Abstract
We report on the development of bioinspired cardiac scaffolds made from electroconductive acid-modified silk fibroin-poly(pyrrole) (AMSF+PPy) substrates patterned with nanoscale ridges and grooves reminiscent of native myocardial extracellular matrix (ECM) topography to enhance the structural and functional properties of cultured human pluripotent stem cells (hPSC)-derived cardiomyocytes. Nanopattern fidelity was maintained throughout the fabrication and functionalization processes, and no loss in conductive behavior occurred due to the presence of the nanotopographical features. AMSF+PPy substrates were biocompatible and stable, maintaining high cell viability over a 21-day culture period while displaying no signs of PPy delamination. The presence of anisotropic topographical cues led to increased cellular organization and sarcomere development, and electroconductive cues promoted a significant improvement in the expression and polarization of connexin 43 (Cx43), a critical regulator of cell-cell electrical coupling. The combination of biomimetic topography and electroconductivity also increased the expression of genes that encode key proteins involved in regulating the contractile and electrophysiological function of mature human cardiac tissue.
Keywords: electroconductivity, nanotopography, silk fibroin, polypyrrole, stem cell-derived cardiomyocyte, cardiac tissue engineering
Graphical Abstract
Imparting electroconductive and nanotopographical cues to biodegradable silk-fibroin films enhanced the maturation of cultured human stem cell-derived cardiomyocytes.
1. Introduction
In recent years there has been an increasing number of studies in which conductive materials are applied as scaffolds for engineering a variety of tissue types.1 Specifically, materials such as graphene,2, 3 carbon nanotubes,4–6 gold,7–9 and polyaniline (PANi),10–12 have all been utilized as promising candidates for cardiac tissue engineering. Polypyrrole (PPy) in particular has been studied extensively due to its ease of synthesis, relatively high conductivity under physiological conditions, ability to be chemically modified to adjust its electrical and biological activity, and well-characterized biocompatibility in vitro and in vivo.13–16 While the brittleness of PPy typically precludes its use as a cell-culture substrate, it can be incorporated within a more pliable polymer to form a composite material better suited for biomedical applications.17–19 For example, previous work has demonstrated the ability to integrate PPy with silk fibroin,20–22 prized for its biocompatibility, robust mechanical properties, low cost, versatility, and biodegradability.23, 24
It has been established that cell behavior within tissues is partially regulated by their surrounding extracellular matrix (ECM) via both biochemical factors and physical interactions with ECM architecture.25–27 Ultrastructural analyses of myocardial ECM indicate that it is comprised of highly-aligned collagen fiber bundles that range in size from a few hundred nanometers to a couple microns in diameter, and that this distinct feature plays a significant role in driving the organization and function of cardiomyocytes, and the organ as a whole.28–31 Specifically, the structural definition of the heart on multiple scales induces the directional propagation of forces and action potentials that then enable synchronous and efficient contractions.32 Inspired by these findings, substrates with anisotropic topography have been utilized to promote not only the polarity and structural development of cardiomyocytes, but their overall maturation as well.28, 33, 34
Human pluripotent stem cell-derived cardiomyocytes (hPSC-CMs) offer tremendous potential for the generation of human cardiac tissues for in vitro drug-screening and disease modeling applications, as well as for in vivo therapies such as those for myocardial infarcts.35–38 However, one of the greatest limitations in using these cells is that they are often phenotypically deficient post-differentiation, resulting in tissues that are not sufficiently representative of the adult human myocardium.39, 40 A primary cause of this observed shortcoming in cultured hPSC-CMs is that conventional cell culture platforms do not fully recapitulate the variety of cues cardiomyocytes are exposed to in their native tissue niche, and the development of new methods that incorporate multiple tissue-specific stimuli remains challenging.
In this study, we developed a facile approach for enhancing the development of hPSC-derived cardiac tissues, in which silk fibroin substrates were first patterned with anisotropic nanotopography that mimicked myocardial ECM. These corrugated silk substrates were subsequently functionalized sulfonic acid groups to facilitate integration of PPy, which imparted electroconductive properties. Nanopattern fidelity was maintained throughout the substrate fabrication process, and the presence of topographical features had no impact on substrate conductivity. It was also found that the nanotopographical and electroconductive cues acted synergistically to promote both tissue organization and the maturation of the contractile and electrical signal conduction capabilities of cultured cardiomyocytes. These results further highlight the benefits of recapitulating the myocardial cell niche in vitro such that physiologically representative cardiac tissues for downstream applications can be feasibly generated.
2. Experimental
2.1. Preparation of Aqueous Silk Fibroin Solutions
Bombyx mori silkworm cocoons were purchased from Oregon Silkworms in Bend, OR. Cocoons were shredded then degummed by boiling in Na2CO3 (20 mM, 4 L per 10 g of shredded cocoons) solution for 60 min, then washed in boiling DI water for 15 min followed by three 15-min washes in room temperature DI water. The fibroin fibers were then allowed to dry overnight in a fume hood. The dry fibers were placed in a beaker and sufficient LiBr (9.3 M) solution was added to make a 20% w/v solution, which was placed in a 60 °C oven until fibers were fully dissolved. This solution was placed in dialysis tubing (Fisherbrand, 3.5 kDa MWCO) and dialyzed for 24 h against nanopure water that was changed 4 times. This solution was then dialyzed against dilute HCl (1 mM) until it reached pH 7. The solution was concentrated by laying the dialysis bag on dry polyethylene glycol (10k MW) until it reached the desired concentration (6–8% w/v) at which point it was passed through a syringe filter (Millex-CV 5.00 μm), and then stored at 4 °C.
2.2. Silk Fibroin Substrate Fabrication
Master molds used to generate nanopatterned silk fibroin films were fabricated using a modification of capillary force lithography (CFL) techniques that have been previously described.28, 33, 41, 42 Briefly, a nanopatterned silicon wafer was coated in polyurethane acrylate (PUA; Minuta Technology, Korea), and a 75 μm thick sheet of polyethylene terephthalate (PET; SKC Inc., Korea) was then pressed down onto the silicon wafer. Ultraviolet (UV) light (λ = 250–400 nm) was then applied for 30 s (dose = 100 mJ/cm2) to give an initial curing. This nanopatterned PUA master mold was then peeled from the silicon wafer and placed under UV overnight to fully cure. A polydimethylsiloxane (PDMS; Sylgard 184, Dow Corning, MI, USA) mold was then produced by pouring PDMS over the PUA master mold and left to cure over a period of 48 h at room temperature before the PUA master and PDMS were subsequently peeled apart.
Prepared silk fibroin solution was then poured into the PDMS mold and left to dry overnight at room temperature before the resulting nanopatterned silk film was peeled from the PDMS mold and soaked in 70% ethanol overnight. Flat silk fibroin substrates were fabricated using an identical process, but with the use of an unpatterned PDMS mold instead.
2.3. Acid Modification of Silk Substrates
Ethanol-cured silk fibroin films were adhered to a Petri dish by applying polyimide tape to their perimeter, taking care to place the corrugated side facing upward. The film was then immersed in borate buffer (20 mL, 100 mM sodium borate, 123 mM sodium chloride, pH 9.5) and placed on ice. Sulfanilic acid (85 mg) and p-toluene sulfonic acid (p-TSA, 380 mg) were dissolved in nanopure water (5 mL) and placed on ice for 10 min. Sodium nitrite (4 M, 160 μL) solution was added to the acid mixture and returned to the ice for 15 min to allow the diazonium salt to form. The solution containing the diazonium salt was then pipetted into the silk solution, mixed and allowed to react on ice for 40 min. The reaction solution was discarded and the silk film was rinsed 3 times with nanopure water. Films with this treatment are referred to as acid-modified silk films (AMSF).
