Abstract
Due to the large numbers of transcription factors (TFs) and cell types, querying binding profiles of all valid TF/cell type pairs is not experimentally feasible. To address this issue, we developed a convolutional-recurrent neural network model, called FactorNet, to computationally impute the missing binding data. FactorNet trains on binding data from reference cell types to make predictions on testing cell types by leveraging a variety of features, including genomic sequences, genome annotations, gene expression, and signal data, such as DNase I cleavage. FactorNet implements several convenient strategies to reduce runtime and memory consumption. By visualizing the neural network models, we can interpret how the model predicts binding. We also investigate the variables that affect cross-cell type accuracy, and offer suggestions to improve upon this field. Our method ranked among the top teams in the ENCODE-DREAM in vivo Transcription Factor Binding Site Prediction Challenge, achieving first place on six of the 13 final round evaluation TF/cell type pairs, the most of any competing team. The FactorNet source code is publicly available, allowing users to reproduce our methodology from the ENCODE-DREAM Challenge.
Keywords: deep learning, transcription factors, ENCODE, DREAM
1. Introduction
High-throughput sequencing has led to a diverse set of methods to interrogate the epigenomic landscape for the purpose of discovering tissue and cell type-specific putative functional elements. Such information provides valuable insights for a number of biological fields, including synthetic biology and translational medicine. Among these methods are ChIP-seq, which applies a large-scale chromatin immunoprecipitation assay that maps in vivo transcription factor (TF) binding sites or histone modifications genome-wide [1], and DNase-seq, which identifies genome-wide locations of open chromatin, or “hotspots”, by sequencing genomic regions sensitive to DNase I cleavage [2, 3]. At deep sequencing depth, DNase-seq can identify TF binding sites, which manifest as dips, or “footprints”, in the digital DNase I cleavage signal [4, 5, 6]. Other studies have shown that cell type-specific functional elements can display unique patterns of motif densities and epigenomic signals [7]. Computational methods can integrate these diverse datasets to elucidate the complex and non-linear combinations of epigenomic markers and raw sequence contexts that underlie functional elements such as enhancers, promoters, and insulators. Some algorithms accomplish this by dividing the entire genome systematically into segments, and then assigning the resulting genome segments into “chromatin states” by applying machine learning methods such as Hidden Markov Models, Dynamic Bayesian Networks, or Self-Organizing Maps [8, 9, 10].
The Encyclopedia of DNA Elements (ENCODE) [11] and NIH Roadmap Epigenomics [12] projects have generated a large number of ChIP-seq and DNase-seq datasets for dozens of different cell and tissue types. Owing to several constraints, including cost, time or sample material availability, these projects are far from completely mapping every mark and sample combination. This disparity is especially large for TF binding profiles because ENCODE has profiled over 600 human biosamples and over 200 TFs, translating to over 120,000 possible pairs of biosamples and TFs, but as of the writing of this article only about 8,000 TF binding profiles are available. Due to the strong correlations between epigenomic markers, computational methods have been proposed to impute the missing datasets. One such imputation method is ChromImpute [13], which applies ensembles of regression trees to impute missing chromatin marks. With the exception of CTCF, ChromImpute does not impute TF binding. Moreover, ChromImpute does not take sequence context into account, which can be useful for predicting the binding sites of TFs like CTCF that are known to have a strong binding motif.
Computational methods designed to predict TF binding include PIQ [14], Centipede [15], and msCentipede [16]. These methods require a collection of motifs and DNase-seq data to predict TF binding sites in a single tissue or cell type. While such an approach can be convenient because the DNase-seq signal for the cell type considered is the only mandatory experimental data, it has several drawbacks. These models are trained in an unsupervised fashion using algorithms such as expectation maximization (EM). The manual assignment of a motif for each TF is a strong assumption that completely ignores any additional sequence contexts such as co-binding, indirect binding, and non-canonical motifs. This can be especially problematic for TFs like REST, which is known to have eight non-canonical binding motifs [17].
More recently, deep neural network (DNN) methods have gained significant traction in the bioinformatics community. DNNs are useful for biological applications because they can efficiently identify complex non-linear patterns from large amounts of feature-rich data. They have been successfully applied to predicting splicing patterns [18], predicting variant deleteriousness [19], and gene expression inference [20]. The convolutional neural network (CNN), a variant of the DNN, has been useful for genomics because it can process raw DNA sequences and the kernels are analogues to position weight matrices (PWMs), which are popular models for describing the sequence-specific binding pattern of TFs. Examples of genomic application of CNNs include DanQ[21], DeepSEA [22], Basset [23], DeepBind [24], and DeeperBind [25]. These methods accept raw DNA sequence inputs and are trained in a supervised fashion to discriminate between the presence and absence of epigenetic markers, including TF binding, open chromatin, and histone modifications. Consequently, these algorithms are not suited to the task of predicting cell type-specific epigenomic markers. Instead, they are typically designed for other tasks such as motif discovery or functional variant annotation. Both DanQ and DeeperBind, unlike the other three CNN methods, also use a recurrent neural network (RNN), another type of DNN, to form a CNN-RNN hybrid architecture that can outperform pure convolutional models. RNNs have been useful in other machine learning applications involving sequential data, including phoneme classification [26], speech recognition [27], machine translation [28], and human action recognition [29]. More recently, CNNs and RNNs have been used for predicting single-cell DNA methylation states [30]
To predict cell type-specific TF binding, we developed FactorNet, which combines elements of the aforementioned algorithms. FactorNet trains a DNN on data from one or more reference cell types for which the TF or TFs of interest have been profiled, and this model can then predict binding in other cell types. The FactorNet model builds upon the DanQ CNN-RNN hybrid architecture by incorporating additional real-valued coordinated-based signals such as DNase-seq signals as features. Our software pipeline includes several convenient utilities to accelerate training and reduce memory consumption. For example, using a combination of the keras builtin utilities and Python wrapper libraries, we developed convenient data generators that can efficiently stream training data directly from standard genomic data formats; thus models can be trained on large datasets without changing memory requirements or producing large intermediate files. In contrast, genomic machine learning methods, such as BoostMe [31] and random forest based model for methylation prediction [32], may limit training to a smaller subset due to memory constraints. Other genomic machine learning methods, such as DeepCpG [30] and DeepSEA [22], inefficiently extract millions of training sequences into the hard drive as HDF5 files before training. Finally, computations are carried out on highly parallelized graphics processing units using the keras deep learning library to make training times tractable.
