target genes were analyzed in reference to the housekeeping gene, beta-actin (Bactin). A list of gene primers (Integrated DNA Technologies, Coralville, IA, USA) and the housekeeping gene, beta-actin, are included in the supplementary files (Table S1).
The CFX96 C1000 Real-Time PCR Detection machine quantifies the amount of PCR product generated by measuring SsoFastEvaGreen dye (Bio-Rad Laboratories, Inc., Hercules, CA) that fluoresces when bound to double-stranded DNA. The qPCR program consisted of an enzyme activation step (95 °C for 1 min), an amplification and quantification program (40 cycles of 95 °C for 10 s, 60 °C for 10 s, single fluorescence reading), a 72 °C for 5 min, a melt curve (65 °C-95 °C heating 0.5 °C/s with continuous fluorescence readings), and a final step at 72 °C for 5 min per the manufacturer’s protocol. All gene expression data were normalized to the housekeeping gene. Relative fold changes were calculated and analyzed using a mathematical model for relative quantification of real-time PCR data developed by Pfaffl [69] and then normalized as a ratio to control group.
DNA methylation analysis
DNA was extracted from frozen whole PND 21 (n = 3 – 7 ovaries/treatment group) ovaries using the AllPrep DNA/RNA Mini Kit (Qiagen, Austin, TX, USA) per the manufacturer’s protocol. DNA was extracted, eluted in 100 μL of EB buffer, and stored in −80 C until further DNA methylation testing. To measure global DNA methylation status, the enzyme-linked immunosorbent assay MethylFlash Methylated DNA 5-mC Quantification Kit (Colorimetric assay, Epigentek Group Inc., Farmingdale, NY, USA) was used according to the manufacturer’s protocol. Briefly, ovarian DNA (100 ng) was added to high affinity strip wells. Methylated DNA was detected using capture and detection antibodies for 5-methyl cytosine (5-mC) and quantified by reading absorbance at 450 nm using a 354 Multiskan Ascent Microplate Reader (Thermo Electron Corp., Shanghai, China). The absolute amount and percentage of methylated DNA were calculated using the absolute quantification method per the manufacturer’s protocol. Briefly, a standard curve was calculated from five known concentrations of methylated DNA (0.5, 1, 2, 5, 10 ng). The slope of the standard curve was quantified and used in the provided formulas per the manufacturer’s protocol to calculate the amount and percentage of methylated DNA (5-mC) in each sample.
Statistical analyses
Data were expressed as the mean ± standard error of the mean (SEM). In all generations, data from multiple female pups originating from the same litter were averaged and combined as n = 1, and data from at least 3 separate litters were used in the analyses. Data were analyzed by comparing treatment groups to control using IBM SPSS version 24 software (SPSS Inc., Chicago, IL, USA). Outliers were removed by the Grubb’s test using GraphPad outlier calculator software (GraphPad Software Inc., La Jolla, CA, USA). Data that were continuous were assessed for normal distribution by Shapiro-Wilk analysis. If data met assumptions of normal distribution and homogeneity of variance, data were analyzed by one-way analysis of variance (ANOVA) followed by Tukey HSD or Dunnett 2-sided post-hoc comparisons. However, if data met assumptions of normal distributions, but not homogeneity of variance, data were analyzed by ANOVA followed by Games-Howell or Dunnett’s T3 post-hoc comparisons. If data were presented as percentages or were not normally distributed, the independent sample Kruskal-Wallis H followed by Mann-Whitney U non-parametric tests were performed. For all comparisons, statistical significance was determined by p-value ≤ 0.05. In instances in which p-values were greater than 0.05, but less than 0.10, data were considered to exhibit a trend towards significance.
Results
The effects of ancestral exposure to DEHP on gene expression in the F3 generation as determined by RNA sequencing and The Database of Annotation, Visualization, and Integrated Discovery Bioinformatics
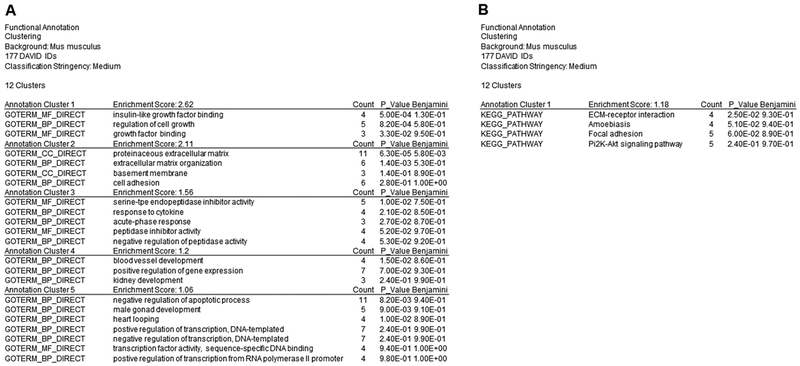
Functional annotation gene clustering analysis via DAVID provided 5 annotation clusters from the “Gene_Ontology” selection (Figure 1A). Within the annotation cluster containing the highest enrichment score was insulin-like growth factor binding, regulation of cell growth, and growth factor binding (Figure 1A). Functional annotation gene clustering from the “Pathway” selection provided 1 annotation cluster (Figure 1B). Within the annotation cluster, extra cellular matrix-receptor interaction, amoebiasis, focal adhesion, and the PI3K-Akt signaling pathway were listed (Figure 1B). Based on these results, subsequent qPCR was conducted to assess the effects of DEHP exposure on ovarian gene expression.
Figure 1.
Data obtained from the RNA sequencing were functionally analyzed using The Database of Annotation, Visualization, and Integrated Discovery (DAVID) Bioinformatics version 6.8. A total of 177 genes were entered into DAVID (false discovery rate < 0.62 and p < 0.007) for functional annotation analysis. “Gene_Ontology” results yield 5 annotation clusters (A) and “Pathways” results yield 1 annotation cluster (B) with a significant enrichment score ≥ 1.
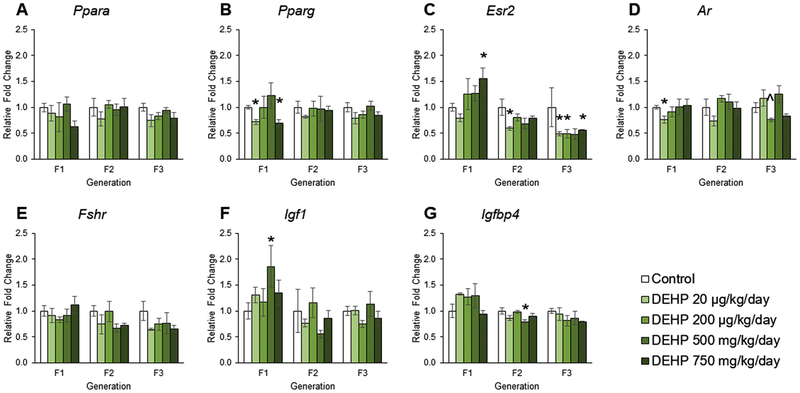
The effects of DEHP exposure on hormone receptors and insulin-like growth factor gene expression in ovaries from the F1 – F3 generations
Several studies suggest that DEHP and its many metabolites act through steroid hormone receptors and peroxisome proliferator-activated receptors (PPAR) [70, 71]. Therefore, the current study examined the effects of prenatal and ancestral exposure to DEHP on the expression of hormone receptors and PPARs. Further, based on the RNA sequencing results, the insulin-like growth factor (IGF) family was examined. In the F1 generation, prenatal exposure to DEHP did not affect the expression of Ppara, Fshr, or Igfbp4 compared to controls (Figures 2A, 2E, and 2G). However prenatal exposure to DEHP decreased the expression of Pparg in the 20 μg/kg/day and 750 mg/kg/day groups, increased Esr2 expression in the 750 mg/kg/day group, decreased the expression of Ar in the 20 μg/kg/day group, and increased the expression of Igfl in the 500 mg/kg/day group compared to controls (Figures 2B, 2C, 2D, and 2F, n = 3 – 5 ovaries/treatment group, p ≤ 0.05). In the F2 generation, exposure to DEHP did not affect the expression of Ppara, Pparg, Ar, Fshr, or Igf1 compared to controls (Figures 2A, 2B, 2D, 2E, and 2F). In contrast, exposure to DEHP decreased the expression of Esr2 in the 20 μg/kg/day group and decreased Igfbp4 expression in the 500 mg/kg/day group compared to controls (Figures 2C and 2G, n = 3 ovaries/treatment group, p ≤ 0.05). In the F3 generation, ancestral exposure to DEHP did not affect the expression of Ppara, Pparg, Fshr, Igf1, or Igfbp4 compared to controls (Figures 2A, 2B, 2E, 2F, and 2G). However ancestral exposure to DEHP decreased the expression of Esr2 in the 20 μg/kg/day, 200 μg/kg/day, and 750 mg/kg/day groups, and decreased the expression of Ar in the 200 μg/kg/day group compared to controls (Figures 2C and 2D, n = 3 – 6 ovaries/treatment group, p ≤ 0.05 but p = 0.071 for Ar in 200 μg/kg/day).