2.4. Deposition of Poly(pyrrole) onto Acid-Modified Silk Substrates
Acid modified films that were still adhered to the Petri dishes were immersed in p-TSA (5 mM) and pyrrole (50 mM) solution. Immediately after the addition of pyrrole, FeCl3 was added to a final concentration of 15 mM and was allowed to polymerize at room temperature for 2 h. Following polymerization, the solution was decanted and the films were rinsed thoroughly with nanopure water. The films were sonicated in nanopure water for 30 s, and then cut away from the Petri dish and stored in ethanol (70% v/v) to ensure sterility. These films are referred to as AMSF+PPy in the remaining text.
2.5. Scanning Electron Microscopy
Substrates were sputter-coated with Au-Pd for 60 s. High resolution imaging of substrate surfaces was achieved using a scanning electron microscope (Sirion XL30, FEI, OR, USA) and an acceleration voltage of 5 kV.
2.6. Atomic Force Microscopy
Topographical profiles of fabricated substrates were obtained using an atomic force microscope (Dimension ICON, Bruker, MA, USA). Tapping mode was used with 1 kHz of scan rate and a 512 × 512 scanning resolution over a 10 μm × 10 μm area of each sample. Measurement of RMS surface roughness was completed using Gwyddion (Czech Metrology Institute, CZ) data analysis software.
2.7. I-V Curve Measurements
I-V curves were recorded using a parameter analyzer (Keithley 4200-SCS, Tektronix, OR, USA) under the conditions of a direct current (DC) voltage sweep ranging from −5 V to 5 V.
2.8. Measurement of Substrate Resistivity
Sheet resistivity of substrates was measured with a four-point resistivity probe (S-304–2, Lucas Labs, CA, USA) mounted with a Signatone SP4–40045TBY tip and powered by a Keithley 2400 Sourcemeter. Values were calculated using the equation , where V (volts) is the voltage measured across the two inner probe electrodes, I (amperes) is the current passed between the two outer probe electrodes, and RS (Ω/sq) is the sheet resistivity.
2.9. Differentiation of hESC-Derived Cardiomyocytes
Cardiomyocyte differentiation from human embryonic stem cells (hESCs) was accomplished using a previously described and established serum-free directed differentiation protocol.43 Undifferentiated RUES-2 hESCs (Rockefeller University, NY, USA) were maintained and expanded on Matrigel-coated culture plates in feeder-free mouse embryonic fibroblast-conditioned media supplemented with 4 ng/mL basic fibroblast growth factor (bFGF; R&D Systems, MN, USA). To induce differentiation of RUES-2 monolayers, the conditioned media was supplemented with 1 μM of the Wnt agonist Chiron 99021 (Tocris Bioscience, MN, USA) for 1 d. This was followed by a change in medium to RPMI 1640/B27 (minus insulin) (Invitrogen, MA, USA) supplemented with 100 ng/mL activin A (R&D Systems). After 18 h of culture, the cells were fed with RPMI/B27 (minus insulin) supplemented with 5 ng/mL bone morphogenetic protein-4 (BMP-4; R&D Systems) and 1 μM Chiron 99021 for 48 h. Cells were then provided with fresh RPMI/B27 (minus insulin) supplemented with 1 μM XAV 939 (Tocris Bioscience), a Wnt antagonist, for 48 h. Seven days after induction, cells were maintained in RPMI/B27 (plus insulin) medium for an additional 7 d before dissociation for flow cytometry analysis. Only differentiation runs that produced a population of cells that were at least 90% cardiac troponin T (cTnT)-positive were used for the experiments described in this study.
2.10. Cell Culture
Prior to cell seeding, fabricated AMSF substrates were incubated at 37 °C in a 60 μg/mL solution of human fibronectin (Invitrogen) for 12 h. For the formation of confluent monolayers, stem cell-derived cardiomyocytes were seeded onto substrates at a density of 2.5 × 105 cells/cm2, while a seeding density of 2.5 × 104 cells/cm2 was used for single-cell imaging experiments. Cultures were maintained in RPMI/B27 (plus insulin) medium that was replaced every other day for a total of 21 d.
2.11. Assessment of Biocompatibility
Stem cell-derived cardiomyocytes were cultured on flat and patterned AMSF and AMSF+PPy substrates, as well as on a standard tissue culture polystyrene (TCPS) control for 21 d. Cells were seeded onto substrates at a density of 2.5 × 105 cells/cm2. Cell viability was then determined using a MTS colorimetric assay (CellTiter96, Promega, WI, USA) according to the manufacturer’s guidelines. Briefly, 20 μL of assay solution reagent was pipetted into a clear-bottom 96-well plate containing the samples and 100 μL of culture medium. The plate was then incubated at 37 °C for 4 h before recording absorbance values at λ = 490 nm with a plate reader.
2.12. Immunostaining and Confocal Fluorescence Microscopy
Cultured cells were fixed with 4% w/v paraformaldehyde in PBS for 10 min at room temperature and were then permeabilized with 0.1% w/v Triton-X 100 (Sigma-Aldrich, MO, USA) in PBS for 5 min at room temperature. For analysis of sarcomeric development, cells were stained for α-actinin (1:200 dilution; Sigma-Aldrich). For analysis of gap junction formation, cells were stained for connexin-43 (1:300 dilution; Sigma-Aldrich) and α-actinin. Mounting medium that included a DAPI counterstain for nuclei was used. Immunostained cells were imaged using confocal microscopy (A1R, Nikon Instruments, NY, USA) at 60x magnification with oil immersion. Analysis of sarcomere lengths, z-band widths, and connexin expression and spatial distribution was conducted using ImageJ software (National Institutes of Health, MD, USA). Analysis of cell orientation was conducted using a MATLAB script that was developed in-house.
2.13. Analysis of Gene Expression
RNA was isolated from cardiomyocytes after 21 d of culture by using the E.Z.N.A. Total RNA Kit I (Omega Bio-Tek, GA, USA) according to the manufacturer’s protocol. Quantity and purity of RNA was determined by 260/280 nm absorbance. The relative expression levels of selected genes were obtained using quantitative reverse-transcription polymerase chain reaction (qRT-PCR) analyses. cDNA of cultured cells in different condition (10 ng) was prepared using the Maxima SYBR Green/ROX qPCR master mix (Thermo Scientific, MA, USA). Reactions were processed by the ABI 7900HT PCR system with the following parameters: 50 °C/2 min and 95 °C/10 min, followed by 40 cycles of 95 °C/15 s and 60°C/1 min. Results were analyzed using SDS 2.3 software, and relative expression was calculated using the comparative Ct method, where GAPDH was designated as the housekeeping gene and cardiomyocytes cultured on TCPS were used as controls. Each sample was run in triplicate reactions for each gene. To examine the maturation of cardiomyocyte contractile machinery, hMYH7 and TNNT2 were used as markers, while GJA1 and SCN5A were used as markers for gap junction and voltage-gated ion channel development, respectively. Primers for all assayed genes were purchased from Bio-Rad Laboratories (CA, USA), and used as provided. Amplicon context sequences for these primers have been listed in Supporting Information Table 1.
2.14. Statistical Analyses
All quantitative data is provided as means ± standard error of the mean (SEM). One-way ANOVA with a Tukey’s post-hoc test was used to analyze data sets that include more than two experimental groups, while a Student’s t-test was used to compare data sets looking at only two variables. In all analyses, a p-value less than 0.05 was considered significant.
