Abstract
Background
Spotted leaf mutants show typical necrotic lesions that appear spontaneously in the absence of any pathogen attack. These mutants are often characterized to exhibit programmed cell death (PCD) and activation of plant defense responses resulting in enhanced disease resistance to multiple pathogens. Here, we reported a novel spotted-leaf mutant, spl40 that showed enhanced disease resistance response.
Results
Initially lesions appeared at leaf tips during seedling stage and gradually covered the whole leaf at the tillering stage. The lesion development was light-dependent. spl40 showed obvious cell death at and around the lesion, and burst of reactive oxygen species (ROS) was accompanied by disturbed ROS scavenging system. Photosynthetic capacity was compromised as evidenced by significant reductions in chlorophyll content, important photosynthesis parameters and downregulated expression of photosynthesis-related genes which ultimately led to poor performance of major agronomic traits. spl40 exhibited enhanced resistance to 14 out of 16 races of bacterial blight pathogen of rice, caused by Xanthomonas oryzae pv. oryzae, most probably though activation of SA and JA signaling pathways, owing to upregulated expression of SA and JA signaling genes, though the exact mechanism remain to be elucidated. The spotted-leaf phenotype was controlled by a novel single recessive nuclear gene. Genetic mapping combined with high throughput sequencing analysis identified Os05G0312000 as the most probable candidate gene. Sequencing of ORF revealed a single SNP change from C to T that resulted in non-synonymous change in amino acid residue from leucine to phenylalanine. Interestingly, the complementation plants did not display lesions before heading but showed lesions at the heading stage and the transgenic T1 progenies could be classified into 3 categories based on their lesion intensity, indicating the complex genetic nature of the spl40 mutation.
Conclusion
The results obtained here clearly show that genes related to defense and PCD were upregulated in accordance with enhanced disease resistance and occurrence of PCD, whereas the photosynthetic capacity and overall ROS homeostasis was compromised in spl40. Our data suggest that a novel spotted-leaf mutant, spl40, would help to elucidate the mechanism behind lesion development involving programmed cell death and associated defense responses.
Electronic supplementary material
The online version of this article (10.1186/s12284-019-0326-6) contains supplementary material, which is available to authorized users.
Keywords: Rice, Spotted-leaf, Programmed cell death, Bacterial blight, Defense response
Background
Plants, during their lifecycle, are exposed to adverse environmental conditions imposed by different biotic and abiotic stresses. To combat against such adversaries, they are equipped with well-developed defense arsenal as a cope-up mechanism. Specifically during the plant innate immune response against pathogen attack, hypersensitive response (HR), a form of programmed cell death, plays an important role (Liu et al. 2005).
Lesion mimic mutants (LMMs) are so called because of the phenotype resembling the pathogen inducible, HR-mediated necrosis, which forms an important resistance mechanism in plants involving recognition of pathogens through a resistance gene-mediated defense system that induces cell death in attacked areas, which prevents further spread of pathogen to the cells in close vicinity of attack (Dangl and Jones 2001; Lam et al. 2001; Qiao et al. 2010). Genetic and molecular data accumulated till date have suggested that cell death in plants associated with HR or developmental processes is genetically programmed (Lorrain et al. 2003), and this cell death is believed to be programmed cell death that plays an important role in plant growth and development (Huang et al. 2010). Several mutants exhibiting spontaneous cell death have been isolated in different plant species like maize (Wang et al. 2013), sorghum (Sindhu et al. 2018), Arabidopsis (Lorrain et al. 2003; Simanshu et al. 2014), barley (Rostoks et al. 2006), rice (Takahashi et al. 1999; Yin et al. 2000; Mori et al. 2007; Wu et al. 2008; Huang et al. 2011; Manosalva et al. 2011; Ma et al. 2012; Zhou et al. 2018)) and wheat (Kamlofski et al. 2007; Tang et al. 2013; Wang et al. 2016). As every new lesion mimic mutant is providing some useful information about plant’s cope-up mechanism at molecular as well as physiological level, the hunt for novel lesion mimic mutants is expected to continue, even at greater pace and efforts. Appearance of lesions in most of the LMM is accompanied by some defense responses like production of reactive oxygen intermediates (ROIs), accumulation of phytoalexin, callose and phenolic compounds, enhanced expression of defense marker genes and elevated levels of signaling compounds like salicylic acid (Mori et al. 2007; Qiao et al. 2010; Chen et al. 2012; Xu et al. 2018). Many LMM with disease resistance phenotype showed the elevated expression of pathogenesis-related proteins like PR1, PR2, PR5, PR10, PBZ1 and PAL, which are the important components of plant defense against pathogens (Wang et al. 2005; Mori et al. 2007; Takahashi et al. 2007; Qiao et al. 2010; Chen et al. 2012).
To date, many LMM or spotted-leaf genes have been cloned and characterized in rice, and most of the mutants show HR-like symptoms, but with various levels of resistance to pathogens and different reaction patterns to environmental factors, indicating the presence of multiple pathways leading to HR in rice. The identified protein products encoded by spotted-leaf genes in rice include ubiquitin ligase (Zeng et al. 2004), heat stress transcription factors (Yamanouchi et al. 2002), zinc finger protein (Wang et al. 2005), clathrin associated adaptor protein complex (Qiao et al. 2010), acyltransferase (Mori et al. 2007) and splicing factor 3 subunit 3 (Chen et al. 2012). These findings indicate that numerous proteins, with distinct functions in multiple signaling pathways, are involved in the regulation of HR cell death and disease resistance (Chen et al. 2012). As the disease resistance relates to expression of defense related genes, HR and immunity, it is expected that studies on LMMs will help to provide more insights into the mechanism underlying these processes (Takahashi et al. 1999; Wu et al. 2008). In a report involving characterization of SPL28, it is showed that the SPL28 gene encodes a clathrin associated adaptor protein complex1, medium subunit μ1 (AP1M1) which appeared to be involved in the regulation of vesicular trafficking, and it was suggested that SPL28 dysfunction caused the formation of hypersensitive response (HR)-like lesions, leading to the initiation of leaf senescence (Qiao et al. 2010). One of the best studied rice LMM spl11, found to encode U-box/Armadillo repeat protein having E3 ubiquitin ligase activity, which showed broad-spectrum disease resistance (Zeng et al. 2004). E3 ubiquitin ligase is a well characterised component of ubiquitin-26 s proteasome system (UPS) which play an important role in the regulation of plant immune responses (Marino et al. 2012). The resistance showed by spl11 to rice blast and bacterial blight is a non-specific resistance which is likely to be regulated by the downstream components of defense pathways, which in turn suggest that there might involve a possible cross-talks between the cell death pathway and defense pathways of blast and bacterial blight resistance (Huang et al. 2010).
Here, we identified a spotted-leaf mutant spl40 from an EMS-induced Zhongjian100 mutant bank. This mutant exhibits necrotic lesions at four leaf stage and showed enhanced resistance to bacterial blight disease. Histochemical staining revealed the ROS accumulation and plant cell death at and around the lesion. The mutant is light sensitive and showed reduced chlorophyll content as well as disturbed photosynthetic capacity and ROS scavenging system. We also compared major agronomic traits as well as different physiological parameters that showed poor performance. The mutation is controlled by a single nuclear recessive gene located on the long arm of chromosome 5. We concluded that spl40 is a novel spotted-leaf mutant with enhanced resistance to multiple races of Xoo probably through activation of JA and SA signaling pathways. This study would facilitate the elucidation of mechanism behind initiation of lesions and HR-induced PCD.
Results
Phenotypic performance of spl40
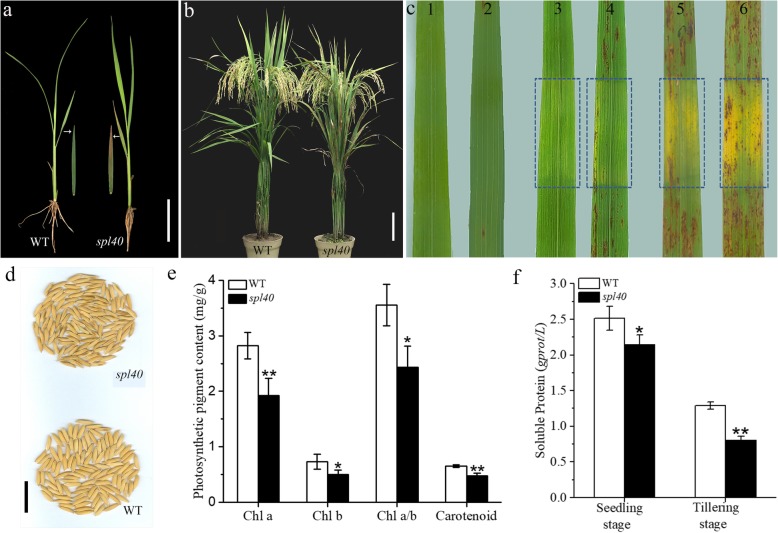
Under the natural summer field conditions, leaves of spl40 largely remained the same as the wild type (WT) till the emergence of the fourth leaf. At four leaf stage, the necrotic spots were initiated from the tip of older leaves (Fig. 1a). Later from tillering to heading, these necrotic spots became more serious and gradually spread through the whole leaf (Fig. 1b). The lesions also appeared on some husks in mutant seeds (Fig. 1d). The major agronomic traits such as number of filled grains per panicle, number of panicle per plant, seed setting rate and 1000-grain weight, were all significantly decreased in spl40 (Table 1).