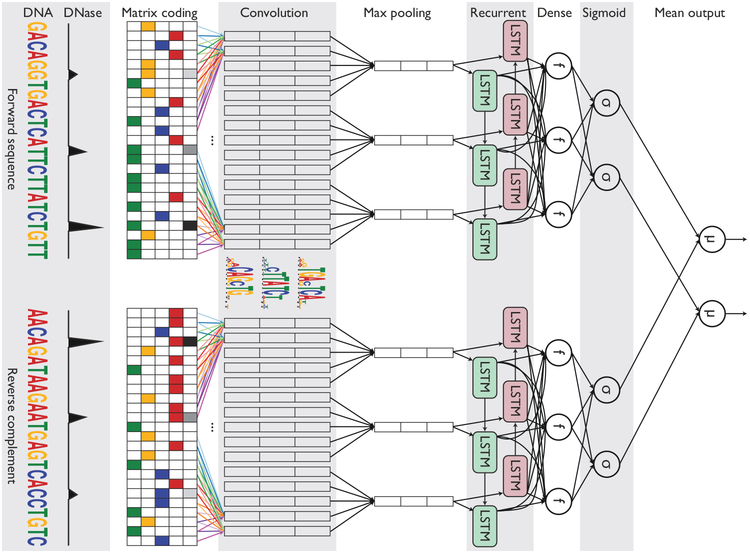
We also extended the DanQ network into a “Siamese” architecture that accounts for reverse complements (Figure 1). This Siamese architecture applies identical networks of shared weights to both strands to ensure that both the forward and reverse complement sequences return the same outputs, essentially halving the total amount of training data, ultimately improving training efficiency and predictive accuracy. Siamese networks are popular among tasks that involve finding similarity or a relationship between two comparable objects, such as signature verification [33] and assessing sentence similarity [34].
Figure 1: Simplified diagram of the FactorNet model.
An input DNA sequence (top) is first one hot encoded into a 4-row bit matrix. Real-valued single-nucleotide signal values (e.g. DNase I cleavage) are concatenated as extra rows to this matrix. A rectifier activation convolution layer transforms the input matrix into an output matrix with a row for each convolution kernel and a column for each position in the input (minus the width of the kernel). Each kernel is effectively a sequence motif. Max pooling downsamples the output matrix along the spatial axis, preserving the number of channels. The subsequent recurrent layer contains long short term memory (LSTM) units connected end-to-end in both directions to capture spatial dependencies between motifs. Recurrent outputs are densely connected to a layer of rectified linear units. The activations are likewise densely connected to a sigmoid layer that nonlinear transformation to yield a vector of probability predictions of the TF binding calls. An identical network, sharing the same weights, is also applied to the reverse complement of the sequence (bottom). Finally, respective predictions from the forward and reverse complement sequences are averaged together, and these averaged predictions are compared via a loss function to the true target vector. Although not pictured, we also include a sequence distributed dense layer between the convolution and max pooling layer to capture higher order motifs.
We submitted the FactorNet model to the ENCODE-DREAM in vivo Transcription Factor Binding Site Prediction Challenge [35], where it ranked among the top teams. The Challenge delivers a crowdsourcing approach to figure out the optimal strategies for solving the problem of TF binding prediction. Although all results discussed in this paper are derived from data in the Challenge, FactorNet is compatible with standard genomic data file formats, can conveniently run on a modern desktop computer, and is therefore readily usable for data outside of the Challenge.
2. Materials and methods
2.1. ENCODE-DREAM Challenge dataset
The ENCODE-DREAM Challenge dataset is comprised of DNase-seq, ChIP-seq, and RNA-seq data from the ENCODE project or The Roadmap Epigenomics Project covering 14 cell types and 32 TFs. All annotations and preprocessing are based on hgl9/GRCh37 release version of the human genome and GENCODE release 19 [36]. Data are restricted to chromosomes X and 1–22. Chromosomes 1, 8 and 21 are set aside exclusively for evaluation purposes and binding data were completely absent for these three chromosomes during the Challenge. TF binding labels are provided at a 200 bp resolution. Specifically, the genome is segmented into 200 bp bins sliding every 50 bp. Each bin is labeled as bound (B), unbound (U) or ambiguously bound (A) depending on the majority label of all nucleotides in the bin. Ambiguous bins overlap peaks that fail to pass the IDR threshold of 5% and are excluded from evaluation. A more complete description of the dataset, including preprocessing details such as peak calling, can be found in the ENCODE-DREAM Challenge website [35].
2.2. Evaluation
The TF binding prediction problem is evaluated as a two-class binary classification task. For each test TF/cell type pair, the following performance measures are computed:
auROC. The area under the receiver operating characteristic curve is a common metric for evaluating classification models. It is equal to the probability that a classifier will rank a randomly chosen positive instance higher than a randomly chosen negative one.
auPR. The area under the precision-recall curve is more appropriate in the scenario of few relevant items, as is the case with TF binding prediction [21]. Unlike the auROC metric, the auPR metric does not take into account the number of true negatives called.
Recall at fixed FDR. The recall at a fixed false discovery rate (FDR) represents a point on the precision-recall curve. Like the auPR metric, this metric is appropriate in the scenario of few relevant items. This metric is often used in applications such as fraud detection in which the goal may be to maximize the recall of true fraudsters while tolerating a given fraction of customers to falsely identify as fraudsters. The ENCODE-DREAM Challenge computes this metric for several FDR values.
