Abstract
Induced pluripotent stem cells (iPSCs) offer an unprecedented opportunity to study human physiology and disease at the cellular level. They also have the potential to be leveraged in the practice of precision medicine, for example, personalized drug testing. This statement comprehensively describes the provenance of iPSC lines, their use for cardiovascular disease modeling, their use for precision medicine, and strategies through which to promote their wider use for biomedical applications. Human iPSCs exhibit properties that render them uniquely qualified as model systems for studying human diseases: they are of human origin, which means they carry human genomes; they are pluripotent, which means that in principle, they can be differentiated into any of the human body’s somatic cell types; and they are stem cells, which means they can be expanded from a single cell into millions or even billions of cell progeny. iPSCs offer the opportunity to study cells that are genetically matched to individual patients, and genome-editing tools allow introduction or correction of genetic variants. Initial progress has been made in using iPSCs to better understand cardiomyopathies, rhythm disorders, valvular and vascular disorders, and metabolic risk factors for ischemic heart disease. This promising work is still in its infancy. Similarly, iPSCs are only just starting to be used to identify the optimal medications to be used in patients from whom the cells were derived. This statement is intended to (1) summarize the state of the science with respect to the use of iPSCs for modeling of cardiovascular traits and disorders and for therapeutic screening; (2) identify opportunities and challenges in the use of iPSCs for disease modeling and precision medicine; and (3) outline strategies that will facilitate the use of iPSCs for biomedical applications. This statement is not intended to address the use of stem cells as regenerative therapy, such as transplantation into the body to treat ischemic heart disease or heart failure.
Keywords: AHA Scientific Statements; models, cardiovascular; precision medicine; stem cells, induced pluripotent
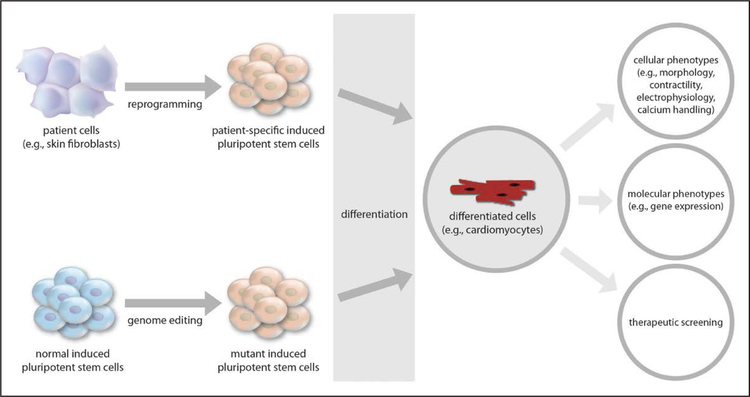
Human pluripotent stem cells (hPSCs) offer an invaluable model system for understanding the genetic basis of human cardiovascular diseases. Unlike nonhuman animal models, hPSCs can be tightly genetically matched to patients with disease. Previously, only in unusual circumstances was it feasible to study primary tissues such as cardiac muscle and blood vessels obtained directly from living patients, and even in such circumstances, the amount of tissue was limited. hPSCs represent a new approach that provides an unprecedented opportunity to study human cells that are matched to the patients of interest. Because they can be expanded into very large numbers and differentiated into a variety of cell types that are relevant to cardiovascular diseases—including cardiomyocytes, vascular endothelial and smooth muscle cells, and hepatocytes—hPSCs can in principle provide a limitless source of material with which to dissect the molecular underpinnings of the patient’s disease process within and beyond the cardiovascular system (Figure 1).
Figure 1.
Induced pluripotent stem cells for cardiovascular disease modeling and precision medicine studies.
hPSCs come in several varieties. Human embryonic stem cells (hESCs), first reported in 1998, are derived directly from human embryos.1,2 Because making each hESC line typically entails the destruction of an embryo, the cell line is not matched to any living person. Arguably, this limits the utility of the hESC line for disease modeling and for precision medicine, because the health of the potential person represented by the source embryo is unknowable. Furthermore, most embryos used for the generation of hESCs have been left over from fertility treatments and, if implanted and taken to term normally, would not yield offspring expected to be strongly predisposed to any particular disease; however, as a result of preimplantation genetic diagnosis, in some cases the embryo is known to carry a specific, highly penetrant disease mutation. When the latter occurs, one can reasonably assume that the hESC line is a good proxy for the disease.
Representing a second type of hPSCs are stem cells derived by somatic cell nuclear transfer, which entails the transfer of a nucleus from a differentiated cell into a denucleated ovum.3 Although this method is technically challenging and rarely used, it does offer a way to produce hPSC lines that are matched to people known to have the relevant disease. In 1 reported case, the hPSC line was made via somatic cell nuclear transfer from a patient with type 1 diabetes mellitus.4
The third type, induced pluripotent stem cells (iPSCs), is now the most abundant and widely used type of hPSCs. In their pioneering work, Yamanaka and colleagues explored the possibility of reprogramming somatic (ie, nongerm) cells into pluripotent stem cells via the heterologous expression of some combination of transcription factors.5 They identified 4 factors (now known as the reprogramming factors or Yamanaka factors)—Oct3/4, Sox2, Klf4, and c-Myc—that could achieve the induction of pluripotency with mouse fibroblasts.5 This discovery was followed by a rapid succession of studies reporting that the Yamanaka factors (or some variation thereof) could successfully reprogram various types of human somatic cells.6–9
Initially, there were concerns that despite also being pluripotent, iPSCs were substantively different than hESCs, which were considered the gold standard in the field. Over the past decade, these concerns have largely subsided. Although there is evidence that iPSCs initially retain some degree of epigenetic memory that reflects the identity of the somatic cell source, which could skew their ability to differentiate into certain cell types, this appears to dissipate as the cells are passaged in culture.10–12 Initial reports suggested that the process of reprogramming itself resulted in aberrant epigenetic signatures in iPSCs that distinguished them from hESCs.13,14 However, a recent study carefully examining this issue in isogenic (shared genetic background) hESCs and iPSCs concluded that transcriptional differences between hESCs and iPSCs are minor and inconsistent, and for all practical purposes, these cell types are molecularly and functionally equivalent.15
OBTAINING iPSCs FOR RESEARCH USE
The use of iPSCs has far exceeded the use of hESCs, because of the unique ability to genetically match iPSCs to living people for whom clinical data might be available. Moreover, hESCs have regulatory restrictions on their use. We outline the means by which investigators can obtain iPSCs from patients with specific traits or diseases, or iPSCs with specific genotypes of interest.
Methods to Derive New iPSC Lines
The first decision to make when generating iPSCs is the cell type to use for reprogramming. Many different somatic cell types have been demonstrated as being competent for the induction of pluripotency. Initially, dermal fibroblasts were a favored source; they could be readily obtained from a patient via a skin punch biopsy, a relatively noninvasive procedure, albeit a surgical procedure with a potential risk of infection of the biopsy site or (more likely) of the biopsied tissue itself. An important development in the field was the ability to perform reprogramming on peripheral blood cells, typically T cells.16–18 Patients could be more easily recruited for a procedure that involves only a routine, nominally uncomfortable, sterile blood draw. It also made it feasible to draw upon peripheral blood cells that previously had been collected for unrelated purposes and placed in long-term storage in a frozen state. Other easily accessible, risk-free cellular sources for reprogramming are renal tubular cells collected from urine samples19 and keratinocytes harvested from plucked hair follicles.20 One consideration in choosing the cellular source is the concern that some types of cells, such as blood cells and dermal fibroblasts, might carry a higher mutational burden because of high turnover rates and exposure to ultraviolet radiation. This could be particularly relevant when deriving iPSCs from older donors, because mutations and other abnormalities appear to occur with increasingly higher frequency with increased age.21
The second choice to make is the method by which to deliver the Yamanaka factors into the cell type. Initial efforts to generate iPSCs entailed the use of an integrating virus (eg, retrovirus or lentivirus) to introduce the factors as DNA transgenes. This led to concerns that constitutive expression of the factors would alter the biology of the iPSCs and interfere with their differentiation into somatic cell types, as well as the fear that undesirable transgenes could be inserted into the genome (eg, oncogenes or tumor suppressor genes). Indeed, the differentiation competency of iPSC lines could be affected by a disproportionate “stemness” load caused by integrated transgenes, which underscores the value of nonintegrative nuclear reprogramming.22 Subsequent efforts focused on the use of transient, integration-free methods of delivering the Yamanaka factors, namely, via Sendai virus (a nonintegrating virus that eventually is lost from newly derived iPSCs as they are passaged),23 episomal plasmid vectors,24,25 minicircle vectors,26 mini-intronic plasmids,27 and messenger RNA transfection.28 A recent comprehensive study comparing the various integration-free methods for the derivation of iPSCs concluded that there was no substantive difference in quality among the iPSC lines generated by the different techniques, but rather that the choice could be safely made based on the needs of the laboratory (Table 1).29 For example, the authors endorsed the use of Sendai virus because of its efficacy in fibroblasts and blood cells, as well as its availability as a commercial reagent, but noted that the unavailability of clinical-grade Sendai virus makes it less useful in generating iPSCs intended for clinical use (eg, transplantation). RNA transfection was noted to be quick and efficient in fibroblasts but was not reported to be effective in blood cells. Episomal vectors were recommended for clinical-grade applications because of the simplicity of the reagents and quick clearance from iPSCs compared with the Sendai virus.
Table 1.
Differences in iPSC Reprogramming Methods
| Feature | RNA Transfection | Sendai Virus | Episomal Vectors | Lentivirus |
|---|---|---|---|---|
| Efficiency (with fibroblasts) | High | Moderate | Low | High |
| Reliability (with fibroblasts) | Moderate | High | High | High |
| Amount of work | High | Low | Low | Very high (requires excision of vector) |
| Aneuploidy rate | Low | Moderate | High | Moderate |
| Number of input cells (fibroblasts) | Low | High | High | Moderate |
| Time to iPSC colony emergence | Low | Moderate | Moderate | Moderate |
| Efficiency (blood cells) | Not reported | High | High | High |
| Reliability (blood cells) | Not reported | High | High | High |
| Special equipment needs | 5% O2 incubator | None | Transfection device and O2 incubator | None |
| Proportion of iPSC lines free of reprogramming reagents by passage 5 | Very high | None | Moderate | None |
| Proportion of iPSC lines free of reprogramming reagents by passage 9–11 | Very high | High | Moderate | None |
iPSC indicates induced pluripotent stem cell.
Adapted from Schlaeger et al29 by permission from Macmillan Publishers Ltd: Nature Biotechnology. Copyright © 2014, Springer Nature.
Upon successful generation of the desired iPSCs, various quality control measures are routinely undertaken. Traditionally, the gold standard to establish pluripotency is to implant the iPSCs into an immunodeficient animal and monitor for formation of teratomas that include tissues of all 3 developmental lineages (ectoderm, mesoderm, and endoderm). Because this is laborious and arguably entails the needless use of animals, newer alternative methods now include formation of embryoid bodies in vitro and assessment that they include cells of the 3 lineages; flow cytometry for markers of pluripotency, such as TRA-1–60 and SSEA-4; and detection of TRA-1–60 and SSEA-5 by nanoparticle-based surface-enhanced Raman scattering technology, which has been demonstrated to be highly sensitive and specific for the identification of pluripotent cells.30 An additional check can be provided through the use of gene expression “scorecards” that evaluate the pluripotency or differentiation capacity of an iPSC line.31,32
Newly made iPSC lines should also be assessed for genomic integrity via cytogenetic karyotyping or array-based virtual karyotyping. The latter has the advantage of potentially detecting copy number changes and microdeletions and microduplications. There has been concern that iPSCs can accumulate mutations during the process of reprogramming,33 which makes them less than perfectly matched to the donor individuals. Furthermore, other studies have established that irrespective of reprogramming, hPSCs propagated for some length of time in culture will unavoidably accumulate mutations.34–36 Routine exome or genome sequencing of newly generated iPSCs is not currently the norm, but as the cost of sequencing continues to fall, sequencing of new iPSC clones will be advisable to assess for any differences from the donor’s germline genome. This might be especially warranted for iPSCs from older donors, in light of the observation that these iPSCs have a higher mutational burden at baseline.21
For iPSC lines that are intended for use for precision medicine applications (ie, to obtain information that will be directly translated to patient care, eg, response to a medication), even more stringent quality control measures are necessary. When they are first being established, iPSC lines should be monitored for clearance of whatever vectors were used for reprogramming (eg, episomal vectors, Sendai virus), and they should not be used until total clearance is rigorously confirmed. For as long as they are in use, iPSC lines need to be continuously monitored for infection, particularly with mycoplasma, which can alter the properties of the cells in ways that might confound phenotypic analyses. Periodic sequencing of iPSC lines as they are propagated in culture should be performed, both to confirm the identity of the cell lines (to ensure they have not inadvertently been mixed with or replaced by other cell lines in the laboratory) and to monitor for new mutations that might affect the properties of the cells. If the iPSC lines are to be used for differentiation into a particular cell type (eg, cardiomyocytes), the lines should initially be evaluated for their propensity to yield the desired cell type before they are committed to use in a study. Use of a poorly differentiating cell line will undermine the goals of the study. It should be recognized that these various quality control measures will add substantially to the costs of an iPSC-based study, and this needs to be proactively considered when planning the budget for the work.
Although it is quite feasible for reprogramming to be performed in an individual laboratory, a number of institutions have established core facilities dedicated to the generation, propagation, quality control, and dissemination of iPSC lines. This allows for a significant reduction of costs and streamlining of the procedure through bulk ordering of reagents and the efficient generation of multiple derivations in parallel by experienced staff. Core facilities now typically charge several thousand US dollars for the generation of iPSC lines from an individual donor, performed over the course of several months—a substantial cost and time investment, but given that only a decade has passed since the discovery of pluripotency induction, this represents remarkable progress.
Availability of Preexisting iPSC Lines
The easiest means by which an investigator can use iPSC lines from patients with specific diseases of interest, or individuals with particular genotypes of interest, is to obtain iPSC lines that have already been made elsewhere. Although this can be achieved via collaboration with other investigators, individual investigators will typically have made at most a few iPSC lines in their own laboratories, which limits the number of lines one can expect to obtain from collaborators. Drawing from the example set by the Jackson Laboratory in serving as a central repository to which investigators can contribute their genetically modified mouse models and thereby make them available to the scientific community, several organizations have been seeking to establish so-called biobanks to serve as repositories for large iPSC collections that are easily accessible to any interested investigators.
In light of the considerable expense of generating iPSC lines, substantial funding is necessary to underwrite the establishment of biobanks. The California Institute of Regenerative Medicine (Oakland, CA) is underwriting the costs of generating 3 iPSC lines each from 3000 patients and healthy individuals, to be stored and distributed by the Coriell Institute for Medical Research (Camden, NJ). These iPSC lines are intended to be broadly representative of a variety of disorders, including cardiovascular diseases, liver diseases, respiratory diseases, Alzheimer disease, autism, and neurodevelopmental disabilities. The National Heart, Lung, and Blood Institute funded the Next Generation Genetic Association Studies (NextGen) Consortium, which is spread across 8 academic institutions in the United States with the goal of generating iPSC lines from >1500 individuals, including those with various cardiovascular, pulmonary, and blood disorders and their healthy control counterparts.37 These cell lines are being stored and distributed by WiCell Research Institute (Madison, WI). On a smaller scale, the National Heart, Lung, and Blood Institute and the California Institute of Regenerative Medicine are funding the Stanford Cardiovascular Institute Biobank, which is generating iPSC lines from individuals with specific cardiovascular disorders such as hypertrophic cardiomyopathy (HCM), dilated cardiomyopathy (DCM), and cardiac rhythm disorders. Other regional efforts, including those sponsored through the Mayo Clinic Center for Regenerative Medicine and linked to dedicated Regenerative Medicine Consult Service portals, are under way to ensure long-term comprehensive repositories that cover a variety of human diseases. Similar international efforts are under way, such as the recently launched European Bank for induced pluripotent Stem Cells.
Genome-Editing Tools to Generate Stem Cell Lines With Specific Genotypes
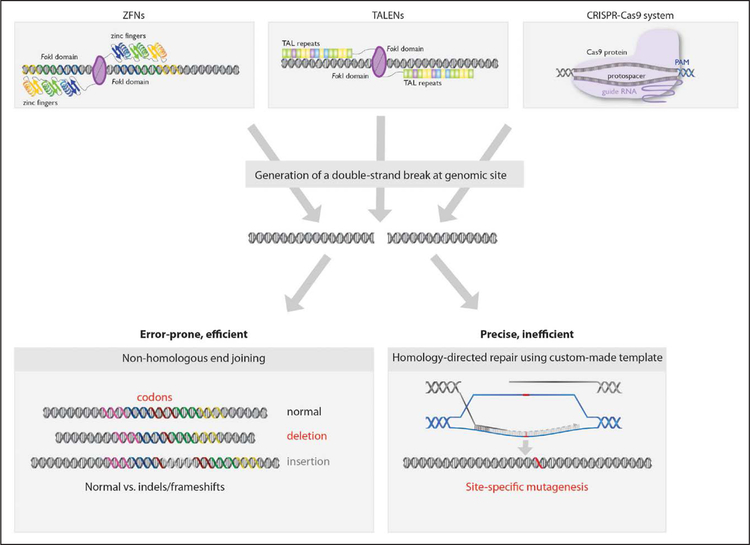
In many cases, a particular disease-associated mutation might be of interest to an investigator but sufficiently rare in a population (and perhaps even unique to 1 individual) that it would be infeasible to recruit an individual with the mutation and generate iPSCs from that person without a relevant iPSC line being available in a biobank. In such cases, genome-editing tools can now introduce specific disease-associated mutations into normal or wild-type iPSC lines from healthy individuals (Figure 1). A variety of tools have been used for this purpose, including adenoviral vectors, zinc-finger nucleases, transcription activator-like effector nucleases (TALENs), and clustered regularly interspaced short palindromic repeats (CRISPR)-associated (Cas) systems (Figure 2).38 The recent introduction of CRISPR-Cas9 to edit mammalian cells has revolutionized the biomedical sciences because of its ease of use, allowing any investigator with basic molecular biology skills to use it, and because of its relatively high efficiency compared with other tools.
Figure 2. Genome-editing tools for use in induced pluripotent stem cells.
Cas indicates CRISPR-associated; CRISPR, clustered regularly interspaced short palindromic repeats; PAM, protospacer adjacent motif; TAL, transcription activator-like; TALENs, transcription activator-like effector nucleases; and ZFN, zinc-finger nuclease.
Regardless of whether zinc-finger nucleases, TALENs, CRISPR-Cas9, or even newer nucleases are used, the aim is to introduce double-strand DNA breaks at desired locations in the genome. The cellular DNA repair machinery uses 1 of 2 mechanisms to restore the integrity of a site of a DNA break, nonhomologous end-joining or homology-directed repair (HDR) (Figure 2). Nonhomologous end-joining is an error-prone process that can introduce insertions or deletions (indels) of various sizes, although such indels are usually a few base pairs in length. HDR is a more precise repair mechanism that uses a homologous template—typically a sister chromatid or chromosome, but alternatively a synthetic, custommade template introduced into the cell by the investigator, allowing for the introduction of specific, desired changes into the genome. In principle, genome editing via HDR provides a means by which any desired disease-associated mutation can be introduced into any iPSC line (or, conversely, a preexisting mutation can be corrected). It can also be used to introduce useful DNA sequences, such as a reporter gene in a desired locus. However, HDR is typically a less efficient mode of genome editing than nonhomologous end-joining, because HDR is restricted to the S and G2 phases of the cell cycle, whereas nonhomologous end-joining occurs throughout the cell cycle. Accordingly, it is easier to knock out genes by introducing frameshift indels into coding sequences than it is to knock in a mutation.