3. Results and Discussion
3.1. Fabrication and characterization of conductive nanopatterned silk fibroin-PPy substrates
To better mimic the nanotopographical features of native tissue ECM and thereby provide a more physiologically-relevant microenvironment for cultured cardiomyocytes, capillary force lithography (CFL) was used to pattern silk fibroin substrates (Figure 1A). CFL techniques allow for the generation of nanoscale features with a greater fidelity and reproducibility than is achievable using methods such as electrospinning44, 45 or directional freezing,46 and can be performed under ambient conditions without the need for a cleanroom. In this study, nanopatterns consisting of aligned grooves and ridges measuring 800 nm × 800 nm × 600 nm (groove width × ridge width × ridge height) were used as these dimensions not only best approximated those of the fiber bundles observed in the ECM, but were also found in a previous study to induce the greatest degree of cellular alignment, hypertrophy, and sarcomere development in cardiomyocytes.33
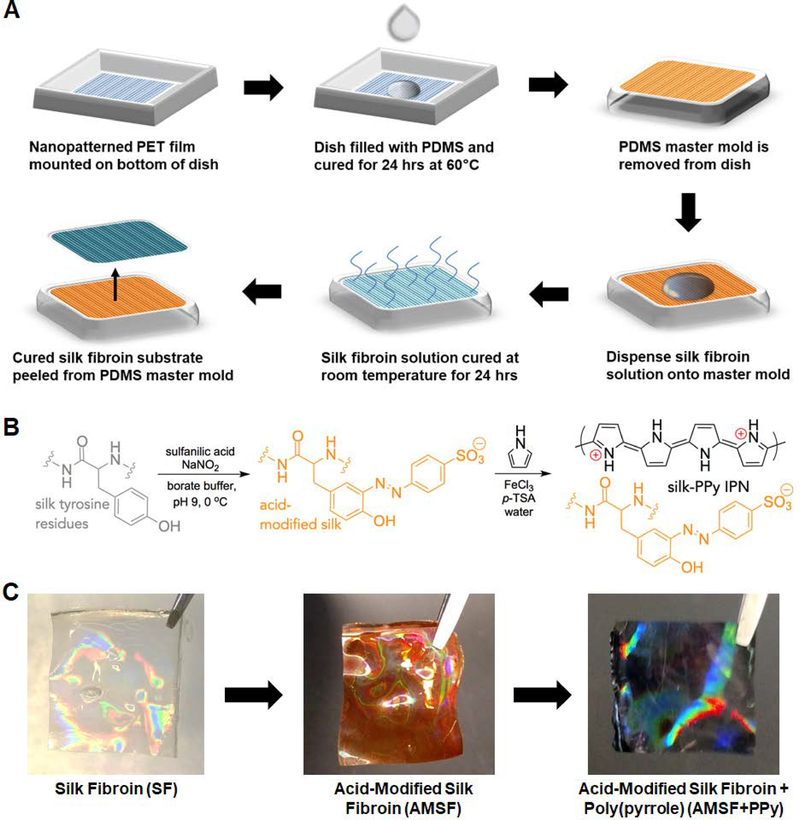
Figure 1. Fabrication of nanopatterned acid-modified silk fibroin and polypyrrole deposition.
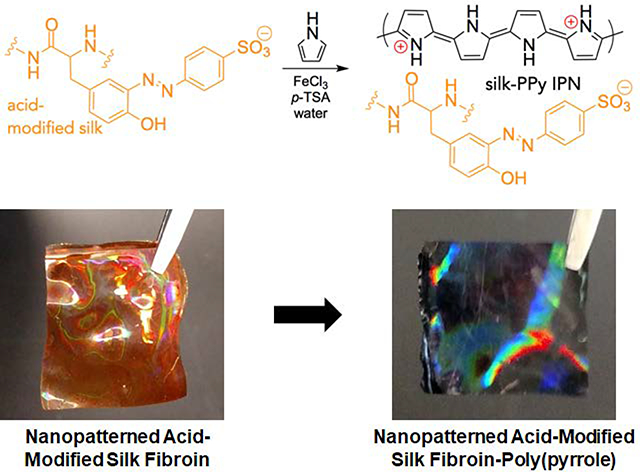
(A) Scheme of the process utilized to generate the master mold and subsequent casting of silk fibroin solution to generate nanopatterned substrates. (B) Description of chemical moieties and reactions that occur during the acid modification and PPy deposition stages. (C) Representative photographs of the nanopatterned silk substrates at each stage. The substrates take on an orange hue after acid modification and turn black after PPy deposition. An iridescent sheen is present throughout. Dimensions of the pictured films are 1 cm × 1 cm.
Fabricated silk films were then acid-modified through a diazonium coupling reaction that introduces sulfonic acid groups covalently bound to the silk protein backbone via the tyrosine residues (Figure 1B). This acid modification of silk fibroin improves the absorption and intercalation of positively-charged PPy into the now negatively-charged silk network, resulting in a significantly more robust interpenetrating-network composite material that resists delamination and maintains stable electroconductive properties in aqueous environments.20 PPy was then deposited onto the AMSF films by simply submerging the films in an aqueous solution of pyrrole, p-TSA and FeCl3 for 2 h. The fabrication, modification, and deposition process, progress could be visually monitored, as the silk substrates changed from colorless to orange in the acid modification step (indicative of azo bond formation between the tyrosine and diazonium salt), and from orange to black in the PPy deposition step (Figure 1C). Further details and chemical characterization of the resulting silk-PPy films are given in previous publications.20, 47
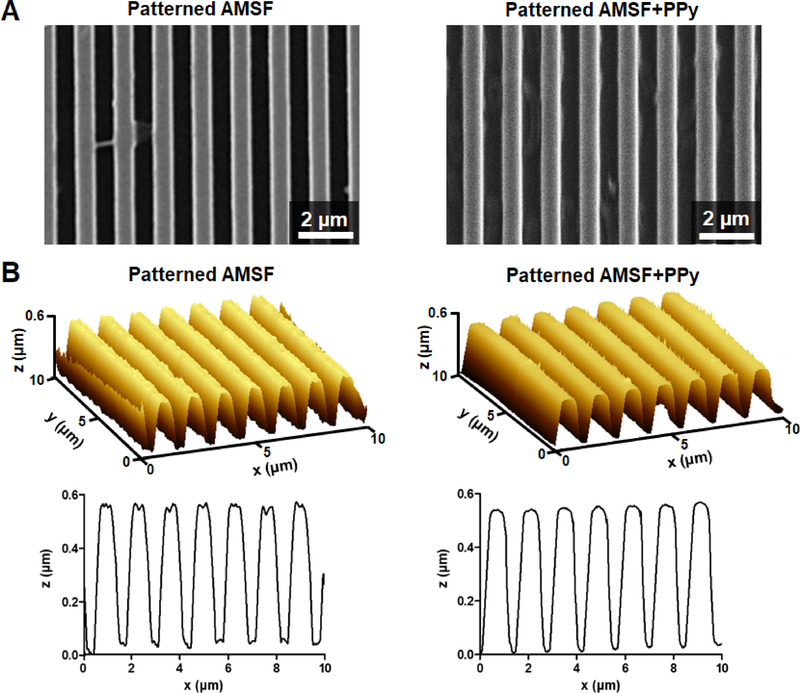
Throughout the reaction steps, it was observed that nanopatterned films maintained the iridescent sheen that is characteristic of patterned materials, indicating at a macroscopic scale that the fidelity of the molded pattern was maintained. This finding was confirmed through SEM and AFM imaging of these substrates, where not only were the patterned grooves and ridges well-defined both after acid modification and PPy deposition (Figure 2A), but the measured dimensions of these features were comparable to the initial design as well (Figure 2B). PPy deposition on AMSF had minimal impact on surface roughness and topographical feature size, with the average RMS roughness value of 8.97 ± 0.21 nm practically negligible compared to the patterned ridges that were 600 nm tall (Supporting Information Figure 1). The nanopatterns did not significantly affect the tensile strength and elastic modulus of the silk-PPy films, as measured values were in the same range as those previously reported for flat films (~7 MPa and ~200 MPa, respectively).20, 47
Figure 2. Characterization of substrate topography.