As illustrated in Figure 1, the FactorNet Siamese architecture operates on both the forward and reverse complement sequences to ensure that both strands return the same outputs during both training and prediction. Although a TF might only physically bind to one strand, this information cannot usually be inferred directly from the peak data. Thus, the same set of labels are assigned to both strands in the evaluation step.
2.3. Features and data preprocessing
FactorNet works directly with standard genomic file formats and requires relatively little preprocessing. FASTA files provides genomic sequences, BED files provide the locations of reference TF binding sites for labels, and bigWig files [37] provide dense, continuous signal data at single-nucleotide resolution. bigWig values are included as extra rows that are appended to the four-row one hot input DNA binary matrix. Training data are streamed using data generators to reduce memory use without impacting the running time. We developed the data generators using a combination of keras [38], pyfasta[39], pybedtools [40], and pyBigWig [41], FactorNet can accept an arbitrary number of bigWig files as input features, and we found the following signals to be highly informative for prediction:
DNase I cleavage. For each cell type, reads from all DNase-seq replicates were trimmed down to first nucleotide on the 5’ end, pooled and normalized to lx coverage using deepTools [42],
35 bp mapability uniqueness. This track quantifies the uniqueness of a 35 bp subsequence on the positive strand starting at a particular base, which is important for distinguishing where in the genome DNase I cuts can be detected. Scores are between 0 and 1, with 1 representing a completely unique sequence and 0 representing a sequence that occurs more than 4 times in the genome. Otherwise, scores between 0 and 1 indicate the inverse of the number of occurrences of that subsequence in the genome. It is available from the UCSC genome browser under the table wgEncodeDukeMapabilityUniqueness35bp.
In addition to sequential features, FactorNet also accepts non-sequential metadata features. At the cell type level, we applied principal component analysis to the inverse hyperbolic sine transformed gene expression levels and extracted the top 8 principal components. Gene expression levels are measured as the average of the fragments per kilobase per million for each gene transcript. At the bin level, we included Boolean features that indicate whether gene annotations (coding sequence, intron, 5’ untranslated region, 3’ untranslated region, and promoter) and CpG islands [43] overlap a given bin. We define a promoter to be the region up to 300 bps upstream and 100 bps downstream from any transcription start site. To incorporate these metadata features as inputs to the model, we append the values to the dense layer of the neural network and insert another dense layer containing the same number of ReLU neurons between the new merged layer and the sigmoid layer (Figure 1)
2.4. ·Training
Our implementation is written in Python, utilizing the Keras 1.2.2 library [38] with the Theano 0.9.0 [44, 45] backend. We used a Linux machine with 32GB of memory and an NVIDIA Titan X Pascal GPU for training.
FactorNet supports single- and multi-task training. Both types of neural network models are trained using the Adam algorithm [46] with a minibatch size of 100 to minimize the mean multi-task binary cross entropy loss function on the training set. We also include dropout [47] to reduce overfitting. One or more chromosomes are set aside as a validation set. Validation loss is evaluated at the end of each training epoch and the best model weights according to the validation loss are saved. Training sequences of constant length centered on each bin are efficiently streamed from the hard drive in parallel to the model training. Random spatial translations are applied in the streaming step as a form of data augmentation. Each epoch, an equal number of positive and negative bins are randomly sampled and streamed for training, but this ratio is an adjustable hyperparameter (see Table SI for a detailed explanation of all hyperparameters). In the case of multi-task training, a bin is considered positive if it is confidently bound to at least one TF. Bins that overlap a blacklisted region [11] are automatically labeled negative and excluded from training.
2.4.1. Single-task training
Single-task training leverages data from multiple cell types by treating bins from all cell types as individually and identically distributed (i.i.d.) records. To make single-task training run efficiently, one bin is allotted per positive peak and these positive bins are included at most once per epoch for training. Ambiguously bound bins are excluded from training. Single-task model training can typically complete in under two hours.
2.4–2. Multi-task training
FactorNet can only perform multi-task training when training on data from a single cell type due to the variation of available binding data for the cell types. For example, the ENCODE-DREAM Challenge provides reference binding data for 15 TFs for GM12878 and 16 TFs for HeLa-S3, but only 8 TFs are shared between the two cell types. Compared to single-task training, multi-task training takes considerably longer to complete due to the larger number of positive bins. At the start of training, positive bins are identified by first segmenting the genome into 200 bins sliding every 50 bp and discarding all bins that fail to overlap at least one confidently bound TF site. Model-task model training can typically complete in two days.
2.5. Ensembling by model averaging
Ensembling is a common strategy for improving classification performance. At the time of the Challenge, we implemented a simple ensembling strategy commonly called “bagging submissions”, which involves averaging predictions from two or more models. Instead of averaging prediction probabilities directly, we first convert the scores to ranks, and then average these ranks. Rank averaging is more appropriate than direct averaging if predictors are not evenly calibrated between 0 and 1, which is often the case with the FactorNet models.
3. Results and Discussion
3.1. Performance varies across transcription factors
Table 1 shows a partial summary of FactorNet cross-cell type performances on a variety of cell type and TF combinations as of the conclusion of the ENCODE-DREAM Challenge. Final rankings in the Challenge are based on performances over 13 TF/cell type pairs. A score combining several primary performance measures is computed for each pair. In addition to the 13 TF/cell type pairs for final rankings, there are 28 TF/cell type “leaderboard” pairs. Competitors can compare performances and receive live updating of their scores for the leaderboard TF/cell type pairs. Scores for the 13 final ranking TF/cell type pairs were not available until the conclusion of the challenge. Our model achieved first place on six of the 13 TF/cell type final ranking pairs, the most of any team.
Table 1: Partial summary of FactorNet cross-cell type performances on the ENCODE-DREAM Challenge data.