The ability to introduce desired mutations into iPSC lines has several potential advantages compared with generating iPSC lines from patients. Except in cases where a patient with the desired mutation is simply not accessible to the investigator, it will typically be much quicker and less expensive to use genome editing than it is to recruit and consent a new patient into a research study, collect somatic cells, undertake reprogramming, and perform quality control and then expansion of iPSC clones through a number of passages before they can be used for studies. The investigator can genome-edit a well-characterized wild-type cell line that has been used for studies previously and for which differentiation protocols have already been tested and optimized rather than take the chance that the newly generated patient-specific iPSC lines may not prove suitable for the desired studies. However, there is a potential shortcoming to this shortcut approach, because it depends on the disease-associated mutation being fully penetrant in the chosen wild-type iPSC line. If the iPSC line does not have a permissive genetic background (ie, if it contains genetic modifiers that mitigate the phenotypic consequences of the mutation), then the genome-edited iPSC line will not properly reflect the disease state. This is presumably not an issue with a patient-derived iPSC line, because the very fact of the patient having the disease indicates that the genetic background is permissive for the phenotypic consequences of the mutation.
Yet another major advantage of genome editing of iPSCs is that it allows more rigorous study designs than the generation of iPSC lines from genetically unmatched disease patients and control individuals, an issue that is discussed in more detail in Study Design.
iPSCs FOR CARDIOVASCULAR DISEASE MODELING
Disease modeling represents perhaps the most productive use of iPSCs to date. A singular advantage of iPSCs is that they are genetically matched to the person from whom they were derived, which makes them ideally suited for the study of diseases that have a strong underlying genetic cause, such as classic monogenic disorders. At the same time, the fact that iPSCs have been reprogrammed from adult cells into a baseline state stripped of many of the epigenetic changes caused by environmental influences during the person’s lifetime allows the investigator to better study the genetic contribution to the disease. This is an important consideration when modeling complex diseases that typically reflect the interplay of multiple genetic and environmental factors. In either case, iPSCs have proven useful in helping researchers better understand how disease genotypes at the genetic level are manifested as phenotypes at the cellular level with respect to the broad disease categories described in this section (Table 2).39
Table 2.
Partial List of Cardiovascular and Related Diseases Modeled With Human Induced Pluripotent Stem Cells or Human Embryonic Stem Cells
| Disease Modeled | Mutated Gene |
|---|---|
| Dilated cardiomyopathy | TTN, TNNT2, LMNA, PLN, DES |
| Duchenne muscular dystrophy | DMD |
| Barth syndrome | TAZ |
| Hypertrophic cardiomyopathy | MYH7 |
| Arrhythmogenic right ventricular dysplasia | PKP2 |
| Left ventricular noncompaction | TBX20, GATA4 |
| Long-QT syndrome type 1 | KCNQ1 |
| Jervell and Lange-Nielsen syndrome | KCNQ1 |
| Long-QT syndrome type 2 | KCNH2 |
| Long-QT syndrome type 3 | SCN5A |
| Timothy syndrome | CACNA1C |
| Catecholaminergic polymorphic ventricular tachycardia type 1 | RYR2 |
| Catecholaminergic polymorphic ventricular tachycardia type 2 | CASQ2 |
| Brugada syndrome | SCN5A |
| Calcific aortic valve | NOTCH1 |
| Williams-Beuren syndrome | ELN |
| Familial pulmonary hypertension | BMPR2 |
| Familial hypercholesterolemia | LDLR, PCSK9 |
| Familial hypobetalipoproteinemia | PCSK9 |
| Tangier disease | ABCA1 |
| Dyslipidemia | SORT1 |
| Maturity-onset diabetes of the young type 2 | GCK |
| Insulin resistance | AKT2 |
| Hypoinsulinemic hypoglycemia and hemihypertrophy | AKT2 |
Cardiomyopathies
Familial cardiomyopathies are among the best characterized of monogenic cardiovascular disorders, with mutations in dozens of genes having been linked to various forms of cardiomyopathy. As such, there has been significant interest in the generation of iPSCs from patients with familial cardiomyopathies and differentiation of the iPSCs into cardiomyocytes (referred to here as iPSC-CMs), as well as early success in gaining new molecular insights into the pathogenesis of the disorders.
DCM is the most common type of cardiomyopathy, with familial DCM cases thought to represent a substantial proportion of the total cases. DCM manifests with ventricular enlargement and thinning and heart failure from a severely compromised ejection fraction. The most commonly mutated gene in familial DCM cases is TTN (titin), followed by LMNA (prelamin-A/C), MYH7 and MYH6 (myosin-7 and −6), SCN5A (sodium channel protein type 5 subunit alpha), MYBPC3 (myosin-binding protein C, cardiac-type), and TNNT2 (troponin T, cardiac muscle).40 In almost all cases, familial DCM is associated with dominant mutations. Perhaps 10% to 20% of familial DCM cases can be attributed to truncating mutations (frameshift, nonsense, or splice site) in TTN, which encodes a component of the sarcomere.41 iPSCs generated from DCM patients with either truncating or missense mutations in TTN, when differentiated into iPSC-CMs and assembled into cardiac microtissues, displayed sarcomere insufficiency, impaired responses to mechanical and β-adrenergic stress, and attenuated growth factor and cell signaling activation.42 Genome-edited wild-type iPSC-CMs into which TTN truncating mutations had been introduced displayed similar phenotypes.42
The most intensively studied familial DCM iPSC lines to date were derived from a family whose affected members harbor a missense R173W mutation in TNNT2, which also encodes a component of the sarcomere. Several iPSC lines were generated from affected family members, and several were generated from unaffected family members as control lines. The mutant iPSC-CMs exhibited abnormal calcium handling, reduced contractility, and myofibrillar disarray, which were exacerbated with β-adrenergic stimulation.43 A deeper molecular characterization of these cell lines revealed increased expression of the phosphodiesterase genes PDE2A and PDE3A via epigenetic activation that was responsible for the compromised β-adrenergic signaling and contractile dysfunction.44
A number of other studies have characterized iPSC-CMs from patients with DCM, either from a primary familial disorder or as part of a genetic syndrome. iPSC-CMs with either a nonsense or missense mutation in LMNA were found to have increased nuclear bleb formation and micronucleation, as well as increased apoptosis on electrical stimulation.45 iPSC-CMs with an in-frame deletion mutation in PLN (phospholamban) showed calcium handling abnormalities that were reversed in iPSC-CMs in which the mutation had been corrected by genome editing.46 A novel DES (desmin) missense mutation found by exome sequencing in a patient with DCM was associated with abnormal desmin aggregations, calcium handling, and response to inotropic stress in the patient’s iPSC-CMs.47 Duchenne muscular dystrophy is a primarily skeletal muscle disorder that is often accompanied by a dilated-type cardiomyopathy, which is often the underlying cause of death in older children surviving past the initial (skeletal-based) stages of the disease. iPSC-CMs from a number of patients with Duchenne muscular dystrophy displayed abnormalities consistent with this disease pathophysiology.48,49 Barth syndrome is a mitochondrial disorder cause by TAZ (tafazzin) mutations that affects both cardiac and skeletal muscle and manifests in part as DCM. iPSC-CMs from 2 patients with either a frame-shift or missense mutation in TAZ exhibited mitochondrial defects, excess levels of reactive oxygen species, abnormal sarcomere assembly, and impaired contractility.50 Genome-edited wild-type iPSC-CMs into which TAZ frameshift mutations were introduced had the same abnormalities as the patient-specific iPSC-CMs.50
Familial HCM is a dominant disorder that manifests as asymmetrical ventricular wall thickening with increased risk of sudden cardiac death (SCD). The most commonly mutated genes in familial HCM cases are sarcomere components, including MYH7, MYBPC3 (myosin-binding protein C, cardiac type), TNNT2, TNNI3 (troponin I, cardiac muscle), and TPM1 (tropomyosin α−1 chain).51 The most intensively studied familial HCM iPSC lines to date were derived from a family whose affected members harbor a missense mutation in MYH7. The iPSC lines were generated from 10 affected and unaffected family members (2 parents and 8 children).52 Besides displaying sarcomere disarray, the mutant iPSC-CMs displayed cellular enlargement and contractile arrhythmia in the setting of abnormal calcium handling, both of which could be normalized by treatment with the calcium channel blocker verapamil. iPSC-CMs with a different MYH7 missense mutation also displayed sarcomere disarray and electrophysiological abnormalities that could be normalized with verapamil treatment.53
Arrhythmogenic right ventricular cardiomyopathy (ARVC), also called arrhythmogenic right ventricular dysplasia, is typically a dominant disorder in which there is gradual, adult-onset fibrofatty replacement of cardiomyocytes, predominantly in the right ventricle but sometimes affecting the left ventricle, with increased risk of SCD. The most commonly mutated genes in ARVC cases are components of a structure involved in cell-to-cell adhesion called desmosome: PKP2 (plakophilin 2), DSG2 (desmoglein 2), DSP (desmoplakin), DSC2 (desmocollin 2), and JUP (junction plakoglobin).54 The most intensively studied ARVC iPSC lines to date were derived from 2 unrelated individuals, one homozygous for a mutation in PKP2 that causes a splicing defect and the other heterozygous for a frame-shift mutation in PKP2.55 Although the iPSC-CMs from these patients exhibited subtle molecular defects in standard differentiation conditions, they did not fully manifest ARVC-related phenotypes until they were exposed to conditions that resulted in metabolic aging, namely, treatment with a cocktail of 5 adipogenic factors, whereupon they displayed increased lipogenesis and apoptosis, as well as abnormal calcium handling. In another study, iPSC-CMs from 2 patients heterozygous for different PKP2 frameshift mutations had increased lipid accumulation and desmosomal distortion in standard differentiation conditions, phenotypes that were intensified on treatment with adipogenic medium.56 These same findings were observed in yet another study with iPSC-CMs from a patient heterozygous for a missense mutation in PKP2.57 Phenotypes observed in iPSC-CMs from patients with PKP2 mutations could be reversed with a small molecule originally identified in a high-throughput screen in a zebrafish model of ARVC.58 A lesson to draw from these studies is that extra stimuli could be needed to fully provoke disease-related phenotypes in the in vitro differentiated cardiomyocytes, particularly with adult-onset diseases such as ARVC. However, caution also must be taken, because the relevance of the extra stimuli to the native human myocardium in the disease setting might be unclear. In addition, these studies also suggest the need to perform more comprehensive side-by-side comparisons between iPSC-CM phenotypes and cardiac abnormalities found in actual donor hearts.
Left ventricular noncompaction, also known as spongiform cardiomyopathy, results when trabeculated structures in the ventricular wall that form during development fail to undergo typical compaction from spongy into solid structures.59 Potential clinical consequences include heart failure and SCD. iPSC-CMs from family members with left ventricular noncompaction who harbored a nonsense mutation in TBX20 (T-box protein 20), compared with iPSC-CMs from unaffected family members, displayed a proliferation defect and abnormal activation of transforming growth factor-β signaling, a finding that was also observed in genetically modified mice.60 iPSC-CMs from patients with delayed-onset cardiomyopathy with noncompaction linked to a missense mutation in GATA4 (GATA binding protein 4) showed impairments in contractility, calcium handling, and metabolic activity.61
Rhythm Disorders
Arrhythmic syndromes are the cardiovascular disorders most extensively studied with iPSC-based models to date, in particular the long-QT syndromes (LQTS). LQTS is marked by delayed repolarization of the heart after contraction, which manifests as an increased QT interval on the ECG and predisposes patients to ventricular arrhythmias and SCD. LQTS is usually inherited in an autosomal dominant manner, although there are a few forms that are inherited in an autosomal recessive manner. Mutations in at least 15 genes have been linked to LQTS, typically affecting the function of potassium, sodium, or calcium channels in cardiomyocytes.62 The 3 most commonly implicated genes, respectively, responsible for LQTS1, LQTS2, and LQTS3, are KCNQ1 (potassium voltage-gated channel subfamily Q member 1), KCNH2 (potassium voltage-gated channel subfamily H member 2, also known as hERG), and SCNA5 (sodium voltage-gated channel α-subunit 5). Intriguingly, these 3 subtypes of LQTS appear to have different triggers for ventricular arrhythmia.63 LQTS1 is triggered by exercise, particularly swimming, whereas LQTS2 is triggered by emotional stress and auditory stimuli (such as alarm clocks). In contrast, arrhythmic events in LQTS3 tend to occur during sleep. These distinct triggers suggest distinct mechanisms by which mutant alleles of the 3 genes predispose to SCD.
In the first published iPSC-based study of an arrhythmic syndrome, iPSCs were derived from affected members of a family with autosomal dominant LQTS1 associated with a missense mutation in KCNQ1.64 Mutant iPSC-CMs displayed prolongation of the action potential duration (APD), as well as increased tachyarrhythmia on exposure to a β-adrenergic agonist, isoproterenol. The mutation was found to confer a dominant-negative impairment of membrane localization of the KCNQ1 channel subunit, resulting in a 70% to 80% reduction in the slowly activating delayed rectifier potassium current (IKs) in the iPSC-CMs. Subsequent studies of iPSC-CMs from LQTS1 patients with KCNQ1 missense or frameshift mutations found similar phenotypes.65,66 iPSC-CMs generated from a patient with Jervell and Lange-Nielsen syndrome, a recessive disorder with severe QT prolongation, congenital bilateral deafness, and high risk of SCD that is usually caused by mutations in KCNQ1, showed the expected pronounced changes in APD, IKs current, and sensitivity to proarrhythmic stressors, as did iPSC-CMs in which genome editing was used to introduce KCNQ1 mutations.67
Numerous studies with iPSC-CMs from patients with LQTS2 associated with missense mutations in KCNH2 have reproducibly shown increased APDs, increased arrhythmogenicity, and decreased rapidly activating delayed rectifier potassium current (IKr), which is sometimes linked to defective trafficking of the hERG protein.68–73 In one study of LQTS2 iPSC-CMs, genetic correction of the KCNH2 mutation reversed the phenotypes,71 and in another study, specific knockdown of the mutant KCNH2 allele with RNA interference reversed the phenotypes, establishing it as a dominant-negative allele.74 In a similar fashion, iPSC-CMs from patients with LQTS3 associated with SCN5A mutations75,76 or Timothy syndrome associated with mutations in CAC-NA1C (calcium voltage-gated channel subunit-αC)77 were found to have increased APDs and either abnormal sodium currents or an abnormal calcium current. Of note, both LQTS1 and LQTS2 have been modeled with iPSC-CMs in which mutant genes were inserted with genome editing, which avoids the need to recruit patients with the particular mutations and to generate new iPSC lines for each patient.78
Another rhythm disorder that has proven amenable to modeling with iPSC-CMs is catecholaminergic polymorphic ventricular tachycardia (CPVT). In CPVT, ventricular arrhythmias are triggered by exercise or emotional stresses in the absence of structural heart disease. Although these arrhythmias are often self-resolving, SCD does occur in many CPVT patients. The most common cause of CPVT is autosomal dominant mutations in RYR2 (ryanodine receptor 2), with the next most common being autosomal recessive mutations in CASQ2 (calsequestrin 2); both genes encode regulators of calcium storage and release in the sarcoplasmic reticulum in cardiomyocytes.79 A large number of studies of iPSC-CMs from CPVT patients with missense mutations in one or the other gene have documented abnormal calcium handling and increased arrhythmogenicity on adrenergic stimulation.80–87
Brugada syndrome is yet another disorder that has been modeled with iPSC-CMs. Brugada syndrome, which like LQTS3 has been linked to mutations in SCN5A but is quite distinct from LQTS3, is a disorder that is associated with characteristic electrocardiographic precordial ST-segment elevation and predisposes to ventricular fibrillation and SCD. iPSC-CMs from patients with Brugada syndrome displayed blunted inward sodium currents, increased triggered activity, and abnormal calcium handling; these cellular phenotypes were rescued with genome editing to correct the causal SCN5A mutation.88
Valvular and Vascular Disorders
Bicuspid aortic valve with calcification of the valve is the most common congenital valvular disorder. A familial form of the disease with severe valve calcification has been linked to heterozygous nonsense mutations in NOTCH1, which encodes a transcriptional regulator.89 iPSCs from patients with NOTCH1 mutations were subjected to genome editing to correct the mutations, followed by differentiation of the isogenic cell lines into endothelial cells.90 When assessed in an in vitro model of vascular shear stress, the mutant iPSC-endothelial cells failed to activate antiosteogenic, anti-inflammatory, and antioxidant pathways that were induced in control iPSC-endothelial cells, consistent with a predisposition towards calcification of the aortic valve.
Supravalvular aortic stenosis can occur in isolation as the result of mutations in ELN (elastin) or as part of Williams-Beuren syndrome, in which a deletion on chromosome 7q11.23 entirely removes the ELN gene.91 iPSCs from a patient with a frameshift mutation in ELN, after differentiation into smooth muscle cells, had defective actin filament bundle formation, a significantly higher proliferation rate, and a higher migration rate in a chemotaxis assay.92 Similar phenotypes, along with impaired vascular tube formation, were observed in iPSC-smooth muscle cells from Williams-Beuren patients,92,93 and the phenotypes could be normalized with treatment with the drug rapamycin.93
Pulmonary arterial hypertension causes progressive right-sided heart failure that is accompanied by substantial morbidity and mortality. A subset of patients with pulmonary arterial hypertension exhibit an inherited form of the disease, with the majority of familial cases linked to mutations in BMPR2 (bone morphogenetic protein receptor 2). iPSC-endothelial cells from patients with familial pulmonary arterial hypertension and BMPR2 mutations displayed reduced adhesion, survival, migration, and angiogenesis compared with iPSC-endothelial cells from either unaffected carriers of BMPR2 mutations or healthy individuals without BMPR2 mutations.94 These findings suggest that iPSC-endothelial cells can model not only familial pulmonary arterial hypertension caused by BMPR2 mutations but also the effects of genetic modifiers of BMPR2 mutations resulting in an absence of disease.
Metabolic Risk Factors for Ischemic Heart Disease
Blood concentrations of lipids, namely, low-density lipoprotein cholesterol (LDL-C) and high-density lipoprotein cholesterol, are well-established risk factors for ischemic heart disease. A number of monogenic lipid disorders have been defined. The best known is autosomal dominant hypercholesterolemia, also known as familial hypercholesterolemia (FH), which can be caused by mutations in LDLR (LDL receptor), APOB (apolipoprotein B), or PCSK9 (proprotein convertase subtilisin/kexin type 9), all of which are important regulators of blood LDL-C levels.95 Two studies generated iPSCs from FH patients with mutations in LDLR and found that differentiated hepatocytes were impaired in LDL particle uptake, consistent with the elevated blood LDL-C levels seen in FH patients.96,97 The second study also found that the patient-derived hepatocytes responded to treatment with a statin drug that increased LDL uptake.97 Another study generated iPSCs from an FH patient with a gain-of-function mutation in PCSK9 and from a patient with familial hypobetalipoproteinemia who had an abnormally low LDL-C level caused by a loss-of-function PCSK9 mutation. As with the LDLR mutant cell lines, the FH iPSC-hepatocytes displayed impaired LDL particle uptake, whereas the familial hypobetalipoproteinemia iPSC-hepatocytes displayed increased LDL particle uptake.98
Tangier disease, also known as hypoalphalipoproteinemia, is marked by abnormally low blood levels of high-density lipoprotein cholesterol caused by recessive mutations in ABCA1 (ATP-binding cassette subfamily A member 1). ABCA1 is the molecule that effluxes cholesterol from cells such as macrophages onto high-density lipoprotein particles and thus is a key mediator of reverse cholesterol transport. iPSCs derived from patients with Tangier disease with missense or nonsense ABCA1 mutations displayed impaired cholesterol efflux on differentiation into macrophages.99 The patient iPSC-macrophages also showed increased expression of proinflammatory cytokines. In a complementary study, genome editing was used to introduce frameshift mutations in the ABCA1 gene to knock out the gene in hPSCs.100 As with the aforementioned Tangier disease patient iPSC-macrophages, ABCA1-knockout differentiated macrophages had both impaired cholesterol efflux and increased expression of cytokines compared with isogenic wild-type differentiated macrophages.