(A) Representative SEM images of nanopatterned substrates with and without PPy. (B) 3D and cross-section AFM profiles illustrate that the fidelity of the nanotopography is well-maintained even after PPy deposition.
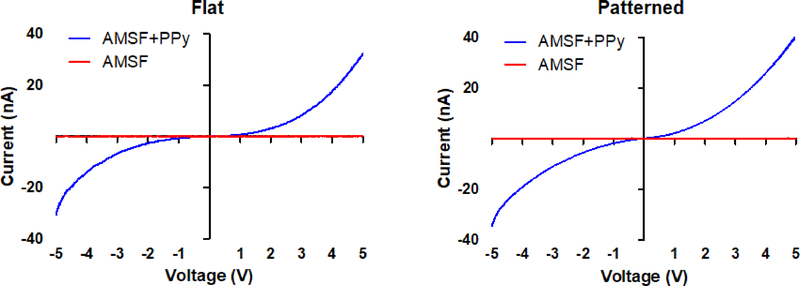
Four-point probe measurements of the sheet resistivity of AMSF substrates with and without PPy indicate that, as expected, the addition of PPy significantly increases substrate conductivity. The sheet resistivity of the unmodified AMSF was greater than 106 Ω/sq, while that of AMSF+PPy was typically in the range of 200-500 Ω/sq (which corresponds to a conductivity of ~1 S/cm). This is further illustrated by current-voltage (I-V) curve measurements in which AMSF+PPy substrates responded to applied voltages and exhibited a non-linear resistive behavior while AMSF-only substrates displayed no response (Figure 3). Additionally, differences in nanotopography had no impact on the overall electrical properties of the silk-PPy composites, as the I-V curves of flat and patterned substrates were virtually identical. Resistivity of the films typically increased by ~3–10 times over 21 days when soaked in aqueous solutions with or without proteins and enzymes,20, 47 but even after degradation the conductivities are still higher than that of the native myocardial tissue (0.0002 to 0.0062 S/cm).48
Figure 3. AMSF+PPy substrates are electrically responsive.
I-V curves of flat and patterned substrates indicate that the electroconductive property of the substrates is unaffected by topographical changes.
The method presented here of fabricating biodegradable substrates possessing both electrical conductivity and biomimetic nanotopography offers some significant advantages over other previously reported techniques. Since electroconductive functionality is achieved by absorbing PPy into the existing silk network in a uniform manner, nanoscale features that are present on the surface are maintained without risk of changes in dimension or shape. This contrasts with other methods, such as chemical vapor deposition (CVD), where the conductive materials are deposited onto patterned surfaces and the resulting coating can be inhomogenous.49–51 Since the PPy itself is not biodegradable, the ability to incorporate it only in the surface of the silk substrate minimizes the amount of PPy necessary to make the composites conductive.18, 19 Furthermore, the relative simplicity of the acid modification, CFL patterning, and PPy conjugation processes obviate the need for complex training and expensive equipment. This drives down cost while improving reproducibility, thereby increasing the translatability of the fabrication method and likelihood of adoption by a variety of end-users.
3.2. Enhanced structural organization and sarcomere development by electroconductive and nanotopographical cues
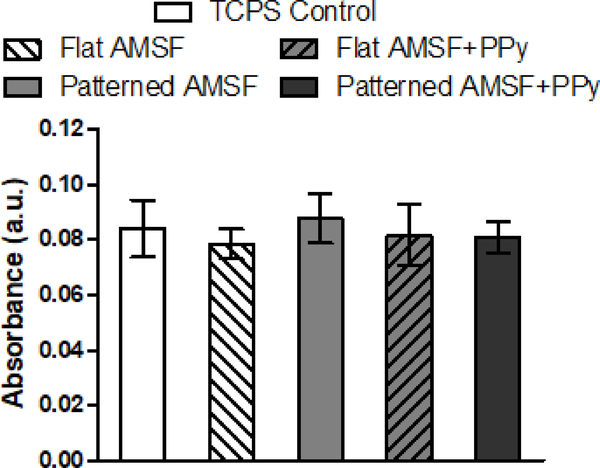
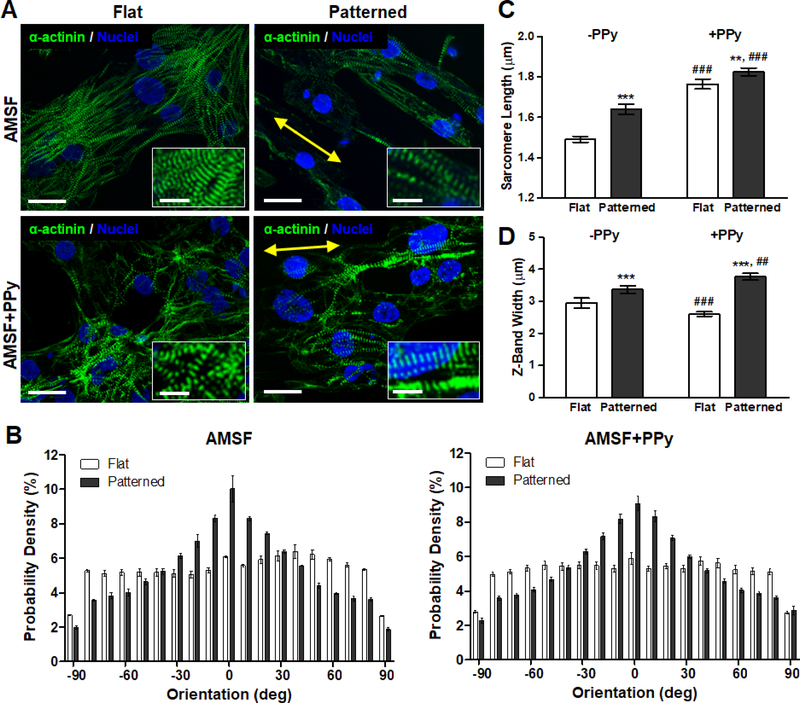
Cardiomyocytes differentiated from human embryonic stem cells were then cultured on flat and patterned AMSF and AMSF+PPy substrates for 21 d before they were fixed and stained for α-actinin to analyze cellular morphology and sarcomeric development. High cell viability was maintained on all substrate types for the entirety of the culture period (Figure 4). Cells cultured on flat substrates were more disorganized than those on patterned substrates which featured elongated cell bodies that appeared to be aligned to the same axis as the underlying nanotopography (Figure 5A). Quantitative analysis of cell orientation revealed that patterned substrates did indeed induce greater anisotropic orientation compared to flat substrates, although overall alignment is slightly reduced on AMSF+PPy substrates (Figure 5B). However, this did not appear to have a negative impact on sarcomeric development, as cardiomyocytes on patterned substrates possessed longer sarcomeres than those cultured on flat substrates (Figure 5C). Furthermore, the conductivity imparted by PPy also induced a significant increase in sarcomere length in cells on both patterned and flat substrates. Cellular alignment due to topographical cues increased z-band width, and although there was a statistically significant increase in z-band width in cells on AMSF+PPy substrates, the impact of electroconductivity on this aspect of muscle development appears to be limited (Figure 5D).