Each final ranking TF/cell type pair is demarcated with a *. For each final ranking TF/cell type pair, we provide, in parentheses, performance scores based on the evaluation pair’s original ChIP-seq fold change signal.
| Factor | Cell type | auROC | auPR | Recall at 50% FDR |
|---|---|---|---|---|
| CTCF* | iPSC | 0.9966 (0.9998) | 0.8608 (0.9794) | 0.9142 (0.9941) |
| CTCF | GM12878 | 0.9968 | 0.8451 | 0.8777 |
| CTCF* | PC-3 | 0.9862 (0.9942) | 0.7827 (0.8893) | 0.7948 (0.9272) |
| ZNF143 | K562 | 0.9884 | 0.6957 | 0.7303 |
| MAX | MCF-7 | 0.9956 | 0.6624 | 0.8290 |
| MAX* | liver | 0.9882 (0.9732) | 0.4222 (0.6045) | 0.3706 (0.6253) |
| EGR1 | K562 | 0.9937 | 0.6522 | 0.7312 |
| EGR1* | liver | 0.9856 (0.9741) | 0.3172 (0.5306) | 0.2164 (0.5257) |
| HNF4A* | liver | 0.9785 (0.9956) | 0.6188 (0.8781) | 0.6467 (0.9291) |
| MAFK | K562 | 0.9946 | 0.6176 | 0.6710 |
| MAFK | MCF-7 | 0.9906 | 0.5241 | 0.5391 |
| GABPA | K562 | 0.9957 | 0.6125 | 0.6299 |
| GABPA* | liver | 0.9860 (0.9581) | 0.4416 (0.5197) | 0.3550 (0.5202) |
| YY1 | K562 | 0.9945 | 0.6078 | 0.7393 |
| TAF1 | HepG2 | 0.9930 | 0.5956 | 0.6961 |
| TAF1* | liver | 0.9892 (0.9657) | 0.4283 (0.4795) | 0.4039 (0.4766) |
| E2F6 | K562 | 0.9885 | 0.5619 | 0.6455 |
| REST | K562 | 0.9958 | 0.5239 | 0.5748 |
| REST* | liver | 0.9800 (0.9692) | 0.4122 (0.5596) | 0.4065 (0.5945) |
| FOXA1* | liver | 0.9862 (0.9813) | 0.4922 (0.6546) | 0.4889 (0.6728) |
| FOXA1 | MCF-7 | 0.9638 | 0.4487 | 0.4613 |
| JUND | H1-hESC | 0.9948 | 0.4098 | 0.3141 |
| JUND* | liver | 0.9765 (0.9825) | 0.2649 (0.6921) | 0.1719 (0.7223) |
| TCF12 | K562 | 0.9801 | 0.3901 | 0.3487 |
| STAT3 | GM12878 | 0.9975 | 0.3774 | 0.3074 |
| NANOG* | iPSC | 0.9885 (0.9876) | 0.3539 (0.6421) | 0.3118 (0.6680) |
| CREB1 | MCF-7 | 0.9281 | 0.3105 | 0.2990 |
| E2F1* | K562 | 0.9574 (0.9888) | 0.2406 (0.6428) | 0.0000 (0.6573) |
| FOXA2* | liver | 0.9773 (0.9932) | 0.2172 (0.7920) | 0.0231 (0.8278) |
All FactorNet were trained using raw DNA sequences and DNase I cleavage signals as input features. In addition to these two features, FactorNet can also incorporate other features, such as mapability and gene annotations. We selected models to make predictions for leaderboard and final ranking TF/cell type pairs according to performance on a held out chromosome validation set in the training cell types. Based on the validation performance, incorporating DNase I features significantly improves performance over a DNA-only model. In contrast, incorporating other features yielded comparatively smaller, but somewhat consistent, improvements (Figure S1).
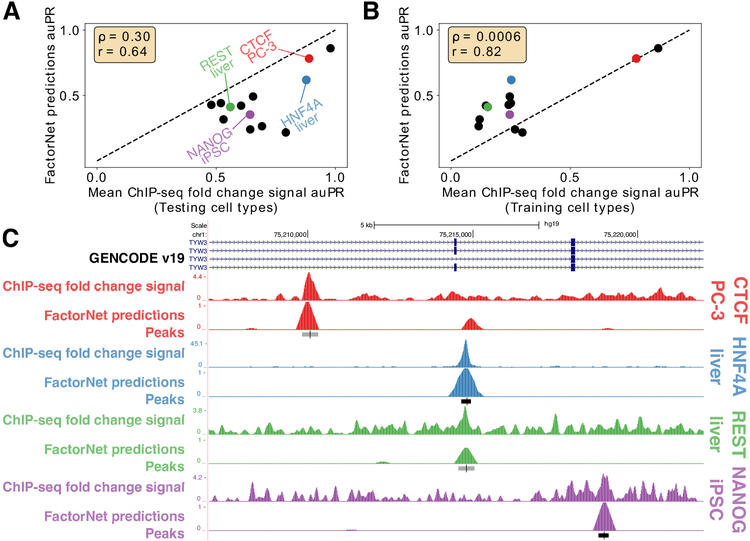
FactorNet typically achieves auROC scores above 97% for most of the TF/cell type pairs, reaching as low as 92.8% for CREB1/MCF-7. auPR scores, in contrast, display a wider range of values, reaching as low as 21.7% for FOXAl/liver and 87.8% for CTCF/iPSC. For some TFs, such as CTCF and ZNF143, the predictions are already accurate enough to be considered useful. Much of the variation in auPR scores can be attributed to noise in the ChIP-seq signal used to generate the evaluation labels, which we demonstrate by building classifiers based on taking the mean in a 200 bp window of the ChIP-seq fold change signal in the testing cell types with respect to input control. Peak calls are derived from the SPP algorithm [48], which uses the fold-change signal and peak shape to score and rank peaks. An additional processing step scores peaks according to an irreproducible discovery rate (IDR), which is a measure of consistency between replicate experiments. Bins are labeled positive if they overlap a peak that meets the IDR threshold of 5%. The IDR scores are not always monotonically associated with the fold-changes. Nevertheless, we expect that performance scores from the fold-change signal classifiers should serve as overly optimistic upper bounds for benchmarking. Commensurate with these expectations, the auPR scores of the FactorNet models are less than, but positively correlative with, the respective auPR scores of the ChIP-seq fold-change signal classifiers (Figure 2A). This pattern does not extend to the auROC scores, and in more than half of the cases the FactorNet auROC scores are greater (Figure S2). These results are consistent with previous studies that showed the auROC can be unreliable and overly optimistic in an imbalanced class setting [49], which is a common occurrence in genomic applications [21], motivating the use of alternative measures like the auPR that ignore the overly abundant true negatives. If instead of using ChIP-seq fold-change signals in the testing cell types, we used signals from the training cell types (or, if multiple training cell types are available for a TF, aggregate fold-change signals), we can get a measure of how consistent TF binding is across cell types. Compared to the training cell type fold-change signal classifiers, FactorNet performs significantly better. Hence, FactorNet predictions are much more accurate than simply assuming binding sites are invariant across cell types.