Figure 2.
The effects of prenatal and ancestral DEHP exposure on steroid hormone receptors and insulin-like growth factors in PND 21 ovaries in the F1 – F3 generations. All gene expression is relative to the housekeeping gene, Bactin, and the relative fold change is normalized to 1 for control. Graphs represent mean ± SEM from 3 – 6 ovaries per treatment group. * p ≤ 0.05 (significant difference compared to control with generation), 0.05 < ^ p < 0.10.
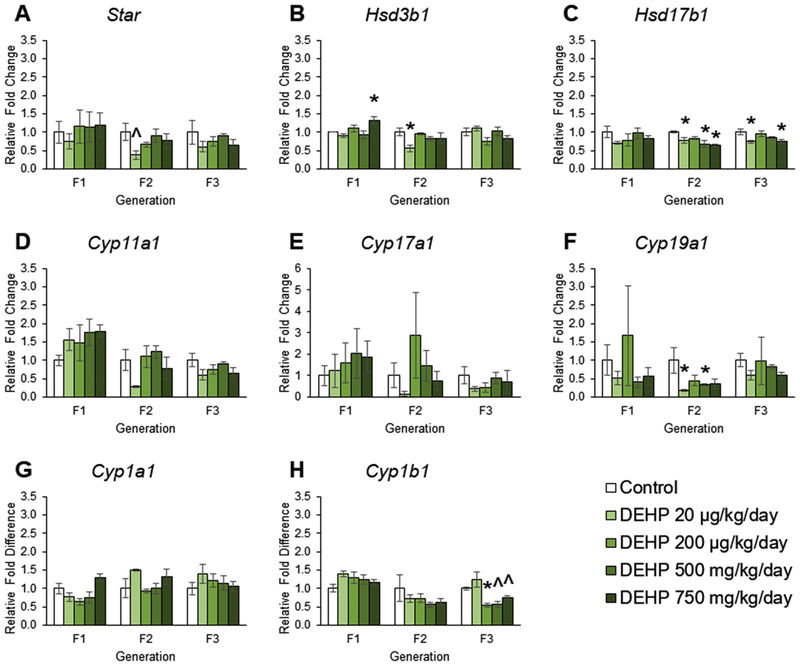
The effects of DEHP on steroidogenic enzyme gene expression in ovaries from the F1 – F3 generations
Our previous work showed that prenatal DEHP exposure dysregulated steroid hormone levels in F2 generations, but not the F1 and F3 generations of mice at PND 21 [24]. The current work was performed to examine estrogen synthesis and expand our knowledge of DEHP dysregulation of steroid hormones by determining if it is due to prenatal or ancestral DEHP effects on expression of steroidogenic enzymes. In the F1 generation, prenatal exposure to DEHP did not affect the expression of Star, Hsd17b1, Cyp11a1, Cyp17a1, Cyp19a1, Cyp1a1, or Cyp1b1 (Figures 3A, 3C, 3D, 3E, 3F, 3G, and 3H), but DEHP at 750 mg/kg/day increased the expression of Hsd3b1 compared to controls (Figure 3B, n = 3 – 5 ovaries/treatment group, p ≤ 0.05). In the F2 generation, exposure to DEHP decreased the expression of Star in the 20 μg/kg/day group, decreased Hsd3b1 expression in the 20 μg/kg/day group, decreased Hsd17b1 expression in the 20 μg/kg/day, 500 mg/kg/day, and 750 mg/kg/day groups, and decreased Cyp19a1 expression in the 20 μg/kg/day and 500 mg/kg/day groups compared to controls (Figures 3A, 3B, 3C, and 3F, n = 3 ovaries/treatment group, p < 0.05, but p = 0.081 for Star in 20 μg/kg/day). In the F3 generation, ancestral exposure to DEHP did not affect the expression of Star, Hsd3b1, Cyp11a1, Cyp17a1, Cyp19a1, or Cyp1a1 compared to controls (Figures 3A, 3B, 3D, 3E, 3F, and 3G), but ancestral exposure decreased the expression of Hsd17b1 in the 20 μg/kg/day and 750 mg/kg/day groups and decreased Cyp1b1 expression in the 200 μg/kg/day, 500 mg/kg/day, and 750 mg/kg/day treatment groups compared to controls (Figures 3C and 3H, n = 3 – 6 ovaries/treatment group, p ≤ 0.05, but p = 0.068 and 0.069 for Cyp1b1 in 500 mg and 750 mg/kg/day, respectfully).
Figure 3.
The effects of prenatal and ancestral DEHP exposure on steroidogenesis and estradiol metabolism in PND 21 ovaries in the F1 – F3 generations. All gene expression is relative to the housekeeping gene, Bactin, and the relative fold change is normalized to 1 for control. Graphs represent mean ± SEM from 3 – 6 ovaries per treatment group. * p ≤ 0.05 (significant difference compared to control with generation), 0.05 < ^ p < 0.10.
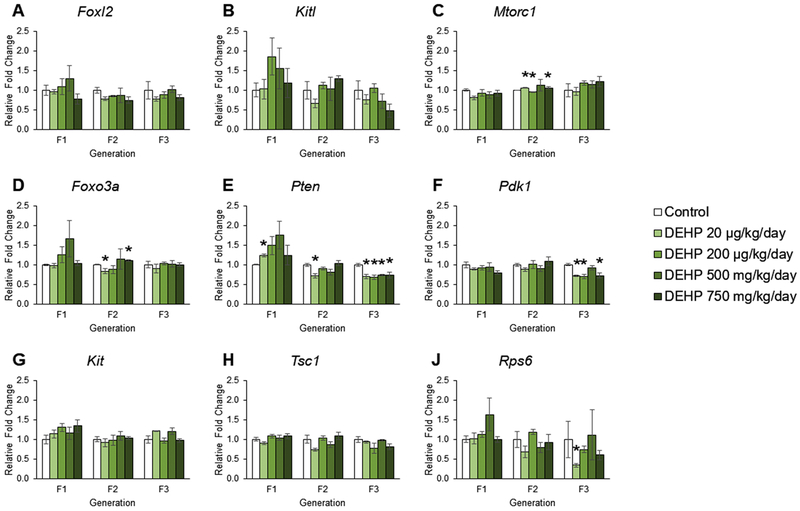
The effects of DEHP exposure on phosphoinositide 3–kinase pathway gene expression ovaries from the F1 – F3 generations
Our previous work showed that ancestral exposure to DEHP accelerated primordial follicle recruitment in the F3 generation, but not in the F1 and F2 generations of mice [24]. Previous studies also indicate that adult exposure to DEHP dysregulates the PI3K pathway, a critical pathway for primordial follicle recruitment [45]. Further, the RNAseq data indicate that ancestral DEHP exposure alter the PI3K pathway (Figure 1). Thus, we examined the effects of prenatal and ancestral exposure to DEHP on the PI3K factors in the F1 – F3 generations. In the F1 generation, prenatal exposure to DEHP did not affect the expression of Foxl2, Kitl, Mtorc1, Foxo3a, Pdk1, Kit, Tsc1, or Rps6 compared to controls (Figures 4A, 4B, 4C, 4D, 4F, 4G, 4H, and 4J). However, prenatal exposure to 20 μg/kg/day of DEHP increased the expression of Pten compared to controls (Figure 4E, n = 3 – 5 ovaries/treatment group, p ≤ 0.05). In the F2 generation, exposure to DEHP did not affect the expression of Foxl2, Kitl, Pdk1, Kit, Tsc1, or Rps6 compared to controls (Figures 4A, 4B, 4F, 4G, 4H, and 4J), but DEHP exposure increased the expression of Mtorc1 at 20 μg/kg/day and 750 mg/kg/day and decreased Mtorc1 expression at 200 μg/kg/day, decreased the expression of Foxo3a in the 20 μg/kg/day group and increased Foxo3a expression in the 750 mg/kg/day group, and decreased the expression of Pten in the 20 μg/kg/day group compared to controls (Figures 4C, 4D, and 4E, n = 3 ovaries/treatment group, p ≤ 0.05). In the F3 generation, ancestral exposure to DEHP did not affect the expression of Foxl2, Kitl, Mtorc1, Foxo3a, Kit, or Tsc1 compared to controls (Figures 4A, 4B, 4C, 4D, 4G, and 4H). Ancestral exposure to DEHP decreased the expression of Pten in all treatment groups, decreased Pdk1 expression in the 20 μg/kg/day, 200 μg/kg/day, and 750 mg/kg/day groups, and decreased Rps6 expression in the 20 μg/kg/day group compared to controls (Figures 4E, 4F, and 4J, n = 3 – 6 ovaries/treatment group, p ≤ 0.05).