Stem cell–based analyses proved useful in clarifying the function of SORT1, a novel regulator of blood LDL-C levels identified by genome-wide association studies. Conflicting studies in mice disagreed on whether hepatic SORT1 decreased or increased lipoprotein particle secretion.101,102 Genome editing was used to knock out the SORT1 gene in hESCs, and SORT1-knockout differentiated hepatocytes displayed increased lipoprotein particle secretion.103 SORT1 also has a role in insulin-stimulated glucose transport, confirmed by the finding that glucose transport was unchanged in SORT1-knockout differentiated adipocytes on stimulation with insulin, in contrast to isogenic wild-type differentiated adipocytes, in which glucose transport was increased.103
Type 2 diabetes mellitus and its attendant insulin resistance are also risk factors for ischemic heart disease. As with other diseases, iPSCs have proven the most useful when modeling monogenic forms of diabetes mellitus. iPSCs derived from patients with maturity-onset diabetes of the young type 2, caused by missense mutations in GCK (glucokinase), were differentiated into pancreatic β-cells and transplanted into mice, whereupon they were found to require higher blood glucose levels to stimulate insulin secretion than control iPSC-derived β-cells.104 This phenotype was reversed when genome editing was used to correct the GCK missense mutations.
A dominant negative missense mutation in AKT2 (V-Akt murine thymoma viral oncogene homolog 2) was found to be responsible for severe insulin resistance and diabetes mellitus,105 whereas a putative gain-of-function missense mutation in AKT2 resulted in hypoinsulinemic hypoglycemia and hemihypertrophy, with clinical signs opposite to those of diabetes mellitus.106 Both of these conditions were modeled in an isogenic series of genome-edited hESCs, without the need to recruit the rare patients with these mutations: insulin resistance via knockout of the AKT2 gene, and hypoinsulinemic hypoglycemia via knockin of the AKT2 gain-of-function missense mutation.103 AKT2-knockout-hepatocytes displayed increased glucose production in response to stimulation with dexamethasone, forskolin, and insulin, whereas AKT2-knockin hESC-hepatocytes displayed decreased glucose production. AKT2-knockout hESC-adipocytes showed impaired insulin-stimulated glucose transport and decreased lipid accumulation, whereas AKT2-knockin hESC-adipocytes showed increased baseline glucose transport, regardless of the presence of insulin, and increased lipid accumulation. These in vitro phenotypes accord well with the metabolic disturbances of the patients. Additionally, AKT2-knockin hESC-adipocytes were observed to have abnormally high secretion of inflammatory molecules, which suggests an unrecognized inflammatory component to hypoinsulinemic hypoglycemia and hemihypertrophy.103
METHODOLOGICAL CONSIDERATIONS IN DISEASE MODELING
Study Design
The vast majority of iPSC-based disease-modeling studies have compared 1 or only a few iPSC lines from patients with a disease with 1 or a few iPSC lines from healthy control individuals. In general, these studies have concluded that the phenotypic differences observed between differentiated disease cells and differentiated control cells are attributable to the disease mutations. However, such conclusions are potentially confounded by other factors that can underlie the observed differences. Most obviously, differences in genetic background are inevitable unless the comparators are identical twins (in which case one would not expect to see any phenotypic differences between the iPSC lines in the first place). Such differences are a particular concern when the disease cells and the control cells are not matched for sex and ethnicity. Some studies attempted to mitigate the confounding by using control cells from unaffected individuals in the same families as the patients from whom the disease cells were derived. Nonetheless, because only ≈50% of the genome is shared between any 2 first-degree relatives, confounding from the genetic differences in the other ≈50% of the genome is still a possibility.
Even if the subjects from whom the iPSCs were derived are well matched, there are a host of other potential confounders that could complicate the conclusions of a study. Factors such as differences in the cellular sources for the iPSCs (eg, skin versus blood versus urine), differences in the methodologies used for the induction of pluripotency (eg, lentivirus versus Sendai virus versus episomes versus RNA transfection), differences in the epigenetic states of the iPSC lines used, differences in the passage numbers and acclimation to cell culture conditions of the iPSC lines, and differences in the capacity for differentiation to the desired cell type all can come into play.
With so many potential confounding factors, studies with unmatched iPSC lines should take steps to minimize their influence. One consideration is the expected effect of the disease-associated mutations under study. If the disease is a dominant monogenic disorder, and the disease mutations are presumed to be highly penetrant and have outsize phenotypic effects, then the signal that results from a mutation might well outweigh the noise that results from the confounding factors. However, if a complex, polygenic disease is being studied, any given disease-associated variant could have a small effect size that is swamped by the confounders. This problem is exacerbated if the disease under study is only partly heritable or very heterogeneous, or if the responsible disease-associated variants are unknown. In such cases, the study would be best served by using large numbers of disease and control iPSC lines so as to balance out the confounding factors and thus sharpen the signal-to-noise ratio. However, it can be challenging to use a large number of lines because of patient scarcity, expense of generating the lines, laboratory capacity, etc. The growing number of biobanks from which many iPSC lines for a particular rare disease are available (eg, HCM) or through which a genetically diverse population cohort of iPSC lines is accessible (to make it possible to identify many lines with a particular common DNA variant) now make it more feasible to conduct large, well-powered studies. One caveat is that many iPSCs derived from patients categorized as “diseased” could be from patients who fall under a variable spectrum of disease (especially in complex forms) and not necessarily end stage; thus, careful side-by-side analysis of iPSC phenotypes to donor tissues would be necessary to determine whether this clinical heterogeneity can be recapitulated in iPSCs. However, if true, this can be advantageous in identifying early drivers of disease.
The recent emergence of efficient genome-editing tools provides alternative study designs that eliminate most of the aforementioned confounders. For instance, genome editing can be used to introduce a disease-associated mutation into a wild-type iPSC line, yielding isogenic cell lines that are matched for origin, acclimation to culture conditions, epigenetic status, differentiation capacity, etc. Any differences observed between wild-type and mutant iPSC lines can be more reliably attributed to the mutation itself, thus establishing a causal connection between genotype and phenotype. However, as mentioned in Genome-Editing Tools, this study design may not be informative if the mutation being modeled is not highly penetrant with a large effect size and if the genetic background of the chosen iPSC line is less permissive (protective) for the disease. Put another way, this study design tests for the sufficiency of the mutation to cause disease phenotypes, regardless of genetic background (Figure 3).
Figure 3. Study designs to assess for sufficiency and necessity of disease variants.
iPSC indicates induced pluripotent stem cell.
A different study design entails genome editing of a patient-specific iPSC line to correct the disease-associated mutation. This is somewhat different from introducing the mutation into a wild-type iPSC line, because it tests for the necessity of the mutation to cause disease phenotypes and does not presume that the mutation is highly penetrant (Figure 3). Because the iPSC line was derived from a patient with the disease, it can be presumed that the genetic background is permissive for the disease. In principle, the most rigorous disease-modeling studies would be those in which genome editing of both wild-type and disease iPSCs is performed to test the sufficiency and necessity of a disease mutation in parallel.
The major shortcoming of the genome-editing approach is that it relies on the investigator having properly identified a disease-associated variant to study. If the genetic cause of a disease remains unclear but the disease clearly segregates within a family in a monogenic fashion, then the use of iPSCs from family members might be more productive in learning about the molecular underpinnings of a disease. If the disease is complex (ie, polygenic), then genome editing might not be informative, because numerous variants would need to be edited to fully reproduce (or fully eliminate) the disease state. Genome editing remains sufficiently arduous that altering >1 variant at a time is technically prohibitive, and editing a single variant might not be instructive because of its relatively small contribution to disease. In addressing a complex disease, use of a large number of well-matched patient and control iPSC lines might instead be the most viable approach.
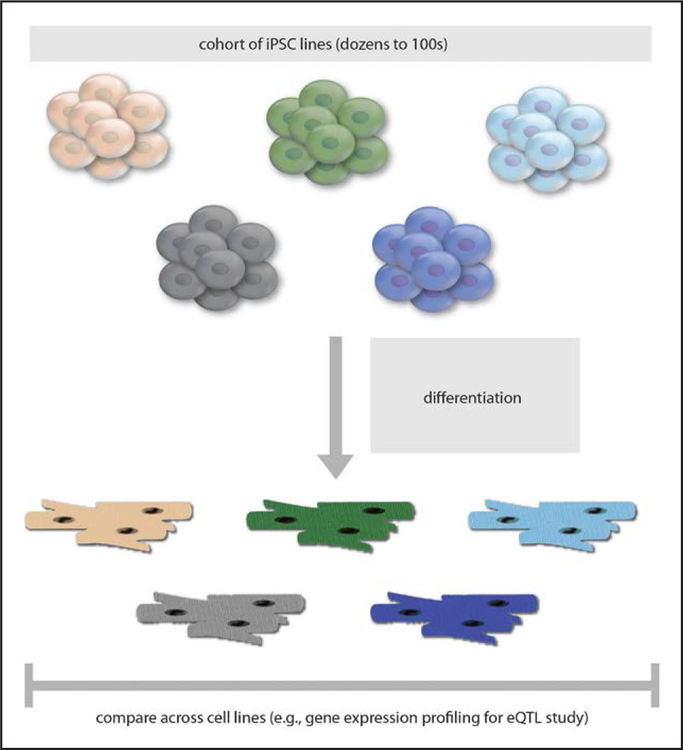
Finally, it is worth noting a study design that is unmoored from any particular disease but that instead seeks to better understand the molecular consequences of naturally occurring genetic variations. In principle, the use of a genetically diverse population cohort of iPSCs numbering in the dozens to hundreds will have sufficient representation of alternate alleles of common genetic variants to identify associations between genotypes and expression levels of nearby genes, known as expression quantitative trait loci (eQTLs) (Figure 4). To date, eQTL studies have been performed with primary tissues obtained from living patients who have undergone surgery or from postmortem cadaveric donors. For obvious reasons, these might not reflect the highest-quality tissues; furthermore, the intact tissues comprise an admixture of multiple cell types, and the obtained material is nonrenewable. By contrast, iPSCs offer an inexhaustible supply of differentiated cells of a single type for use in eQTL studies. Such studies could yield new insights not only into the tissue-specific regulation of gene expression but possibly into other molecular mechanisms, such as the regulation of RNA transcript splicing. One recent study used a population cohort of iPSC-hepatocytes to perform eQTL studies and discovered several novel causal variants and causal genes involved in lipid metabolism.107
Figure 4. Study design using a cohort of iPSC lines.
eQTL indicates expression quantitative trait loci; and iPSC, induced pluripotent stem cell.
Differentiation Protocols
An enormously attractive property of iPSCs is that they can be differentiated in vitro into a variety of lineages— in theory, into all of the existing cell types in the human body. The previous section on cardiovascular disease modeling described studies that used the following differentiated cell types to assess disease-related phenotypes: cardiomyocytes, endothelial cells, smooth muscle cells, hepatocytes, macrophages, and adipocytes. Although such studies can prove informative, they can also have the limitations inherent in the differentiation protocols, namely, the lack of consistency, lack of purity, and lack of maturity of the differentiated cells.
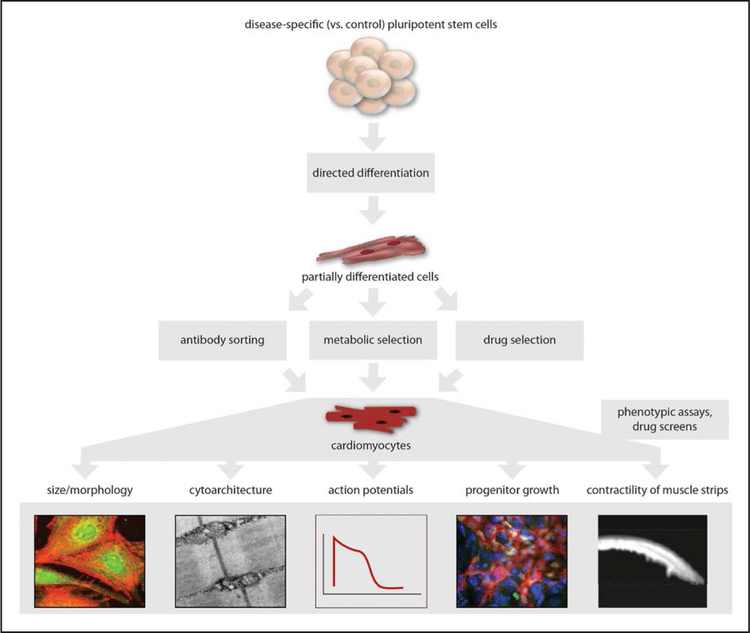
This is illustrated by the continuing efforts to develop and optimize protocols to yield pure, mature cardiomyocytes from hPSCs (Figure 5). Initial protocols involved the dispersion of hPSCs and culturing in suspension, followed by spontaneous aggregation of the cells into embryoid bodies. As a result of spontaneous differentiation of cells in the embryoid bodies into different lineages, some proportion of the cells take on the properties of cardiomyocytes and can then be isolated and studied.108,109 iPSCs have proven to be just as amenable to embryoid body–based differentiation as hESCs.110,111 However, embryoid body–based differentiation is labor intensive and has low efficiency with respect to the proportion of functional cardiomyocytes, as well as significant sample-to-sample variability. This has prompted exploration of protocols in which hPSCs are differentiated in a 2-dimensional monolayer format, which greatly simplifies the process.112 An important conceptual innovation was the use of growth factors or protein inhibitors to direct the differentiation of hPSCs into cardiomyocytes, rather than relying on the hPSCs to spontaneously differentiate into cardiomyocytes.112–116 A widely used monolayer-based differentiation protocol incorporates glycogen synthase kinase-3β and Wnt inhibitors, yielding cardiomyocytes with an efficiency as high as >90%.117 A more recent protocol simplifies the process even further by combining glycogen synthase kinase-3β and Wnt inhibitors with a 3-component basal medium (as opposed to previous protocols that used a basal medium with >20 components).118 In principle, simplification of differentiation conditions should foster more sample-to-sample consistency and reproducibility.
Figure 5.
Cardiomyocyte differentiation.
Another approach to generating differentiated cardiomyocytes in large numbers is to convert hPSCs into multipotent cardiac progenitor cells (CPCs) that in turn have the capacity to be differentiated into cardiomyocytes, smooth muscle cells, and endothelial cells.113,119,120 CPCs have the ability to proliferate in culture, which in principle makes it possible to greatly expand a CPC source from which cardiomyocytes (and the 2 other cell types) can then be made at large scale. Exploration of directed differentiation conditions that permit the conversion of CPCs into a pure population of cardiomyocytes, rather than a mix of 3 cell types, is a work in progress. The recent demonstration of the ability to directly reprogram mouse fibroblasts into expandable CPCs,121,122 without the need for an intermediate iPSC stage, points to the possibility of using human “iCPCs” instead of iPSCs for disease modeling and precision medicine studies in the future, although rigorous quality control measures for the CPCs would need to be established.
Despite these advances in cardiomyocyte differentiation, the lack of purity and lack of maturity remain obstacles. For some studies, mixed populations of cardiomyocytes and noncardiomyocytes might be adequate, as in cases when one can visually inspect for spontaneously beating cells and focus one’s analyses on just those cells. However, more generally, highly pure preparations of cardiomyocytes are desirable and optimal. One strategy is to use a cardiomyocyte-specific membrane protein with which to deploy antibodies for fluorescence-activated cell sorting or magnetic-activated cell sorting to separate cardiomyocytes from other cells, followed by replating of the purified population of cardiomyocytes. Signal-regulatory protein-α (SIRPA) and vascular cell adhesion molecule 1 (VCAM1) have been identified as 2 such cardiomyocyte markers, and cell sorting with antibodies against either of these proteins resulted in >95% pure populations of functional cardiomyocytes.123,124 A fluorescent dye that labels mitochondria and tetramethylrhodamine methyl ester perchlorate can be similarly used to sort cardiomyocytes from noncardiomyocytes.125 However, a disadvantage to this strategy is that it requires additional manipulation of the cells, namely, dispersion from the culture dish followed by sorting through a machine and then replating. Not only is this labor intensive, especially when working with a large number of samples, it also can adversely affect the quality of the cells.
An ideal approach would be to purify the cardiomyocytes while still in the dish. In one study, lentiviral vectors were used to introduce antibiotic resistance genes driven by cardiomyocyte-specific promoters into hPSCs.126 In a test case, drug selection in the dish after differentiation of infected cells yielded 96% pure cardiomyocytes. In another study, an antibiotic resistance gene was inserted by homologous recombination directly into the endogenous MYH6 locus such that the gene would be coexpressed with MYH6 in cardiomyocytes, with drug selection resulting in >98% purity.127 In yet another study with a more sophisticated approach, genome editing of hPSCs was used to incorporate a quiescent transgene harboring an antibiotic resistance gene.128 Differentiated cells were treated with a lentivirus expressing Cre recombinase from a muscle-specific promoter to potentiate the antibiotic resistant gene only in cardiomyocytes. Drug selection in the dish improved the yield of cardiomyocytes from 80% to >99%. A nongenetic strategy to purify iPSC-CMs in the dish takes advantage of their ability to metabolize lactate, in contrast to undifferentiated cells; prolonged incubation with glucose-depleted, lactate-rich medium increases the yield of iPSC-CMs to as high as 99%.129 However, even when purity is achieved with any of these strategies, heterogeneity remains in the iPSC-CMs; differentiated cardiomyocytes are typically a mixture of cells, some of which most closely resemble ventricular cells, with others resembling atrial cells and still others resembling nodal cells.109 Obtaining a pure population of just 1 of these cardiomyocyte subtypes is a challenge that remains to be solved, although some progress has been made.114,130–135 For example, a recently reported protocol can generate nodal-like cells of >80% purity.135
Although the strategies outlined above can successfully purify in vitro differentiated iPSC-CMs, the cells display important differences from authentic adult human cardiomyocytes and are more similar to fetal human cardiomyocytes (Table 3).136–139 Whereas adult cardiomyocytes are long and cylindrical in shape, differentiated cells are more rounded and much shorter, with a smaller length-to-width ratio.140 The adult cells are multinucleated, whereas differentiated cells tend to be mononuclear, like fetal cardiomyocytes.141 Furthermore, differentiated cells have less well-developed sarcoplasmic reticulum and lack transverse tubules, which results in calcium handling that relies more on flux through the sarcolemma rather than the sarcoplasmic reticulum, which resembles fetal cardiomyocytes.142,143 With respect to metabolism, iPSC-CMs have fewer mitochondria and accordingly depend more on glycolysis rather than β-oxidation of fatty acids for energy.55,125,144 In general, iPSC-CMs have higher resting membrane potentials (ranging from −30 to −75 mV) than adult ventricular cardiomyocytes (−85 to −90 mV) but slower depolarization speeds.136,137 The gene expression profile of differentiated cells most closely resembles that of fetal cardiomyocytes and is quite distinct from that of adult cardiomyocytes with respect to calcium handling and cardiac ion channel genes.145–147
Table 3.