Fig. 1.
Spotted leaf phenotype of spl40. a Lesions appear at 4-leaf stage (shown detached 3rd leaf with lesions) (bar = 5 cm). b WT and spl40 at maturity stage (Scale bar = 10 cm). c Effect of light on lesion formation under the natural condition (1–2) Zhongjian100 and spl40 before shading. (2–3) Zhongjian100 and spl40 shaded for 7 days. (5) spl40 re-instated for 5 d. (6) spl40 re-instated for 15 d. Shaded areas are boxed. d Lesion phenotype on matured seeds (bar = 2 cm). e Photosynthetic pigment contents at tillering stage. f Soluble protein content at seedling and tillering stage. Values are means ± SD (n = 3); ** indicates significance at P ≤ 0.01 and * indicates significance at P ≤ 0.05 by Student’s t test
Table 1.
Agronomic performance of spl40
| Material | Plant height (cm) | No. tiller/Plant | No. Panicle/plant | No. filled grain/panicle | Seed-setting rate (%) | 1000-grain weight (g) |
|---|---|---|---|---|---|---|
| WT | 103.3 ± 4.2 | 23.3 ± 1.2 | 23 ± 2.0 | 85.7 ± 2.5 | 81.4 ± 6.2 | 21 ± 0.3 |
| spl40 | 105 ± 2.3 | 21.7 ± 1.2 | 20.7 ± 3.6* | 80.3 ± 2.1* | 57.3 ± 8.9** | 18.3 ± 0.3** |
Values are means ± SD (n = 3)
** indicates significance at P ≤ 0.01 and * indicates significance at P ≤ 0.05 by Student’s t test
Photosynthetic pigment content was measured to check the effect of lesion on it. The levels of chlorophyll a, chlorophyll b, carotenoid and Chl a/b ratio all were significantly reduced in spl40 compared with WT (Fig. 1e). Furthermore, there was a significant decrease in the soluble protein content in spl40 at the seedling and tillering stages compared to WT (Fig. 1f). Altogether, this data suggested that the mutation-induced formation of necrotic lesions had negative effect on photosynthetic pigment content as well as soluble protein content, which might be the cause of poor agronomic performance of the mutant.
Lesion initiation is light-dependent
A number of lesion mimic mutants show light-dependent initiation of lesions (Huang et al. 2010; Chen et al. 2018). To determine the effect of light on lesion initiation of spl40, a second leaf from both WT and spl40 was covered with 2–3 cm aluminium foil for 7 days at the tillering stage. The mutant leaf was shaded well before lesions spread to the whole leaf. The results showed that in case of spl40, the shaded area had nothing but very few lesions at the edge while unshaded area showed presence of numerous lesions (Fig. 1c). Later, upon re-instating the light for 5 days resulted in the initiation of lesions which were subsequently increased in number after 15 days (Fig. 1c). WT showed no obvious changes before and after the shading treatment (Fig. 1c). These results confirmed that the initiation of lesions in spl40 was light-dependent.
Altered photosynthetic capacity in spl40
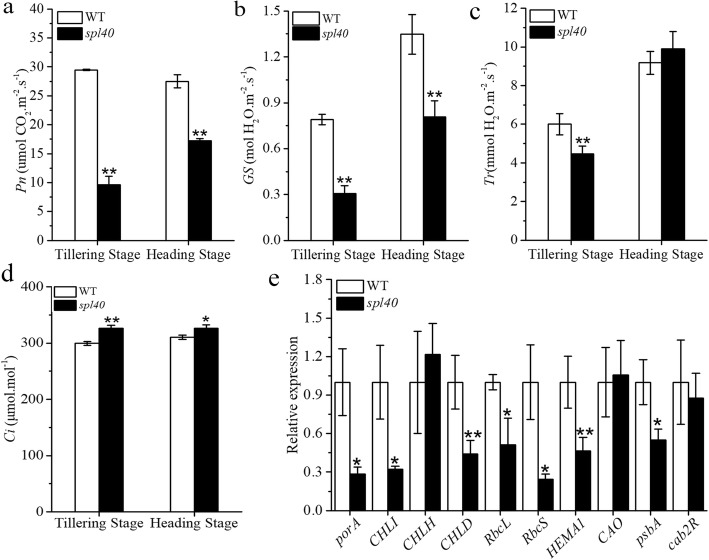
As shown above there was a reduction in the photosynthetic pigment content in the spl40, presence of the lesions might also lead to changes in the photosynthetic capacity. We measured the photosynthesis parameters at the tillering and heading stage in spl40 and WT. It was found that there was a significant reduction the net photosynthetic rate (Pn) and stomatal conductance (GS) in spl40 compared to WT at both stages (Fig. 2a, b). In case of transpiration rate (Tr), spl40 showed significant reduction compared to WT at the tillering stage, whereas it was similar at the heading stage (Fig. 2c). Intercellular CO2 concentration (Ci) was significantly increased in spl40 compared to WT at both stages (Fig. 2d). Altogether, the photosynthetic capacity was impaired in spl40 which might be the result of reductions in photosynthetic pigment content and this ultimately led to poor agronomic performance of spl40.
Fig. 2.
Photosynthesis parameters of leaves at the tillering and heading stages. a Net photosynthetic rate (Pn). b Stomatal conductance (Gs). c Transpiration rate (Tr). d Intercellular CO2 concentration (Ci). e Expression profile of photosynthesis related genes in spl40 and WT at tillering stage. Values are means ± SD (n = 3); ** indicates significance at P ≤ 0.01 and * indicates significance at P ≤ 0.05 by Student’s t test
To clarify whether the impaired photosynthetic capacity was stemmed from changes at genetic level, we measured the expression of different photosynthesis related genes in the mutant and WT at the tillering stage by qRT-PCR. The expression level of most of the photosythesis related genes was found to be significantly reduced in spl40 compared to WT (Fig. 2e). This shows that the mutation in spl40 has resulted in reduced expression of photosynthesis related genes which was in accordance with reduced photosynthetic pigment content and altered photosynthetic capacity.
ROS accumulation and cell death occur in spl40
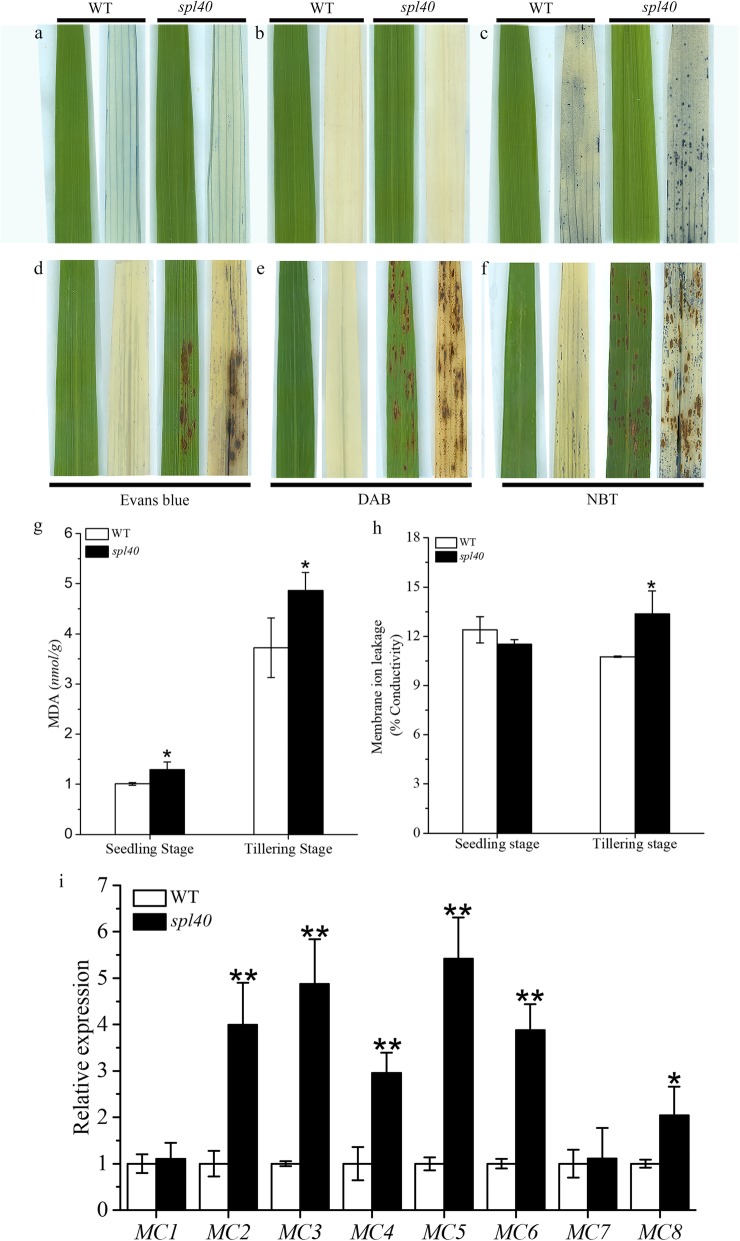
To determine the occurrence of cell death as well as the accumulation of ROS at or around the lesions, we carried out histochemical staining with spl40 and WT leaves both before the lesion development and after the lesion development. Staining with evans blue, which is an indicator of irreversible membrane damage or cell death (Liu et al. 2008), showed presence of blue stained cells in spl40 leaves with lesion, whereas there was no staining observed before the lesion development (Fig. 3a, d). On the contrary, WT leaves were devoid of staining at both time points. To confirm the membrane damage and cell death, we further measured the levels of malonaldehyde (MDA) and membrane ion leakage. The mutant showed significantly higher levels of malonaldehyde content compared to WT both at the seedling and tillering stage (Fig. 3g), whereas membrane ion leakage was significantly higher in spl40 than WT only at the tillering stage (Fig. 3h). Taken together these results suggested that indeed the cell death occurred in the leaves with lesions.