Figure 4.
The effects of prenatal and ancestral DEHP exposure on the phosphoinositide 3-kinase pathway in PND 21 ovaries in the F1 – F3 generations. All gene expression is relative to the housekeeping gene, Bactin, and the relative fold change is normalized to 1 for control. Graphs represent mean ± SEM from 3 – 6 ovaries per treatment group. * p ≤ 0.05 (significant difference compared to control with generation), 0.05 < ^ p < 0.10.
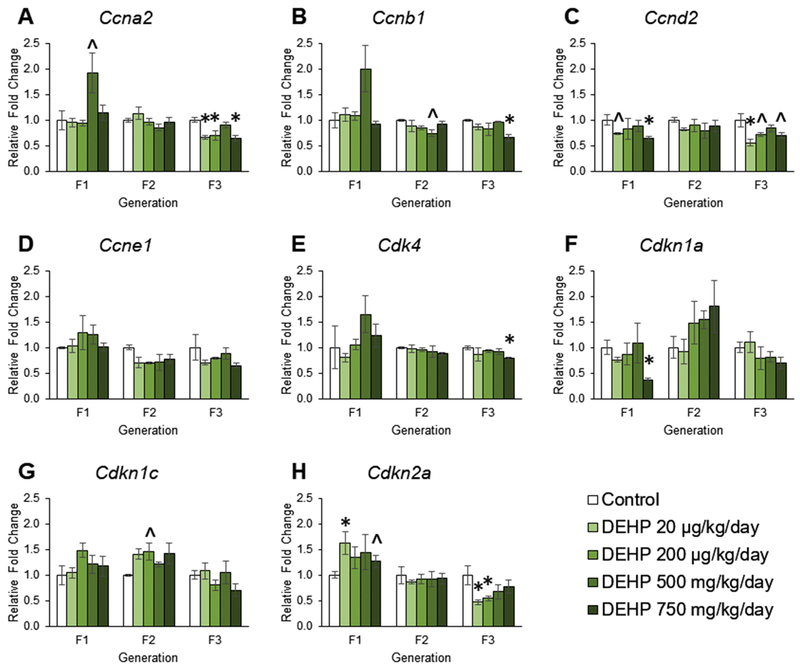
The effects of DEHP exposure on cell cycle regulator gene expression in ovaries from the F1 – F3 generations
Our previous work showed the prenatal exposure to DEHP dysregulated folliculogenesis at PND 21 in all three generations of mice [24]. Folliculogenesis is regulated by many factors, but the cell cycle regulators are heavily involved in cell proliferation and follicle growth [49, 72]. The RNAseq data suggested that ancestral exposure to DEHP affected regulators of growth (Figure 1). Therefore, we measured the mRNA expression levels of cyclins, cyclin dependent kinases, and cyclin dependent kinase inhibitors. In the F1 generation, prenatal exposure to DEHP increased the expression of Ccna2 in the 500 mg/kg/day group, decreased the expression of Ccnd2 in the 750 mg/kg/day group, decreased the expression of Cdkn1a in the 750 mg/kg/day group, increased the expression of Cdkn1c in the 200 μg/kg/day group, and increased the expression of Cdkn2a expression in the 20 μg/kg/day, 200 μg/kg/day, and 750 mg/kg/day groups compared to controls (Figure 5A, 5C, 5F, 5G, and 5H n = 3 – 6 ovaries/treatment group, p ≤ 0.05, but p = 0.101 for Ccnd2 in 20 μg/kg/day, p = 0.086 for Cdkn1c in 200 μg/kg/day, and p = 0.086 for Cdkn2a for 200 μg/kg/day). Prenatal exposure to DEHP did not affect the expression of Ccnb1, Ccne1, or Cdk4 compared to controls (Figures 5B, 5E, and 5D). In the F2 generation, exposure to DEHP did not affect the expression of Ccna2, Ccnd2, Ccne1, Cdk4, Cdkn1a, Cdkn1c, or Cdkn2a compared to controls (Figures 5A, 5C, 5D, 5E, 5F, 5G, and 5H). However, exposure to DEHP at 500 mg/kg/day decreased the expression of Ccnb1 compared to controls, but it was borderline statistically significant (Figure 5B, n = 3 ovaries/treatment group, p = 0.057). In the F3 generation, ancestral exposure to DEHP decreased the expression of Ccna2 in the 20 μg/kg/day, 200 μg/kg/day, and 750 mg/kg/day groups, decreased Ccnb1 expression in the 750 mg/kg/day group, decreased Ccnd2 expression in the 20 μg/kg/day, 200 μg/kg/day, and 750 mg/kg/day groups, decreased Cdk4 expression in the 750 mg/kg/day group, and decreased the expression of Cdkn2a in the 20 μg/kg/day and 200 μg/kg/day groups compared to controls (Figures 5A, 5B, 5C, 5E, and 5H n = 3 – 6 ovaries/treatment group, p ≤ 0.05, but p = 0.084 and 0.060 for Ccnd2 in 200 μg/kg/day and 750 mg/kg/day, respectfully). Further, in the F3 generation, ancestral exposure to DEHP did not affect the expression of Ccne1, Dckn1a, or Cdkn1c compared to controls (Figures 5D, 5F, and 5G).
Figure 5.
The effects of prenatal and ancestral DEHP exposure on cell cycle regulators in PND 21 ovaries in the F1 – F3 generations. All gene expression is relative to the housekeeping gene, Bactin, and the relative fold change is normalized to 1 for control. Graphs represent mean ± SEM from 3 – 6 ovaries per treatment group. * p ≤ 0.05 (significant difference compared to control with generation), 0.05 < ^ p < 0.10.
The effects of DEHP exposure on apoptosis and oxidative stress pathway gene expression in ovaries from the F1 – F3 generations
Our previous study showed the prenatal exposure to DEHP decreased the percentage of atretic follicles in the F1 generation [24]. The B-cell lymphomas/leukemia-2 (Bcl-2) family includes inhibitors and promoters of apoptosis in the ovary [47]. The balance of promotors and inhibitors of apoptosis is critical for the healthy development and maintenance follicular cells. The Bcl-2 family has been shown to directly regulate apoptosis in the ovary [47, 73]. In addition, oxidative stress is an imbalance of pro-oxidant molecules and anti-oxidant defenses and the balance of these factors are critical for adequate growth and development of follicles [48]. Thus, we examined the effects of prenatal and ancestral exposure to DEHP on the expression of the Bcl-2 family and oxidative stress factors in the ovary of the F1 – F3 generations.