Differences Between iPSC-CMs and Adult Human Cardiomyocytes
| iPSC-CMs | Adult Cardiomyocytes | |
|---|---|---|
| Morphology | ||
| Shape | Round or polygonal | Rod and elongated |
| Size | 20–30 μm | 150 μm |
| Nuclei per cell | Mononucleated | ≈25% multinucleated |
| Multicellular organization | Disorganized | Polarized |
| Sarcomere appearance | Disorganized | Organized |
| Sarcomere length | Shorter (≈1.6 μm) | Longer (≈2.2 μm) |
| Sarcomeric protein – MHC | β>α | β>>α |
| Sarcomeric protein – titin | N2BA | N2B |
| Sarcomeric protein – troponin I | ssTnI | cTnI |
| Sarcomere units – H zones and A bands | Formed after prolonged differentiation | Formed |
| Sarcomere units – M bands and T tubules | Absent | Present |
| Distribution of gap junctions | Circumferential | Polarized to intercalated disks |
| Electrophysiology | ||
| Resting membrane potential | ≈ –60 mV | ≈ –90 mV |
| Upstroke velocity | ≈50 V/s | ≈250 V/s |
| Amplitude | Small | Large |
| Spontaneous automaticity | Exhibited | Absent |
| Hyperpolarization-activated pacemaker (If) | Present | Absent |
| Sodium (INa) | Low | High |
| Inward rectifier potassium (IK1) | Low or absent | High |
| Transient outward potassium current (Ito) | Inactivated | Activated |
| Conduction velocity | Slower (≈0.1 m/s) | Faster (0.3–1.0 m/s) |
| Calcium handling | ||
| Ca2+ transient | Inefficient | Efficient |
| Amplitudes of Ca2+ transient | Small and decrease with pacing | Increase with pacing |
| Excitation-contraction coupling | Slow | Fast |
| Contractile force | ≈nN range/cell | ≈μN range/cell |
| Ca2+-handling proteins (CASQ2, RyR2, and PLN) | Low or absent | Normal |
| Force-frequency relationship | Positive | Negative |
| Mitochondrial bioenergetics | ||
| Mitochondrial number | Low | High |
| Mitochondrial volume | Low | High |
| Mitochondrial structure | Irregular distribution, perinuclear | Regular distribution, aligned |
| Mitochondrial proteins – DRP1 and OPA1 | Low | High |
| Metabolic substrate | Glycolysis (glucose) | Oxidative (fatty acid) |
| Adrenergic signaling | ||
| Responses to β-adrenergic stimulation | Lack of inotropic reaction | Inotropic reaction |
| Cardiac α-adrenergic receptor ADRA1A | Absent | Present |
cTnI indicates cardiac troponin I; iPSC-CMs, induced pluripotent stem cell–derived cardiomyocytes; MHC, myosin heavy chain; and ssTnI, slow skeletal troponin I.
Modified from Sayed et al139 with permission from Elsevier. Copyright © 2016, Elsevier.
A number of strategies have been attempted to increase the maturity of iPSC-CMs and make them more similar to adult cardiomyocytes, although a definitive solution to the problem remains a vigorously pursued goal in the field. One strategy has been to simply maintain the cells for a lengthier time in culture. Compared with iPSC-CMs differentiated and maintained for 20 to 40 days, cells maintained for 80 to 120 days were increased in size and anisotropy and had greater myofibril density and alignment, visible sarcomeres, a much larger proportion of multinucleated cells, hyperpolarized maximum diastolic potentials, increased action potential amplitudes, and faster upstroke velocities.148 In a separate study, iPSC-CMs maintained for a full year accrued structural characteristics of mature cells.149
Altering the stiffness or morphology of the growth substrate for iPSC-CMs is an increasingly popular approach. One study found that differentiated cardiomyocytes maintained on polyacrylamide hydrogels with increasing degrees of stiffness attained morphologies more similar to adult cells.150 Another study found that iPSC-CMs maintained on stiffer polyacrylamide hydrogels generated increase contractile stress.151 Yet another study found that differentiated cardiomyocytes most resembled adult cardiomyocytes with respect to contractile activity, myofibril alignment, electrophysiology, direction of calcium flow, organization of mitochondria, and presence of transverse tubules when the cells were maintained on polyacrylamide hydrogels of stiffness similar to that found in myocardial tissue and micropatterned to have a 7:1 length-to-width ratio.152 Finally, iPSC-CMs maintained on an easy-to-prepare Matrigel mattress had rod-shaped morphologies and increased sarcomere length, contractility, and expression of maturation markers.153
Other methods of inducing maturity include electrical stimulation,154,155 treatment with pharmacological agents such as phenylephrine and angiotensin II,156 and overexpression of certain genes or microRNAs.157–159 For example, heterologous overexpression of miR-499 and miR-1 enhanced the differentiation of iPSC-CMs and facilitated electrophysiological maturation of the cells,157,158 and overexpression of Kir2.1 also promoted electrophysiological maturation.159
Some of the shortcomings of iPSC-CMs in the dish can be overcome by fabricating the cells into engineered 3-dimensional structures.155,160–165 In addition to displaying characteristics that suggest a greater degree of maturity and of similarity to adult cardiomyocytes, iPSC-CMs within these engineered myocardial structures allow for more authentic disease modeling by permitting the analysis of characteristics that are difficult to assess in a dish, including cytoskeletal architecture, force generation, and arrhythmogenesis. In one example, individual iPSC-CMs were seeded on micropatterned fibronectin rectangles designed to mimic the length-to-width ratio of adult cardiomyocytes, providing the investigators with a better way to assess sarcomere disorganization in TAZ-mutant iPSC-CMs.50 The same investigators used a “heart-on-chip” platform in which iPSC-CMs were seeded onto thin elastomers micropatterned with fibronectin lines and supported by glass coverslips, thus fabricating muscular thin film tissue constructs in which the impaired contractility of TAZ-mutant iPSC-CMs could be observed and precisely measured.50 In another study, 3-dimensional tissues were generated by combining iPSC-CMs with human mesenchymal stem cells along with collagen and fibrinogen, which were then embedded within 3-dimensional micropatterned matrices that incorporated cantilevers to directly measure forces generated by the tissues.42 This platform demonstrated that tissues made from TTN-mutant iPSC-CMs had decreased contractile function and were impaired in their response to β-adrenergic stimulation.
Although we have focused on iPSC-CMs in this section, the same limitations and considerations—lack of consistency, lack of purity, lack of maturity, and lack of ability to fully model disease in the dish—apply to all in vitro differentiated cell types. For example, the deficiencies of iPSC-hepatocytes with respect to metabolic functions are well documented.166 A large cohort of iPSC-hepatocytes that were subjected to whole-genome gene expression profiling were found to be quite distinct from both primary human hepatocytes and whole-liver samples; the iPSC-hepatocytes faithfully modeled some hepatocyte-specific eQTLs but failed to appropriately model other eQTLs.107 Just as with iPSC-CMs, the past decade has seen substantial effort being devoted to overcoming these obstacles for each particular differentiated cell type.
Need for Biological Replicates
The vagaries of iPSC generation, single-cell clonal expansion, propagation of cells in culture for numerous passages, imperfect differentiation protocols, and even stochastic variation between environment conditions in side-by-side wells on the same dish can all introduce confounders into even the most careful, rigorous iPSC-based study designs. This makes it mandatory to use biological replicates—ideally, many biological replicates—when using iPSCs for disease modeling.
We have already discussed in Study Design how the use of unmatched iPSC lines from different individuals makes a study susceptible to confounders, and how the best remedy is to use a large number of cell lines in each experimental group. Yet even if the cell lines being compared are isogenic cell lines generated by genome editing, it is still problematic to draw conclusions from a comparison of single lines representing each experimental group (eg, a single wild-type line versus a single knockout line) for several reasons. First, genome-editing tools carry the risk of off-target effects such as cleavage and mutagenesis (indels) at sites in the genome other than the desired target site. Off-target effects present a potential confounding factor that could account for any difference observed between single cell lines. Although initial studies with genome-editing tools (especially CRISPR-Cas9) in transformed cultured cell lines suggested that they might incur a high frequency of off-target effects, subsequent studies using whole-genome sequencing in hESC and iPSC clones targeted with genome-editing tools indicated that for any given clone, the number of off-target mutations was very few, perhaps zero.34–36 However, these same studies brought to light a second issue of arguably greater concern, which is the possibility that the individual clones could carry a large number (dozens to hundreds) of unique single-nucleotide variants. Such variants are not expected to result from the use of genome-editing tools (which preferentially produce indels), which suggests that single-nucleotide variants arise spontaneously in culture in different cells at different times. This means that any 2 clonal lines expanded from single cells within the same stock will almost certainly differ with respect to some variants and thus are not truly isogenic, which raises the possibility of a confounding factor. The third issue arises from the practice of expanding clonal lines from single cells, a process that inevitably exerts selection pressure on the cells and thereby alters the cells in unpredictable ways—if not at the genetic level, then quite possibly at the epigenetic level. The resulting differences between cell lines, whether with respect to proliferation rate, capacity for differentiation into the desired cell type, or some other factors, can also act as confounders.
With all of these potential confounders in play, the best way to ensure that any observed phenotypic differences between experimental groups are authentic is the use of multiple clonal cell lines per experimental group. If one uses 2 cell lines per experimental group, it becomes unlikely that the same off-target effects, single-nucleotide variants, or selective pressures will manifest in both cell lines within a single group and not in any of the cell lines in the other group(s); this becomes extremely unlikely if one uses ≥3 cell lines per group.
In some cases, it might not be feasible to generate multiple cell lines per experimental group. For example, knocking in a variant into 1 or both alleles of an iPSC line via HDR-based genome editing, to generate heterozygous or homozygous cells, might occur with such low efficiency that one would only obtain a single clone with the desired genotype despite screening hundreds of clones.107 Even generating that single clone might entail multiple steps: knocking in the variant on one allele, single-cell expansion of the heterozygous clone, knocking in the variant on the other allele, and single-cell expansion of the homozygous clone. With so many manipulations of the cells and passages in culture, one should be greatly concerned about confounding factors. In such cases, one way to ensure that any observed phenotypic differences are genuinely attributable to the genetic variant is to attempt to rescue the phenotype with a complementary genetic alteration, such as by using genome editing to restore the original genotype, or taking an alternative approach (eg, if an introduced variant is thought to result in loss of function of the target gene, to heterologously express the wild-type gene).103,167 If the phenotypic differences are successfully reversed, then they can be confidently attributed to the genetic variant.
A final consideration is variability within a single clonal iPSC line. Even when grown in the same dish, cells in different wells in the dish can experience subtly different conditions that result in discernible phenotypic effects. These differences can be particularly accentuated with the process of in vitro differentiation, which, as described previously, can vary significantly in efficiency from sample to sample and result in inconsistently heterogeneous cellular mixtures (eg, undifferentiated cells with differentiated cells, or less mature cells with more mature cells).107,168 With these confounding factors in play, one should generally avoid making comparisons between single samples of cell lines. Rather, it is best to use multiple samples for each clonal cell line, which can be undertaken by simply growing and differentiating the samples in multiple wells in the same dish or in multiple dishes in the same incubator and then processing the samples in parallel. In principle, the greater the concern for inconsistency in growth and differentiation conditions, the greater the number of replicates should be used for each clonal cell line.
iPSCs FOR PRECISION MEDICINE
Use of iPSCs to Perform Patient-Specific Therapeutic Screening
One important goal of precision medicine is to prescribe the right dose of the right drug for the right patient at the right time. A substantial amount of energy has been devoted to pharmacogenomics, defined by the US Food and Drug Administration as the study of variations of DNA and RNA characteristics that are related to drug response. A number of pharmacogenomic applications related to cardiovascular drugs such as warfarin, clopidogrel, and statins are being explored.169 Such applications rely on genotyping of 1 or a few DNA variants that have been found to be strongly associated with either a beneficial response or an adverse response to the medication in question. Arguably, this is an overly simplistic approach to achieving the goal of pharmacogenomics; as with most clinical traits and diseases, it is likely that a large number of genomic loci or many factors in one’s genetic background could influence the response to the medication. By virtue of having a perfectly matched genetic background, patient-specific iPSCs can provide a model system that integrates all of the genomic loci involved in the response to the medication. Thus, iPSCs offer an opportunity to better test the efficacy and safety of the medication in vitro without exposing a patient to risk. Indeed, it should be possible to assess the effects of several alternative medications in an iPSC-based model and thereby choose the most appropriate medication for the particular patient, rather than prescribing the medications to patients in general without knowing how each would react and determining the best choice only by trial and error.170
However, the utility of iPSC-based models depends on differentiated cells such as iPSC-CMs being faithful proxies for the tissues in the living human being. The investigation of whether iPSC-based models will prove to be sufficiently predictive to warrant their use in clinical decision making is still in its infancy, but initial studies show some promise. In one study, iPSC-CMs generated from patients with LQTS or HCM were observed to be more susceptible to the arrhythmogenic effects of cisapride, a medication that was withdrawn from the US market because of side effects such as ventricular arrhythmias and SCD in patients with conditions such as heart failure and QT prolongation secondary to other medications.171 In another study, 2 potent blockers of IKs displayed signs of increased toxicity in iPSC-CMs from a patient with LQTS2 compared with control iPSC-CMs.172 In yet another study, iPSC-CMs from patients who had demonstrated extreme sotalol-induced QT prolongation exhibited larger increases in field potential durations than iPSC-CMs from patients who showed minimal QT prolongation with sotalol.173
In an example that underscores the potential of using iPSC-CMs for personalized treatment, iPSC-CMs were generated from a patient with LQTS and frequent ventricular arrhythmias who had been found to have 2 candidate causal variants, 1 in KCNH2 and 1 in SCN5A. Electrophysiological analysis of the iPSC-CMs revealed sodium channel current defects but no potassium channel current defects, which indicates that the SCN5A variant was responsible for the patient’s LQTS. The defect was ameliorated with treatment with the sodium channel blocker mexiletine, an effect that was accentuated by increased pacing of the iPSC-CMs but not by addition of a second sodium channel blocker, flecainide.75 The effects observed in the iPSC-CMs suggested that the optimal treatment for the patient would be to prescribe mexiletine alone and to set a high pacemaker rate (via the patient’s implantable cardioverter-defibrillator). Indeed, this was the treatment that had been empirically found to best control the patient’s arrhythmias.
Cancer patients who receive the chemotherapeutic agent doxorubicin are at a significant risk for developing cardiomyopathy, particularly with higher doses of the drug, but the mechanistic basis for this drug toxicity is unclear, and there is currently no way to predict which patients will experience this serious adverse consequence upon receiving the drug. iPSC-CMs were generated from 8 breast cancer patients who had received doxorubicin, 4 of whom had developed cardiomyopathy and 4 of whom had not.174 Upon treatment with doxorubicin in vitro, iPSC-CMs from cardiomyopathy patients had reduced viability compared with those from noncardiomyopathy patients. The former also displayed impaired mitochondrial and metabolic function, impaired calcium handling, decreased antioxidant pathway activity, and increased reactive oxygen species production. Thus, the patient-specific iPSC-CMs were able to recapitulate the vulnerability of the patients to doxorubicin cardiac toxicity, and one could envision using iPSC-CMs to guide the choice of using doxorubicin versus adjusting its dosage to tailor the optimal chemotherapeutic regimen.
In similar fashion, iPSC-CMs from 13 individuals were screened with 21 US Food and Drug Administration–approved tyrosine kinase inhibitors, used to treat various types of cancer, for cardiotoxicity. The results were used to generate a cardiac safety index for the tyrosine kinase inhibitors.175 Bioinformatics analysis of transcriptomic profiles of iPSC-CMs has been used to predict patient-specific drug-induced cardiotoxicity.176 Going beyond drug toxicity, iPSC-CMs might also be used to assess a patient’s risk of adverse outcomes from other types of environmental exposures; for example, iPSC-CMs have been used as an in vitro model for coxsackievirus B3–induced myocarditis.177
Although it might not presently be feasible to routinely generate iPSCs and differentiated cells for every patient, given the effort and expense involved, the examples described above suggest that judicious use of iPSC-based models could be valuable in complex patient cases, particularly when they involve genetic disorders in which the causal mechanism is unclear or when there is a substantial risk that administration of a medication will have grave consequences. Another possible future use of patient-specific iPSCs is to perform drug screening with differentiated cells, to choose among a panel of established medications or even identify novel candidate drugs to treat the patient’s condition.
Use of iPSCs to Define Patient-Specific Mechanisms
One of the challenges of studying complex cardiovascular disorders is that different pathophysiological mechanisms can result in similar clinical outcomes. Mutations in different genes or even different mutations in the same gene can have can different consequences at the molecular level but still have the same downstream phenotypic consequences. Patient-specific iPSCs offer the opportunity to dissect the mechanisms directly relevant to the patients’ mutations. As related in Use of iPSCs to Perform Patient-Specific Therapeutic Screening, this can be useful for the personalization of therapy, but it can also reap rewards with respect to basic science investigations.
One example of the use of iPSCs to define patientspecific mechanisms is the aforementioned studies on the missense R173W mutation in TNNT2.43,44 Derived from patients with DCM linked to this mutation, iPSC-CMs were found to have epigenetic activation of phosphodiesterase gene expression, a hitherto unrecognized mechanism that plausibly contributes to DCM pathophysiology via the modulation of β-adrenergic signaling. Detailed studies of the iPSC-CMs suggested that the R173W mutant troponin T protein is altered in such a way as to aberrantly localize in the nucleus and interact with the histone demethylase proteins KDM1A and KDM5A, presumably resulting in chromatin remodeling at the genomic sites of the implicated phosphodiesterase genes.
Whether this mechanistic pathway is generalizable to all cases of familial DCM related to TNNT2 mutations or, even more broadly, to cases of DCM linked to mutations in other genes, remains to be determined. The ability to generate iPSCs from many DCM individuals with many different gene mutations will be invaluable in determining the prevalence of epigenetic activation of phosphodiesterase genes among DCM patients, as well as in elucidating alternative pathogenetic DCM-associated mechanisms. A deeper understanding of all of the potential mechanisms that can result in DCM might lead to a better appreciation of DCM subtypes, with patients being assigned to subtypes based on molecular phenotypes, whether measured in iPSC-CMs or other proxy tissues. Each DCM subtype might well require its own distinct clinical management strategy in order for patients to be optimally treated, which involves not just choice of medication but other interventions such as lifestyle changes, implantation of devices to prevent ventricular arrhythmias, and pacemaker settings to optimize long-term cardiac function. The same can be said for other phenotypically complex inherited cardiovascular disorders such as ARVC and HCM. Thus, iPSC-based studies could inform precision medicine in ways that go far beyond pharmacogenomics. Indeed, the individualized nature of iPSC-based platforms enables the decoding of patient-specific, cell-autonomous disease development. Evolution from product-focused disease modeling to process-focused disease discovery brings with it the opportunity to understand prephenotypic disease onset and lays the groundwork for targeted individualized therapeutics.178
Use of iPSCs to Functionally Annotate Patient-Specific DNA Variants
Variants of uncertain significance (VUSs) are emerging as a significant challenge in clinical genetics, one with enormous implications for precision medicine. With many patients now undergoing exome or genome sequencing, a trend that is sure to increase exponentially in the coming years, some are being found to have novel variants in genes linked to monogenic cardiovascular disorders such as the cardiomyopathies and rhythm disorders described in previous sections. This poses a dilemma to clinical providers as to whether and how to counsel and manage their patients with these findings. Cardiomyopathy and rhythm disorder genes present a special challenge in light of the current recommendation from the American College of Medical Genetics that potentially pathogenic variants in these genes be reported to patients, irrespective of their preference and even if the variants were discovered incidentally from testing related to noncardiovascular conditions.179 The knowledge can be troublesome, because there is still no reliable means by which to distinguish pathogenic variants from benign variants with respect to gene function. Computational methods to predict pathogenicity have performed poorly in discriminating among established pathogenic and benign mutations,180 and attempts to use population-based methods to determine pathogenicity (ie, searching public exome databases to determine whether the variants are absent in the general population, which suggests they could be pathogenic) have resulted in patients being incorrectly advised that they have pathogenic mutations in cardiomyopathy genes.181
Adding to the complexity of the problem is the fact that variants in the same gene can give rise to very different diseases. For example, MYH7, MYBPC3, and TNNT2, the genes most commonly found to be mutated in familial HCM, are also mutated in substantial proportions of familial DCM cases. Different variants in SCN5A have been linked to LQTS3, Brugada syndrome, idiopathic ventricular fibrillation, sick sinus syndrome, and even familial DCM. For such genes, a VUS presents not only the question of whether it is pathogenic or benign but also the question of which disease it might cause in the patient, with serious implications for the management of the patient.