Figure 2: Performance and ChIP-seq signal varies across TF/cell-type pairs.
Scatterplots comparing auPR scores between FactorNet predictions and mean ChIP-seq fold change signal within a 200 bp window from either the testing cell type (A) or training cell type (B) (if multiple training cell types are available for a TF, then the aggregate ChIP-seq signals from multiple cell types are used). Each marker corresponds to one of the 13 final ranking TF/cell type pairs. Spearman (ρ) and Pearson (r) correlations are displayed. (C) Genome browser [50] screenshot displays the ChIP-seq fold change signal, FactorNet predictions, and peak calls for four TF/cell type pairs in the TYW3 locus. Confidently bound regions are more heavily shaded than ambiguously bound regions.
We can also visualize the FactorNet predictions as genomic signals that can be viewed alongside the ChIP-seq signals and peak calls (Figure 2C). Higher FactorNet prediction values tend to coalesce around called peaks, forming peak-like shapes in the prediction signal that resemble the signal peaks in the original ChIP-seq signal. The visualized signals also demonstrate the differences in signal noise across the ChIP-seq datasets. The NANOG/iPSC ChIP-seq dataset, for example, displays a large amount of signal outside of peak regions, unlike the HNF4A/liver ChIP-seq dataset which has most of its signal focused in peak regions.
The ENCODE-DREAM challenge data, documentation, and results can be found on the Challenge homepage: https://www.synapse.org/ENCODE. We also provide comparisons to other top ENCODE-DREAM competitors and existing published methods in the Supplementary Files.
3.2. Interpreting neural network models
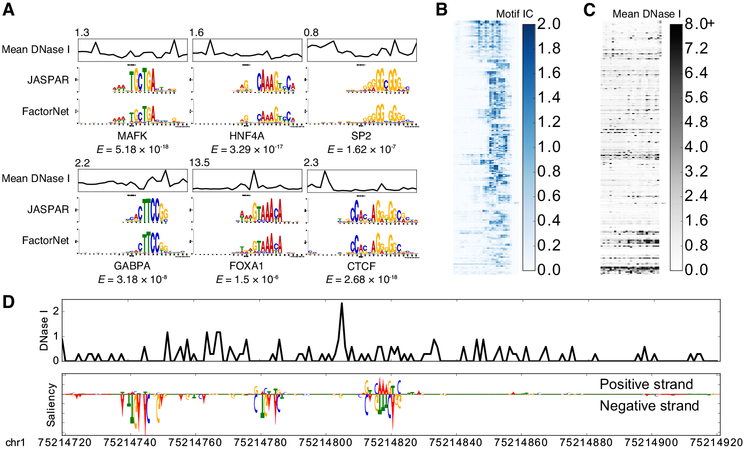
Using the same heuristic from DeepBind [24] and DanQ [21], we visualized several kernels from a HepG2 multi-task model as sequence logos by aggregating subsequences that activate the kernels (Figure 3A). The kernels significantly match motifs associated with the target TFs. Furthermore, the aggregated DNase I signals also inform us of the unique “footprint” signatures the models use to identify true binding sites at single-nucleotide resolution. After visualizing and aligning all the kernels, we confirmed that the model learned a variety of motifs (Figure 3B). A minority of kernels display very little sequence specificity while recognizing regions of high chromatin accessibility (Figure 3C).
Figure 3: Visually interpreting FactorNet models.
(A) Network kernels from a HepG2 multi-task FactorNet model are converted to sequence logos and aligned with motifs from JASPAR [51] using TOMTOM [52]. Mean normalized DNase I cleavage signals and their maximum values are displayed above the aligned logos. E-values measure similarity between query and target motifs, corrected for multiple hypothesis testing. All kernels are converted to sequence logos and aligned with RSAT [53]. The heatmaps are ordered by this alignment and colored according to the motif information content (IC) (B) or mean DNase I cleavage signal (C) at each nucleotide position. (D) Normalized liver DNase I cleavage signal and saliency maps of aligned stranded sequences centered on the summit of a liver HNF4A peak in the TYW3 locus (Figure 2C). Negative gradients are converted to zeros. We visualized saliency maps with the DeepLIFT visualizer [54].
Saliency maps are another common technique of visualizing neural network models [55]. To generate a saliency map, we compute the gradient of the output category with respect to the input sequence. By visualizing the saliency maps of a genomic sequence, we can identify the parts of the sequence the neural network finds most relevant for predicting binding, which we interpret as sites of TF binding at single-nucleotide resolution. Using a liver HNF4A peak sequence and HNF4A predictor model as an example, the saliency map highlights a subsequence overlapping the summit that strongly matches the known canonical HNF4A motif, as well as two putative binding sites upstream of the summit on the reverse complement (Figure 3D). More examples of FactorNet saliency maps can be found in the kipoi github repository [56].