In the F1 generation, prenatal exposure to DEHP did not affect the expression of Bcl2, Bax, Bad, Casp3, Casp8, Catalase, Gpx, or Gsr compared to controls (Figures S1A, S1B, S1C, S1E, S1F, S1H, S1J, and S1K). In contrast, prenatal exposure to DEHP at 750 mg/kg/day decreased the ratio of Bax/Bcl2 and DEHP at 500 mg/kg/day increased the expression of Bok compared to controls, but it was borderline statistically significant (Figures S1D and S1G, n = 3 – 5 ovaries/treatment group, p ≤ 0.05 and p = 0.076 for Bok). In the F2 generation, exposure to DEHP did not affect the expression of Bcl2, Bax, Bax/Bcl2 ratio, Bok, Catalase, or Gsr compared to controls (Figures S1A, S1B, S1D, S1E, S1H, and S1K). However, exposure to 20 μg/kg/day of DEHP decreased the expression of Bad, Casp3, and Casp8, and exposure to 500 mg/kg/day of DEHP increased the expression of Gpx compared to controls (Figures S1C, S1E, S1F, and S1J, n = 3 ovaries/treatment group, p ≤ 0.05). In the F3 generation, ancestral exposure to DEHP decreased the expression of Bcl2 in all treatment groups, increased the expression of Bax/Bcl2 ratio in the 20μg/kg/day, 200 μg/kg/day, and 500 mg/kg/day groups, decreased the expression of Casp3 in the 20 μg/kg/day, 200 μg/kg/day, and 500 mg/kg/day groups, decreased the expression of Casp8 in all treatment groups, decreased the expression of Bok in the 20 μg/kg/day group, decreased the expression of Gpx in the 750 mg/kg/day group, and decreased the expression of Gsr in the 20 μg/kg/day, 200 μg/kg/day, and 750 mg/kg/day groups compared to controls (Figures S1A, S1D, S1E, S1F, S1G, S1J, and S1K, n = 3 – 6 ovaries/treatment group, p < 0.05, but p = 0.071 for Casp8 in 750 mg/kg/day, p = 0.088 for Bok in 20 μg/kg/day, and p = 0.067 for Gsr in 200 μg/kg/day).
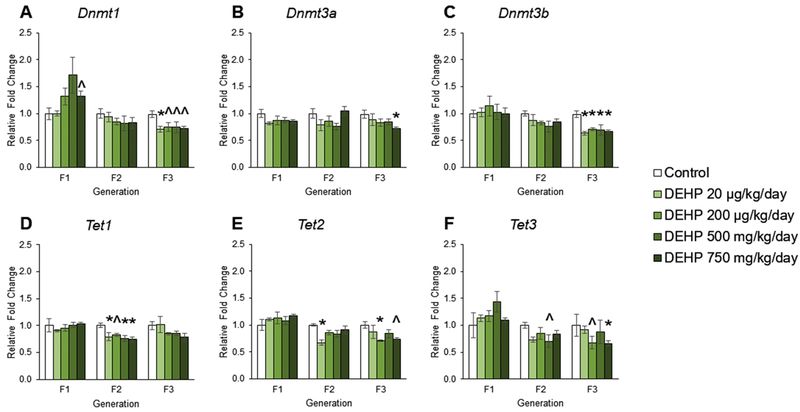
The effects of DEHP exposure on DNA methyltransferases and ten-eleven translocation enzyme gene expression in ovaries from the F1 – F3 generations
Although previous studies demonstrate that DEHP exposure causes transgenerational inheritance of ovarian dysfunction [24, 74, 75], the DNA methylation mediators underlying these changes have not been well studied. Therefore, we examined the expression levels Dnmt and Tet in the F1 – F3 generations. In the F1 generation, prenatal exposure to DEHP did not affect the expression of Dnmt3a, Dnmt3b, Tet1, Tet2, and Tet compared to controls (Figures 6B, 6C, 6D, 6E, and 6F). However, prenatal exposure to DEHP at 750 mg/kg/day increased the expression of Dnmt1 compared to controls, but it was borderline statistically significant (Figure 6A, n = 3 – 5 ovaries/treatment group, p = 0.068). In the F2 generation, exposure to DEHP did not affect the expression of Dnmt1, Dnmt3a, or Dnmt3b compared to controls (Figures 6A, 6B, and 6C). In contrast, DEHP exposure decreased the expression of Tet1 in all groups, decreased the expression of Tet2 in the 20 μg/kg/day group, and decreased the expression of Tet3 in the 500 mg/kg/day group compared to control, but it was borderline statistically significant (Figures 6D, 6E, and 6F, n = 3 ovaries/treatment group, p ≤ 0.05, but p = 0.085 for Tet3 in 500 mg/kg/day). In the F3 generation, ancestral exposure to DEHP decreased the expression of Dnmt1 in all groups compared to control, but some of the decreases were borderline statistically significant (Figure 6A, n = 3 – 6 ovaries/treatment group, p ≤ 0.05, but p = 0.097 for 200 μg/kg/day, p = 0.095 for 500 mg/kg/day, and p = 0.059 for 750 mg/kg/day). Further, ancestral exposure to DEHP decreased the expression of Dnmt3a in the 750 mg/kg/day group, decreased the expression of Dnmt3b in all groups, decreased Tet2 expression in the 200 μg/kg/day and 750 mg/kg/day groups, and decreased Tet3 expression in the 200 μg/kg/day and 750 mg/kg/day groups compared to controls (Figures 6B, 6C, 6E, and 6F, n = 3 – 6 ovaries/treatment group, p ≤ 0.05, but p = 0.064 for Tet2 in 750 mg/kg/day and p = 0.063 for Tet3 in 200 μg/kg/day). In the F3 generation, ancestral exposure to DEHP did not affect the expression of Tet1 compared to controls (Figure 6D).
Figure 6.
The effects of prenatal and ancestral DEHP exposure on DNA methyltransferases and ten-eleven translocation enzymes in PND 21 ovaries in the F1 – F3 generations. All gene expression is relative to the housekeeping gene, Bactin, and the relative fold change is normalized to 1 for control. Graphs represent mean ± SEM from 3 – 6 ovaries per treatment group. * p ≤ 0.05 (significant difference compared to control with generation), 0.05 < ^ p < 0.10.
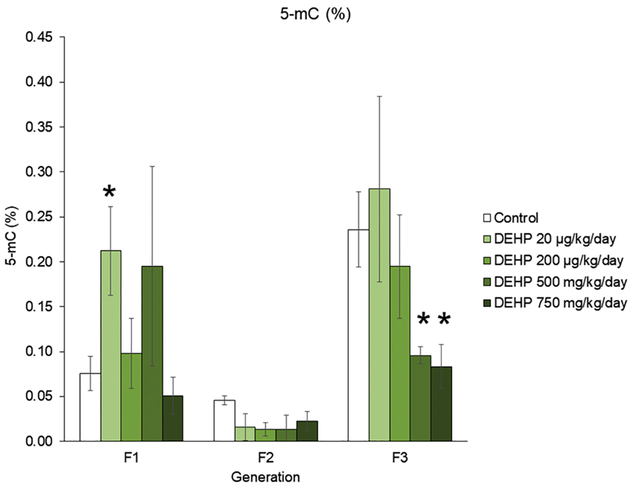
The effects of DEHP exposure on DNA methylation percentage in whole ovaries from the F1 – F3 generations
Previous studies determined that prenatal exposure to DEHP causes both multigenerational and transgenerational inheritance in ovarian dysfunction [23–25]. Transgenerational inheritance is thought to be mediated by epigenetic mechanisms, and DNA methylation is a commonly studied epigenetic mechanism. Therefore, the current study measured the percentage of 5-mC in the whole ovary in the F1 – F3 generations. In the F1 generation, prenatal exposure to 20 μg/kg/day of DEHP increased the percentage of 5-mC compared to controls and in the F3 generation, ancestral exposure to 500 mg/kg/day and 750 mg/kg/day of DEHP decreased the percentage of 5-mC in the whole ovary compared to controls (Figure 7, n = 3 – 7 ovaries/treatment group, p ≤ 0.05).
Figure 7.
The effects of prenatal and ancestral DEHP exposure on the percentage of 5-mC in whole ovaries at PND 21 in the F1 – F3 generations. Graphs represent mean ± SEM from 3 – 7 ovaries per treatment group. * p ≤ 0.05 (significant difference compared to control with generation).
Discussion
Our previous study showed that prenatal and ancestral exposure to DEHP disrupted sex steroid hormone levels in the F1 and F2 generations, disrupted ovarian follicle counts in the F1 – F3 generations, and altered select reproductive outcomes in the F1 – F3 generations [23, 24]. Our current study provides additional information on the multigenerational and transgenerational effects of DEHP exposure on the ovary. We show that prenatal exposure to DEHP disrupts the expression of the sex steroid hormone synthesis pathway, factors in the PI3K pathway, steroid hormone receptors, DNA demethylation processes, and DNA methylation in the F1 and the F2 generations of the ovary. Further, we show that ancestral exposure to DEHP disrupts the expression of estrogen metabolism, the PI3K pathway, cell cycle regulators, apoptosis and oxidative stress factors, estrogen receptor beta, DNA methylation and demethylation factors, and DNA methylation in the F3 ovary. This study provides potential mechanisms and pathways explaining how both prenatal and ancestral exposure to DEHP disrupt ovarian functions in the F1–F3 generations of mice.