What is needed to fulfill the promise of precision medicine for patients in light of the discovery of VUSs is an efficient and reliable method of functionally annotating the variants. This would allow clinical providers to properly advise patients as to whether their particular variants put them at risk for disease and, ideally, which disease, as well as to recommend a proper course of action. It would also guide providers in deciding whether to offer to test family members for the same variants. iPSCs could potentially provide a platform for the functional annotation of variants. Unlike biochemical assays or heterologous expression of mutant genes in cultured transformed cells, iPSCs could be differentiated into the relevant cell type (eg, iPSC-CMs for cardiomyopathy gene variants) and more authentically recapitulate the phenotypic consequences of a given variant.
In principle, one could derive an iPSC line from a patient with a VUS, differentiate it into the relevant cell type, and compare its phenotypes with a collection of iPSCs from patients with the disease of concern and from healthy individuals. This might help to determine whether the patient is at risk for the disease based on the patient’s genetic background. Although this outcome would certainly be useful, this approach has several shortcomings. First, it will be challenging to generate enough iPSC lines to keep pace with the large number of patients who will be found to have VUSs in cardiovascular disease genes. Second, even if a patient’s iPSC line suggests a predisposition for the disease, it does not establish whether the variant in question is a driving factor for the disease and thus needs to be screened in family members. Third, there is the possibility that the patient’s iPSC line might turn out to be a poor differentiator with respect to the relevant cell type.
Genome editing enables a more systematic approach to the problem of VUSs. One can envision a standardized platform in which a well-characterized, high-quality (with respect to differentiation), wild-type iPSC line would be used for the insertion of many VUSs. All known variants in a gene (cataloged in clinical databases such as ClinVar) could be compared in a series of isogenic cell lines, thus removing many potential confounders and isolating the effects of the variants. Upon establishing and calibrating the platform with respect to the known variants (ie, being able to classify variants based on their pathogenicity and the disease to which they predispose, if any), novel variants discovered in patients could be introduced into the platform and then appropriately classified. Although it might not be possible to predict with 100% reliability whether a given variant will definitely cause a disease in a given person (given that genetic background and environmental factors will likely be involved), the functional annotation would certainly be more reliable than any existing approaches, such as computational methods.
In the long term, one could envision this kind of platform being integrated into genetics clinics such that any patient who undergoes clinical sequencing and is found to have a known variant in a cardiovascular disease gene could be given a functional interpretation of the variant immediately or, if found to have a novel variant, could be given a functional readout within a few months, which would then be used to inform the patient’s prognosis and management.
PROMOTING THE USE OF iPSCs FOR BIOMEDICAL APPLICATIONS
Establishing and Facilitating Access to Biobanks
As described in Methods to Derive New iPSC Lines, the generation of large numbers of iPSC lines is not a trivial undertaking and at present requires a substantial amount of funding. One way in which to amortize the investment of generating cell lines is to make them widely and easily available to the scientific community, so that many investigators can make use of the same cell lines rather than needing to generate cell lines of their own. This will be especially helpful when investigators wish to perform large-scale studies using dozens or even hundreds of cell lines. The establishment of iPSC biobanks is a welcome trend in this direction.
The National Heart, Lung, and Blood Institute has invested $80 million in its 5-year NextGen program,37 which has resulted in the generation and banking of ≈1500 iPSC lines. The California Institute of Regenerative Medicine has invested a comparably large amount of money to generate and bank 9000 iPSC lines from 3000 people. The New York Stem Cell Foundation seeks to build a biobank of 2500 iPSC lines. A number of smaller biobank collections have been underwritten by various international, federal, and state organizations. Collectively, these efforts represent a promising start toward the goal of making genetically diverse cohorts of iPSC lines available to the scientific community.
An additional important step for funding agencies will be to encourage grant recipients to routinely collect and freeze peripheral blood cells from any patients recruited for their studies, even if the generation of iPSCs is not the primary intent of the studies. As long as the consent forms signed by recruited patients cover the possibility of the generation of iPSC lines from their cells, the frozen samples will be available for iPSC generation indefinitely and could potentially be used by investigators in the future, upon request. Although such collections could and should be maintained by individual local institutions, they could also be maintained on a national scale. The National Institutes of Health’s Precision Medicine Initiative Cohort Program has the goal of recruiting 1 million people nationwide and collecting an extensive amount of medical data, as well as millions of biospecimens that will be stored in a biobank at Mayo Clinic. It is unrealistic to expect that iPSC lines will ever be made from all 1 million participants. However, as genetic and clinical data become available to investigators, it could be feasible for an investigator to select specific individuals with genotypes and phenotypes of interest and request that iPSC lines be generated from those individuals’ stored biospecimens, with measures taken to ensure their anonymity.
Collectively, the emerging concept of stem cell “theranostics” incorporates the triad of disease modeling, cellular diagnostics, and personalized therapeutics.182 The workflow of disease modeling starts with patient-derived iPSC biobanks to enable molecular profiling and characterization of defining traits of health and disease. Cellular diagnostics then builds on disease-specific biomarkers to resolve the genotype-phenotype relationships that could offer insight into the innate capacity of individuals to be treated. Personalized therapeutics is based on the knowledge of dysfunctional tissues, which provides the rationale to target the underlying mechanism of disease.
Development of Technologies to Scale Up Production and Differentiation of iPSC Lines
One way in which to more rapidly populate large iPSC biobanks and to empower research making use of large cohorts of iPSCs is to foster the development of technologies to produce iPSC lines at a large scale and to differentiate a large number of iPSC lines in parallel. Such efforts are underway and can be expected to greatly accelerate progress in the field.
The generation of iPSC lines can be made more cost-effective and efficient by simply optimizing the manual steps routinely used to reprogram patient cells. In one report, such optimization enabled a single staff member to generate 20 to 40 iPSC lines at a time, making it feasible for a single small laboratory to generate a few hundred iPSC lines in a year.183 Other efforts have focused instead on automation. One such attempt resulted in the development of a modular, robotic platform that can perform all of the steps in the process of reprogramming cells from skin biopsy samples; a key feature of this platform entailed the high throughput, pooled selection of polyclonal pluripotent cells derived from each patient sample, which resulted in the iPSCs displaying less line-to-line variation (with respect to global gene expression) than iPSCs resulting from single-cell selection and expansion.184 The platform was also designed to perform automated differentiations, although the purity of iPSC-CMs was typically around 50%. It was estimated that the automated platform could further increase throughput by 10- to 12-fold and decrease reagent costs by 5- to 6-fold compared with the most optimized manual reprogramming strategy reported to date.183
A number of automated systems have been used to maintain hPSCs in 2-dimensional culture conditions, either in flasks or in a 96-well format, the latter being particularly useful for drug screens or testing differentiation conditions.185–187 An important innovation has been the ability to maintain and expand hPSCs in suspension culture, which allows for large-scale growth of the cells in a more cost-effective manner than with flasks and plates.188,189 Another key step forward has been the development of platforms to differentiate hPSCs into cardiomyocytes with reasonably high yield (surpassing 80% purity) in suspension culture.190–193 The use of simulated microgravity increased the purity even further.194 With continued progress, it might soon be possible to produce hundreds of patient-specific iPSC-CM samples for use in single large-scale experiments with little need for manual intervention. Furthermore, many of these innovations can be readily applied to other differentiated cell types relevant to cardiovascular diseases.
Incorporation of iPSCs Into Drug Development Pipelines to Assess for Toxicity
It has been extensively documented that many drug candidates that had appeared to be safe in preclinical cellular and animal models displayed unacceptable toxicities when ultimately used in human beings; one example is increased risk of ventricular arrhythmias, which has been involved in 28% of drug withdrawals from the market in the United States.195,196 This perhaps speaks to the lack of fidelity of standard preclinical models to important aspects of human physiology, such as cardiac electrophysiology.
Drug candidates are often tested in Chinese hamster ovary (CHO) cells and human embryonic kidney (HEK) cells overexpressing the hERG channel, which contributes the rapidly activating delayed rectifier potassium current (IKr) and is a primary mediator of drug-induced QT prolongation and ventricular arrhythmias. These cell lines are popular because they are easy to maintain in culture, are straightforward to use, and lend themselves to high-throughput drug screening; however, these cell lines are quite distinct from human cardiomyocytes and do not recapitulate important aspects of their physiology (hence the need to overexpress the hERG ion channel in the first place). The lack of expression of ion channels other than hERG has resulted in incorrect assessments of drug effects. For example, verapamil acts on both potassium and calcium channels, with countervailing effects that render the drug neutral overall in both human cardiomyocytes and iPSC-CM models.197 However, in a system in which only the hERG channel is overexpressed, verapamil produces a result that is interpreted as false-positive toxicity.198 It is also notable that the aforementioned cell lines are aneuploid and thus could be dysregulated in their responses to drugs for that reason.199
Although testing of drug candidates in preclinical animal models might appear to make up for the shortcomings of CHO and HEK cultured cell lines, in fact these animals also have significant disadvantages related to major differences in cardiac physiology compared with humans. The mouse heart naturally beats about 9 times faster than the human heart, with correspondingly briefer APDs, and dependent on different ion channels.200 The IKr current, which plays an important role in human cardiomyocytes and is involved in drug-induced QT prolongation, has a negligible role in mouse cardiomyocytes. Genes involved in sarcomere structure and function and calcium handling differ in their expression patterns between the species, which can result in major differences in disease pathophysiology. Although larger animals more closely resemble humans with respect to cardiac physiology, they are not as readily bred and maintained as mice, which makes them less suitable for candidate drug assessment. Moreover, even larger animals can differ profoundly from humans in their responses to drugs.201
In principle, primary human cardiomyocytes would be the optimal model with which to test for drug toxicities; however, they are difficult to procure and to maintain in culture and do not lend themselves to high-throughput screening. By comparison, iPSC-CMs are advantageous in all of these respects. As discussed in Differentiation Protocols, although iPSC-CMs have characteristics that mark them as immature relative to adult human cardiomyocytes, they nonetheless have many shared properties that make them appropriate for use in preclinical drug testing as an alternative to currently used cellular and animal models. For instance, in contrast to hERG-overexpressing cell lines, iPSC-CMs, which express hERG at similar levels as adult human cardiomyocytes,171 correctly determined verapamil to be nontoxic with respect to QT prolongation.197 Alfuzosin, a drug that can cause QT prolongation via its action on sodium channels rather than hERG and thus is scored as nontoxic in hERG-overexpressing cell lines,202 was correctly found to be toxic in iPSC-CMs.197
In 1 study, iPSC-CMs were used to screen 131 different drugs at 6 different concentrations for cardiotoxicity using a 384-well format, which demonstrated the ability to use iPSC-CMs in high-throughput assays.203 In another study, iPSC-CMs were used to assess 51 previously characterized compounds for their effects on cardiomyocyte contraction and were found to provide a sensitivity of 87% and specificity of 70%.204 A more recent study by investigators at the US Food and Drug Administration confirmed the potential utility of iPSC-CMs for the prediction of proarrhythmic risk of drugs.205 As iPSC-CMs gain broader recognition as an advantageous system for the testing of candidate drugs, they are starting to be used in drug discovery efforts. For example, a liposomal formulation of doxorubicin, an agent with well-known cardiotoxic effects (as related in Use of iPSCs to Perform Patient-Specific Therapeutic Screening), was tested on iPSC-CMs, leading to limitation of delivery of the drug into cardiomyocytes that prevented signs of toxicity.206 These findings informed the decision to advance the formulation to phase I clinical trials.
A good case can be made for the routine incorporation of iPSC-CMs into drug development pipelines now, if not as a replacement for some of the currently used preclinical models, then at least as an additional, complementary model. A particular advantage to iPSC-CMs is that they reflect the genetic backgrounds of the individuals from whom they were generated. Because responses to drugs can vary widely depending on particular genetic variants, it would be advantageous to use a panel of iPSC lines from a diverse group of individuals who reflect a diversity of genotypes. It would then be feasible to assess for differences in safety and, if relevant, efficacy across genetic subgroups within the target population. A drug that displayed evidence of toxicity with some but not all iPSC lines would likely be disqualified, whereas a drug that displayed evidence of efficacy with some but not all iPSC lines could be further investigated to define which individual patients would benefit from it versus which would not. Thus, incorporating iPSC-CMs early into drug development pipelines could greatly empower precision medicine.
ACKNOWLEDGMENT
The authors are grateful to Amy N. Thomas at Stanford University for her assistance in preparing the figures for this statement.
DISCLOSURES
Writing Group Disclosures
| Writing Group Member | Employment | Research Grant | Other Research Support | Speakers’ Bureau/Honoraria | Expert Witness | Ownership Interest | Consultant/Advisory Board | Other |
|---|---|---|---|---|---|---|---|---|
| Kiran Musunuru | University of Pennsylvania Cardiovascular Institute | None | None | None | None | None | None | None |
| Farah Sheikh | University of California-San Diego | None | None | None | None | None | None | None |
| Rajat M. Gupta | Harvard University | None | None | None | None | None | None | None |
| Steven R. Houser | Temple University School of Medicine Cardiovascular Research Center | None | None | None | None | None | None | None |
| Kevin O. Maher | Emory University, Children’s Healthcare of Atlanta | None | None | None | None | None | None | None |
| David J. Milan | Massachusetts General Hospital | None | None | None | None | None | None | None |
| Andre Terzic | Mayo Clinic Center for Regenerative Medicine | None | None | None | None | None | None | None |
| Joseph C. Wu | Stanford University School of Medicine, Stanford Cardiovascular Institute | NIH* | None | None | None | None | None | None |
This table represents the relationships of writing group members that may be perceived as actual or reasonably perceived conflicts of interest as reported on the Disclosure Questionnaire, which all members of the writing group are required to complete and submit. A relationship is considered to be “significant” if (a) the person receives $10 000 or more during any 12-month period, or 5% or more of the person’s gross income; or (b) the person owns 5% or more of the voting stock or share of the entity, or owns $10 000 or more of the fair market value of the entity. A relationship is considered to be “modest” if it is less than “significant” under the preceding definition.
Significant.
Reviewer Disclosures
| Reviewer | Employment | Research Grant | Other Research Support | Speakers’ Bureau/Honoraria | Expert Witness | Ownership Interest | Consultant/Advisory Board | Other |
|---|---|---|---|---|---|---|---|---|
| Piero Anversa | Cardiocentro Ticino (Switzerland) | None | None | None | None | None | None | None |
| Joshua M. Hare | University of Miami | NIH* | None | None | None | Vestion* | Vestion*; Longeveron* | None |
| Steven P. Jones | University of Louisville | NIH (cell therapy grant)† | None | None | None | None | None | None |
| Yibing Qyang | Yale University | None | None | None | None | None | None | None |
This table represents the relationships of reviewers that may be perceived as actual or reasonably perceived conflicts of interest as reported on the Disclosure Questionnaire, which all reviewers are required to complete and submit. A relationship is considered to be “significant” if (a) the person receives $10 000 or more during any 12-month period, or 5% or more of the person’s gross income; or (b) the person owns 5% or more of the voting stock or share of the entity, or owns $10 000 or more of the fair market value of the entity. A relationship is considered to be “modest” if it is less than “significant” under the preceding definition.
Modest.
Significant.
FOOTNOTES
The American Heart Association makes every effort to avoid any actual or potential conflicts of interest that may arise as a result of an outside relationship or a personal, professional, or business interest of a member of the writing panel. Specifically, all members of the writing group are required to complete and submit a Disclosure Questionnaire showing all such relationships that might be perceived as real or potential conflicts of interest.
This statement was approved by the American Heart Association Science Advisory and Coordinating Committee on September 29, 2017, and the American Heart Association Executive Committee on December 11, 2017. A copy of the document is available at http://professional.heart.org/statements by using either “Search for Guidelines & Statements” or the “Browse by Topic” area. To purchase additional reprints, call 843-216-2533 or e-mail kelle.ramsay@wolterskluwer.com.
The American Heart Association requests that this document be cited as follows: Musunuru K, Sheikh F, Gupta RM, Houser SR, Maher KO, Milan DJ, Terzic A, Wu JC; on behalf of the American Heart Association Council on Functional Genomics and Translational Biology; Council on Cardiovascular Disease in the Young; and Council on Cardiovascular and Stroke Nursing. Induced pluripotent stem cells for cardiovascular disease modeling and precision medicine: a scientific statement from the American Heart Association. Circ Genom Precis Med. 2018;11:e000043. DOI: 10.1161/HCG.0000000000000043.
Expert peer review of AHA Scientific Statements is conducted by the AHA Office of Science Operations. For more on AHA statements and guidelines development, visit http://professional.heart.org/statements. Select the “Guidelines & Statements” drop-down menu, then click “Publication Development.”
Permissions: Multiple copies, modification, alteration, enhancement, and/or distribution of this document are not permitted without the express permission of the American Heart Association. Instructions for obtaining permission are located at http://www.heart.org/HEARTORG/General/Copyright-Permission-Guidelines_UCM_300404_Article.jsp. A link to the “Copyright Permissions Request Form” appears on the right side of the page.
Circ Genom Precis Med is available at http://circgenetics.ahajournals.org.