Figure 4. AMSF and AMSF+PPy substrates are biocompatible.
Colorimetric MTS assays of cardiomyocytes cultured for 21 days show that acid-modification and the presence of PPy have no negative impact on cell viability. Viability is presented as normalized absorbance values.
Figure 5. Nanotopographical and electroconductive cues enhance structural organization and contractile apparatus development.
(A) Representative images of cardiomyocytes fluorescently stained for α-actinin (green) and nuclei (blue). Cells on nanopatterned substrates exhibit elongated and aligned morphologies. Yellow arrows indicate the direction of the nanopattern. Scale bars: 25 μm; inset scale bars: 10 μm. (B) Anisotropic nanopatterns induce cell orientation along the axis of the pattern, indicated here by 0°, whereas cells are randomly oriented on flat substrates. (C) Sarcomere length is significantly increased due to nanopatterning and PPy. (D) Z-band width is greatly influenced by topography but is only slightly increased on nanopatterned substrates due to PPy. All quantitative data are presented as means ± SEM, n≥10 different cultures. **p < 0.01, ***p < 0.001 (flat vs. patterned; Student’s t-test); ##p < 0.01, ###p < 0.001 (−PPy vs. +PPy; Student’s t-test).
Given the importance of tissue structure on the functional capabilities of the myocardium in vivo, it is critical that this cellular organization is recapitulated within engineered myocardial tissues. Our results confirm the benefits of using bioinspired nanotopographies to achieve this goal, as not only was the overall anisotropic alignment of cultured cardiomyocytes enhanced by topography-mediated contact guidance cues, but this alignment also led to significant improvements in sarcomere organization and development. The sarcomere is the fundamental unit for cardiomyocyte contraction, and cells that possess longer sarcomeres are generally capable of producing greater contractile forces.52 In relaxed adult human cardiomyocytes, sarcomere lengths are typically approximately 2.2 μm,53 whereas sarcomere lengths in immature hPSC-CMs have been reported to be approximately 33% shorter at 1.65 μm.54 In this study, by simply inducing an elongated morphology in cells, average sarcomere lengths could be increased by 10%. For cells cultured on the electroconductive AMSF+PPy substrates, a 23% increase was observed, with average sarcomere length at a near physiological value of 1.83 μm. The lateral alignment of multiple myofibrils is also important, as this allows for synchronous contractions of sarcomeres over larger length-scales, thereby enabling cardiac tissue to produce strong twitch forces.55 Correspondingly, more mature cardiomyocytes possess larger z-band widths. The maturation benefits of promoting of cellular alignment is once again evident as these morphological changes led to a greater number of sarcomeres to be in register, thus leading to significantly increased z-band widths in anisotropically organized cells.
3.3. Modulation of gap junction formation and localization by electroconductive and nanotopographical cues
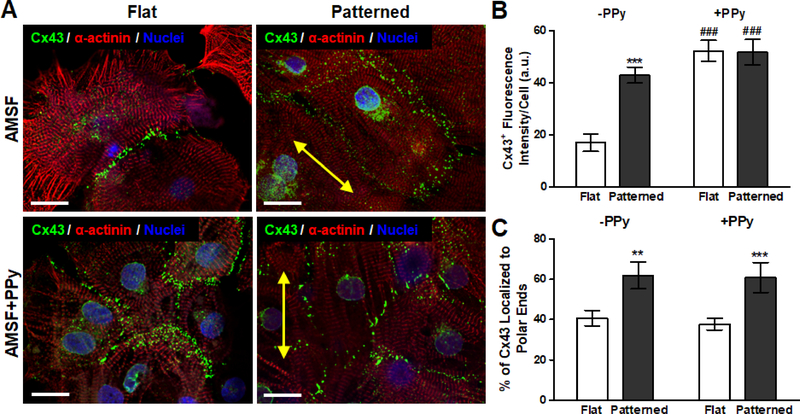
Cardiomyocytes were also stained for connexin 43 (Cx43), a gap junction protein critical for regulating the electrical coupling of cardiomyocytes within the mammalian myocardium (Figure 6A). Quantitative analysis of Cx43 expression based on immunostaining revealed that while overall expression on a per cell basis was not significantly different between cells cultured on flat and patterned AMSF+PPy substrates, their expression of Cx43 was significantly greater than that of cells cultured on patterned AMSF, which were in turn was greater than that of cells cultured on flat AMSF (Figure 6B). Localization of Cx43 was then determined by quantifying the amount of Cx43 in the polar regions of each cell as a percentage of total expression.56 It was found that while nanotopographical cues induced a greater degree of Cx43 polarization, electroconductive cues had no impact on protein distribution (Figure 6C).
Figure 6. Modulation of gap junction protein expression and localization due to electroconductivity and anisotropic nanotopography.
(A) Representative images of cardiomyocytes fluorescently stained for α-actinin (red), Cx43 (green), and nuclei (blue). Yellow arrows indicate the direction of the nanopattern. Scale bars: 25 μm. (B) While bioinspired nanotopography significantly increased Cx43 expression, the advantage of topographical cues is nearly nullified in the presence of electroconductive cues as PPy elicited a similar degree of Cx43 expression on both flat and patterned substrates. (C) Cx43 polarization is not significantly affected by electroconductive cues as topography dictated protein localization patterns. All quantitative data are presented as means ± SEM, n≥10 different cultures. **p < 0.01, ***p < 0.001 (flat vs. patterned; Student’s t-test); ###p < 0.001 (-PPy vs. +PPy; Student’s t-test).