In this study, mice were orally exposed daily to DEHP starting at embryonic day 10.5 and ending at birth. During this exposure window, primordial germ cells in the fetus (F2 generation) migrate to the genital ridge and undergo mitosis, meiosis, methylation, and demethylation processes [76–79]. We anticipate that this window of exposure targets epigenetic inheritance and likely causes transgenerational inheritance in the F3 generation [79]. This window of exposure is critical because the F1, F2, and F3 generations receive exposure at different developmental windows. The F1 generation is exposed as a developing pup, and therefore, the hypothalamus-pituitary-gonadal axis is directly exposed to DEHP. The F2 generation is exposed as the developing germ cells in the gonad. The F3 generation is not directly exposed to DEHP and thus, it is the first generation to experience transgenerational inheritance. Each generation is exposed to DEHP at different developmental time points; therefore, we anticipated and observed that the effects of DEHP on ovarian functions would be different in each generation.
Endocrine disruptors, such as DEHP, cause nonmonotonic dose responses such as sigmoid, U-shaped, or inverted-U-shaped curves [80, 81]. Although the mechanisms behind such nonmonotonic effects are not fully understood, they may be due to receptor type and abundance in specific cells or tissues [82], receptor down-regulation and desensitization [83, 84], and endocrine feedback loops [85, 86]. Therefore, it is not surprising that the majority of the results indicate that DEHP exposure did not have a linear dose response effect on gene expression in the ovary. Further, in each generation, baseline differences in control groups were present. Baseline differences in control groups between generations is a phenomenon that has been observed in numerous transgenerational studies [23–25, 75, 87]. Although it is not clear what causes baseline changes between control groups, it may be due to differences in the timing of exposure and/or age of mice [87].
Data from RNA sequencing were analyzed using DAVID. Of the 177 genes, themes and pathways were identified. Genes were selected for further qPCR analysis as a mean to verify sequencing data. Interestingly, some themes including regulation of cell growth, insulin-like growth factor binding, and PI3K-Akt signaling pathway were identified. Previous work in the Flaws’ laboratory has determined that exposures to toxicants such as DEHP and bisphenol A disrupts genes associated with cell growth, insulin-like growth factor, and the PI3K-Akt signaling pathway in the ovary [45, 88, 89]. Thus, numerous signaling pathways were analyzed.
Our results indicate that DEHP exposure affected the expression of Pparg, Ar, and Esr2 in the F1 generation. These finding are in contrast to a previous study that showed that DEHP exposure repressed Esr1 gene expression via PPARα-dependent pathways in a multigenerational manner [90]. We were surprised that ancestral DEHP exposure did not change Ppar expression in the F3 generation in our study because it is thought that the endocrine disrupting effects of DEHP are mediated through PPAR action [90–93]. However, most notably in the F3 generation, ancestral DEHP exposure caused significant decreases in Esr2 in our study. Given that Esr2 is important for regulatory effects of estrogens on granulosa proliferation, it is possible that DEHP-induced decreases in Esr2 may lead to many of the observed gene expression changes in the F3 generation [94–96].
Our current results show that DEHP exposure disrupted expression of steroidogenic enzymes in the F2 and F3 generations, but not in the F1 generation. According to our previous study, prenatal DEHP exposure did not affect serum 17β-estradiol levels in the F1 generation at PND 21 [24]. Therefore, it is not surprising that prenatal DEHP exposure did not significantly affect the expression of sex steroid hormone synthesis enzymes in the F1 generation. However, in the F2 generation, our previous study showed that prenatal exposure to DEHP borderline decreased serum 17β-estradiol levels and increased serum progesterone levels in the 20 μg/kg/day treatment group compared to control [24]. In our current study, DEHP exposure at 20 μg/kg/day decreased the expression of Star, Hsd3b1, Hsd17b1, and Cyp19a1 in the F2 generation and this decrease in enzymes correlates well with the previously observed serum sex steroid hormone levels. Likely, the increase in serum progesterone level is due to the decrease of enzymes necessary to further biotransform it to androgens and estrogens. Further, the decrease of Hsd17b1 and Cyp19a1 likely leads to a decrease in serum 17β-estradiol levels because these two enzymes biotransform estrone and testosterone into 17β-estradiol, respectfully [97]. Finally, in the F3 generation, we observed a decrease in Hsd17b1 expression with DEHP exposure, which is important for biotransforming androstenedione into testosterone and estrone into 17β-estradiol [97]. However, in the previous study, we did not observe a serum sex steroid hormone change in response to DEHP exposure [24].
Interestingly, our steroidogenic enzyme gene expression results are in contrast with another study that exposed mice to DEHP during an early developmental time period. Specifically, Pocar et al. perinatally dosed mice throughout gestation and lactation with low doses of DEHP and observed decreases in steroidogenic enzyme expression in the F1 generation and not the F2 or F3 generations [75]. The reason why our results and Pocar et al. vary may be due to the many differences between the experiments. In our study, we dosed animals only during the second half of gestation and our doses included 20 μg/kg/day – 750 mg/kg/day, whereas Pocar et al. dosed animals throughout lactation and gestation with 50 μg/kg/day and 5 mg/kg/day [75]. Additional studies that examine the direct effects of DEHP exposure on steroidogenesis show mixed results, demonstrating that the route, timing, and dose of DEHP greatly contribute to DEHP-induced effects [23, 24, 51, 98].
Results from our study indicate that prenatal and ancestral DEHP exposure disrupted the PI3K pathway in the F1, F2, and F3 generations. In the F1 generation, prenatal DEHP exposure increased Pten expression in the PI3K pathway, but it did not affect other factors in the pathway. Pten is a gene that encodes the PI3K negative regulator; if deleted, the entire pool of primordial follicles activates [42]. Therefore, an increase in Pten expression suggests primordial follicle quiescence. Interestingly, in our previous study, we observed data supporting primordial follicle quiescence in the F1 generation [24]. Our previous data also showed that at PND 21, ancestral DEHP exposure decreased primordial follicle numbers [24]. A correlated decrease in Pten, Pdk1, and Rps6 expression correlates well with decreased primordial follicle numbers because decreased Pten expression suggests that primordial follicles activate and continue folliculogenesis [42]. Further, decreased Pdk1 and Rps6 expression decreases primordial follicle survival [99]. Therefore, decreased expression of these factors supports previously published follicle count numbers in the F3 generation [24] and provides a potential mechanism for follicle count disruption observed at PND 21.
Prenatal and ancestral exposure to DEHP significantly decreased gene expression of cell cycle regulators in the F1 and F3 generations. In somatic cells, the cell cycle is made of four phases, with different cyclin-dependent kinases and cyclins to control the cell cycle [100]. Cyclin A2 is expressed during the S phase and is critical for DNA replication [49]. Cyclin B1 is necessary for cell cycle progression through mitosis [49]. Cyclin D2 binding to CDK4 is a critical positive regulator for ovarian granulosa cell proliferation in response to follicle-stimulating hormone (FSH) [72, 101]. In the F1 generation, it is likely that prenatal DEHP exposure inhibits cell cycle progression by decreasing promotors of the cell cycle such as Ccnd2 and increasing the expression of inhibitors of cell cycle such as Cdkn2a and Cdkn1c [102, 103]. Although the expression of Cdkn1a, another cell cycle inhibitor, was decreased in the F1 generation, it is likely that it was not biologically significant enough to counteract the expression of the other inhibitors. Further, the effects of prenatal DEHP exposure on cell cycle regulators is somewhat similar to studies that directly exposed the ovary to DEHP. Direct exposure to DEHP in vitro increased the expression of Ccna2, Ccnb1, Ccnd2, Cdk4, and Ccne1 after 72 hours of exposure [50]. In the F3 generation, DEHP-induced decreases in cyclins and cyclin-dependent kinase suggest that ancestral DEHP exposure reduces cell cycle progression and proliferation, likely causes cells to undergo cell cycle arrest.