REFERENCES
- 1.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts [published correction appears in Science. 1998;282:1827]. Science 1998;282:1145–1147. [DOI] [PubMed] [Google Scholar]
- 2.Shamblott MJ, Axelman J, Wang S, Bugg EM, Littlefield JW, Donovan PJ, Blumenthal PD, Huggins GR, Gearhart JD. Derivation of pluripotent stem cells from cultured human primordial germ cells. Proc Natl Acad Sci U S A 1998;95:13726–13731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tachibana M, Amato P, Sparman M, Gutierrez NM, Tippner-Hedges R, Ma H, Kang E, Fulati A, Lee HS, Sritanaudomchai H, Masterson K, Larson J, Eaton D, Sadler-Fredd K, Battaglia D, Lee D, Wu D, Jensen J, Patton P, Gokhale S, Stouffer RL, Wolf D, Mitalipov S. Human embryonic stem cells derived by somatic cell nuclear transfer. Cell 2013;153:1228–1238. doi: 10.1016/j.cell.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamada M, Johannesson B, Sagi I, Burnett LC, Kort DH, Prosser RW, Paull D, Nestor MW, Freeby M, Greenberg E, Goland RS, Leibel RL, Solomon SL, Benvenisty N, Sauer MV, Egli D. Human oocytes reprogram adult somatic nuclei of a type 1 diabetic to diploid pluripotent stem cells. Nature 2014;510:533–536. doi: 10.1038/nature13287. [DOI] [PubMed] [Google Scholar]
- 5.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 6.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 7.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA. Induced pluripotent stem cell lines derived from human somatic cells. Science 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 8.Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, Lerou PH, Lensch MW, Daley GQ. Reprogramming of human somatic cells to pluripotency with defined factors. Nature 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 9.Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T, Okita K, Mochiduki Y, Takizawa N, Yamanaka S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- 10.Ghosh Z, Wilson KD, Wu Y, Hu S, Quertermous T, Wu JC. Persistent donor cell gene expression among human induced pluripotent stem cells contributes to differences with human embryonic stem cells. PLoS One 2010;5:e8975. doi: 10.1371/journal.pone.0008975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, Kim J, Aryee MJ, Ji H, Ehrlich LI, Yabuuchi A, Takeuchi A, Cunniff KC, Hongguang H, McKinney-Freeman S, Naveiras O, Yoon TJ, Irizarry RA, Jung N, Seita J, Hanna J, Murakami P, Jaenisch R, Weissleder R, Orkin SH, Weissman IL, Feinberg AP, Daley GQ. Epigenetic memory in induced pluripotent stem cells. Nature 2010;467:285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Polo JM, Liu S, Figueroa ME, Kulalert W, Eminli S, Tan KY, Apostolou E, Stadtfeld M, Li Y, Shioda T, Natesan S, Wagers AJ, Melnick A, Evans T, Hochedlinger K. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat Biotechnol 2010;28:848–855. doi: 10.1038/nbt.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lister R, Pelizzola M, Kida YS, Hawkins RD, Nery JR, Hon G, Antosiewicz-Bourget J, O’Malley R, Castanon R, Klugman S, Downes M, Yu R, Stewart R, Ren B, Thomson JA, Evans RM, Ecker JR. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature 2011;471:68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruiz S, Diep D, Gore A, Panopoulos AD, Montserrat N, Plongthongkum N, Kumar S, Fung HL, Giorgetti A, Bilic J, Batchelder EM, Zaehres H, Kan NG, Schöler HR, Mercola M, Zhang K, Belmonte JC Izpisua. Identification of a specific reprogramming-associated epigenetic signature in human induced pluripotent stem cells. Proc Natl Acad Sci U S A 2012;109:16196–16201. doi: 10.1073/pnas.1202352109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choi J, Lee S, Mallard W, Clement K, Tagliazucchi GM, Lim H, Choi IY, Ferrari F, Tsankov AM, Pop R, Lee G, Rinn JL, Meissner A, Park PJ, Hochedlinger K. A comparison of genetically matched cell lines reveals the equivalence of human iPSCs and ESCs. Nat Biotechnol 2015;33:1173–1181. doi: 10.1038/nbt.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seki T, Yuasa S, Oda M, Egashira T, Yae K, Kusumoto D, Nakata H, Tohyama S, Hashimoto H, Kodaira M, Okada Y, Seimiya H, Fusaki N, Hasegawa M, Fukuda K. Generation of induced pluripotent stem cells from human terminally differentiated circulating T cells. Cell Stem Cell 2010;7:11–14. doi: 10.1016/j.stem.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 17.Loh YH, Hartung O, Li H, Guo C, Sahalie JM, Manos PD, Urbach A, Heffner GC, Grskovic M, Vigneault F, Lensch MW, Park IH, Agarwal S, Church GM, Collins JJ, Irion S, Daley GQ. Reprogramming of T cells from human peripheral blood. Cell Stem Cell 2010;7:15–19. doi: 10.1016/j.stem.2010.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Staerk J, Dawlaty MM, Gao Q, Maetzel D, Hanna J, Sommer CA, Mostoslavsky G, Jaenisch R. Reprogramming of human peripheral blood cells to induced pluripotent stem cells. Cell Stem Cell 2010;7:20–24. doi: 10.1016/j.stem.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou T, Benda C, Duzinger S, Huang Y, Li X, Li Y, Guo X, Cao G, Chen S, Hao L, Chan YC, Ng KM, Ho JC, Wieser M, Wu J, Redl H, Tse HF, Grillari J, Grillari-Voglauer R, Pei D, Esteban MA. Generation of induced pluripotent stem cells from urine. J Am Soc Nephrol 2011;22:1221–1228. doi: 10.1681/ASN.2011010106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aasen T, Raya A, Barrero MJ, Garreta E, Consiglio A, Gonzalez F, Vassena R, Bilić J, Pekarik V, Tiscornia G, Edel M, Boué S, Belmonte JC Izpisúa. Efficient and rapid generation of induced pluripotent stem cells from human keratinocytes. Nat Biotechnol 2008;26:1276–1284. doi: 10.1038/nbt.1503. [DOI] [PubMed] [Google Scholar]
- 21.Sardo V Lo, Ferguson W, Erikson GA, Topol EJ, Baldwin KK, Torkamani A. Influence of donor age on induced pluripotent stem cells. Nat Biotechnol 2017;35:69–74. doi: 10.1038/nbt.3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martinez-Fernandez A, Nelson TJ, Reyes S, Alekseev AE, Secreto F, Perez-Terzic C, Beraldi R, Sung HK, Nagy A, Terzic A. iPS cell-derived cardiogenicity is hindered by sustained integration of reprogramming transgenes. Circ Cardiovasc Genet 2014;7:667–676. doi: 10.1161/CIRCGENETICS.113.000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fusaki N, Ban H, Nishiyama A, Saeki K, Hasegawa M. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc Jpn Acad Ser B Phys Biol Sci 2009;85:348–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu J, Hu K, Smuga-Otto K, Tian S, Stewart R, Slukvin II, Thomson JA. Human induced pluripotent stem cells free of vector and transgene sequences. Science 2009;324:797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okita K, Matsumura Y, Sato Y, Okada A, Morizane A, Okamoto S, Hong H, Nakagawa M, Tanabe K, Tezuka K, Shibata T, Kunisada T, Takahashi M, Takahashi J, Saji H, Yamanaka S. A more efficient method to generate integration-free human iPS cells. Nat Methods 2011;8:409–412. doi: 10.1038/nmeth.1591. [DOI] [PubMed] [Google Scholar]
- 26.Jia F, Wilson KD, Sun N, Gupta DM, Huang M, Li Z, Panetta NJ, Chen ZY, Robbins RC, Kay MA, Longaker MT, Wu JC. A nonviral minicircle vector for deriving human iPS cells. Nat Methods 2010;7:197–199. doi: 10.1038/nmeth.1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Diecke S, Lu J, Lee J, Termglinchan V, Kooreman NG, Burridge PW, Ebert AD, Churko JM, Sharma A, Kay MA, Wu JC. Novel codon-optimized mini-intronic plasmid for efficient, inexpensive, and xeno-free induction of pluripotency. Sci Rep 2015;5:8081. doi: 10.1038/srep08081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Warren L, Manos PD, Ahfeldt T, Loh YH, Li H, Lau F, Ebina W, Mandal PK, Smith ZD, Meissner A, Daley GQ, Brack AS, Collins JJ, Cowan C, Schlaeger TM, Rossi DJ. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell 2010;7:618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schlaeger TM, Daheron L, Brickler TR, Entwisle S, Chan K, Cianci A, DeVine A, Ettenger A, Fitzgerald K, Godfrey M, Gupta D, McPherson J, Malwadkar P, Gupta M, Bell B, Doi A, Jung N, Li X, Lynes MS, Brookes E, Cherry AB, Demirbas D, Tsankov AM, Zon LI, Rubin LL, Feinberg AP, Meissner A, Cowan CA, Daley GQ. A comparison of non-integrating reprogramming methods. Nat Biotechnol 2015;33:58–63. doi: 10.1038/nbt.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han J, Qian X, Wu Q, Jha R, Duan J, Yang Z, Maher KO, Nie S, Xu C. Novel surface-enhanced Raman scattering-based assays for ultra-sensitive detection of human pluripotent stem cells. Biomaterials 2016;105:66–76. doi: 10.1016/j.biomaterials.2016.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bock C, Kiskinis E, Verstappen G, Gu H, Boulting G, Smith ZD, Ziller M, Croft GF, Amoroso MW, Oakley DH, Gnirke A, Eggan K, Meissner A. Reference maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell 2011;144:439–452. doi: 10.1016/j.cell.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Müller FJ, Schuldt BM, Williams R, Mason D, Altun G, Papapetrou EP, Danner S, Goldmann JE, Herbst A, Schmidt NO, Aldenhoff JB, Laurent LC, Loring JF. A bioinformatic assay for pluripotency in human cells. Nat Methods 2011;8:315–317. doi: 10.1038/nmeth.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gore A, Li Z, Fung HL, Young JE, Agarwal S, Antosiewicz-Bourget J, Canto I, Giorgetti A, Israel MA, Kiskinis E, Lee JH, Loh YH, Manos PD, Montserrat N, Panopoulos AD, Ruiz S, Wilbert ML, Yu J, Kirkness EF, Belmonte JC Izpisua, Rossi DJ, Thomson JA, Eggan K, Daley GQ, Goldstein LS, Zhang K. Somatic coding mutations in human induced pluripotent stem cells. Nature 2011;471:63–67. doi: 10.1038/nature09805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Veres A, Gosis BS, Ding Q, Collins R, Ragavendran A, Brand H, Erdin S, Cowan CA, Talkowski ME, Musunuru K. Low incidence of off-target mutations in individual CRISPR-Cas9 and TALEN targeted human stem cell clones detected by whole-genome sequencing. Cell Stem Cell 2014;15:27–30. doi: 10.1016/j.stem.2014.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suzuki K, Yu C, Qu J, Li M, Yao X, Yuan T, Goebl A, Tang S, Ren R, Aizawa E, Zhang F, Xu X, Soligalla RD, Chen F, Kim J, Kim NY, Liao HK, Benner C, Esteban CR, Jin Y, Liu GH, Li Y, Belmonte JC Izpisua. Targeted gene correction minimally impacts whole-genome mutational load in human-disease-specific induced pluripotent stem cell clones. Cell Stem Cell 2014;15:31–36. doi: 10.1016/j.stem.2014.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith C, Gore A, Yan W, Abalde-Atristain L, Li Z, He C, Wang Y, Brodsky RA, Zhang K, Cheng L, Ye Z. Whole-genome sequencing analysis reveals high specificity of CRISPR/Cas9 and TALEN-based genome editing in human iPSCs. Cell Stem Cell 2014;15:12–13. doi: 10.1016/j.stem.2014.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sweet DJ. iPSCs meet GWAS: the NextGen Consortium. Cell Stem Cell 2017;20:417–418. doi: 10.1016/j.stem.2017.03.020. [DOI] [PubMed] [Google Scholar]
- 38.Musunuru K Genome editing of human pluripotent stem cells to generate human cellular disease models. Dis Model Mech 2013;6:896–904. doi: 10.1242/dmm.012054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matsa E, Ahrens JH, Wu JC. Human induced pluripotent stem cells as a platform for personalized and precision cardiovascular medicine. Physiol Rev 2016;96:1093–1126. doi: 10.1152/physrev.00036.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hershberger RE, Morales A. Dilated cardiomyopathy overview. In: Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A, Bean LJH, Bird TD, Fong CT, Mefford HC, Smith RJH, Stephens K, eds. GeneReviews [Internet] Seattle, WA: University of Washington, Seattle; 1993–2016. Posted July 27, 2007 [updated September 24, 2015]. [Google Scholar]
- 41.Herman DS, Lam L, Taylor MR, Wang L, Teekakirikul P, Christodoulou D, Conner L, DePalma SR, McDonough B, Sparks E, Teodorescu DL, Cirino AL, Banner NR, Pennell DJ, Graw S, Merlo M, Di Lenarda A, Sinagra G, Bos JM, Ackerman MJ, Mitchell RN, Murry CE, Lakdawala NK, Ho CY, Barton PJ, Cook SA, Mestroni L, Seidman JG, Seidman CE. Truncations of titin causing dilated cardiomyopathy. N Engl J Med 2012;366:619–628. doi: 10.1056/NEJMoa1110186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hinson JT, Chopra A, Nafissi N, Polacheck WJ, Benson CC, Swist S, Gorham J, Yang L, Schafer S, Sheng CC, Haghighi A, Homsy J, Hubner N, Church G, Cook SA, Linke WA, Chen CS, Seidman JG, Seidman CE. Titin mutations in iPS cells define sarcomere insufficiency as a cause of dilated cardiomyopathy. Science 2015;349:982–986. doi: 10.1126/science.aaa5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun N, Yazawa M, Liu J, Han L, Sanchez-Freire V, Abilez OJ, Navarrete EG, Hu S, Wang L, Lee A, Pavlovic A, Lin S, Chen R, Hajjar RJ, Snyder MP, Dolmetsch RE, Butte MJ, Ashley EA, Longaker MT, Robbins RC, Wu JC. Patient-specific induced pluripotent stem cells as a model for familial dilated cardiomyopathy. Sci Transl Med 2012;4:130ra47. doi: 10.1126/scitranslmed.3003552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu H, Lee J, Vincent LG, Wang Q, Gu M, Lan F, Churko JM, Sallam KI, Matsa E, Sharma A, Gold JD, Engler AJ, Xiang YK, Bers DM, Wu JC. Epigenetic regulation of phosphodiesterases 2A and 3A underlies compromised β-adrenergic signaling in an iPSC model of dilated cardiomyopathy. Cell Stem Cell 2015;17:89–100. doi: 10.1016/j.stem.2015.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Siu CW, Lee YK, Ho JC, Lai WH, Chan YC, Ng KM, Wong LY, Au KW, Lau YM, Zhang J, Lay KW, Colman A, Tse HF. Modeling of lamin A/C mutation premature cardiac aging using patient‐specific induced pluripotent stem cells. Aging (Albany NY) 2012;4:803–822. doi: 10.18632/aging.100503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karakikes I, Stillitano F, Nonnenmacher M, Tzimas C, Sanoudou D, Termglinchan V, Kong CW, Rushing S, Hansen J, Ceholski D, Kolokathis F, Kremastinos D, Katoulis A, Ren L, Cohen N, Gho JM, Tsiapras D, Vink A, Wu JC, Asselbergs FW, Li RA, Hulot JS, Kranias EG, Hajjar RJ. Correction of human phospholamban R14del mutation associated with cardiomyopathy using targeted nucleases and combination therapy. Nat Commun 2015;6:6955. doi: 10.1038/ncomms7955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tse HF, Ho JC, Choi SW, Lee YK, Butler AW, Ng KM, Siu CW, Simpson MA, Lai WH, Chan YC, Au KW, Zhang J, Lay KW, Esteban MA, Nicholls JM, Colman A, Sham PC. Patient-specific induced-pluripotent stem cells-derived cardiomyocytes recapitulate the pathogenic phenotypes of dilated cardiomyopathy due to a novel DES mutation identified by whole exome sequencing. Hum Mol Genet 2013;22:1395–1403. doi: 10.1093/hmg/dds556. [DOI] [PubMed] [Google Scholar]
- 48.Dick E, Kalra S, Anderson D, George V, Ritso M, Laval SH, Barresi R, Aartsma-Rus A, Lochmüller H, Denning C. Exon skipping and gene transfer restore dystrophin expression in human induced pluripotent stem cells-cardiomyocytes harboring DMD mutations. Stem Cells Dev 2013;22:2714–2724. doi: 10.1089/scd.2013.0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin B, Li Y, Han L, Kaplan AD, Ao Y, Kalra S, Bett GC, Rasmusson RL, Denning C, Yang L. Modeling and study of the mechanism of dilated cardiomyopathy using induced pluripotent stem cells derived from individuals with Duchenne muscular dystrophy. Dis Model Mech 2015;8:457–466. doi: 10.1242/dmm.019505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang G, McCain ML, Yang L, He A, Pasqualini FS, Agarwal A, Yuan H, Jiang D, Zhang D, Zangi L, Geva J, Roberts AE, Ma Q, Ding J, Chen J, Wang DZ, Li K, Wang J, Wanders RJ, Kulik W, Vaz FM, Laflamme MA, Murry CE, Chien KR, Kelley RI, Church GM, Parker KK, Pu WT. Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart-on-chip technologies. Nat Med 2014;20:616–623. doi: 10.1038/nm.3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cirino AL, Ho C. Hypertrophic cardiomyopathy overview. In: Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A, Bean LJH, Bird TD, Fong CT, Mefford HC, Smith RJH, Stephens K, eds. GeneReviews [Internet] Seattle, WA: University of Washington, Seattle; 1993–2016. Posted August 5, 2008 [updated January 16, 2014]. [PubMed] [Google Scholar]
- 52.Lan F, Lee AS, Liang P, Sanchez-Freire V, Nguyen PK, Wang L, Han L, Yen M, Wang Y, Sun N, Abilez OJ, Hu S, Ebert AD, Navarrete EG, Simmons CS, Wheeler M, Pruitt B, Lewis R, Yamaguchi Y, Ashley EA, Bers DM, Robbins RC, Longaker MT, Wu JC. Abnormal calcium handling properties underlie familial hypertrophic cardiomyopathy pathology in patient-specific induced pluripotent stem cells. Cell Stem Cell 2013;12:101–113. doi: 10.1016/j.stem.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Han L, Li Y, Tchao J, Kaplan AD, Lin B, Li Y, Mich-Basso J, Lis A, Hassan N, London B, Bett GC, Tobita K, Rasmusson RL, Yang L. Study familial hyper-trophic cardiomyopathy using patient-specific induced pluripotent stem cells. Cardiovasc Res 2014;104:258–269. doi: 10.1093/cvr/cvu205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McNally E, MacLeod H, Dellefave-Castillo L. Arrhythmogenic right ventricular dysplasia/cardiomyopathy. In: Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A, Bean LJH, Bird TD, Fong CT, Mefford HC, Smith RJH, Stephens K, eds. GeneReviews [Internet] Seattle, WA: University of Washington, Seattle; 1993–2016. Posted April 18, 2005 [updated January 9, 2014]. [Google Scholar]