Fig. 3.
Histochemical staining of spl40 and WT. a and d Evans blue staining: a before lesion development. d after lesion development. b and e DAB staining: b before lesion development. e after lesion development. c and f NBT staining: c before lesion development. f after lesion development. g Malonaldehyde (MDA) content at seedling and tillering stage. h Membrane ion leakage rate at seedling and tillering stage. i Expression analysis of PCD-related genes at tillering stage. Values are means ± SD (n = 3); ** indicates significance at P ≤ 0.01 and * indicates significance at P ≤ 0.05 by Student’s t test
Further, we checked whether the occurrence of cell death in spl40 was also accompanied by burst of reactive oxygen species (ROS). DAB staining was carried out to determine the H2O2 accumulation. The red brown precipitate was observed on spl40 leaves with lesion (Fig. 3e), whereas no precipitate was observed on leaves without lesion (Fig. 3b). WT leaves remained clear without any precipitate both before and after the lesion development stage. NBT staining, which is the indicator of O2− accumulation, showed blue stained regions in spl40 leaves both before and after lesion development (Fig. 3c, f). Leaves with lesion had higher precipitate than leaves without lesion. WT leaves also showed some positive staining but it was significantly less compared to spl40. These results confirmed that the occurrence of cell death was accompanied by burst of ROS.
Later, we analysed the expression pattern of different rice metacaspase (OsMC) genes, which are the important regulators of programmed cell death (Huang et al. 2015). The results showed that expression levels of OsMC2, OsMC3, OsMC4, OsMC5, OsMC6 and OsMC8 were significantly increased compared to WT (Fig. 3i). While the expression levels of OsMC1 and OsMC7 were similar in spl40 and WT. Together, our results further demonstrated that the mutation caused PCD to occur with concurrent increase in the expression of rice metacaspase genes in mutant.
Perturbed ROS scavenging system in spl40
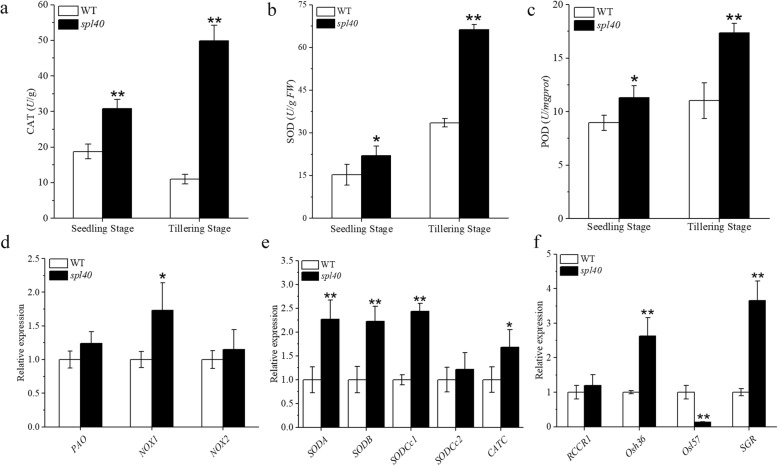
Burst of reactive oxygen species (ROS) is commonly associated with perturbed ROS scavenging system. To check this possibility, we measured the activities ROS scavenging enzymes in spl40 and WT leaves. The activities of CAT, SOD and POD all were significantly increased in spl40 compared to WT both at the seedling and tillering stage (Fig. 4a–c). This clearly showed that burst of ROS resulted in perturbed ROS scavenging system in spl40. To further confirm this conclusion, we carried out the expression analysis of different ROS-homeostasis related genes. NOX (NOX1 and NOX2) and PAO genes encode the major ROS generating enzymes NADPH oxidase (NOX) and polyamine oxidase (PAO), respectively (Lee et al. 2018). Expression of NOX1 was significantly increased in the mutant compared to WT (Fig. 4d). As spl40 exhibited enhanced ROS accumulation, we next examined the expression of ROS scavenging genes. Expression levels of SODA, SODB, SODCc1 and CATC were significantly higher in the mutant than WT (Fig. 4e). Subsequently, we also measured the expression of important senescence related genes. spl40 showed significantly enhanced expression of Osh36 and SGR compared to WT, whereas the expression of Osl57 was significantly decreased in the mutant (Fig. 4f). Our results demonstrated that the mutation led to increased activity of ROS scavenging enzymes along with varied changes in the expression of ROS-generating, ROS-scavenging and senescence related genes, which contributed to overall perturbed ROS homeostasis in the mutant.
Fig. 4.
ROS scavenging enzymes and associated genes. a–c Enzymatic activities of CAT, SOD and POD at seedling and tillering stage. SOD, superoxide dismutase; CAT, catalase; POD, peroxidase. d Expression levels of genes encoding ROS-generating enzymes. e Expression levels of ROS detoxification-related genes. f Expression levels of senescence-related genes. Values are means ± SD (n = 3); ** indicates significance at P ≤ 0.01 and * indicates significance at P ≤ 0.05 by Student’s t test
Enhanced resistance to bacterial blight pathogen
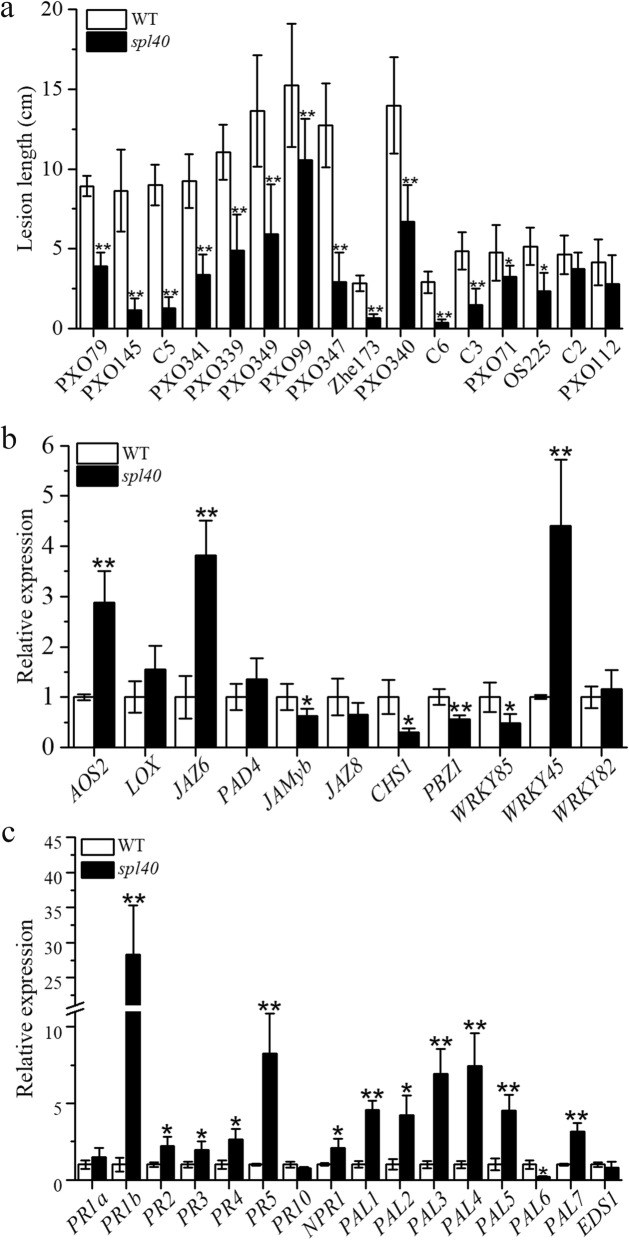
Formation of lesions in rice LMM often lead to activation of defense responses that results in enhanced resistance to one or more pathogens (Wu et al. 2008; Feng et al. 2013; Zhang et al. 2018). We carried out the disease resistance assessment with 16 races of Xanthomonas oryzae pv. oryzae under the natural field conditions by leaf clipping method (Kauffman 1973). Top second leaf of spl40, WT and susceptible control IR24 were inoculated with Xoo pathogen at the tillering stage. The results showed that spl40 exhibited significantly enhanced resistance to races PXO79, PXO145, C5, PXO339, PXO341, PXO349, PXO99, PXO347, Zhe173, PXO340, C6, C3 (P ≤ 0.01), PXO71 and OS225 (P ≤ 0.05) compared to WT, while resistance response to races C2 and PXO112 was similar in both spl40 and WT (Fig. 5a). The IR24 showed susceptible reaction to all the races tested. The results suggested that spl40 is a mutant with enhanced resistance to multiple races of Xoo.
Fig. 5.