DEHP exposure disrupted DNA methylation in the ovaries in each generation. In the F1 generation, prenatal DEHP exposure increased Dnmt expression and increased the percentage of 5-mC. Increased Dnmt1 expression supports increased 5-mC in the ovary because Dnmt1 is important for translating DNMT1, the maintenance DNA methyltransferase. Prenatal DEHP exposure increased Dnmt expression in testicular Leydig cells, increased methylation in promoter regions of steroidogenic transcription factors, and decreased gene expression of steroidogenic enzymes in the F1 generation of rats [104]. In the F2 generation, DEHP exposure did not affect DNA methylation percentage, but it significantly decreased Tet expression. These data suggest that prenatal DEHP exposure modulates DNA demethylation pathways, but not to a large enough degree for the decrease to significantly affect 5-mC percentage in the ovary. In the F3 generation, ancestral DEHP exposure decreased Dnmt, Tet, and 5-mC expression. It is likely that ancestral DEHP exposure decreases Dnmt, subsequently decreasing 5-mC in the ovary, and that DEHP-induced changes in 5-mC percentage in the F1 and F3 generations may contribute to some of the DEHP-induced changes in expression [40, 105–107]. However, additional studies are necessary to determine if global 5-mC translates to altered methylation in promoters of transcripts for critical ovarian functions.
In summary, our observations indicate that prenatal and ancestral DEHP exposure causes differential gene expression in multiple pathways necessary for healthy ovarian function in the F1, F2, and F3 generations. Further, our study suggests that DEHP-induced DNA methylation may underly some of the transgenerational effects of DEHP. However, our studies focus on the expression of mRNA and not protein levels in the ovary and therefore, caution should be taken when interpreting these results and comparing them to studies with protein expression. Therefore, future studies should measure protein expression of the genes and examine the specific epigenetic mechanisms underlying the transgenerational effects of DEHP exposure. Finally, the metabolic and pharmacokinetic differences between mice and humans are not clear for DEHP [108, 109], therefore, the unknowns in species differences in metabolism may contribute to uncertainty in the species specific effects of DEHP.
Supplementary Material
Highlights.
Prenatal DEHP exposure disrupts gene expression in the F1, F2, and F3 ovary.
Prenatal DEHP exposure disrupts Dnmt in the F1 and F3 generations.
Prenatal DEHP exposure decreases Tet in F2 and F3 generation.
DEHP increases DNA methylation in the F1, but decreases it in the F3 generation.
Acknowledgments
We would like to thank the Flaws laboratory members for their help and support. This work was supported by the Billie A. Field Fellowship in Reproductive Biology (SR); National Institutes of Health [P01 ES 022848 (JAF), F31 ES030467 (SR), and T32 ES007326 (SR)]; and the Environmental Protection Agency [RD83 543401 (JAF)].
Grants/Fellowships
This work was supported by the Billie A. Field Fellowship in Reproductive Biology (SR); National Institutes of Health [P01 ES 022848 (JAF), F31 ES030467 (SR), and T32 ES007326 (SR)]; and the Environmental Protection Agency [RD83 543401 (JAF)].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interest statement
The authors have no conflicts of interest to disclose.
References
- 1.ATSDR, Toxicological Profile for Di(2-ethylhexyl) Phthalate. U.S. Department of Health and Human Services, 2002: p. 176–213. [Google Scholar]
- 2.Hannon PR and Flaws JA, The effects of phthalates on the ovary. Front Endocrinol (Lausanne), 2015. 6: p. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Erythropel HC, et al. , Leaching of the plasticizer di(2-ethylhexyl)phthalate (DEHP) from plastic containers and the question of human exposure. Appl Microbiol Biotechnol, 2014. 98(24): p. 9967–81. [DOI] [PubMed] [Google Scholar]
- 4.Heudorf U, Mersch-Sundermann V, and Angerer J, Phthalates: toxicology and exposure. Int J Hyg Environ Health, 2007. 210(5): p. 623–34. [DOI] [PubMed] [Google Scholar]
- 5.Koch HM and Calafat AM, Human body burdens of chemicals used in plastic manufacture. Philos Trans R Soc Lond B Biol Sci, 2009. 364(1526): p. 2063–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wormuth M, et al. , What are the sources of exposure to eight frequently used phthalic acid esters in Europeans? Risk Anal, 2006. 26(3): p. 803–24. [DOI] [PubMed] [Google Scholar]
- 7.Helm D, Correlation between production amounts of DEHP and daily intake. Sci Total Environ, 2007. 388(1–3): p. 389–91. [DOI] [PubMed] [Google Scholar]
- 8.Silva MJ, et al. , Exposure to di-2-ethylhexyl terephthalate in a convenience sample of U.S. adults from 2000 to 2016. Arch Toxicol, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hogberg J, et al. , Phthalate diesters and their metabolites in human breast milk, blood or serum, and urine as biomarkers of exposure in vulnerable populations. Environ Health Perspect, 2008. 116(3): p. 334–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kato K, et al. , Mono(2-ethyl-5-hydroxyhexyl) phthalate andmono-(2-ethyl-5-oxohexyl) phthalate as biomarkers for human exposure assessment to di-(2-ethylhexyl) phthalate. Environ Health Perspect, 2004. 112(3): p. 327–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Silva MJ, et al. , Measurement of eight urinary metabolites of di(2-ethylhexyl) phthalate as biomarkers for human exposure assessment. Biomarkers, 2006. 11(1): p. 1–13. [DOI] [PubMed] [Google Scholar]
- 12.Latin G, et al. , Exposure to Di(2-ethylhexyl)phthalate in humans during pregnancy. A preliminary report. Biol Neonate, 2003. 83(1): p. 22–4. [DOI] [PubMed] [Google Scholar]
- 13.Patel S, et al. , Effects of Endocrine-Disrupting Chemicals on the Ovary. Biol Reprod, 2015. 93(1): p. 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rattan S, et al. , Exposure to endocrine disruptors during adulthood: consequences for female fertility. J Endocrinol, 2017. 233(3): p. R109–r129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Araki A, et al. , Association between maternal exposure to di(2-ethylhexyl) phthalate and reproductive hormone levels in fetal blood: the Hokkaido study on environment and children’s health. PLoS One, 2014. 9(10): p. e109039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin LC, et al. , Associations between maternal phthalate exposure and cord sex hormones in human infants. Chemosphere, 2011. 83(8): p. 1192–9. [DOI] [PubMed] [Google Scholar]
- 17.Marie C, Vendittelli F, and Sauvant-Rochat M-P, Obstetrical outcomes and biomarkers to assess exposure to phthalates: A review. Environment International, 2015. 83: p. 116–136. [DOI] [PubMed] [Google Scholar]
- 18.Watkins DJ, et al. , In utero and peripubertal exposure to phthalates and BPA in relation to female sexual maturation. Environ Res, 2014. 134: p. 233–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hart R, et al. , The influence of antenatal exposure to phthalates on subsequent female reproductive development in adolescence: a pilot study. Reproduction, 2014. 147(4): p. 379–90. [DOI] [PubMed] [Google Scholar]