- 55.Kim C, Wong J, Wen J, Wang S, Wang C, Spiering S, Kan NG, Forcales S, Puri PL, Leone TC, Marine JE, Calkins H, Kelly DP, Judge DP, Chen HS. Studying arrhythmogenic right ventricular dysplasia with patient-specific iPSCs. Nature 2013;494:105–110. doi: 10.1038/nature11799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Caspi O, Huber I, Gepstein A, Arbel G, Maizels L, Boulos M, Gepstein L. Modeling of arrhythmogenic right ventricular cardiomyopathy with human induced pluripotent stem cells. Circ Cardiovasc Genet 2013;6:557–568. doi: 10.1161/CIRCGENETICS.113.000188. [DOI] [PubMed] [Google Scholar]
- 57.Ma D, Wei H, Lu J, Ho S, Zhang G, Sun X, Oh Y, Tan SH, Ng ML, Shim W, Wong P, Liew R. Generation of patient-specific induced pluripotent stem cell-derived cardiomyocytes as a cellular model of arrhythmogenic right ventricular cardiomyopathy. Eur Heart J 2013;34:1122–1133. doi: 10.1093/eurheartj/ehs226. [DOI] [PubMed] [Google Scholar]
- 58.Asimaki A, Kapoor S, Plovie E, Arndt A Karin, Adams E, Liu Z, James CA, Judge DP, Calkins H, Churko J, Wu JC, MacRae CA, Kléber AG, Saffitz JE. Identification of a new modulator of the intercalated disc in a zebrafish model of arrhythmogenic cardiomyopathy. Sci Transl Med 2014;6:240ra74. doi: 10.1126/scitranslmed.3008008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Arbustini E, Favalli V, Narula N, Serio A, Grasso M. Left ventricular non-compaction: a distinct genetic cardiomyopathy? J Am Coll Cardiol 2016;68:949–966. doi: 10.1016/j.jacc.2016.05.096. [DOI] [PubMed] [Google Scholar]
- 60.Kodo K, Ong SG, Jahanbani F, Termglinchan V, Hirono K, InanlooRahatloo K, Ebert AD, Shukla P, Abilez OJ, Churko JM, Karakikes I, Jung G, Ichida F, Wu SM, Snyder MP, Bernstein D, Wu JC. iPSC-derived cardiomyocytes reveal abnormal TGF-β signalling in left ventricular non-compaction cardiomyopathy. Nat Cell Biol 2016;18:1031–1042. doi: 10.1038/ncb3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ang YS, Rivas RN, Ribeiro AJS, Srivas R, Rivera J, Stone NR, Pratt K, Mohamed TMA, Fu JD, Spencer CI, Tippens ND, Li M, Narasimha A, Radzinsky E, Moon-Grady AJ, Yu H, Pruitt BL, Snyder MP, Srivastava D. Disease model of GATA4 mutation reveals transcription factor cooperativity in human cardiogenesis. Cell 2016;167:1734–1749.e22. doi: 10.1016/j.cell.2016.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Alders M, Christiaans I. Long QT syndrome. In: Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A, Bean LJH, Bird TD, Fong CT, Mefford HC, Smith RJH, Stephens K, eds. GeneReviews [Internet] Seattle, WA: University of Washington, Seattle; 1993–2016. Posted February 20, 2003 [updated June 18, 2015]. [Google Scholar]
- 63.Schwartz PJ, Priori SG, Spazzolini C, Moss AJ, Vincent GM, Napolitano C, Denjoy I, Guicheney P, Breithardt G, Keating MT, Towbin JA, Beggs AH, Brink P, Wilde AA, Toivonen L, Zareba W, Robinson JL, Timothy KW, Corfield V, Wattanasirichaigoon D, Corbett C, Haverkamp W, Schulze-Bahr E, Lehmann MH, Schwartz K, Coumel P, Bloise R. Genotype-phenotype correlation in the long-QT syndrome: gene-specific triggers for life-threatening arrhythmias. Circulation 2001;103:89–95. [DOI] [PubMed] [Google Scholar]
- 64.Moretti A, Bellin M, Welling A, Jung CB, Lam JT, Bott-Flügel L, Dorn T, Goedel A, Höhnke C, Hofmann F, Seyfarth M, Sinnecker D, Schömig A, Laugwitz KL. Patient-specific induced pluripotent stem-cell models for long-QT syndrome. N Engl J Med 2010;363:1397–1409. doi: 10.1056/NEJMoa0908679. [DOI] [PubMed] [Google Scholar]
- 65.Egashira T, Yuasa S, Suzuki T, Aizawa Y, Yamakawa H, Matsuhashi T, Ohno Y, Tohyama S, Okata S, Seki T, Kuroda Y, Yae K, Hashimoto H, Tanaka T, Hattori F, Sato T, Miyoshi S, Takatsuki S, Murata M, Kurokawa J, Furukawa T, Makita N, Aiba T, Shimizu W, Horie M, Kamiya K, Kodama I, Ogawa S, Fukuda K. Disease characterization using LQTS-specific induced pluripotent stem cells. Cardiovasc Res 2012;95:419–429. doi: 10.1093/cvr/cvs206. [DOI] [PubMed] [Google Scholar]
- 66.Ma D, Wei H, Lu J, Huang D, Liu Z, Loh LJ, Islam O, Liew R, Shim W, Cook SA. Characterization of a novel KCNQ1 mutation for type 1 long QT syndrome and assessment of the therapeutic potential of a novel IKs activator using patient-specific induced pluripotent stem cell-derived cardiomyocytes. Stem Cell Res Ther 2015;6:39. doi: 10.1186/s13287-015-0027-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang M, D’Aniello C, Verkerk AO, Wrobel E, Frank S, Oostwaard D Ward-van, Piccini I, Freund C, Rao J, Seebohm G, Atsma DE, Schulze-Bahr E, Mummery CL, Greber B, Bellin M. Recessive cardiac phenotypes in induced pluripotent stem cell models of Jervell and Lange-Nielsen syndrome: disease mechanisms and pharmacological rescue. Proc Natl Acad Sci U S A 2014;111:E5383–E5392. doi: 10.1073/pnas.1419553111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Itzhaki I, Maizels L, Huber I, Zwi-Dantsis L, Caspi O, Winterstern A, Feldman O, Gepstein A, Arbel G, Hammerman H, Boulos M, Gepstein L. Modelling the long QT syndrome with induced pluripotent stem cells. Nature 2011;471:225–229. doi: 10.1038/nature09747. [DOI] [PubMed] [Google Scholar]
- 69.Matsa E, Rajamohan D, Dick E, Young L, Mellor I, Staniforth A, Denning C. Drug evaluation in cardiomyocytes derived from human induced pluripotent stem cells carrying a long QT syndrome type 2 mutation. Eur Heart J 2011;32:952–962. doi: 10.1093/eurheartj/ehr073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lahti AL, Kujala VJ, Chapman H, Koivisto AP, Pekkanen-Mattila M, Kerkelä E, Hyttinen J, Kontula K, Swan H, Conklin BR, Yamanaka S, Silvennoinen O, Aalto-Setälä K. Model for long QT syndrome type 2 using human iPS cells demonstrates arrhythmogenic characteristics in cell culture. Dis Model Mech 2012;5:220–230. doi: 10.1242/dmm.008409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bellin M, Casini S, Davis RP, D’Aniello C, Haas J, Oostwaard D Ward-van, Tertoolen LG, Jung CB, Elliott DA, Welling A, Laugwitz KL, Moretti A, Mummery CL. Isogenic human pluripotent stem cell pairs reveal the role of a KCNH2 mutation in long-QT syndrome. EMBO J 2013;32:3161–3175. doi: 10.1038/emboj.2013.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mehta A, Sequiera GL, Ramachandra CJ, Sudibyo Y, Chung Y, Sheng J, Wong KY, Tan TH, Wong P, Liew R, Shim W. Re-trafficking of hERG reverses long QT syndrome 2 phenotype in human iPS-derived cardiomyocytes. Cardiovasc Res 2014;102:497–506. doi: 10.1093/cvr/cvu060. [DOI] [PubMed] [Google Scholar]
- 73.Jouni M, Si-Tayeb K, Es-Salah-Lamoureux Z, Latypova X, Champon B, Caillaud A, Rungoat A, Charpentier F, Loussouarn G, Baró I, Zibara K, Lemarchand P, Gaborit N. Toward personalized medicine: using cardiomyocytes differentiated from urine-derived pluripotent stem cells to recapitulate electrophysiological characteristics of type 2 long QT syndrome. J Am Heart Assoc 2015;4:e002159. doi: 10.1161/JAHA.115.002159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Matsa E, Dixon JE, Medway C, Georgiou O, Patel MJ, Morgan K, Kemp PJ, Staniforth A, Mellor I, Denning C. Allele-specific RNA interference rescues the long-QT syndrome phenotype in human-induced pluripotency stem cell cardiomyocytes. Eur Heart J 2014;35:1078–1087. doi: 10.1093/eurheartj/eht067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Terrenoire C, Wang K, Tung KW, Chung WK, Pass RH, Lu JT, Jean JC, Omari A, Sampson KJ, Kotton DN, Keller G, Kass RS. Induced pluripotent stem cells used to reveal drug actions in a long QT syndrome family with complex genetics. J Gen Physiol 2013;141:61–72. doi: 10.1085/jgp.201210899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ma D, Wei H, Zhao Y, Lu J, Li G, Sahib NB, Tan TH, Wong KY, Shim W, Wong P, Cook SA, Liew R. Modeling type 3 long QT syndrome with cardiomyocytes derived from patient-specific induced pluripotent stem cells. Int J Cardiol 2013;168:5277–5286. doi: 10.1016/j.ijcard.2013.08.015. [DOI] [PubMed] [Google Scholar]
- 77.Yazawa M, Hsueh B, Jia X, Pasca AM, Bernstein JA, Hallmayer J, Dolmetsch RE. Using induced pluripotent stem cells to investigate cardiac phenotypes in Timothy syndrome. Nature 2011;471:230–234. doi: 10.1038/nature09855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang Y, Liang P, Lan F, Wu H, Lisowski L, Gu M, Hu S, Kay MA, Urnov FD, Shinnawi R, Gold JD, Gepstein L, Wu JC. Genome editing of isogenic human induced pluripotent stem cells recapitulates long QT phenotype for drug testing. J Am Coll Cardiol 2014;64:451–459. doi: 10.1016/j.jacc.2014.04.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Napolitano C, Priori SG, Bloise R. Catecholaminergic polymorphic ventricular tachycardia. In: Pagon RA, Adam MP, Ardinger HH, Wallace SE, Ame-miya A, Bean LJH, Bird TD, Fong CT, Mefford HC, Smith RJH, Stephens K, eds. GeneReviews [Internet] Seattle, WA: University of Washington, Seattle; 1993–2016. Posted October 14, 2004 [updated March 6, 2014]. [Google Scholar]
- 80.Fatima A, Xu G, Shao K, Papadopoulos S, Lehmann M, Arnáiz-Cot JJ, Rosa AO, Nguemo F, Matzkies M, Dittmann S, Stone SL, Linke M, Zechner U, Beyer V, Hennies HC, Rosenkranz S, Klauke B, Parwani AS, Haverkamp W, Pfitzer G, Farr M, Cleemann L, Morad M, Milting H, Hescheler J, Saric T. In vitro modeling of ryanodine receptor 2 dysfunction using human induced pluripotent stem cells. Cell Physiol Biochem 2011;28:579–592. doi: 10.1159/000335753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jung CB, Moretti A, y Schnitzler M Mederos, Iop L, Storch U, Bellin M, Dorn T, Ruppenthal S, Pfeiffer S, Goedel A, Dirschinger RJ, Seyfarth M, Lam JT, Sinnecker D, Gudermann T, Lipp P, Laugwitz KL. Dantrolene rescues arrhythmogenic RYR2 defect in a patient-specific stem cell model of catecholaminergic polymorphic ventricular tachycardia. EMBO Mol Med 2012;4:180–191. doi: 10.1002/emmm.201100194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Itzhaki I, Maizels L, Huber I, Gepstein A, Arbel G, Caspi O, Miller L, Belhassen B, Nof E, Glikson M, Gepstein L. Modeling of catecholaminergic polymorphic ventricular tachycardia with patient-specific human-induced pluripotent stem cells. J Am Coll Cardiol 2012;60:990–1000. doi: 10.1016/j.jacc.2012.02.066. [DOI] [PubMed] [Google Scholar]
- 83.Kujala K, Paavola J, Lahti A, Larsson K, Pekkanen-Mattila M, Viitasalo M, Lahtinen AM, Toivonen L, Kontula K, Swan H, Laine M, Silvennoinen O, Aalto-Setälä K. Cell model of catecholaminergic polymorphic ventricular tachycardia reveals early and delayed afterdepolarizations. PLoS One 2012;7:e44660. doi: 10.1371/journal.pone.0044660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang XH, Haviland S, Wei H, Sarić T, Fatima A, Hescheler J, Cleemann L, Morad M. Ca2+ signaling in human induced pluripotent stem cell-derived cardiomyocytes (iPS-CM) from normal and catecholaminergic polymorphic ventricular tachycardia (CPVT)-afflicted subjects. Cell Calcium 2013;54:57–70. doi: 10.1016/j.ceca.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Di Pasquale E, Lodola F, Miragoli M, Denegri M, Avelino-Cruz JE, Buonocore M, Nakahama H, Portararo P, Bloise R, Napolitano C, Condorelli G, Priori SG. CaMKII inhibition rectifies arrhythmic phenotype in a patient-specific model of catecholaminergic polymorphic ventricular tachycardia. Cell Death Dis 2013;4:e843. doi: 10.1038/cddis.2013.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Novak A, Barad L, Lorber A, Gherghiceanu M, Reiter I, Eisen B, Eldor L, Itskovitz-Eldor J, Eldar M, Arad M, Binah O. Functional abnormalities in iPSC-derived cardiomyocytes generated from CPVT1 and CPVT2 patients carrying ryanodine or calsequestrin mutations. J Cell Mol Med 2015;19:2006–2018. doi: 10.1111/jcmm.12581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Novak A, Barad L, Zeevi-Levin N, Shick R, Shtrichman R, Lorber A, Itskovitz-Eldor J, Binah O. Cardiomyocytes generated from CPVTD307H patients are arrhythmogenic in response to β-adrenergic stimulation. J Cell Mol Med 2012;16:468–482. doi: 10.1111/j.1582-4934.2011.01476.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liang P, Sallam K, Wu H, Li Y, Itzhaki I, Garg P, Zhang Y, Vermglinchan V, Lan F, Gu M, Gong T, Zhuge Y, He C, Ebert AD, Sanchez-Freire V, Churko J, Hu S, Sharma A, Lam CK, Scheinman MM, Bers DM, Wu JC. Patient-specific and genome-edited induced pluripotent stem cell-derived cardiomyocytes elucidate single-cell phenotype of Brugada syndrome. J Am Coll Cardiol 2016;68:2086–2096. doi: 10.1016/j.jacc.2016.07.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Garg V, Muth AN, Ransom JF, Schluterman MK, Barnes R, King IN, Gross-feld PD, Srivastava D. Mutations in NOTCH1 cause aortic valve disease. Nature 2005;437:270–274. doi: 10.1038/nature03940. [DOI] [PubMed] [Google Scholar]
- 90.Theodoris CV, Li M, White MP, Liu L, He D, Pollard KS, Bruneau BG, Srivastava D. Human disease modeling reveals integrated transcriptional and epigenetic mechanisms of NOTCH1 haploinsufficiency. Cell 2015;160:1072–1086. doi: 10.1016/j.cell.2015.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Merla G, Brunetti-Pierri N, Piccolo P, Micale L, Loviglio MN. Supravalvular aortic stenosis: elastin arteriopathy. Circ Cardiovasc Genet 2012;5:692–696. doi: 10.1161/CIRCGENETICS.112.962860. [DOI] [PubMed] [Google Scholar]
- 92.Ge X, Ren Y, Bartulos O, Lee MY, Yue Z, Kim KY, Li W, Amos PJ, Bozkulak EC, Iyer A, Zheng W, Zhao H, Martin KA, Kotton DN, Tellides G, Park IH, Yue L, Qyang Y. Modeling supravalvular aortic stenosis syndrome with human induced pluripotent stem cells. Circulation 2012;126:1695–1704. doi: 10.1161/CIRCULATIONAHA.112.116996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kinnear C, Chang WY, Khattak S, Hinek A, Thompson T, de Carvalho Rodrigues D, Kennedy K, Mahmut N, Pasceri P, Stanford WL, Ellis J, Mital S. Modeling and rescue of the vascular phenotype of Williams-Beuren syndrome in patient induced pluripotent stem cells. Stem Cells Transl Med 2013;2:2–15. doi: 10.5966/sctm.2012-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gu M, Shao NY, Sa S, Li D, Termglinchan V, Ameen M, Karakikes I, Sosa G, Grubert F, Lee J, Cao A, Taylor S, Ma Y, Zhao Z, Chappell J, Hamid R, Austin ED, Gold JD, Wu JC, Snyder MP, Rabinovitch M. Patient-specific iPSC-derived endothelial cells uncover pathways that protect against pulmonary hypertension in BMPR2 mutation carriers. Cell Stem Cell 2017;20:490–504. doi: 10.1016/j.stem.2016.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Youngblom E, Knowles JW. Familial hypercholesterolemia. In: Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A, Bean LJH, Bird TD, Fong CT, Mefford HC, Smith RJH, Stephens K, eds. GeneReviews [Internet] Seattle, WA: University of Washington, Seattle; 1993–2016. Posted January 2, 2014. [Google Scholar]
- 96.Rashid ST, Corbineau S, Hannan N, Marciniak SJ, Miranda E, Alexander G, Huang-Doran I, Griffin J, Ahrlund-Richter L, Skepper J, Semple R, Weber A, Lomas DA, Vallier L. Modeling inherited metabolic disorders of the liver using human induced pluripotent stem cells. J Clin Invest 2010;120:3127–3136. doi: 10.1172/JCI43122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cayo MA, Cai J, DeLaForest A, Noto FK, Nagaoka M, Clark BS, Collery RF, Si-Tayeb K, Duncan SA. JD induced pluripotent stem cell-derived hepatocytes faithfully recapitulate the pathophysiology of familial hypercholesterolemia. Hepatology 2012;56:2163–2171. doi: 10.1002/hep.25871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Si-Tayeb K, Idriss S, Champon B, Caillaud A, Pichelin M, Arnaud L, Lemarchand P, Le May C, Zibara K, Cariou B. Urine-sample-derived human induced pluripotent stem cells as a model to study PCSK9-mediated autosomal dominant hypercholesterolemia. Dis Model Mech 2016;9:81–90. doi: 10.1242/dmm.022277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhang H, Xue C, Shah R, Bermingham K, Hinkle CC, Li W, Rodrigues A, Tabita-Martinez J, Millar JS, Cuchel M, Pashos EE, Liu Y, Yan R, Yang W, Gosai SJ, VanDorn D, Chou ST, Gregory BD, Morrisey EE, Li M, Rader DJ, Reilly MP. Functional analysis and transcriptomic profiling of iPSC-derived macrophages and their application in modeling Mendelian disease. Circ Res 2015;117:17–28. doi: 10.1161/CIRCRESAHA.117.305860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gupta RM, Meissner TB, Cowan CA, Musunuru K. Genome-edited human pluripotent stem cell-derived macrophages as a model of reverse cholesterol transport: brief report. Arterioscler Thromb Vasc Biol 2016;36:15–18. doi: 10.1161/ATVBAHA.115.305956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Musunuru K, Strong A, Frank-Kamenetsky M, Lee NE, Ahfeldt T, Sachs KV, Li X, Li H, Kuperwasser N, Ruda VM, Pirruccello JP, Muchmore B, Prokunina-Olsson L, Hall JL, Schadt EE, Morales CR, Lund-Katz S, Phillips MC, Wong J, Cantley W, Racie T, Ejebe KG, Orho-Melander M, Melander O, Koteliansky V, Fitzgerald K, Krauss RM, Cowan CA, Kathiresan S, Rader DJ. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature 2010;466:714–719. doi: 10.1038/nature09266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kjolby M, Andersen OM, Breiderhoff T, Fjorback AW, Pedersen KM, Madsen P, Jansen P, Heeren J, Willnow TE, Nykjaer A. Sort1, encoded by the cardiovascular risk locus 1p13.3, is a regulator of hepatic lipoprotein export. Cell Metab 2010;12:213–223. doi: 10.1016/j.cmet.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 103.Ding Q, Lee YK, Schaefer EA, Peters DT, Veres A, Kim K, Kuperwasser N, Motola DL, Meissner TB, Hendriks WT, Trevisan M, Gupta RM, Moisan A, Banks E, Friesen M, Schinzel RT, Xia F, Tang A, Xia Y, Figueroa E, Wann A, Ahfeldt T, Daheron L, Zhang F, Rubin LL, Peng LF, Chung RT, Musunuru K, Cowan CA. A TALEN genome-editing system for generating human stem cell-based disease models. Cell Stem Cell 2013;12:238–251. doi: 10.1016/j.stem.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hua H, Shang L, Martinez H, Freeby M, Gallagher MP, Ludwig T, Deng L, Greenberg E, Leduc C, Chung WK, Goland R, Leibel RL, Egli D. iPSC-derived β cells model diabetes due to glucokinase deficiency. J Clin Invest 2013;123:3146–3153. doi: 10.1172/JCI67638. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 105.George S, Rochford JJ, Wolfrum C, Gray SL, Schinner S, Wilson JC, Soos MA, Murgatroyd PR, Williams RM, Acerini CL, Dunger DB, Barford D, Umpleby AM, Wareham NJ, Davies HA, Schafer AJ, Stoffel M, O’Rahilly S, Barroso I. A family with severe insulin resistance and diabetes due to a mutation in AKT2. Science 2004;304:1325–1328. doi: 10.1126/science.1096706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hussain K, Challis B, Rocha N, Payne F, Minic M, Thompson A, Daly A, Scott C, Harris J, Smillie BJ, Savage DB, Ramaswami U, De Lonlay P, O’Rahilly S, Barroso I, Semple RK. An activating mutation of AKT2 and human hypoglycemia. Science 2011;334:474. doi: 10.1126/science.1210878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pashos EE, Park Y, Wang X, Raghavan A, Yang W, Abbey D, Peters DT, Arbelaez J, Hernandez M, Kuperwasser N, Li W, Lian Z, Liu Y, Lv W, Lytle-Gabbin SL, Marchadier DH, Rogov P, Shi J, Slovik KJ, Stylianou IM, Wang L, Yan R, Zhang X, Kathiresan S, Duncan SA, Mikkelsen TS, Morrisey EE, Rader DJ, Brown CD, Musunuru K. Large, diverse population cohorts of hiPSCs and derived hepatocyte-like cells reveal functional genetic variation at blood lipid-associated loci. Cell Stem Cell 2017;20:558–570.e10. doi: 10.1016/j.stem.2017.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kehat I, Kenyagin-Karsenti D, Snir M, Segev H, Amit M, Gepstein A, Livne E, Binah O, Itskovitz-Eldor J, Gepstein L. Human embryonic stem cells can differentiate into myocytes with structural and functional properties of cardiomyocytes. J Clin Invest 2001;108:407–414. doi: 10.1172/JCI12131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.He JQ, Ma Y, Lee Y, Thomson JA, Kamp TJ. Human embryonic stem cells develop into multiple types of cardiac myocytes: action potential characterization. Circ Res 2003;93:32–39. doi: 10.1161/01.RES.0000080317.92718.99. [DOI] [PubMed] [Google Scholar]