Evaluation of disease resistance to Xanthomonas oryzae pv. oryzae and expression of defense genes involved in JA and SA signaling pathway. a Lesion lengths after Xoo inoculation at 20 DPI. b Expression analysis of JA signaling pathway genes. c Expression analysis of SA signaling pathway genes. Values are means ± SD (A n = 9; B and C n = 3); ** indicates significance at P ≤ 0.01 and * indicates significance at P ≤ 0.05 by Student’s t test
To ascertain whether enhanced disease resistance response was stemmed from elavated expression of defence related genes, we examined their expression by real-time quantitative PCR analysis. These defense genes mainly consisted of those involved in SA and JA signalling and/or biosynthesis. Expression level of JA-signalling genes AOS2, WRKY45 and JAZ6 was significantly higher, while the expression of JAMyb, CHS1, PBZ1 and WRKY85 was singificantly reduced, in spl40 compared to WT (Fig. 5b). Most of the SA-signalling genes, PR1b, PR2, PR3, PR4, PR5, NPR1, PAL1, PAL2, PAL3, PAL4, PAL5 and PAL7 were highly expressed in spl40 compared to WT (Fig. 5c). This clearly showed that the elavated expression of SA signalling genes as well as some JA signalling genes might have resulted in enhanced resistance to Xoo in spl40.
Genetic control and physical mapping of spl40 candidate gene
To determine the genetic control of the mutation, a cross was made between spl40 and the wild type Zhongjian100. F1 progenies showed normal phenotype similar to WT suggesting that mutation was recessive in nature. Further, 423 F2 progenies were screened for genetic segregation and found that 308 individuals exhibited normal phenotype, while 115 individuals exhibited mutant phenotype. Thus, segregation pattern fitted to 3:1 Mendelian ratio (χ2 = 0.62 < χ20.05 = 3.84). Collectively, this suggested that spotted-leaf phenotype of the spl40 was governed by a single recessive nuclear gene.
An F2 population derived from a cross between spl40 and Nipponbare (Japonica) was used to map the locus responsible for spl40 mutant phenotype. The spotted-leaf phenotype was found to be linked with two simple sequence repeat (SSR) markers RM17952 and RM18522 on chromosome 5 by bulk segregant analysis. To fine map the locus, 1266 F2 individuals with spl40 phenotype were genotyped and the mutation was finally delimited to a 523 kb interval between markers InDel1 and RM18379 (Fig. 6a).
Fig. 6.
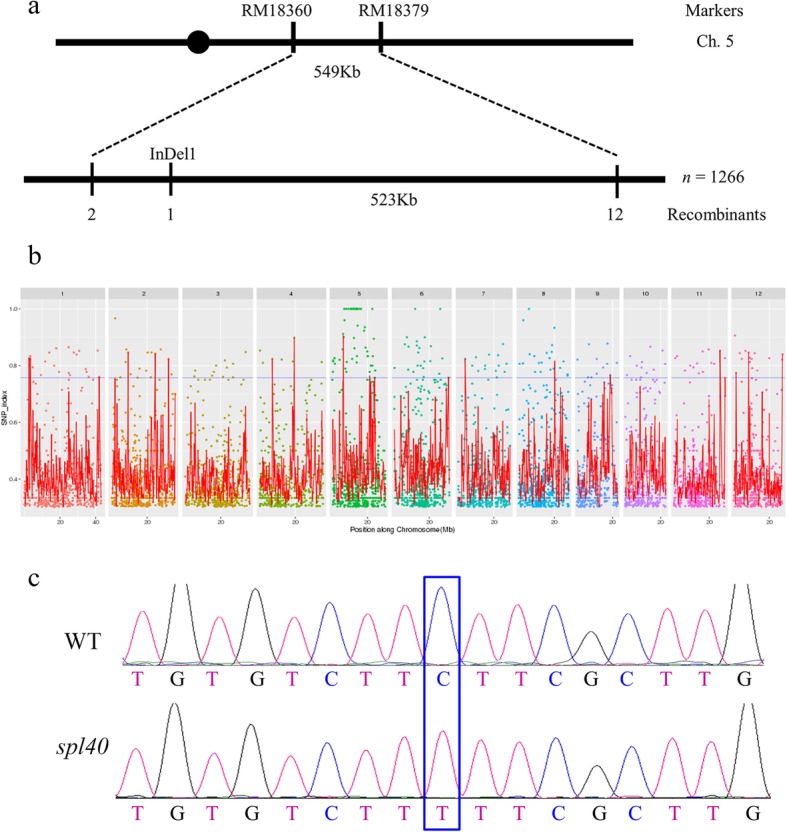
MutMap assisted mapping of spl40 gene on the long arm of chromosome 5. a Fine mapping of the candidate region of spl40. b Manhattan plot depicting the distribution of SNPs along 12 rice chromosomes. c Confirmation of the spl40 mutation by sequencing
The WT DNA and the pooled DNA from backcross F2 individuals were subjected to high throughput sequencing, and the results showed that more than five thousand SNPs were detected between WT and the mutant (Fig. 6b). Out of these, a total of 15 SNPs were identified with SNP index of 1 on 3 chromosomes. Among them, two were located in the 523 kb candidate region, out of which only one was present in the genic region (Fig. 6b). This resultant SNP was localized in the 10th exon Os05G0312000 which was of non-synonymous nature. A single base substitution from C to T occurred at position 2092 in the coding sequence that resulted in change of amino acid residue from leucine to phenylalanine. The mutation was confirmed by sequencing the exonic region of Os05G0312000 between the mutant and WT (Fig. 6c).
Functional complementation with SPL40
The WT allele was transferred into the spl40 mutant background via Agrobacterium-mediated plant transformation to validate the candidate gene. At the tillering stage, when spl40 showed obvious spotted-leaf phenotype, the WT and T0 complementation plants did not show lesion mimic phenotype. Surprisingly, as the complementation plants entered the heading stage, brown lesions started to appear on the older leaves. At this stage spl40 had severe spotted-leaf phenotype, while WT was normal. Further, T1 progenies were grown and checked for lesion mimic phenotype (Fig. 7a). It was found that, in all the lines, T1 plants showed different degree of phenotype severity. Based on the severity of phenotype we grouped the T1 plants in three categories viz., lesion mimic type, intermediate type and wild type (Fig. 7b). But in all the lines, T1 plants displayed complex segregation pattern (Fig. 7c). We carried out genotyping of randomly selected T1 plants representing each category to check for the transgene of WT allele by sequencing the region of SPL40 that contains the mutation. We found that all the progenies showed heterozygous nature at the mutation site (Additional file 1: Table S1). Furthermore, no significance difference in the frequencies of WT allele among the three categories of T1 plants was detected (Additional file 2: Figure S1). The results indicate that the genetic nature of spl40 is very complex and further work is needed to understand its genetic behavior.
Fig. 7.
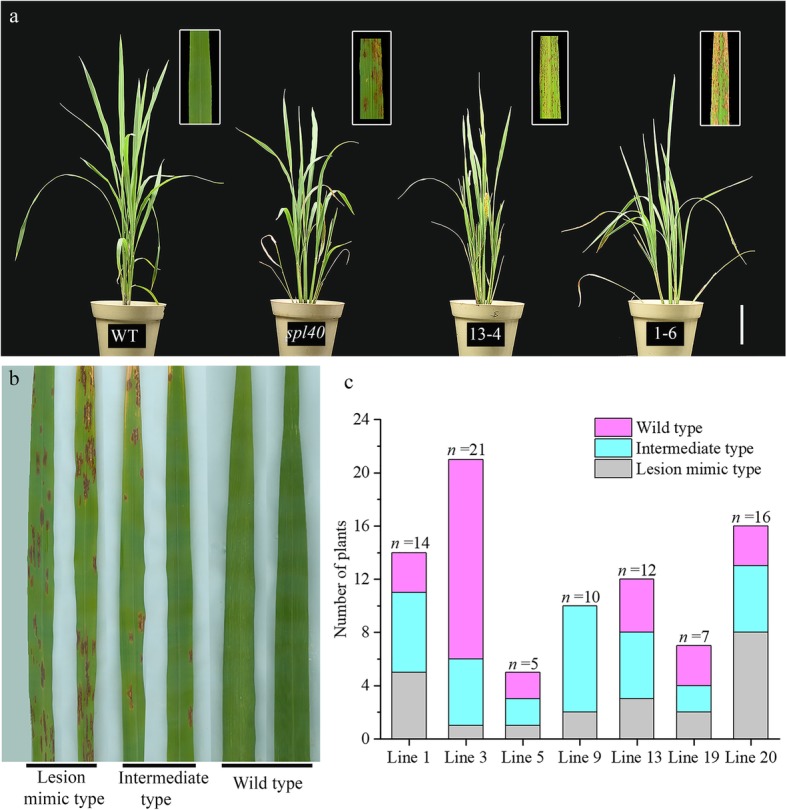
T1 transgenic complementation plants. a Phenotype of WT, spl40 and T1 complementation plants (13–4 and 1–6). b T1 leaves representing three phenotypic categories viz., lesion mimic type, intermediate type and wild type. c Distribution of T1 plants in three phentoypic categories
Discussion
Spotted-leaf mutants are generally considered as LMM mutants, owing to the nature of cell necrosis they show similar to that caused by hypersensitive response (HR), a type of programmed cell death (PCD) induced by pathogen invasion (Walbot et al. 1983). In present study, we have identified a novel spotted-leaf mutant spl40 that showed lesion development similar to the HR. The mutation was controlled by a single nuclear recessive gene located on the long arm of chromosome 5. In rice, a number of spotted-leaf mutants are affected in agronomic as well as physiological processes/traits (Huang et al. 2011; Feng et al. 2013; Chen et al. 2018). For example, Spl24, was affected in major physiological and agronomic traits like photosynthetic pigment content, plant height, seed setting rate and 1000-grain weight (Chen et al. 2018). Similar to this, spl40 mutant showed decrease in photosynthetic pigment content and content of soluble protein, as well as decrease in important agronomic parameters like number of filled grains per panicle, number of panicle per plant, seed setting rate and 1000-grain weight.