- 20.Machtinger R, et al. , Urinary concentrations of biomarkers of phthalates and phthalate alternatives and IVF outcomes. Environ Int, 2018. 111: p. 23–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Skinner MK, Epigenetic transgenerational inheritance. Nat Rev Endocrinol, 2016. 12(2): p. 68–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Skinner MK, What is an epigenetic transgenerational phenotype? F3 or F2. Reprod Toxicol, 2008. 25(1): p. 2–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brehm E, et al. , Prenatal Exposure to Di(2-Ethylhexyl) Phthalate Causes Long-Term Transgenerational Effects on Female Reproduction in Mice. Endocrinology, 2018. 159(2): p. 795–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rattan S, et al. , Prenatal exposure to di(2-ethylhexyl) phthalate disrupts ovarian function in a transgenerational manner in female mice. Biol Reprod, 2018. 98(1): p. 130–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rattan S, et al. , Di(2-Ethylhexyl) Phthalate Exposure During Prenatal Development Causes Adverse Transgenerational Effects on Female Fertility in Mice. Toxicol Sci, 2018. 163(2): p. 420–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Otterdijk SD and Michels KB, Transgenerational epigenetic inheritance in mammals: how good is the evidence? Faseb j, 2016. 30(7): p. 2457–65. [DOI] [PubMed] [Google Scholar]
- 27.Rubin H, Etymology of epigenetics. Science, 2001. 294(5551): p. 2477–8. [DOI] [PubMed] [Google Scholar]
- 28.Zama AM and Uzumcu M, Epigenetic effects of endocrine-disrupting chemicals on female reproduction: an ovarian perspective. Front Neuroendocrinol, 2010. 31(4): p. 420–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Angarica VE and Del Sol A, Bioinformatics Tools for Genome-Wide Epigenetic Research. Adv Exp Med Biol, 2017. 978: p. 489–512. [DOI] [PubMed] [Google Scholar]
- 30.McCarthy MM and Rissman EF, Chapter 52 - Epigenetics of Reproduction, in Knobil and Neill’s Physiology of Reproduction (Fourth Edition). 2015, Academic Press: San Diego: p. 2439–2501. [Google Scholar]
- 31.Okano M, et al. , DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell, 1999. 99(3): p. 247–57. [DOI] [PubMed] [Google Scholar]
- 32.Kohli RM and Zhang Y, TET enzymes, TDG and the dynamics of DNA demethylation. Nature, 2013. 502(7472): p. 472–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okano M, Xie S, and Li E, Cloning and characterization o f a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat Genet, 1998. 19(3): p. 219–20. [DOI] [PubMed] [Google Scholar]
- 34.Szyf M, The role of DNA methyltransferase 1 in growth control. Front Biosci, 2001. 6:p. D599–609. [DOI] [PubMed] [Google Scholar]
- 35.Rountree MR, Bachman KE, and Baylin SB, DNMT1 binds HDAC2 and a new corepressor, DMAP1, to form a complex at replication foci. Nat Genet, 2000. 25(3): p. 269–77. [DOI] [PubMed] [Google Scholar]
- 36.Robertson KD, et al. , DNMT1 forms a complex with Rb, E2F1 andHDAC1 and represses transcription from E2F-responsive promoters. Nat Genet, 2000. 25(3): p. 338–42. [DOI] [PubMed] [Google Scholar]
- 37.Ito S, et al. , Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature, 2010. 466(7310): p. 1129–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Putiri EL, et al. , Distinct and overlapping control of 5-methylcytosine and 5- hydroxymethylcytosine by the TET proteins in human cancer cells. Genome biology, 2014. 15(6): p. R81–R81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stenz L, et al. , Testicular Dysgenesis Syndrome and Long-Lasting Epigenetic Silencing of Mouse Sperm Genes Involved in the Reproductive System after Prenatal Exposure to DEHP. PLoS One, 2017. 12(1): p. e0170441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen CH, et al. , Association between fetal exposure to phthalate endocrine disruptor and genome-wide DNA methylation at birth. Environ Res, 2018. 162: p. 261–270. [DOI] [PubMed] [Google Scholar]
- 41.Richardson KA, et al. , Di (2-ethylhexyl) phthalate (DEHP) alters proliferation and uterine gland numbers in the uteri of adult exposed mice. Reprod Toxicol, 2018. 77: p. 70–79. [DOI] [PubMed] [Google Scholar]
- 42.Zheng W, et al. , Functional roles of the phosphatidylinositol 3-kinases (PI3Ks) signaling in the mammalian ovary. Mol Cell Endocrinol, 2012. 356(1–2): p. 24–30. [DOI] [PubMed] [Google Scholar]
- 43.Stokoe D, The phosphoinositide 3-kinase pathway and cancer. Expert Rev Mol Med, 2005. 7(10): p. 1–22. [DOI] [PubMed] [Google Scholar]
- 44.Engelman JA, Luo J, and Cantley LC, The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet, 2006. 7(8): p. 606–19. [DOI] [PubMed] [Google Scholar]
- 45.Hannon PR, Peretz J, and Flaws JA, Daily exposure to Di(2-ethylhexyl) phthalate alters estrous cyclicity and accelerates primordial follicle recruitment potentially via dysregulation of the phosphatidylinositol 3-kinase signaling pathway in adult mice. Biol Reprod, 2014. 90(6): p. 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hannon PR, et al. , Mono(2-Ethylhexyl) Phthalate Accelerates Early Folliculogenesis and Inhibits Steroidogenesis in Cultured Mouse Whole Ovaries and Antral Follicles. Biol Reprod, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hussein MR, Apoptosis in the ovary: molecular mechanisms. Hum Reprod Update, 2005. 11(2): p. 162–77. [DOI] [PubMed] [Google Scholar]
- 48.Agarwal A, et al. , The effects of oxidative stress on female reproduction: a review. Reproductive biology and endocrinology : RB&E, 2012. 10: p. 49–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grana X and Reddy EP, Cell cycle control in mammalian cells: role of cyclins, cyclin dependent kinases (CDKs), growth suppressor genes and cyclin-dependent kinase inhibitors (CKIs). Oncogene, 1995. 11(2): p. 211–9. [PubMed] [Google Scholar]
- 50.Hannon PR, et al. , Di(2-ethylhexyl) phthalate inhibits antral follicle growth, induces atresia, and inhibits steroid hormone production in cultured mouse antral follicles. Toxicol Appl Pharmacol, 2015. 284(1): p. 42–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hannon PR, Niermann S, and Flaws JA, Acute Exposure to Di(2-Ethylhexyl) Phthalate in Adulthood Causes Adverse Reproductive Outcomes Later in Life and Accelerates Reproductive Aging in Female Mice. Toxicological Sciences, 2016. 150(1): p. 97–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Doyle TJ, et al. , Transgenerational effects of di-(2-ethylhexyl) phthalate on testicular germ cell associations and spermatogonial stem cells in mice. Biol Reprod, 2013. 88(5):p. 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Niermann S, et al. , Prenatal exposure to di-(2-ethylhexyl) phthalate (DEHP) affects reproductive outcomes in female mice. Reprod Toxicol, 2015. 53: p. 23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.USEPA, Di(2-ethylhexyl)phthalate (DEHP); CASRN117-81-7 Integrated Risk Information System (IRIS) Chemical Assessment Summary; 1988. [Google Scholar]
- 55.Hirshfield AN, Development of Follicles in the Mammalian Ovary, in International Review of Cytology, Jeon KW and Friedlander M, Editors. 1991, Academic Press; p. 43–101. [DOI] [PubMed] [Google Scholar]
- 56.Pepling ME, From primordial germ cell to primordial follicle: mammalian female germ cell development. Genesis, 2006. 44(12): p. 622–32. [DOI] [PubMed] [Google Scholar]
- 57.Menke DB, Koubova J, and Page DC, Sexual differentiation of germ cells in XX mouse gonads occurs in an anterior-to-posterior wave. Developmental Biology, 2003. 262(2): p. 303–312. [DOI] [PubMed] [Google Scholar]
- 58.Durcova-Hills G and Capel B, Development of germ cells in the mouse. Curr Top Dev Biol, 2008. 83: p. 185–212. [DOI] [PubMed] [Google Scholar]
- 59.TEAM, R.C. R: A language and environment for ststiscal compuing. 2017; Available from: http://www.R-project.org/.