3.3. Example: applying FactorNet to predict E2F1 binding
Many variables can affect the accuracy of cross-cell prediction accuracy. In addition to the type of model used, other competitors have noted the importance of preprocessing and training strategies to counteract the effects of batch effects and overfitting. For example, DNase-seq data widely varies in terms of sequencing depth and signal-to-noise ratio (SNR) across the cell types, which we measure as the fraction of reads that fall into conservative peaks (FRiP) (Figure S3A). Notably, liver displays the lowest SNR with a FRiP score of 0.05, which is consistent with its status as a primary tissue; all other cell types are cultured cell lines. Some ENCODE-DREAM competitors proposed normalization steps to correct for the differences in DNase-seq data across cell types. Batch effects, which occur because measurements are affected by laboratory conditions, reagent lots, and personnel differences, can also negatively impact accuracy. Due to batch effects and biological differences between cell types, a model trained on a reference cell type may overfit on any technical or biological biases present in that sample and thus fail to generalize to a new cell type. In the cases where a TF has multiple reference cell types to train on, some competitors propose training exclusively on one cell type (ideally the cell type that is most “compatible” with the testing cell type), whereas another competitor used a cross cell-type cross-validation early stopping training strategy to improve cross-cell type generalizability. To demonstrate the flexibility and utility of FactorNet, we incorporate similar strategies into the FactorNet model to yield improved binding prediction for the TF E2F1.
For the ENCODE-DREAM Challenge, the TF E2F1 has two reference cell types for training, GM12878 and HeLa-S3, and one cell type for final round blind evaluation, K562. Reference binding data for other TFs are available for both GM12878 and HeLa-S3, including GABPA, ZNF143, and TAF1. To quantify the errors induced by batch effects present in the different datasets, FactorNet can train on one cell type and validate against another cell type (Figure S3B). We surmise that some of the batch effects that cause discrepancies between a training cell type and a validation cell type include differences in DNase-seq quality, ChIP-seq sequencing (e.g. single-end 36 bp vs. paired-end 100 bp), or antibodies. For E2F1, the GM12878 and HeLa-S3 E2F1 ChIP-seq datasets were generated using two different antibodies: ENCAB0370HX and ENCAB000AFU, respectively. The K562 E2F1 ChIP-seq dataset was generated using the antibodies ENCAB0370HX and ENCAB851KCY, the former of which was also used for GM12878. As expected, a model trained exclusively on GM12878 data is more accurate than a model trained exclusively on HeLa-S3 data (Figure S3C–D). Given that ChIP-seq signal noise can significantly influence the accuracy of predictions (Figure 2), we propose that future data generation efforts should use protocol improvements such as ChIP-exo[57], CUT&RUN[58], or higher quality antibodies to complement the development of prediction models. Protocols across experiments should also be as uniform as possible.
We also compare single-task and multi-task models for E2F1 binding. Several deep learning methods, including DeepSEA [22] and Basset [23], primarily use multi-task training, which involves assigning multiple labels, corresponding to different chromatin markers, to the same DNA sequence. The authors of these methods propose that the multi-task training improves efficiency and performance. FactorNet supports both types of training. To the best of our knowledge, neither single-task nor multi-task training confers any particular advantage in terms of accuracy. For the K562/E2F1 cross-cell prediction, the GM12878 single-task model outperformed GM12878 multi-task model (Figure S3C). In contrast, for the NANOG/iPSC cross-cell type prediction, the Hl-hESC multi-task model outperformed the Hl-hESC single-task model (Figure S4). Nevertheless, ensembling single- and multi-task models together is an effective method of improving performance. In both the NANOG and E2F1 examples, the cross-cell type performance of the single-task and multi-task ensemble models significantly outclasses the performances reported at the conclusion of the Challenge, demonstrating the potential for FactorNet to readily adapt improved training heuristics.
4. Conclusion
FactorNet is a flexible framework that lends itself to a variety of future research avenues. FactorNet’s open source code, documentation, and adherence to standardized file formats ensures its utility in the bioinformatics community. For example, integrating attention mechanisms [59] into the FactorNet neural network model may improve accuracy and interpretability. Furthermore, FactorNet can readily accept other genomic signals that were not included as part of the Challenge but are likely relevant to TF binding prediction, such as conservation and methylation. Along these same lines, if we were to refine our preprocessing strategies for the DNase-seq data, we can easily incorporate these improved features into our model as long as the data are available as bigWig files [37]. Other sources of open chromatin information, such as ATAC-seq [60] and FAIRE-seq [61], can also be used to replace or complement the existing DNase-seq data. Consequently, FactorNet is not limited to any single preprocessing pipeline. In addition, FactorNet is not necessarily constrained to only TF binding predictions. If desired, users can provide the BED files of positive intervals to train models for predicting other markers, such as histone modifications. As more epigenomic datasets are constantly added to data repositories, FactorNet is already in a prime position to integrate both new and existing datasets.
Supplementary Material
Open source method, FactorNet, for predicting cell type-specific transcription factor binding
One of the top performing methods in the ENCODE-DREAM Challenge
Transcription factor binding prediction problem is far from solved
Acknowledgments
We thank the ENCODE-DREAM challenge organizers for providing the opportunity to test and improve our method. We also thank David Knowles for helping with generating gene expression metadata features.
This work was supported by the National Institute of Biomedical Imaging and Bioengineering, National Research Service Award (EB009418) from the University of California, Irvine, Center for Complex Biological Systems and the National Science Foundation Graduate Research Fellowship under Grant No. (DGE-1321846). Any opinion, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Software availability
Source code is available at the github repository http://github.com/ucicbcl/FactorNet. In addition to the source code, the github repository contains all models and data used for the ENCODE-DREAM Challenge. FactorNet is also available through the kipoi model zoo [56].
Conflict of interest statement.
None declared.