Till now, many LMMs have shown to exhibit light dependent appearance of lesions (Wang et al. 2015; Xiao et al. 2015). The rice LIL1 mutant shows remarkable decrease in lesions when the light was intercepted by covering the leaf with aluminium foil (Zhou et al. 2017). Similarly, both rice Spl24 and HM47 spotted-leaf mutants also exhibit light-induced formation of lesions under natural light conditions (Feng et al. 2013; Chen et al. 2018). In spl40, lesion development was suppressed in the absence of light and appeared again when the light was re-instated. Though, at this point it is unknown which light dependent pathway is responsible for the phenomenon, but it is certain that light acts as a trigger for lesion development in spl40.
Lesions formed in spotted-leaf mutants are similar to the HR lesions caused upon pathogen attack. HR in response to pathogen attack involves rapid death of plant cells in vicinity of pathogen infection to prevent the further spread (Lorrain et al. 2003). Evans blue staining, an indicator of cell death, showed the occurrence of cell death after the lesion development in spl40. This was supported by the elevated expression of OsMC genes that are important regulators of PCD in plants. ROS signaling plays an important role in HR, and ROS generation is closely associated with localized cell death (Zurbriggen et al. 2010). As indicated by DAB and NBT staining, spl40 mutant showed burst of ROS like H2O2 and superoxide (O2−). Number of reports suggests that ROS can function as a regulator of cell death (Jabs et al. 1996; Overmyer et al. 2000; Mateo 2004; Danon et al. 2005; Li et al. 2013; Kaurilind et al. 2015). Arabidopsis lsd1 mutants exhibited impaired control of cell death in the absence of pathogen and, superoxide was necessary and sufficient to initiate lesion formation (Jabs et al. 1996). Superoxide accumulated before the onset of cell death and subsequently in live cells adjacent to spreading lsd1 lesions. Another Arabidopsis mutant, rcd1 also showed superoxide dependent propagation of lesions (Overmyer et al. 2000). In rcd1, ethylene dependent cellular superoxide accumulation occurred ahead of expanding lesions before visible symptoms appear and exogenous application of ethylene increased superoxide-dependent cell death. We speculate that higher superoxide accumulation in spl40 before the onset of lesion might be acting as a trigger to induce cell death. NOX1, encoding NADPH oxidase which is one of the main ROS source, was highly expressed in spl40. Plants detoxify the harmful effects of hyper-accumulated ROS using ROS scavenging system comprising of important enzymes like CAT, SOD and POD. In this study, the activities of ROS scavenging enzymes were significantly higher in spl40 mutant. Elevated expression of genes involved in ROS scavenging enzyme synthesis was in concurrent with higher activity of ROS scavenging enzymes. ROS burst along with increased activities of ROS scavenging enzymes indicate that the overall ROS homeostasis was disturbed in spl40 mutant. In addition to this, senescence related genes were differentially expressed in the mutant as well. More importantly, the SGR gene was highly expressed in spl40, which is known to involve in chlorophyll degradation (Lee et al. 2018). This indicates that the activation of chlorophyll degradation genes might be responsible for decrease in chlorophyll level in spl40.
Spotted-leaf mutants often show enhanced resistance to pathogen infection along with constitutive expression of defense response genes (Yin et al. 2000; Mizobuchi et al. 2002; Wu et al. 2008; Xu et al. 2018). In a rice spl21 mutant, upregulated expression of pathogenesis-related genes and increased level of jasmonic acid leads to enhanced resistance to bacterial blight pathogen Xanthomonas oryzae pv. oryzae (Xu et al. 2018). In present study, spl40 mutant showed enhanced resistance to 14 races of this pathogen. This was supported by upregulated expression of important genes involved in JA and SA signalling pathways. Therefore we conclude that spl40 confers enhanced resistance to bacterial blight pathogen through activation of SA and JA metabolic pathway. But the specific mechanism and the factors involved in the resistance remain to be identified, and this will lead to the future course of this research.
MutMap is a powerful tool that employs whole genome resequencing approach to rapidly identify the causal mutations (Abe et al. 2012; Fekih et al. 2013; Takagi et al. 2013, 2015). Genetic mapping combined with MutMap analysis revealed that Os05G0312000 harbored a single base substitution in the 10th exon that resulted in change in the amino acid residue from leucine to phenylalanine. Although, we could not test the expression of large number ORFs present in the mapping region (about 101 ORFs in 523 kb region), based on high-throughput sequencing results we considered that Os05G0312000 is the most likely candidate gene and decided to carry out functional complementation test. Functional complementation with WT allele was carried out and the T0 transgenic plants did not develop the phenotype till the tillering stage, while spl40 showed the phenotype at the seedling (four leaf) stage. Unexpectedly, the lesion mimic phenotype was observed at the beginning of the heading stage in the T0 plants. Further, T1 progenies consisted of three categories based on the phenotype viz., lesion mimic type, intermediate type and wild type. Though, through functional complementation, we confirmed that Os05G0312000 was likely the correct candidate gene, its genetic nature seems complex. In silico search in Rice Annotation Project Database (http://rice.plantbiology.msu.edu/) showed that Os05G0312000 is predicted to function as a subunit (Med5_1) of the Mediator complex in Arabidopsis. Mediator is a master transcriptional regulator in eukaryotes that serves as a molecular bridge between gene-specific transcription factors bound at enhancers, and RNA polymerase II (RNA pol II) (Flanagan et al. 1991; Carlsten et al. 2013; Buendía-Monreal and Gillmor 2016). It consist of 25 subunits in yeast and approximately 34 subunits in plants, which reside in four distinct modules, termed head, middle, tail and CDK8/kinase (Samanta and Thakur 2015). Bäckström et al. (2007) successfully carried out the purifcation of Mediator complex in Arabidopsis and showed that specific plant Mediator subunits were linked to the regulation of specialized processes such as the control of cell proliferation and the regulation of flowering time in response to light quality. In plants, Mediator has been found to play important roles in diverse aspects of plant life right from growth and development upto the response against biotic and abiotic stresses (Samanta and Thakur 2015). In the present study, we figure that dysfunction of the Mediator indirectly leads to disturbed ROS homeostasis and upregulated expression of cell death related genes which might finally contribute to the occurrence of cell death in spl40. However, the T1 heterozygotes with a similar expression level of WT allele displaying various intensity of lesions are yet to be further invertigated. Furthermore, the complicated genetic nature of spl40 is also evident from the fact that an allelic Nipponbare mutant spl40NIP with single base substitution in the same locus (Os05G0312000) exhibited different phenotype compared with spl40 (Additional file 2: Figure S2). We are considering that the locus may possess complicated biological function possibly associated with genotypes because of the diverse phenotypes. Our laboratory is currently examining the differences in background by exploring the differences in spl40 allele in different cultivars.
Conclusions
Our study demonstrated that spl40 was a novel spotted leaf mutant compromised in number of traits such as poor agronomic performance with altered photosynthetic capacity, HR-like cell death and disturbed ROS homeostasis. In addition, spl40 showed enhanced disease resistance to bacterial blight and immunity-associated senescence. This study would facilitate the efforts to unravel the mechanism of lesion development involving programmed cell death and defense responses.
Methods
Plant materials and growth conditions
A spotted-leaf mutant spl40 was isolated from an ethyl methane sulfonate (EMS)-induced mutant bank of Zhongjian100, an indica rice. The mutant, Zhongjian-100 (wild-type, WT) and F2 populations were grown in the paddy field in summer of 2016 at China National Rice Research Institute (CNRRI) in Hangzhou, Zhejiang, China. The back-cross F2 population derived from the cross spl40/Zhongjian-100 was used for genetic analysis and the spl40/Nipponbare derived F2 population was used for gene mapping. The WT and spl40 were grown for evaluation of agronomic traits including plant height, tiller number, number of panicle/plant, number of filled grain/panicle, seed setting rate and 1000-grain weight. Agronomic traits were recorded at maturity stage on three individual plants, and the means from three replicates were used for analysis.
Histochemical analysis
Leaf samples from WT and spl40 were collected at the tillering stage. Evans blue staining and nitro blue tetrazolium (NBT) staining was performed to determine the occurrence of cell death and accumulation of superoxide (O2−) (Liu et al. 2008; Qiao et al. 2010). The detection of H2O2 was carried out by using 3, 3′-Diaminobenzidine (DAB) staining as previously described (Thordal-Christensen et al. 1997).
Shading experiment
At the tillering stage, the top second leaf of WT and spl40 was used to carry out the shading experiment. The mutant leaves in which lesions have started to appear at the tip, were shaded with a piece of 2–3 cm foil for 7 days under natural light conditions. The foil was then removed and light was reinstated for 5–15 days to investigate the influence of natural light on the initiation of lesions. Leaves were photographed by a scanner.