- 60.Huber W, et al. , Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods, 2015. 12(2): p. 115–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Robinson MD and Oshlack A, A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol, 2010. 11(3): p. R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Robinson MD, McCarthy DJ, and Smyth GK, edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics, 2010. 26(1): p. 139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Leek JT and Storey JD, Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet, 2007. 3(9): p. 1724–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Leek JT and Storey JD, A general framework for multiple testing dependence. Proc Natl Acad Sci U S A, 2008. 105(48): p. 18718–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lun AT, Chen Y, and Smyth GK, It’s DE-licious: A Recipe for Differential Expression Analyses of RNA-seq Experiments Using Quasi-Likelihood Methods in edgeR. Methods Mol Biol, 2016. 1418: p. 391–416. [DOI] [PubMed] [Google Scholar]
- 66.Benjamini Y and Hochberg Y, Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. Series B (Methodological), 1995. 57(1): p. 289–300. [Google Scholar]
- 67.Huang da W, Sherman BT, and Lempicki RA, Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc, 2009. 4(1): p. 44–57. [DOI] [PubMed] [Google Scholar]
- 68.Huang da W, Sherman BT, and Lempicki RA, Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res, 2009. 37(1): p. 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pfaff MW, A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res, 2001. 29(9): p. e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Engel A, et al. , Agonistic and antagonistic effects of phthalates and their urinary metabolites on the steroid hormone receptors ERalpha, ERbeta, and AR. Toxicol Lett, 2017. 277: p. 54–63. [DOI] [PubMed] [Google Scholar]
- 71.Corton JC and Lapinskas PJ, Peroxisomeproliferator-activated receptors: mediators of phthalate ester-induced effects in the male reproductive tract? Toxicol Sci, 2005. 83(1): p. 4–17. [DOI] [PubMed] [Google Scholar]
- 72.Sicinski P, et al. , Cyclin D2 is an FSH-responsive gene involved in gonadal cell proliferation and oncogenesis. Nature, 1996. 384(6608): p. 470–474. [DOI] [PubMed] [Google Scholar]
- 73.Flaws JA, et al. , Effect of bcl-2 on the primordial follicle endowment in the mouse ovary. Biol Reprod, 2001. 64(4): p. 1153–9. [DOI] [PubMed] [Google Scholar]
- 74.Nilsson E, et al. , Environmentally Induced Epigenetic Transgenerational Inheritance of Ovarian Disease. PLoS One, 2012. 7(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pocar P, et al. , Maternal exposure to di(2-ethylhexyl)phthalate (DEHP) promotes the transgenerational inheritance of adult-onset reproductive dysfunctions through the female germline in mice. Toxicology and Applied Pharmacology, 2017. 322: p. 113–121. [DOI] [PubMed] [Google Scholar]
- 76.Wear HM, McPike MJ, and Watanabe KH, From primordial germ cells to primordial follicles: a review and visual representation of early ovarian development in mice. J Ovarian Res, 2016. 9(1): p. 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ramathal C, Reijo Pera RA, and Chavez SL, Chapter 6 - Preimplantation Embryo Development and Primordial Germ Cell Lineage Specification, in Knobil and Neill’s Physiology of Reproduction (Fourth Edition). 2015, Academic Press: San Diego: p. 233–265. [Google Scholar]
- 78.Reik W and Surani MA, Germline and Pluripotent Stem Cells. Cold Spring Harb Perspect Biol, 2015. 7(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Anway MD and Skinner MK, Epigenetic transgenerational actions of endocrine disruptors. Endocrinology, 2006. 147(6 Suppl): p. S43–9. [DOI] [PubMed] [Google Scholar]
- 80.Conolly RB and Lutz WK, Nonmonotonic dose-response relationships: mechanistic basis, kinetic modeling, and implications for risk assessment. Toxicol Sci, 2004. 77(1): p. 151–7. [DOI] [PubMed] [Google Scholar]
- 81.Gore AC, et al. , EDC-2: The Endocrine Society’s Second Scientific Statement on Endocrine-Disrupting Chemicals. Endocrine Reviews, 2015. 36(6): p. E1–E150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Katzenellenbogen BS, et al. , Estrogen receptors: selective ligands, partners, and distinctive pharmacology. Recent Prog Horm Res, 2000. 55: p. 163–93; discussion 194–5. [PubMed] [Google Scholar]
- 83.Ismail A and Nawaz Z, Nuclear hormone receptor degradation and gene transcription: an update. IUBMB Life, 2005. 57(7): p. 483–90. [DOI] [PubMed] [Google Scholar]
- 84.Freedman NJ and Lefkowitz RJ, Desensitization o f G protein-coupled receptors. Recent Prog Horm Res, 1996. 51: p. 319–51; discussion 352–3. [PubMed] [Google Scholar]
- 85.McKenna NJ, Chapter 9 - Gonadal Steroid Action, in Knobil and Neill’s Physiology of Reproduction (Fourth Edition). 2015, Academic Press: San Diego: p. 313–333. [Google Scholar]
- 86.Vandenberg LN, Non-monotonic dose responses in studies of endocrine disrupting chemicals: bisphenol a as a case study. Dose Response, 2014. 12(2): p. 259–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Krishnan K, et al. , Maternal care modulates transgenerational effects of endocrine-disrupting chemicals on offspring pup vocalizations and adult behaviors. Horm Behav, 2019. 107: p. 96–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Berger A, et al. , The effects of in utero bisphenol A exposure on the ovaries in multiple generations of mice. Reprod Toxicol, 2016. 60: p. 39–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhou C and Flaws JA, Effects of an environmentally relevant phthalate mixture on cultured mouse antral follicles. Toxicol Sci, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kawano M, et al. , Peroxisome proliferator-activated receptor alpha mediates di-(2-ethylhexyl) phthalate transgenerational repression of ovarian Esr1 expression in female mice. Toxicol Lett, 2014. 228(3): p. 235–40. [DOI] [PubMed] [Google Scholar]
- 91.Huang Q, et al. , The Inflammation Response to DEHP through PPARgamma in Endometrial Cells. Int J Environ Res Public Health, 2016. 13(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hayashi Y, Ito Y, and Nakajima T, Effects of exposure to Di(2-ethylhexyl)phthalate during fetal period on next generation. Nihon Eiseigaku Zasshi, 2014. 69(2): p. 86–91. [DOI] [PubMed] [Google Scholar]
- 93.Desvergne B, Feige JN, and Casals-Casas C, PPAR-mediated activity of phthalates: A link to the obesity epidemic? Mol Cell Endocrinol, 2009. 304(1–2): p. 43–8. [DOI] [PubMed] [Google Scholar]
- 94.Drummond AE and Fuller PJ, The importance of ERbeta signalling in the ovary. J Endocrinol, 2010. 205(1): p. 15–23. [DOI] [PubMed] [Google Scholar]
- 95.Krege JH, et al. , Generation and reproductive phenotypes of mice lacking estrogen receptor beta. Proc Natl Acad Sci U S A, 1998. 95(26): p. 15677–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dupont S, et al. , Effect of single and compound knockouts of estrogen receptors alpha (ERalpha) and beta (ERbeta) on mouse reproductive phenotypes. Development, 2000. 127(19): p. 4277–91. [DOI] [PubMed] [Google Scholar]
- 97.Auchus RJ, Chapter 8 - Human Steroid Biosynthesis, in Knobil and Neill’s Physiology of Reproduction (Fourth Edition). 2015, Academic Press: San Diego: p. 295–312. [Google Scholar]
- 98.Lai FN, et al. , Di (2-ethylhexyl) phthalate impairs steroidogenesis in ovarian follicular cells of prepuberal mice. Arch Toxicol, 2017. 91(3): p. 1279–1292. [DOI] [PubMed] [Google Scholar]
- 99.Reddy P, et al. , PDK1 signaling in oocytes controls reproductive aging and lifespan by manipulating the survival of primordial follicles. Hum Mol Genet, 2009. 18(15): p. 24. [DOI] [PubMed] [Google Scholar]
- 100.De Clercq A and Inze D, Cyclin-dependent kinase inhibitors in yeast, animals, and plants: a functional comparison. Crit Rev Biochem Mol Biol, 2006. 41(5): p. 293–313. [DOI] [PubMed] [Google Scholar]
- 101.Xiong Y, Zhang H, and Beach D, D type cyclins associate with multiple protein kinases and the DNA replication and repair factor PCNA. Cell, 1992. 71(3): p. 505–14. [DOI] [PubMed] [Google Scholar]
- 102.Jiao Y, Feng Y, and Wang X, Regulation of Tumor Suppressor Gene CDKN2A and Encoded p16-INK4a Protein by Covalent Modifications. Biochemistry (Mosc), 2018. 83(11): p. 1289–1298. [DOI] [PubMed] [Google Scholar]
- 103.Lee MH, Reynisdottir I, and Massague J, Cloning of p57KIP2, a cyclin-dependent kinase inhibitor with unique domain structure and tissue distribution. Genes Dev, 1995. 9(6): p. 639–49. [DOI] [PubMed] [Google Scholar]
- 104.Sekaran S and Jagadeesan A, In utero exposure tophthalate downregulates critical genes in Leydig cells of F1 male progeny. J Cell Biochem, 2015. 116(7): p. 1466–77. [DOI] [PubMed] [Google Scholar]
- 105.Drobna Z, et al. , Transgenerational Effects of Bisphenol A on Gene Expression and DNA Methylation of Imprinted Genes in Brain. Endocrinology, 2018. 159(1): p. 132–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Elmhiri G, et al. , DNA methylation and potential multigenerational epigenetic effects linked to uranium chronic low-dose exposure in gonads of males and females rats. Toxicol Lett, 2018. 282: p. 64–70. [DOI] [PubMed] [Google Scholar]
- 107.Gebhard C, et al. , General transcription factor binding at CpG islands in normal cells correlates with resistance to de novo DNA methylation in cancer cells. Cancer Res, 2010. 70(4): p. 1398–407. [DOI] [PubMed] [Google Scholar]
- 108.Johnson KJ, Heger NE, and Boekelheide K, Of mice and men (and rats): phthalate-induced fetal testis endocrine disruption is species-dependent. Toxicol Sci, 2012. 129(2): p. 235–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ito Y, et al. , Species and inter-individual differences in metabolic capacity of di(2-ethylhexyl)phthalate (DEHP) between human and mouse livers. Environmental Health and Preventive Medicine, 2014. 19(2): p. 117–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.