Taken together, these results provide interesting insights into the impact of electroconductivity on gap junction formation. Culturing of cardiomyocytes on electroconductive substrates leads to an overall increase in the amount of Cx43 expressed per cell, and this result confirms previous observations reported in literature.7, 51 At the same time, it is apparent that while anisotropic cellular organization as a result of nanotopography also increases expression in cells, it plays an even more significant role in concentrating Cx43 at the polar junctions of adjacent cells. This finding highlights the critical importance of organizing cardiomyocytes in a physiological manner, as it is this polarization of Cx43 that enables the rapid and efficient propagation of action potentials in a uniaxial manner that subsequently leads to the contraction in a concerted direction that is required for the proper function of myocardial tissue.57, 58 Indeed, abundant expression of Cx43 and its polar localization is one of the hallmarks of fully differentiated ventricular myocytes in vivo.59
3.4. Synergistic effects of nanotopography and electroconductivity on expression of genetic markers for cardiac maturation
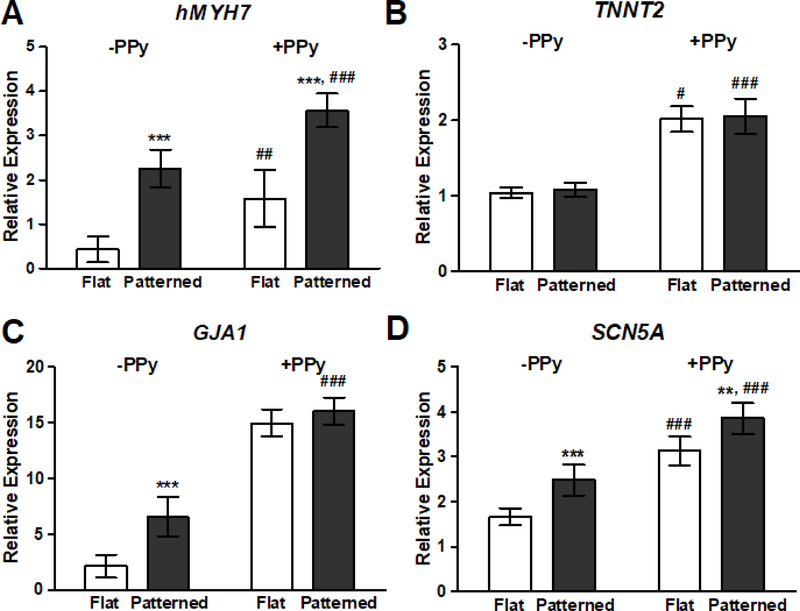
After the 21-day culture period, RNA was collected from cultured hPSC-CMs and the expression levels of representative markers for cardiomyocyte development were quantitatively analyzed with qRT-PCR. Genes were selected such that the impact of substrate properties on both contractile and electrical function could be examined at the tissue level. Cardiomyocytes on patterned substrates saw a significantly increased expression of hMYH7, which codes for β-myosin heavy chain (β-MHC), the predominant isoform of MHC in the adult human ventricular myocardium.60 hMYH7 expression was further increased in cells cultured on AMSF+PPy substrates compared to their counterparts on non-conductive substrates (Figure 7A). On the other hand, expression of cardiac troponin T2 (TNNT2), a key regulator of myocyte contraction, was unaffected by substrate topography and only showed increases in expression when cells were exposed to PPy (Figure 7B). In agreement with the aforementioned observations from the immunohistochemical analysis of gap junction development via Cx43, electroconductivity appears to be a more significant driving factor compared to nanotopography when it comes to regulating the expression of (GJA1) (Figure 7C). Similar to the trends noted for hMYH7 expression, topographical and electroconductive cues both promoted the upregulation of the voltage-gated sodium channel NaV1.5 (SCN5A), with the greatest expression levels seen in cells that were cultured on AMSF+PPy substrates (Figure 7D). NaV1.5 generates the fast sodium current which regulates the cardiac action potential; therefore, the degree of its expression has a significant effect on cardiac conduction velocity.61
Figure 7. Nanotopography and electroconductivity synergistically induced the largest increases in genetic markers for cardiac maturation.
qRT-PCR analysis of the relative expression levels of: (A) β-myosin heavy chain (hMYH7), (B) cardiac troponin T2 (TNNT2), (C) connexin 43 (GJA1), and (D) NaV1.5 (SCN5A). Presence of biomimetic nanotopography significantly increased gene expression, with the exception of TNNT2. Cardiomyocytes cultured on electroconductive silk substrates exhibited an increased expression of all assayed genes when compared to cells on non-conductive substrates. All data are presented as means ± SEM, n≥6 different cultures, **p < 0.01, ***p < 0.001 (flat vs. patterned; Student’s t-test); #p < 0.05, ##p < 0.01, ###p < 0.001 (-PPy vs. +PPy; Student’s t-test).
It has become increasingly clear that mechanical and topographical cues play a crucial role in the modulation of cellular epigenetics and function. These cell-matrix interactions are largely mediated by integrins, which are cell surface receptors that regulate a variety of downstream signaling cascades that affect cell morphology, proliferation, differentiation, migration, and gene expression.62 Integrins typically cluster together to form focal adhesion complexes, and the frequency, shape, and distribution of this clustering is greatly influenced by topographical features at the sub-micron level.63 Focal adhesion kinase (FAK) localizes at these complexes and acts in conjunction with integrins to transduce extracellular mechanical signals to dictate cellular processes. FAK and FAK-associated signaling has been implicated in regulating cardiogenesis via adhesive and topographical cues in vitro, 34, 64, 65 and the role of both integrins and FAK in cardiac organogenesis and maturation is so critical, that ablation or disruption of their function has been shown to result in embryonic mortality in vivo.66–68 Thus, by mimicking native ECM topography with the substrates developed in this study, we were able to leverage these mechanotransductive processes to impart beneficial maturation effects on cultured hPSC-CMs.
It is also evident that electroconductive cues have a profound effect on almost all major aspects of cardiomyocyte development. While there is an abundance of evidence that conductive materials drive the maturation of myocytes, the exact mechanisms and pathways involved have yet to be fully elucidated. One hypothesis suggests that close proximity of cell membranes to conductive surfaces induces a hyperpolarization of the membrane resting potential, which has been demonstrated to be an important factor in promoting the cardiomyocyte maturation process.69 It has also been suggested that the ability for conducting materials to not only improve the adsorption of proteins, but also induce a more bioactive conformation of these proteins, allows for a greater presentation of maturation-promoting biochemical cues.70, 71 The myocardium is an electrically active tissue with an inherent conductivity reported to range from 0.02 to 0.62 S/m and is permeated with conductive Purkinje fibers that assist with action potential propagation.48 Thus, the presence of an electrically-responsive material in the cell culture microenvironment could serve to reproduce the extracellular voltage gradients and electrical fields that are present in vivo and are known to significantly influence developmental processes.72
4. Conclusion
By using nanopatterned AMSF+PPy substrates for culturing hPSC-CMs, we have demonstrated the ability to harness the combined benefits of electroconductivity and bioinspired nanotopography for enhancing cardiomyocyte development. In addition to adopting an anisotropic morphology, cultured cardiomyocytes exhibited markedly improved sarcomere and gap junction development and organization. The pro-maturation effects of the substrates were also reflected in the increased expression levels of genes associated with proteins that are critical for proper cardiac tissue excitation-contraction function. Corresponding improvements in functional capability, such as force generation and action potential conduction velocity, remain to be investigated in future studies. The results presented here add to a growing body of evidence that the overall maturation of a variety of cell types, especially cardiac and skeletal myocytes, can be drastically improved when biomimetic nanotopography and electrical cues are both present in the culture microenvironment.50, 51, 73–76 In addition to the beneficial effects of the substrates on cell maturation, the ability to obtain defined nanoscale features and electroconductive properties in a pliable, biodegradable, and biocompatible platform also greatly expands the potential for this composite material to be used in a variety of in vivo applications, such as for engineering tissue patches and implantable devices.77–79
Supplementary Material
Acknowledgements
This work was supported by National Institutes of Health grants R01 HL135143 and R21 EB020132 (to D.-H.K.), and a National Science Foundation grant DMR-1411292 (to A.R.M.). The authors would like to thank Prof. Seungkeun Choi at the University of Washington Bothell for his assistance with gathering I-V curve data. Part of this work was conducted at the Molecular Analysis Facility, a National Nanotechnology Coordinated Infrastructure site at the University of Washington which is supported in part by the National Science Foundation (grant ECC-1542101), the University of Washington, the Molecular Engineering & Sciences Institute, the Clean Energy Institute, and the National Institutes of Health.
Footnotes
Conflicts of Interest
Deok-Ho Kim is a co-founder and scientific board member at NanoSurface Biomedical Inc.