Measurement of physico-biochemical parameters
The contents of Chlorophyll a (Chl a), chlorophyll b (Chl b) and carotenoid were determined according to He et al. (2018). The contents of malonaldehyde (MDA) and soluble protein, and the enzymatic activities of peroxidase (POD), superoxide dismutase (SOD) and catalase (CAT) were measured following the manufacturer’s instructions (Nanjing Jiancheng Bioengineering Institute, Nanjing, China). Enzymatic activity analysis of POD, SOD, CAT and measurements of soluble protein and MDA content were carried out at the seedling and tillering stages. Photosynthesis related parameters such as net photosynthesis rate (Pn), stomatal conductance (Gs), intercellular CO2 concentration (Ci) and transpiration rate (Tr) were measured in the field by using portable L-6400XT (LI_COR, Lincoln, NB, USA). Means of three replicates were used for analysis. The membrane ion leakage measurement was carried out according to He et al. (2018). The means from three measurement were used for analysis.
RNA extraction and gene expression analysis
The total RNA was isolated from the leaves of spl40, T1 complementary plants and WT using NucleoZOL Reagent Kit according to the manufacturer’s instructions (MACHERY-NAGEL, Düren, Germany). RNA samples were treated with DNase (Promega), and first strand copy of DNA (cDNA) was synthesized using 1 μg of RNA. Real-time fluorescent quantitative PCR was carried out using SYBR Premix ExTag™II (Tli RNaseH plus) master mix (Takara, Kusatsu, Japan) and performed on Thermal Cycler Dice® Real Time System (Takara, Kusatsu, Japan). The rice Ubiquitin was used as the internal control to normalize expression levels. Three biological repeats were used to obtain the final results. The relevant sequences of primer sets used in different experiments are listed in Additional file 1: Table S2. To determine the expression level of WT allele in the three categories of T1 complementary plants (WT, intermediate, and lesion mimic type), the m40-D primer pairs (m40-D F: GCTGCATGCTCTCATATC, m40-D R: GGAAATGAGTCAATATACA) located at exon 9 and exon 10 respectively were used to amplify the cDNA region covering the mutation site, and the RT-PCR products were cloned into the vector pMD18-T. A total of 75 colonies (3 replicates with 25 colonies/replicate) from each category of T1 plants were selected and used for sequencing. The frequncy of WT allele was used to determine the expression level of WT allele in T1 plants. Means of three biological replicates were subjected to one-way ANOVA followed by the Duncan multiple range test.
Disease resistance evaluation
Rice plants of spl40, WT and susceptible control IR24 were inoculated with 16 races (PXO79, PXO145, C5, PXO339, PXO341, PXO349, PXO99, PXO347, Zhe173, PXO340, C6, C3, PXO71, OS225, C2 and PXO112) of Xanthomonas oryzae pv. oryzae (Xoo) by the leaf-clipping method (Kauffman 1973) at the tillering stage. The bacterial cultures were separately suspended in distilled water and adjusted to OD600 = 1.0 for inoculation. Pathogen inoculation was carried out on fully expanded second youngest leaf. Three individual plants and three leaves per plant were inoculated for each race. Disease evaluation was carried out 20 days after inoculation by measuring the legion length. Means from nine leaves were used for analysis.
Genetic analysis and gene mapping
The F1 plants from the cross between spl40/Zhongjian100 were grown in the paddy field at CNRRI to determine the genetic nature (dominant/recessive) of spl40. The subsequent F2 individuals derived from selfing were used for segregation analysis. To map the mutation, a population derived from the cross spl40/Nipponbare was used. Equal amount of leaf tissues from 10 WT and 10 mutant plants were collected for DNA extraction to make the respective DNA pools. Bulk segregant analysis using the DNA pools were used to rapidly map the mutation. The F2 individuals with mutant phenotype were used for fine mapping of the mutation. Plant genomic DNA was isolated according to the previous protocol (Edwards et al. 1991). Simple sequence repeat (SSR) markers were obtained from the website (http://www.gramene.org/). For the insertion/deletion (InDel) markers, the genomic seqeunces of japonica cultivar Nipponbare and indica cultivar 9311 from the public databases RGP (http://rgp.dna.affrc.go.jp/E/toppage.html), Gramene (http://gramene.org/genome_ browser/index.html) and the Gene Research Center of the Chinese Academy of Sciences (http://rice.genomics.org.cn/rice/index2.jsp) were compared and markers were designed by using NCBI Primer-BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast/). The primers were synthesized by Sangon Biotech Co. Ltd. (Shanghai, China). PCR reaction and detection were carried out as described previously (Feng et al. 2013). The relevant primer sequences for gene mapping are listed in Additional file 1: Table S3.
For the high thoughput sequencing by a modified MutMap approach, spl40 was back-crossed to Zhongjian100 and then self crossed to generate BC1F2 population. The WT DNA and the pooled DNA of 25 mutant type individuals were used for high throughput sequencing analysis with 25 average read depth to determine SNPs between WT and the mutant (AnoRoad Genome, Beijing). The SNP index is calculated based on the frequency of mutant type SNP and the SNP with index of 1 is considered as the candidate SNP (Abe et al. 2012).
Vector contruction for functional complementation
The genomic DNA sequence containing the entire coding sequence SPL40 (12,419-bp), including 1966-bp upstream and 1392-bp downstream sequence was divided into four parts so as to clone it into pCAMBIA1300 vector. The four fragments were amplified using respective primers (Additional file 1: Table S4) and cloned into pMD19T(simple) vector. After sequencing, the fragments were sequentially cloned into the pCAMBIA1300 vector. Trangenic plants were developed via Agrobacterium-mediated plant transformation and regeneration (Hiei and Komari 2008).
Additional files
Table S1. Genotype of the transgene of WT allele in T1 progenies from three lines. Table S2. RT-PCR primers. Table S3. Markers used for fine mapping and sequencing of spl40 locus. Table S4. Primers used for making complementation construct. (DOCX 31 kb)
Figure S1. Frequency of WT SPL40 allele in the T1 generation of complementary plants. Wild type (W), intermediate type (I) and Lesion mimic type(L). Values are means ± SD of three biological repeats and re subjected to one way ANOVA followed by the Duncan multiple range test. Figure S2. Phenotype of spl40 and spl40NIP. (a) Phenotype of spl40; (b) Phenotype of spl40NIP. (PDF 255 kb)
Acknowledgements
Not applicable.
Abbreviations
- APX
Ascorbate peroxidase
- CAT
Catalase
- Chl a
Chlorophyll a
- Chl b
Chlorophyll b
- Ci
Intercellular CO2 concentration
- DAB
3,3′-Diaminobenzidine
- Gs
Stomatal conductance
- HR
Hypersensitive response
- InDel
Insertion/deletion marker
- JA
Jasmonic acid
- LMM
Lesion mimic mutant
- MDA
Malonaldehyde
- NBT
Nitro blue tetrazolium
- NOX
NADPH oxidase
- ORF
Open reading frame
- PAO
Polyamine oxidase
- PCD
Programmed cell death
- Pn
Net photosynthesis rate
- POD
Peroxidase
- ROS
Reactive oxygen species
- SA
Salicylic acid
- SNP
Single nucleotide polymorphism
- SOD
Superoxide dismutase
- spl40
Spotted-leaf mutant 40
- SSR
Simple sequence repeat marker
- Tr
Transpiration rate
- WT
Wild type
- Xoo
Xanthomonas oryzae pv. oryzae
Authors’ contributions
XZ and JW conceived and designed the research, AS, XS, ZC and TC performed the experiments, AS, CZ, XW and XZ carried out the data analysis, AS, ST and JW wrote and revised the manuscript. All authors read and approved the manuscript.
Funding
This work was supported by the Ministry of Science and Technology of China (2016YFD0101104) and China National Transgenic Program (2016ZX08001–002).
Availability of data and materials
Not applicable.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Atul Prakash Sathe, Email: atulsathe@163.com.
Xiaona Su, Email: suxiaona1990@163.com.
Zheng Chen, Email: chenzheng7@126.com.
Ting Chen, Email: chenting218@163.com.
Xiangjing Wei, Email: weixiangjing@caas.cn.
Shaoqing Tang, Email: sqtang@126.com.
Xiao-bo Zhang, Phone: 86-571-63370295, Email: hello.xiaobo@163.com.
Jian-li Wu, Phone: 86-571-63370326, Email: beishangd@163.com.