References
- [1].Johnson DS, Mortazavi A, Myers RM, Wold B, Genome-wide mapping of in vivo protein-dna interactions, Science 316 (2007) 1497–502. [DOI] [PubMed] [Google Scholar]
- [2].Crawford G et al. , Genome-wide mapping of dnase hypersensitive sites using massively parallel signature sequencing (mpss), Genome Res 16 (2006) 123–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].John S, Sabo PJ, Canfield TK, Lee K, Vong S, Weaver M, Wang H, Vierstra J, Reynolds AP, Thurman RE, et al. , Genome-scale mapping of dnase i hypersensitivity, Current protocols in molecular biology (2013) 21–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Hesselberth JR, Chen X, Zhang Z, Sabo PJ, Sandstrom R, Reynolds AP, Thurman RE, Neph S, Kuehn MS, Noble WS, Fields S, Stamatoyannopoulos JA, Global mapping of protein-dna interactions in vivo by digital genomic footprinting, Nat Methods 6 (2009) 283–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Boyle AP, Song L, Lee B-K, London D, Keefe D, Birney E, Iyer VR, Crawford GE, Furey TS, High-resolution genome-wide in vivo footprinting of diverse transcription factors in human cells, Genome research 21 (2011) 456–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Neph S et al. , An expansive human regulatory lexicon encoded in transcription factor footprints, Nature 489 (2012) 83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Quang DX, Erdos MR, Parker SCJ, Collins FS, Motif signatures in stretch enhancers are enriched for disease-associated genetic variants, Epigenetics Chromatin 8 (2015) 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ernst J, Kellis M, Chromhmm: automating chromatin-state discovery and characterization, Nature methods 9 (2012) 215–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hoffman MM, Buske OJ, Wang J, Weng Z, Bilmes JA, Noble WS, Unsupervised pattern discovery in human chromatin structure through genomic segmentation, Nature methods 9 (2012) 473–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Mortazavi A, Pepke S, Jansen C, Marinov GK, Ernst J, Kellis M, Hardison RC, Myers RM, Wold BJ, Integrating and mining the chromatin landscape of cell-type specificity using self-organizing maps, Genome research 23 (2013) 2136–2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].ENCODE Project Consortium, An integrated encyclopedia of dna elements in the human genome, Nature 489 (2012) 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Roadmap Epigenomics Consortium et al. , Integrative analysis of 111 reference human epigenomes, Nature 518 (2015) 317–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ernst J, Kellis M, Large-scale imputation of epigenomic datasets for systematic annotation of diverse human tissues, Nat Biotechnol 33 (2015) 364–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Sherwood RI, Hashimoto T, O’Donnell CW, Lewis S, Barkal AA, van Hoff JP, Karun V, Jaakkola T, Gifford DK, Discovery of directional and nondirectional pioneer transcription factors by modeling dnase profile magnitude and shape, Nat Biotechnol 32 (2014) 171–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Pique-Regi R, Degner JF, Pai AA, Gaffney DJ, Gilad Y, Pritchard JK, Accurate inference of transcription factor binding from dna sequence and chromatin accessibility data, Genome Res 21 (2011) 447–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Raj A, Shim H, Gilad Y, Pritchard JK, Stephens M, mscentipede: Modeling heterogeneity across genomic sites and replicates improves accuracy in the inference of transcription factor binding, PLoS One 10 (2015) e0138030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Quang D, Xie X, Extreme: an online em algorithm for motif discovery, Bioinformatics 30 (2014) 1667–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Leung MKK, Xiong HY, Lee LJ, Frey BJ, Deep learning of the tissue-regulated splicing code, Bioinformatics 30 (2014) il21–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Quang D, Chen Y, Xie X, Dann: a deep learning approach for annotating the pathogenicity of genetic variants, Bioinformatics 31 (2015) 761–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Chen Y, Li Y, Narayan R, Subramanian A, Xie X, Gene expression inference with deep learning, Bioinformatics (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Quang D, Xie X, Danq: a hybrid convolutional and recurrent deep neural network for quantifying the function of dna sequences, Nucleic Acids Res 44 (2016) e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Zhou J, Troyanskaya OG, Predicting effects of noncoding variants with deep learning-based sequence model, Nat Methods 12 (2015) 931–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Kelley DR, Snoek J, Rinn JL, Basset: learning the regulatory code of the accessible genome with deep convolutional neural networks, Genome Res 26 (2016) 990–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Alipanahi B, Delong A, Weirauch MT, Frey BJ, Predicting the sequence specificities of dna- and rna-binding proteins by deep learning, Nat Biotechnol 33 (2015) 831–8. [DOI] [PubMed] [Google Scholar]
- [25].Hassanzadeh HR, Wang MD, Deeperbind: Enhancing prediction of sequence specificities of dna binding proteins, in: Bioinformatics and Biomedicine (BIBM), 2016. IEEE International Conference on, IEEE, pp. 178–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Graves A, Schmidhuber J, Framewise phoneme classification with bidirectional lstm and other neural network architectures, Neural Networks 18 (2005) 602–610. [DOI] [PubMed] [Google Scholar]
- [27].Graves A, Jaitly N, Mohamed A-R, Hybrid speech recognition with deep bidirectional lstm, in: Automatic Speech Recognition and Understanding, 2013. IEEE Workshop on, pp. 273–278. [Google Scholar]
- [28].Sundermeyer M, Alkhouli T, Wuebker J, Ney H, Translation modeling with bidirectional recurrent neural networks, in: Proceedings of the Conference on Empirical Methods on Natural Language Processing, October. [Google Scholar]
- [29].Zhu W, Lan C, Xing J, Li Y, Shen L, Zeng W, Xie X, Co-occurrence feature learning for skeleton based action recognition using regularized deep lstm networks, The 30th AAAI Conference on Artificial Intelligence (AAAI-16) (2016). [Google Scholar]
- [30].Angermueller C, Lee HJ, Reik W, Stegle O, Deepcpg: accurate prediction of single-cell dna methylation states using deep learning, Genome biology 18 (2017) 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Zou LS, Erdos MR, Taylor DL, Chines PS, Varshney A, Parker SC, Collins FS, Didion JP, Boostme accurately predicts dna methylation values in whole-genome bisulfite sequencing of multiple human tissues, BMC genomics 19 (2018) 390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Zhang W, Spector TD, Deloukas P, Bell JT, Engelhardt BE, Predicting genome-wide dna methylation using methylation marks, genomic position, and dna regulatory elements, Genome biology 16 (2015) 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Bromley J, Bentz JW, Bottou L, Guyon I, LeCun Y, Moore C, Sackinger E, Shah R, Signature verification using a “siamese” time delay neural network, IJPRAI 7 (1993) 669–688. [Google Scholar]
- [34].Mueller J, Thyagarajan A, Siamese recurrent architectures for learning sentence similarity., in: AAAI, pp. 2786–2792. [Google Scholar]
- [35].Encode-dream challenge description, https://www.synapse.org/ENCODE, Accessed: 2018-10-08.