References
- 1.Guo BL, Glavas L and Albertsson AC, Prog Polym Sci, 2013, 38, 1263–1286. [Google Scholar]
- 2.Lee TJ, Park S, Bhang SH, Yoon JK, Jo I, Jeong GJ, Hong BH and Kim BS, Biochem. Biophys. Res. Commun, 2014, 452, 174–180. [DOI] [PubMed] [Google Scholar]
- 3.Shin SR, Zihlmann C, Akbari M, Assawes P, Cheung L, Zhang KZ, Manoharan V, Zhang YS, Yuksekkaya M, Wan KT, Nikkhah M, Dokmeci MR, Tang XW and Khademhosseini A, Small, 2016, 12, 3677–3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martinelli V, Cellot G, Toma FM, Long CS, Caldwell JH, Zentilin L, Giacca M, Turco A, Prato M, Ballerini L and Mestroni L, ACS Nano, 2013, 7, 5746–5756. [DOI] [PubMed] [Google Scholar]
- 5.Shin SR, Jung SM, Zalabany M, Kim K, Zorlutuna P, Kim SB, Nikkhah M, Khabiry M, Azize M, Kong J, Wan KT, Palacios T, Dokmeci MR, Bae H, Tang XW and Khademhosseini A, ACS Nano, 2013, 7, 2369–2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu Y, Wang L, Guo B and Ma PX, ACS Nano, 2017, 11, 5646–5659. [DOI] [PubMed] [Google Scholar]
- 7.You JO, Rafat M, Ye GJC and Auguste DT, Nano Lett, 2011, 11, 3643–3648. [DOI] [PubMed] [Google Scholar]
- 8.Fleischer S, Shevach M, Feiner R and Dvir T, Nanoscale, 2014, 6, 9410–9414. [DOI] [PubMed] [Google Scholar]
- 9.Shevach M, Maoz BM, Feiner R, Shapira A and Dvir T, J. Mater. Chem. B, 2013, 1, 5210–5217. [DOI] [PubMed] [Google Scholar]
- 10.Borriello A, Guarino V, Schiavo L, Alvarez-Perez MA and Ambrosio L, J. Mater. Sci. Mater. Med, 2011, 22, 1053–1062 [DOI] [PubMed] [Google Scholar]
- 11.Wang L, Wu Y, Hu T, Guo B and Ma PX, Acta Biomater, 2017, 59, 68–81. [DOI] [PubMed] [Google Scholar]
- 12.Dong RN, Zhao X, Guo BL and Ma PX, ACS Appl. Mater. Interfaces, 2016, 8, 17138–17150. [DOI] [PubMed] [Google Scholar]
- 13.Guimard NK, Gomez N and Schmidt CE, Prog. Polym. Sci, 2007, 32, 876–921.. [Google Scholar]
- 14.Wang XD, Gu XS, Yuan CW, Chen SJ, Zhang PY, Zhang TY, Yao J, Chen F and Chen G, J. Biomed. Mater. Res. A, 2004, 68a, 411–422. [DOI] [PubMed] [Google Scholar]
- 15.Balint R, Cassidy NJ and Cartmell SH, Acta Biomater, 2014, 10, 2341–2353. [DOI] [PubMed] [Google Scholar]
- 16.Kai D, Prabhakaran MP, Jin GR and Ramakrishna S, J. Biomed. Mater. Res. A, 2011, 99a, 376–385. [DOI] [PubMed] [Google Scholar]
- 17.Spearman BS, Hodge AJ, Porter JL, Hardy JG, Davis ZD, Xu T, Zhang XY, Schmidt CE, Hamilton MC and Lipke EA, Acta Biomater, 2015, 28, 109–120. [DOI] [PubMed] [Google Scholar]
- 18.Kaur G, Adhikari R, Cass P, Bown M and Gunatillake P, RSC Adv, 2015, 5, 37553–37567. [Google Scholar]
- 19.Hardy JG, Lee JY and Schmidt CE, Curr. Opin. Biotechnol, 2013, 24, 847–854. [DOI] [PubMed] [Google Scholar]
- 20.Romero IS, Schurr ML, Lally JV, Kotlik MZ and Murphy AR, ACS Appl. Mater. Interfaces, 2013, 5, 553–564. [DOI] [PubMed] [Google Scholar]
- 21.Aznar-Cervantes S, Roca MI, Martinez JG, Meseguer-Olmo L, Cenis JL, Moraleda JM and Otero TF, Bioelectrochemistry, 2012, 85, 36–43. [DOI] [PubMed] [Google Scholar]
- 22.Hardy JG, Khaing ZZ, Xin S, Tien LW, Ghezzi CE, Mouser DJ, Sukhavasi RC, Preda RC, Gil ES, Kaplan DL and Schmidt CE, J. Biomater. Sci. Polym. Ed, 2015, 26, 1327–1342. [DOI] [PubMed] [Google Scholar]
- 23.Wang YZ, Kim HJ, Vunjak-Novakovic G and Kaplan DL, Biomaterials, 2006, 27, 6064–6082. [DOI] [PubMed] [Google Scholar]
- 24.Omenetto FG and KapLan DL, Nat. Photonics, 2008, 2, 641–643. [Google Scholar]
- 25.Bissell MJ, Hall HG and Parry G, J. Theor. Biol, 1982, 99, 31–68. [DOI] [PubMed] [Google Scholar]
- 26.LaNasa SM and Bryant SJ, Acta Biomater, 2009, 5, 2929–2938. [DOI] [PubMed] [Google Scholar]
- 27.Kim DH, Provenzano PP, Smith CL and Levchenko A, J. Cell Biol, 2012, 197, 351–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim DH, Lipke EA, Kim P, Cheong R, Thompson S, Delannoy M, Suh KY, Tung L and Levchenko A, Proc. Natl. Acad. Sci. U. S. A, 2010, 107, 565–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Genet M, Lee LC, Nguyen R, Haraldsson H, Acevedo-Bolton G, Zhang Z, Ge L, Ordovas K, Kozerke S and Guccione JM, J. Appl. Physiol, 2014, 117, 142–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sosnovik DE, Wang R, Dai G, Reese TG and Wedeen VJ, J. Cardiovasc. Magn. Reson, 2009, 11, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Streeter DD and Bassett DL, Anat. Rec, 1966, 155, 503–511. [Google Scholar]
- 32.Thavandiran N, Nunes SS, Xiao Y and Radisic M, Stem Cell Res. Ther, 2013, 4, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carson D, Hnilova M, Yang XL, Nemeth CL, Tsui JH, Smith AST, Jiao A, Regnier M, Murry CE, Tamerler C and Kim DH, ACS Appl. Mater. Interfaces, 2016, 8, 21923–21932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim DH, Kshitiz, Smith RR, Kim P, Ahn EH, Kim HN, Marban E, Suh KY and Levchenko A, Integr. Biol, 2012, 4, 1019–1033. [DOI] [PubMed] [Google Scholar]
- 35.Liu WH, Chang YL, Lo WL, Li HY, Hsiao CW, Peng CH, Chiou SH, Ma HI and Chen SJ, Cell Transplant, 2015, 24, 2185–2195. [DOI] [PubMed] [Google Scholar]
- 36.Acimovic I, Vilotic A, Pesl M, Lacampagne A, Dvorak P, Rotrekl V and Meli AC, BioMed Res. Int, 2014, 2014, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lancaster MA and Knoblich JA, Science, 2014, 345, 1247125. [DOI] [PubMed] [Google Scholar]