References
- Abe A, Kosugi S, Yoshida K, Natsume S, Takagi H, Kanzaki H, Matsumura H, Yoshida K, Mitsuoka C, Tamiru M, Innan H, Cano L, Kamoun S, Terauchi R. Genome sequencing reveals agronomically important loci in rice using MutMap. Nat Biotechnol. 2012;30:174–178. doi: 10.1038/nbt.2095. [DOI] [PubMed] [Google Scholar]
- Bäckström S, Elfving N, Nilsson R, Wingsle G, Björklund S. Purification of a plant mediator from Arabidopsis thaliana identifies PFT1 as the Med25 subunit. Mol Cell. 2007;26:717–729. doi: 10.1016/j.molcel.2007.05.007. [DOI] [PubMed] [Google Scholar]
- Buendía-Monreal M, Gillmor CS. Mediator: a key regulator of plant development. Dev Biol. 2016;419:7–18. doi: 10.1016/j.ydbio.2016.06.009. [DOI] [PubMed] [Google Scholar]
- Carlsten JOP, Zhu X, Gustafsson CM. The multitalented mediator complex. Trends Biochem Sci. 2013;38:531–537. doi: 10.1016/j.tibs.2013.08.007. [DOI] [PubMed] [Google Scholar]
- Chen X, Hao L, Pan J, Zheng X, Jiang G, Jin Y, Gu Z, Qian Q, Zhai W, Ma B. SPL5, a cell death and defense-related gene, encodes a putative splicing factor 3b subunit 3 (SF3b3) in rice. Mol Breed. 2012;30:939–949. doi: 10.1007/s11032-011-9677-4. [DOI] [Google Scholar]
- Chen Z, Chen T, Sathe AP, He Y, Zhang XB, Wu JL. Identification of a novel semi-dominant spotted-leaf mutant with enhanced resistance to Xanthomonas oryzae pv. oryzae in rice. Int J Mol Sci. 2018;19:3766. doi: 10.3390/ijms19123766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl JL, Jones JD. Plant pathogens and integrated defence responses to infection. Nature. 2001;411:826–833. doi: 10.1038/35081161. [DOI] [PubMed] [Google Scholar]
- Danon A, Miersch O, Felix G, RGL o d C, Apel K. Concurrent activation of cell death-regulating signaling pathways by singlet oxygen in Arabidopsis thaliana. Plant J. 2005;41:68–80. doi: 10.1111/j.1365-313X.2004.02276.x. [DOI] [PubMed] [Google Scholar]
- Edwards K, Johnstone C, Thompson C. A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 1991;19:1991. doi: 10.1093/nar/19.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekih R, Takagi H, Tamiru M, Abe A, Natsume S, Yaegashi H, Sharma S, Sharma S, Kanzaki H, Matsumura H, Saitoh H, Mitsuoka C, Utsushi H, Uemura A, Kanzaki E, Kosugi S, Yoshida K, Csno L, Kamoun S, Terauchi R. MutMap+: genetic mapping and mutant identification without crossing in rice. PLoS One. 2013;8:1–10. doi: 10.1371/journal.pone.0068529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng BH, Yang Y, Shi YF, Shen HC, Wang HM, Huang QN, Xu X, Lü XG, Wu JL. Characterization and genetic analysis of a novel rice spotted-leaf mutant HM47 with broad-spectrum resistance to Xanthomonas oryzae pv. oryzae. J Integr Plant Biol. 2013;55:473–483. doi: 10.1111/jipb.12021. [DOI] [PubMed] [Google Scholar]
- Flanagan PM, Kelleher RJ, Sayre MH, Tschochner S, Kornberg RD. A mediator required for activation of RNA polymerase II transcription in vitro. Nature. 1991;354:436–438. doi: 10.1038/350436a0. [DOI] [PubMed] [Google Scholar]
- He Y, Zhang Z, Li L, Tang S, Wu JL. Genetic and physio-biochemical characterization of a novel premature senescence leaf mutant in rice (Oryza sativa L.) Int J Mol Sci. 2018;19:2339. doi: 10.3390/ijms19082339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiei Y, Komari T. Agrobacterium-mediated transformation of rice using immature embryos or calli induced from mature seed. Nat Protoc. 2008;3:824–834. doi: 10.1038/nprot.2008.46. [DOI] [PubMed] [Google Scholar]
- Huang L, Zhang H, Hong Y, Liu S, Li D, Song F. Stress-responsive expression, subcellular localization and protein–protein interactions of the rice metacaspase family. Int J Mol Sci. 2015;16:16216–16241. doi: 10.3390/ijms160716216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang QN, Shi YF, Yang Y, Feng BH, Wei YL, Chen J, Baraoidan M, Leung H, Wu JL. Characterization and genetic analysis of a light- and temperature-sensitive spotted-leaf mutant in rice. J Integr Plant Biol. 2011;53:671–681. doi: 10.1111/j.1744-7909.2011.01056.x. [DOI] [PubMed] [Google Scholar]
- Huang QN, Yang Y, Shi YF, Chen J, Wu JL. Spotted-leaf mutants of rice (Oryza sativa) Rice Sci. 2010;17:247–256. doi: 10.1016/S1672-6308(09)60024-X. [DOI] [Google Scholar]
- Jabs T, Dietrich RA, Dangl JL. Initiation of runaway cell death in an Arabidopsis mutant by extracellular superoxide. Science (80) 1996;273:1853 LP-1856. doi: 10.1126/science.273.5283.1853. [DOI] [PubMed] [Google Scholar]
- Kamlofski CA, Antonelli E, Bender C, Jaskelioff M, Danna CH, Ugalde R, Acevedo A. A lesion-mimic mutant of wheat with enhanced resistance to leaf rust. Plant Pathol. 2007;56:46–54. doi: 10.1111/j.1365-3059.2006.01454.x. [DOI] [Google Scholar]
- Kauffman HE. An improved technique for evaluating resistance of rice varieties to Xanthomonas oryzae. Plant Dis Rep. 1973;57:537–541. [Google Scholar]
- Kaurilind E, Xu E, Brosché M. A genetic framework for H2O2 induced cell death in Arabidopsis thaliana. BMC Genomics. 2015;16:1–17. doi: 10.1186/s12864-015-1964-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam E, Kato N, Lawton M. Programmed cell death, mitochondria and the plant hypersensitive response. Nature. 2001;411:848–853. doi: 10.1038/35081184. [DOI] [PubMed] [Google Scholar]
- Lee D, Lee G, Kim B, Jang S, Lee Y, Yu Y, Seo J, Kim S, Lee YH, Lee J, Kim S, Koh HJ. Identification of a Spotted Leaf Sheath gene involved in early senescence and defense response in rice. Front Plant Sci. 2018;9:1–12. doi: 10.3389/fpls.2018.01274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Chen L, Mu J, Zuo J. LESION SIMULATING DISEASE1 interacts with catalases to regulate hypersensitive cell death in Arabidopsis. Plant Physiol. 2013;163:1059–1070. doi: 10.1104/pp.113.225805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Wang Y, Xu J, Su T, Liu G, Ren D. Ethylene signaling is required for the acceleration of cell death induced by the activation of AtMEK5 in Arabidopsis. Cell Res. 2008;18:422–432. doi: 10.1038/cr.2008.2. [DOI] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Czymmek K, Tallóczy Z, Levine B, Dinesh-Kumar SP. Autophagy regulates programmed cell death during the plant innate immune response. Cell. 2005;121:567–577. doi: 10.1016/j.cell.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Lorrain S, Vailleau F, Balagué C, Roby D. Lesion mimic mutants: keys for deciphering cell death and defense pathways in plants? Trends Plant Sci. 2003;8:263–271. doi: 10.1016/S1360-1385(03)00108-0. [DOI] [PubMed] [Google Scholar]
- Ma JY, Chen SL, Zhang JH, Dong YJ, Teng S. Identification and genetic mapping of a lesion mimic mutant in rice. Rice Sci. 2012;19:1–7. doi: 10.1016/S1672-6308(12)60013-4. [DOI] [Google Scholar]
- Manosalva PM, Bruce M, Leach JE. Rice 14-3-3 protein (GF14e) negatively affects cell death and disease resistance. Plant J. 2011;68:777–787. doi: 10.1111/j.1365-313X.2011.04728.x. [DOI] [PubMed] [Google Scholar]
- Marino D, Peeters N, Rivas S. Ubiquitination during plant immune signaling. Plant Physiol. 2012;160:15–27. doi: 10.1104/pp.112.199281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateo A. LESION SIMULATING DISEASE 1 is required for acclimation to conditions that promote excess excitation energy. Plant Physiol. 2004;136:2818–2830. doi: 10.1104/pp.104.043646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizobuchi R, Hirabayashi H, Kaji R, Nishizawa Y, Satoh H, Ogawa T, Okamoto M. Differential expression of disease resistance in rice lesion-mimic mutants. Plant Cell Rep. 2002;21:390–396. doi: 10.1007/s00299-002-0525-1. [DOI] [Google Scholar]
- Mori M, Tomita C, Sugimoto K, Hasegawa M, Hayashi N, Dubouzet JG, Ochiai H, Sekimoto H, Hirochika H, Kikuchi S. Isolation and molecular characterization of a Spotted leaf 18 mutant by modified activation-tagging in rice. Plant Mol Biol. 2007;63:847–860. doi: 10.1007/s11103-006-9130-y. [DOI] [PubMed] [Google Scholar]