- [36].Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, et al. , Gen-code: the reference human genome annotation for the encode project, Genome research 22 (2012) 1760–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Kent WJ, Zweig AS, Barber G, Hinrichs AS, Karolchik D, Bigwig and bigbed: enabling browsing of large distributed datasets, Bioinformatics 26 (2010) 2204–2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Chollet F, et al. , Keras, https://github.com/fchollet/keras, 2015.
- [39].Shirley MD, Ma Z, Pedersen BS, Wheelan SJ, Efficient” pythonic” access to FASTA files using pyfaidx, Technical Report, PeerJ PrePrints, 2015.
- [40].Dale RK, Pedersen BS, Quinlan AR, Pybedtools: a flexible python library for manipulating genomic datasets and annotations, Bioinformatics 27 (2011) 3423–3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Ramirez F, Ryan DP, Gruning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dundar F, Manke T, deeptools2: a next generation web server for deep-sequencing data analysis, Nucleic acids research 44 (2016) W160–W165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Ramirez F, Dundar F, Diehl S, Gruning BA, Manke T, deeptools: a flexible platform for exploring deep-sequencing data, Nucleic acids research 42 (2014) W187–W191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Gardiner-Garden M, Frommer M, CpG islands in vertebrate genomes, Journal of molecular biology 196 (1987) 261–282. [DOI] [PubMed] [Google Scholar]
- [44].Bastien F, Lamblin P, Pascanu R, Bergstra J, Goodfellow IJ, Bergeron A, Bouchard N, Bengio Y, Theano: new features and speed improvements, Deep Learning and Unsupervised Feature Learning NIPS 2012 Workshop, 2012. [Google Scholar]
- [45].Bergstra J, Breuleux O, Bastien F, Lamblin P, Pascanu R, Desjardins G, Turian J, Warde-Farley D, Bengio Y, Theano: a cpu and gpu math expression compiler, in: Proceedings of the Python for scientific computing conference, volume 4, Austin, TX, p. 3. [Google Scholar]
- [46].Kingma D, Ba J, Adam: A method for stochastic optimization, arXiv preprint arXiv:1412.6980 (2014). [Google Scholar]
- [47].Srivastava N, Hinton G, Krizhevsky A, Sutskever I, Salakhutdinov R, Dropout: A simple way to prevent neural networks from overfitting, The Journal of Machine Learning Research 15 (2014) 1929–1958. [Google Scholar]
- [48].Kharchenko PV, Tolstorukov MY, Park PJ, Design and analysis of chip-seq experiments for dna-binding proteins, Nature biotechnology 26 (2008) 1351–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Saito T, Rehmsmeier M, The precision-recall plot is more informative than the roc plot when evaluating binary classifiers on imbalanced datasets, PloS one 10 (2015) e0118432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D, The human genome browser at ucsc, Genome research 12 (2002) 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Mathelier A et al. , JASPAR 2016: a major expansion and update of the open-access database of transcription factor binding profiles, Nucleic Acids Research 44 (2016) D110–D115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Gupta S, Stamatoyannopoulos JA, Bailey TL, Noble WS, Quantifying similarity between motifs, Genome Biol 8 (2007) R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Medina-Rivera A, Defrance M, Sand O, Herrmann C, Castro-Mondragon JA, Delerce J, Jaeger S, Blanchet C, Vincens P, Caron C, Staines DM, Contreras-Moreira B, Artufel M, Charbonnier-Khamvongsa L, Hernandez C, Thieffry D, Thomas-Chollier M, van Helden J, Rsat 2015: Regulatory sequence analysis tools, Nucleic Acids Res 43 (2015) W50–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Shrikumar A, Greenside P, Kundaje A, Learning important features through propagating activation differences, arXiv preprint arXiv:1704.02685 (2017). [Google Scholar]
- [55].Simonyan K, Vedaldi A, Zisserman A, Deep inside convolutional networks: Visualising image classification models and saliency maps, arXiv preprint arXiv:1312.6034 (2013). [Google Scholar]
- [56].Avsec Z, Kreuzhuber R, Israeli J, Xu N, Cheng J, Shrikumar A, Banerjee A, Kim DS, Urban L, Kundaje A, Stegle O, Gagneur J, Kipoi: accelerating the community exchange and reuse of predictive models for genomics, bioRxiv (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Rhee HS, Pugh BF, Comprehensive genome-wide protein-dna interactions detected at single-nucleotide resolution, Cell 147 (2011) 1408–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Skene PJ, Henikoff S, An efficient targeted nuclease strategy for high-resolution mapping of dna binding sites, Elife 6 (2017) e21856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Bahdanau D, Cho K, Bengio Y, Neural machine translation by jointly learning to align and translate, arXiv preprint arXiv:1409.0473 (2014). [Google Scholar]
- [60].Buenrostro JD, Wu B, Chang HY, Greenleaf WJ, Atac-seq: A method for assaying chromatin accessibility genome-wide, Current protocols in molecular biology (2015) 21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Giresi PG, Kim J, McDaniell RM, Iyer VR, Lieb JD, Faire (formaldehyde-assisted isolation of regulatory elements) isolates active regulatory elements from human chromatin, Genome research 17 (2007) 877–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Thorvaldsdóttir H, Robinson JT, Mesirov JP, Integrative genomics viewer (igv): high-performance genomics data visualization and exploration, Briefings in bioinformatics 14 (2013) 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.