- 38.Smith AST, Macadangdang J, Leung W, Laflamme MA and Kim DH, iotechnol. Adv, 2017, 35, 77–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laflamme MA and Murry CE, Nature, 2011, 473, 326–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gherghiceanu M, Barad L, Novak A, Reiter I, Itskovitz-Eldor J, Binah O and Popescu LM, J. Cell. Mol. Med, 2011, 15, 2539–2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Macadangdang J, Lee HJ, Carson D, Jiao A, Fugate J, Pabon L, Regnier M, Murry C and Kim DH, J. Vis. Exp, 2014, 50039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsui JH, Janebodin K, Ieronimakis N, Yama DMP, Yang HS, Chavanachat R, Hays AL, Lee H, Reyes M and Kim DH, ACS Nano, 2017, 11, 11954–11968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu WZ, Van Biber B and Laflamme MA, Methods Mol. Biol, 2011, 767, 419–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang XH, Baughman CB and Kaplan DL, Biomaterials, 2008, 29, 2217–2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meinel AJ, Kubow KE, Klotzsch E, Garcia-Fuentes M, Smith ML, Vogel V, Merkle HP and Meinel L, Biomaterials, 2009, 30, 3058–3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stoppel WL, Hu DJ, Domian IJ, Kaplan DL and Black LD, Biomed. Mater, 2015, 10, 034105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Larson JD, Fengel CV, Bradshaw NP, Romero IS, Leger JM and Murphy AR, Mater. Chem. Phys, 2017, 186, 67–74. [Google Scholar]
- 48.Roth BJ, IEEE Trans. Biomed. Eng, 1997, 44, 326–328. [DOI] [PubMed] [Google Scholar]
- 49.Yu QK, Jauregui LA, Wu W, Colby R, Tian JF, Su ZH, Cao HL, Liu ZH, Pandey D, Wei DG, Chung TF, Peng P, Guisinger NP, Stach EA, Bao JM, Pei SS and Chen YP, Nat. Mater, 2011, 10, 443–449. [DOI] [PubMed] [Google Scholar]
- 50.Yang HS, Lee B, Tsui JH, Macadangdang J, Jang SY, Im SG and Kim DH, Adv. Healthc. Mater, 2016, 5, 137–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith AST, Yoo H, Yi H, Ahn EH, Lee JH, Shao G, Nagornyak E, Laflamme MA, Murry CE and Kim DH, Chem. Commun, 2017, 53, 7412–7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hibberd MG and Jewell BR, J. Physiol, 1982, 329, 527–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bird SD, Doevendans PA, van Rooijen MA, Brutel A de la Riviere R Hassink J, Passier R and Mummery CL, Cardiovasc. Res, 2003, 58, 423–434. [DOI] [PubMed] [Google Scholar]
- 54.Lundy SD, Zhu WZ, Regnier M and Laflamme MA, Stem Cells Dev, 2013, 22, 1991–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rodriguez AG, Rodriguez ML, Han SJ, Sniadecki NJ and Regnier M, Integr. Biol, 2013, 5, 1366–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Salameh A, Wustmann A, Karl S, Blanke K, Apel D, Rojas-Gomez D, Franke H, Mohr FW, Janousek J and Dhein S, Circ. Res, 2010, 106, 1592–1602. [DOI] [PubMed] [Google Scholar]
- 57.Roell W, Lewalter T, Sasse P, Tallini YN, Choi BR, Breitbach M, Doran R, Becher UM, Hwang SM, Bostani T, von Maltzahn J, Hofmann A, Reining S, Eiberger B, Gabris B, Pfeifer A, Welz A, Willecke K, Salama G, Schrickel JW, Kotlikoff MI and Fleischmann BK, Nature, 2007, 450, 819–824. [DOI] [PubMed] [Google Scholar]
- 58.Bukauskas FF, Jordan K, Bukauskiene A, Bennett MV, Lampe PD, Laird DW and Verselis VK, Proc. Natl. Acad. Sci. U. S. A, 2000, 97, 2556–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fromaget C, el Aoumari A and Gros D, Differentiation, 1992, 51, 9–20. [DOI] [PubMed] [Google Scholar]
- 60.Morkin E, Microsc. Res. Tech, 2000, 50, 522–531. [DOI] [PubMed] [Google Scholar]
- 61.Park DS and Fishman GI, Circulation, 2011, 123, 904–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ross RS and Borg TK, Circ. Res, 2001, 88, 1112–1119. [DOI] [PubMed] [Google Scholar]
- 63.Biggs MJP, Richards RG and Dalby MJ, Nanomedicine, 2010, 6, 619–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hakuno D, Takahashi T, Lammerding J and Lee RT, J. Biol. Chem, 2005, 280, 39534–39544. [DOI] [PubMed] [Google Scholar]
- 65.Simpson DG, Terracio L, Terracio M, Price RL, Turner DC and Borg TK, J. Cell. Physiol, 1994, 161, 89–105. [DOI] [PubMed] [Google Scholar]
- 66.Stephens LE, Sutherland AE, Klimanskaya IV, Andrieux A, Meneses J, Pedersen RA and Damsky CH, Genes Dev, 1995, 9, 1883–1895. [DOI] [PubMed] [Google Scholar]
- 67.Baudoin C, Goumans MJ, Mummery C and Sonnenberg A, Genes Dev, 1998, 12, 1202–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Peng X, Wu XY, Druso JE, Wei HJ, Park AYJ, Kraus MS, Alcaraz A, Chen J, Chien S, Cerione RA and Guan JL, Proc. Natl. Acad. Sci. U. S. A, 2008, 105, 6638–6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.van Vliet P, de Boer TP, van der Heyden MAG, El Tamer MK, Sluijter JPG, Doevendans PA and Goumans MJ, Stem Cell Rev. Rep, 2010, 6, 178–185. [DOI] [PubMed] [Google Scholar]
- 70.Molino PJ, Higgins MJ, Innis PC, Kapsa RMI and Wallace GG, Langmuir, 2012, 28, 8433–8445. [DOI] [PubMed] [Google Scholar]
- 71.Wan AMD, Inal S, Williams T, Wang K, Leleux P, Estevez L, Giannelis EP, Fischbach C, Malliaras GG and Gourdon D, J. Mater. Chem. B, 2015, 3, 5040–5048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McCaig CD, Rajnicek AM, Song B and Zhao M, Physiol. Rev, 2005, 85, 943–978. [DOI] [PubMed] [Google Scholar]
- 73.Ku SH, Lee SH and Park CB, Biomaterials, 2012, 33, 6098–6104. [DOI] [PubMed] [Google Scholar]
- 74.Chen MC, Sun YC and Chen YH, Acta Biomater, 2013, 9, 5562–5572. [DOI] [PubMed] [Google Scholar]
- 75.Thrivikraman G, Mallik PK and Basu B, Biomaterials, 2013, 34, 7073–7085. [DOI] [PubMed] [Google Scholar]
- 76.Heidi Au HT, Cui B, Chu ZE, Veres T and Radisic M, Lab Chip, 2009, 9, 564–575. [DOI] [PubMed] [Google Scholar]
- 77.Wendel JS, Ye L, Zhang PY, Tranquillo RT and Zhang JYJ, Tissue Eng. Part A, 2014, 20, 1325–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dvir T, Timko BP, Brigham MD, Naik SR, Karajanagi SS, Levy O, Jin HW, Parker KK, Langer R and Kohane DS, Nat. Nanotechnol, 2011, 6, 720–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhou J, Chen J, Sun HY, Qiu XZ, Mou YC, Liu ZQ, Zhao YW, Li X, Han Y, Duan CM, Tang RY, Wang CL, Zhong W, Liu J, Luo Y, Xing M and Wang CY, Sci. Rep, 2014, 4, 3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.