- Overmyer K, Tuominen H, Kettunen R, Betz C, Langebartels C, Sandermann H, Kangasjärvi J. Ozone-sensitive Arabidopsis rcd1 mutant reveals opposite roles for ethylene and jasmonate signaling pathways in regulating superoxide-dependent cell death. Plant Cell. 2000;12:1849–1862. doi: 10.2307/3871197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Y, Jiang W, Lee J, Park B, Choi MS, Piao R, Woo MO, Roh JH, Han L, Paek NC, Seo HS, Koh HJ. SPL28 encodes a clathrin-associated adaptor protein complex 1, medium subunit μ1 (AP1M1) and is responsible for spotted leaf and early senescence in rice (Oryza sativa) New Phytol. 2010;185:258–274. doi: 10.1111/j.1469-8137.2009.03047.x. [DOI] [PubMed] [Google Scholar]
- Rostoks N, Schmierer D, Mudie S, Drader T, Brueggeman R, Caldwell DG, Waugh R, Kleinhofs A. Barley necrotic locus nec1 encodes the cyclic nucleotide-gated ion channel 4 homologous to the Arabidopsis HLM1. Mol Gen Genomics. 2006;275:159–168. doi: 10.1007/s00438-005-0073-9. [DOI] [PubMed] [Google Scholar]
- Samanta S, Thakur JK. Importance of mediator complex in the regulation and integration of diverse signaling pathways in plants. Front Plant Sci. 2015;6:1–16. doi: 10.3389/fpls.2015.00757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simanshu DK, Zhai X, Munch D, Hofius D, Markham JE, Bielawski J, Bielawska A, Malinina L, Molotkovsky JG, Mundy JW, Patel DJ, Brown RE. Arabidopsis accelerated cell death 11, ACD11, is a ceramide-1-phosphate transfer protein and intermediary regulator of phytoceramide levels. Cell Rep. 2014;6:388–399. doi: 10.1016/j.celrep.2013.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sindhu A, Janick-buckner D, Buckner B, Gray J, Zehr U, Dilkes BP, Johal GS. Propagation of cell death in dropdead1, a sorghum ortholog of the maize lls1 mutant. PLoS One. 2018;13(9):e0201359. doi: 10.1371/journal.pone.0201359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi H, Tamiru M, Abe A, Yoshida K, Uemura A, Yaegashi H, Obara T, Oikawa K, Utsushi H, Kanzaki E, Mitsuoka C, Natsume S, Kosugi S, Kanzaki H, Matsumura H, Urasaki N, Kamoun S, Terauchi R. MutMap accelerates breeding of a salt-tolerant rice cultivar. Nat Biotechnol. 2015;33:445–449. doi: 10.1038/nbt.3188. [DOI] [PubMed] [Google Scholar]
- Takagi H, Uemura A, Yaegashi H, Tamiru M, Abe A, Mitsuoka C, Utsushi H, Natsume S, Kanzaki H, Matsumura H, Saitoh H, Yoshida K, Cano LM, Kamoun S, Terauchi R. MutMap-gap: whole-genome resequencing of mutant F2 progeny bulk combined with de novo assembly of gap regions identifies the rice blast resistance gene Pii. New Phytol. 2013;200:276–283. doi: 10.1111/nph.12369. [DOI] [PubMed] [Google Scholar]
- Takahashi A, Agrawal GK, Yamazaki M, Onosato K, Miyao A, Kawasaki T, Shimamoto K, Hirochika H. Rice Pti1a negatively regulates RAR1-dependent defense responses. Plant Cell. 2007;19:2940–2951. doi: 10.1105/tpc.106.047142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi A, Kawasaki T, Henmi K, Shii K, Kodama O, Satoh H, Shimamoto K. Lesion mimic mutants of rice with alterations in early signaling events of defense. Plant J. 1999;17:535–545. doi: 10.1046/j.1365-313X.1999.00405.x. [DOI] [PubMed] [Google Scholar]
- Tang C, Wang X, Duan X, Wang X, Huang L, Kang Z. Functions of the lethal leaf-spot 1 gene in wheat cell death and disease tolerance to Puccinia striiformis. J Exp Bot. 2013;64:2955–2969. doi: 10.1093/jxb/ert135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thordal-Christensen H, Zhang Z, Wei Y, Collinge DB. Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J. 1997;11:1187–1194. doi: 10.1046/j.1365-313X.1997.11061187.x. [DOI] [Google Scholar]
- Walbot V, Hoisington DA, Neuffer MG. Disease lesion mimic mutations. In: Kosuge T, Meredith CP, Hollaender A, Wilson CM, editors. Genetic engineering of plants. Basic life sciences. Boston: Springer; 1983. [Google Scholar]
- Wang F, Wu W, Wang D, Yang W, Sun J, Liu D, Zhang A. Characterization and genetic analysis of a novel light-dependent lesion mimic mutant, lm3, showing adult-plant resistance to powdery mildew in common wheat. PLoS One. 2016;11(5):e0155358. doi: 10.1371/journal.pone.0155358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Ye B, Yin J, Yuan C, Zhou X, Li W, He M, Wang J, Chen W, Qin P, Ma B, Wang Y, Li S, Chen X. Characterization and fine mapping of a light-dependent leaf lesion mimic mutant 1 in rice. Plant Physiol Biochem. 2015;97:44–51. doi: 10.1016/j.plaphy.2015.09.001. [DOI] [PubMed] [Google Scholar]
- Wang L, Han S, Zhong S, Wei H, Zhang Y, Zhao Y, Liu B. Characterization and fine mapping of a necrotic neaf mutant in maize (Zea mays L.) J Genet Genomics. 2013;40:307–314. doi: 10.1016/j.jgg.2013.04.004. [DOI] [PubMed] [Google Scholar]
- Wang L, Pei Z, Tian Y, He C. OsLSD1, a rice zinc finger protein, regulates programmed cell death and callus differentiation. Mol Plant-Microbe Interact. 2005;18:375–384. doi: 10.1094/MPMI-18-0375. [DOI] [PubMed] [Google Scholar]
- Wu C, Bordeos A, Madamba MRS, Baraoidan M, Ramos M, Wang GL, Leach JE, Leung H. Rice lesion mimic mutants with enhanced resistance to diseases. Mol Gen Genomics. 2008;279:605–619. doi: 10.1007/s00438-008-0337-2. [DOI] [PubMed] [Google Scholar]
- Xiao GQ, Zhang HW, Lu XY, Huang RF. Characterization and mapping of a novel light-dependent lesion mimic mutant lmm6 in rice (Oryza sativa L.) J Integr Agric. 2015;14:1687–1696. doi: 10.1016/S2095-3119(14)60975-8. [DOI] [Google Scholar]
- Xu X, Chen Z, Shi YF, Wang HM, He Y, Shi L, Chen T, Wu JL, Zhang XB. Functional inactivation of OsGCNT induces enhanced disease resistance to Xanthomonas oryzae pv. oryzae in rice. BMC Plant Biol. 2018;18:1–15. doi: 10.1186/s12870-018-1489-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanouchi U, Yano M, Lin H, Ashikari M, Yamada K. A rice spotted leaf gene, Spl7, encodes a heat stress transcription factor protein. Proc Natl Acad Sci U S A. 2002;99:7530–7535. doi: 10.1073/pnas.112209199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Z, Chen J, Zeng L, Goh M, Leung H, Khush GS, Wang GL. Characterizing rice lesion mimic mutants and identifying a mutant with broad-spectrum resistance to rice blast and bacterial blight. Mol Plant-Microbe Interact. 2000;13:869–876. doi: 10.1094/MPMI.2000.13.8.869. [DOI] [PubMed] [Google Scholar]
- Zeng LR, Qu S, Bordeos A, Yang C, Baraoidan M, Yan H, Xie Q, Nahm BH, Leung H, Wang GL. Spotted leaf11, a negative regulator of plant cell death and defense, encodes a U-box/armadillo repeat protein endowed with E3 ubiquitin ligase activity. Plant Cell. 2004;16:2795–2808. doi: 10.1105/tpc.104.025171.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XB, Feng BH, Wang HM, Xu X, Shi YF, He Y, Chen Z, Sathe AP, Shi L, Wu JL. A substitution mutation in OsPELOTA confers bacterial blight resistance by activating the salicylic acid pathway. J Integr Plant Biol. 2018;60:160–172. doi: 10.1111/jipb.12613. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Zhang Z, Liu T, Gao B, Xiong X. Identification and map-based cloning of the light-induced lesion mimic mutant 1 (LIL1) gene in rice. Front Plant Sci. 2017;8:1–11. doi: 10.3389/fpls.2017.02122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Liao H, Chern M, Yin J, Chen Y, Wang J, Zhu X, Chen Z, Yuan C, Zhao W, Wang J, Li W, He M, Ma B, Wang J, Qin P, Chen W, Wang Y, Liu J, Qian Y, Wang W, Wu X, Li P, Zhu L, Li S, Ronald PC, Chen X. Loss of function of a rice TPR-domain RNA-binding protein confers broad-spectrum disease resistance. Proc Natl Acad Sci U S A. 2018;115:3174–3179. doi: 10.1073/pnas.1705927115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurbriggen MD, Carrillo N, Hajirezaei MR. ROS signaling in the hypersensitive response. Plant Signal Behav. 2010;5:393–396. doi: 10.4161/psb.5.4.10793. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Genotype of the transgene of WT allele in T1 progenies from three lines. Table S2. RT-PCR primers. Table S3. Markers used for fine mapping and sequencing of spl40 locus. Table S4. Primers used for making complementation construct. (DOCX 31 kb)
Figure S1. Frequency of WT SPL40 allele in the T1 generation of complementary plants. Wild type (W), intermediate type (I) and Lesion mimic type(L). Values are means ± SD of three biological repeats and re subjected to one way ANOVA followed by the Duncan multiple range test. Figure S2. Phenotype of spl40 and spl40NIP. (a) Phenotype of spl40; (b) Phenotype of spl40NIP. (PDF 255 kb)
Data Availability Statement
Not applicable.