- 110.Zwi L, Caspi O, Arbel G, Huber I, Gepstein A, Park IH, Gepstein L. Cardiomyocyte differentiation of human induced pluripotent stem cells. Circulation 2009;120:1513–1523. doi: 10.1161/CIRCULATIONAHA.109.868885. [DOI] [PubMed] [Google Scholar]
- 111.Zhang J, Wilson GF, Soerens AG, Koonce CH, Yu J, Palecek SP, Thomson JA, Kamp TJ. Functional cardiomyocytes derived from human induced pluripotent stem cells. Circ Res 2009;104:e30–e41. doi: 10.1161/CIRCRESAHA.108.192237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Laflamme MA, Chen KY, Naumova AV, Muskheli V, Fugate JA, Dupras SK, Reinecke H, Xu C, Hassanipour M, Police S, O’Sullivan C, Collins L, Chen Y, Minami E, Gill EA, Ueno S, Yuan C, Gold J, Murry CE. Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nat Biotechnol 2007;25:1015–1024. doi: 10.1038/nbt1327. [DOI] [PubMed] [Google Scholar]
- 113.Kattman SJ, Witty AD, Gagliardi M, Dubois NC, Niapour M, Hotta A, Ellis J, Keller G. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell 2011;8:228–240. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 114.Zhang Q, Jiang J, Han P, Yuan Q, Zhang J, Zhang X, Xu Y, Cao H, Meng Q, Chen L, Tian T, Wang X, Li P, Hescheler J, Ji G, Ma Y. Direct differentiation of atrial and ventricular myocytes from human embryonic stem cells by alternating retinoid signals. Cell Res 2011;21:579–587. doi: 10.1038/cr.2010.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Elliott DA, Braam SR, Koutsis K, Ng ES, Jenny R, Lagerqvist EL, Biben C, Hatzistavrou T, Hirst CE, Yu QC, Skelton RJ, Oostwaard D, Lim SM, Khammy O, Li Ward-van X, Hawes SM, Davis RP, Goulburn AL, Passier R, Prall OW, Haynes JM, Pouton CW, Kaye DM, Mummery CL, Elefanty AG, Stanley EG. NKX2–5(eGFP/w) hESCs for isolation of human cardiac progenitors and cardiomyocytes. Nat Methods 2011;8:1037–1040. doi: 10.1038/nmeth.1740. [DOI] [PubMed] [Google Scholar]
- 116.Hudson J, Titmarsh D, Hidalgo A, Wolvetang E, Cooper-White J. Primitive cardiac cells from human embryonic stem cells. Stem Cells Dev 2012;21:1513–1523. doi: 10.1089/scd.2011.0254. [DOI] [PubMed] [Google Scholar]
- 117.Lian X, Hsiao C, Wilson G, Zhu K, Hazeltine LB, Azarin SM, Raval KK, Zhang J, Kamp TJ, Palecek SP. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc Natl Acad Sci U S A 2012;109:E1848–E1857. doi: 10.1073/pnas.1200250109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Burridge PW, Matsa E, Shukla P, Lin ZC, Churko JM, Ebert AD, Lan F, Diecke S, Huber B, Mordwinkin NM, Plews JR, Abilez OJ, Cui B, Gold JD, Wu JC. Chemically defined generation of human cardiomyocytes. Nat Methods 2014;11:855–860. doi: 10.1038/nmeth.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yang L, Soonpaa MH, Adler ED, Roepke TK, Kattman SJ, Kennedy M, Henckaerts E, Bonham K, Abbott GW, Linden RM, Field LJ, Keller GM. Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature 2008;453:524–528. doi: 10.1038/nature06894. [DOI] [PubMed] [Google Scholar]
- 120.Cao N, Liang H, Huang J, Wang J, Chen Y, Chen Z, Yang HT. Highly efficient induction and long-term maintenance of multipotent cardiovascular progenitors from human pluripotent stem cells under defined conditions. Cell Res 2013;23:1119–1132. doi: 10.1038/cr.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lalit PA, Salick MR, Nelson DO, Squirrell JM, Shafer CM, Patel NG, Saeed I, Schmuck EG, Markandeya YS, Wong R, Lea MR, Eliceiri KW, Hacker TA, Crone WC, Kyba M, Garry DJ, Stewart R, Thomson JA, Downs KM, Lyons GE, Kamp TJ. Lineage reprogramming of fibroblasts into proliferative induced cardiac progenitor cells by defined factors. Cell Stem Cell 2016;18:354–367. doi: 10.1016/j.stem.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhang Y, Cao N, Huang Y, Spencer CI, Fu JD, Yu C, Liu K, Nie B, Xu T, Li K, Xu S, Bruneau BG, Srivastava D, Ding S. Expandable cardiovascular progenitor cells reprogrammed from fibroblasts. Cell Stem Cell 2016;18:368–381. doi: 10.1016/j.stem.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Dubois NC, Craft AM, Sharma P, Elliott DA, Stanley EG, Elefanty AG, Gramolini A, Keller G. SIRPA is a specific cell-surface marker for isolating cardiomyocytes derived from human pluripotent stem cells. Nat Biotechnol 2011;29:1011–1018. doi: 10.1038/nbt.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Uosaki H, Fukushima H, Takeuchi A, Matsuoka S, Nakatsuji N, Yamanaka S, Yamashita JK. Efficient and scalable purification of cardiomyocytes from human embryonic and induced pluripotent stem cells by VCAM1 surface expression. PLoS One 2011;6:e23657. doi: 10.1371/journal.pone.0023657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hattori F, Chen H, Yamashita H, Tohyama S, Satoh YS, Yuasa S, Li W, Yamakawa H, Tanaka T, Onitsuka T, Shimoji K, Ohno Y, Egashira T, Kaneda R, Murata M, Hidaka K, Morisaki T, Sasaki E, Suzuki T, Sano M, Makino S, Oikawa S, Fukuda K. Nongenetic method for purifying stem cell-derived cardiomyocytes. Nat Methods 2010;7:61–66. doi: 10.1038/nmeth.1403. [DOI] [PubMed] [Google Scholar]
- 126.Kita-Matsuo H, Barcova M, Prigozhina N, Salomonis N, Wei K, Jacot JG, Nelson B, Spiering S, Haverslag R, Kim C, Talantova M, Bajpai R, Calzolari D, Terskikh A, McCulloch AD, Price JH, Conklin BR, Chen HS, Mercola M. Lentiviral vectors and protocols for creation of stable hESC lines for fluorescent tracking and drug resistance selection of cardiomyocytes. PLoS One 2009;4:e5046. doi: 10.1371/journal.pone.0005046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ma J, Guo L, Fiene SJ, Anson BD, Thomson JA, Kamp TJ, Kolaja KL, Swanson BJ, January CT. High purity human-induced pluripotent stem cell-derived cardiomyocytes: electrophysiological properties of action potentials and ionic currents. Am J Physiol Heart Circ Physiol 2011;301:H2006–H2017. doi: 10.1152/ajpheart.00694.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gantz JA, Palpant NJ, Welikson RE, Hauschka SD, Murry CE, Laflamme MA. Targeted genomic integration of a selectable floxed dual fluorescence reporter in human embryonic stem cells. PLoS One 2012;7:e46971. doi: 10.1371/journal.pone.0046971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Tohyama S, Hattori F, Sano M, Hishiki T, Nagahata Y, Matsuura T, Hashimoto H, Suzuki T, Yamashita H, Satoh Y, Egashira T, Seki T, Muraoka N, Yamakawa H, Ohgino Y, Tanaka T, Yoichi M, Yuasa S, Murata M, Suematsu M, Fukuda K. Distinct metabolic flow enables large-scale purification of mouse and human pluripotent stem cell-derived cardiomyocytes. Cell Stem Cell 2013;12:127–137. doi: 10.1016/j.stem.2012.09.013. [DOI] [PubMed] [Google Scholar]
- 130.Zhu WZ, Xie Y, Moyes KW, Gold JD, Askari B, Laflamme MA. Neuregulin/ ErbB signaling regulates cardiac subtype specification in differentiating human embryonic stem cells. Circ Res 2010;107:776–786. doi: 10.1161/CIRCRESAHA.110.223917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Fu JD, Jiang P, Rushing S, Liu J, Chiamvimonvat N, Li RA. Na+/Ca2+ exchanger is a determinant of excitation-contraction coupling in human embryonic stem cell-derived ventricular cardiomyocytes. Stem Cells Dev 2010;19:773–782. doi: 10.1089/scd.2009.0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bizy A, Guerrero-Serna G, Hu B, Ponce-Balbuena D, Willis BC, Zarzoso M, Ramirez RJ, Sener MF, Mundada LV, Klos M, Devaney EJ, Vikstrom KL, Herron TJ, Jalife J. Myosin light chain 2-based selection of human iPSC-derived early ventricular cardiac myocytes. Stem Cell Res 2013;11:1335–1347. doi: 10.1016/j.scr.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Karakikes I, Senyei GD, Hansen J, Kong CW, Azeloglu EU, Stillitano F, Lieu DK, Wang J, Ren L, Hulot JS, Iyengar R, Li RA, Hajjar RJ. Small molecule-mediated directed differentiation of human embryonic stem cells toward ventricular cardiomyocytes. Stem Cells Transl Med 2014;3:18–31. doi: 10.5966/sctm.2013-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Josowitz R, Lu J, Falce C, D’Souza SL, Wu M, Cohen N, Dubois NC, Zhao Y, Sobie EA, Fishman GI, Gelb BD. Identification and purification of human induced pluripotent stem cell-derived atrial-like cardiomyocytes based on sarcolipin expression. PLoS One 2014;9:e101316. doi: 10.1371/journal.pone.0101316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Protze SI, Liu J, Nussinovitch U, Ohana L, Backx PH, Gepstein L, Keller GM. Sinoatrial node cardiomyocytes derived from human pluripotent cells function as a biological pacemaker. Nat Biotechnol 2017;35:56–68. doi: 10.1038/nbt.3745. [DOI] [PubMed] [Google Scholar]
- 136.Robertson C, Tran DD, George SC. Concise review: maturation phases of human pluripotent stem cell-derived cardiomyocytes. Stem Cells 2013;31:829–837. doi: 10.1002/stem.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Yang X, Pabon L, Murry CE. Engineering adolescence: maturation of human pluripotent stem cell-derived cardiomyocytes. Circ Res 2014;114:511–523. doi: 10.1161/CIRCRESAHA.114.300558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Karakikes I, Ameen M, Termglinchan V, Wu JC. Human induced pluripotent stem cell-derived cardiomyocytes: insights into molecular, cellular, and functional phenotypes. Circ Res 2015;117:80–88. doi: 10.1161/CIRCRESAHA.117.305365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Sayed N, Liu C, Wu JC. Translation of human-induced pluripotent stem cells: from clinical trial in a dish to precision medicine. J Am Coll Cardiol 2016;67:2161–2176. doi: 10.1016/j.jacc.2016.01.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Snir M, Kehat I, Gepstein A, Coleman R, Itskovitz-Eldor J, Livne E, Gepstein L. Assessment of the ultrastructural and proliferative properties of human embryonic stem cell-derived cardiomyocytes. Am J Physiol Heart Circ Physiol 2003;285:H2355–H2363. doi: 10.1152/ajpheart.00020.2003. [DOI] [PubMed] [Google Scholar]
- 141.Smolich JJ. Ultrastructural and functional features of the developing mammalian heart: a brief overview. Reprod Fertil Dev 1995;7: 451–461. [DOI] [PubMed] [Google Scholar]
- 142.Itzhaki I, Schiller J, Beyar R, Satin J, Gepstein L. Calcium handling in embryonic stem cell-derived cardiac myocytes: of mice and men. Ann N Y Acad Sci 2006;1080:207–215. doi: 10.1196/annals.1380.017. [DOI] [PubMed] [Google Scholar]
- 143.Lieu DK, Liu J, Siu CW, McNerney GP, Tse HF, Abu-Khalil A, Huser T, Li RA. Absence of transverse tubules contributes to non-uniform Ca(2+) wavefronts in mouse and human embryonic stem cell-derived cardiomyocytes. Stem Cells Dev 2009;18:1493–1500. doi: 10.1089/scd.2009.0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Rana P, Anson B, Engle S, Will Y. Characterization of human-induced pluripotent stem cell-derived cardiomyocytes: bioenergetics and utilization in safety screening. Toxicol Sci 2012;130:117–131. doi: 10.1093/toxsci/kfs233. [DOI] [PubMed] [Google Scholar]
- 145.Cao F, Wagner RA, Wilson KD, Xie X, Fu JD, Drukker M, Lee A, Li RA, Gambhir SS, Weissman IL, Robbins RC, Wu JC. Transcriptional and functional profiling of human embryonic stem cell-derived cardiomyocytes. PLoS One 2008;3:e3474. doi: 10.1371/journal.pone.0003474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Synnergren J, Akesson K, Dahlenborg K, Vidarsson H, Améen C, Steel D, Lindahl A, Olsson B, Sartipy P. Molecular signature of cardiomyocyte clusters derived from human embryonic stem cells. Stem Cells 2008;26:1831–1840. doi: 10.1634/stemcells.2007-1033. [DOI] [PubMed] [Google Scholar]
- 147.Xu XQ, Soo SY, Sun W, Zweigerdt R. Global expression profile of highly enriched cardiomyocytes derived from human embryonic stem cells. Stem Cells 2009;27:2163–2174. doi: 10.1002/stem.166. [DOI] [PubMed] [Google Scholar]
- 148.Lundy SD, Zhu WZ, Regnier M, Laflamme MA. Structural and functional maturation of cardiomyocytes derived from human pluripotent stem cells. Stem Cells Dev 2013;22:1991–2002. doi: 10.1089/scd.2012.0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kamakura T, Makiyama T, Sasaki K, Yoshida Y, Wuriyanghai Y, Chen J, Hattori T, Ohno S, Kita T, Horie M, Yamanaka S, Kimura T. Ultrastructural maturation of human-induced pluripotent stem cell-derived cardiomyocytes in a long-term culture. Circ J 2013;77:1307–1314. [DOI] [PubMed] [Google Scholar]
- 150.Jacot JG, Kita-Matsuo H, Wei KA, Chen HS, Omens JH, Mercola M, McCulloch AD. Cardiac myocyte force development during differentiation and maturation. Ann N Y Acad Sci 2010;1188:121–127. doi: 10.1111/j.1749-6632.2009.05091.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hazeltine LB, Simmons CS, Salick MR, Lian X, Badur MG, Han W, Delgado SM, Wakatsuki T, Crone WC, Pruitt BL, Palecek SP. Effects of substrate mechanics on contractility of cardiomyocytes generated from human pluripotent stem cells. Int J Cell Biol 2012;2012:508294. doi: 10.1155/2012/508294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ribeiro AJ, Ang YS, Fu JD, Rivas RN, Mohamed TM, Higgs GC, Srivastava D, Pruitt BL. Contractility of single cardiomyocytes differentiated from pluripotent stem cells depends on physiological shape and substrate stiffness. Proc Natl Acad Sci U S A 2015;112:12705–12710. doi: 10.1073/pnas.1508073112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Feaster TK, Cadar AG, Wang L, Williams CH, Chun YW, Hempel JE, Bloodworth N, Merryman WD, Lim CC, Wu JC, Knollmann BC, Hong CC. Matrigel mattress: a method for the generation of single contracting human-induced pluripotent stem cell-derived cardiomyocytes. Circ Res 2015;117:995–1000. doi: 10.1161/CIRCRESAHA.115.307580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Chan YC, Ting S, Lee YK, Ng KM, Zhang J, Chen Z, Siu CW, Oh SK, Tse HF. Electrical stimulation promotes maturation of cardiomyocytes derived from human embryonic stem cells. J Cardiovasc Transl Res 2013;6:989–999. doi: 10.1007/s12265-013-9510-z. [DOI] [PubMed] [Google Scholar]
- 155.Nunes SS, Miklas JW, Liu J, Aschar-Sobbi R, Xiao Y, Zhang B, Jiang J, Massé S, Gagliardi M, Hsieh A, Thavandiran N, Laflamme MA, Nanthakumar K, Gross GJ, Backx PH, Keller G, Radisic M. Biowire: a platform for maturation of human pluripotent stem cell-derived cardiomyocytes. Nat Methods 2013;10:781–787. doi: 10.1038/nmeth.2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Földes G, Mioulane M, Wright JS, Liu AQ, Novak P, Merkely B, Gorelik J, Schneider MD, Ali NN, Harding SE. Modulation of human embryonic stem cell-derived cardiomyocyte growth: a testbed for studying human cardiac hypertrophy? J Mol Cell Cardiol 2011;50:367–376. doi: 10.1016/j.yjmcc.2010.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Wilson KD, Hu S, Venkatasubrahmanyam S, Fu JD, Sun N, Abilez OJ, Baugh JJ, Jia F, Ghosh Z, Li RA, Butte AJ, Wu JC. Dynamic microRNA expression programs during cardiac differentiation of human embryonic stem cells: role for miR-499. Circ Cardiovasc Genet 2010;3:426–435. doi: 10.1161/CIRCGENETICS.109.934281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Fu JD, Rushing SN, Lieu DK, Chan CW, Kong CW, Geng L, Wilson KD, Chiamvimonvat N, Boheler KR, Wu JC, Keller G, Hajjar RJ, Li RA. Distinct roles of microRNA-1 and −499 in ventricular specification and functional maturation of human embryonic stem cell-derived cardiomyocytes. PLoS One 2011;6:e27417. doi: 10.1371/journal.pone.0027417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Lieu DK, Fu JD, Chiamvimonvat N, Tung KC, McNerney GP, Huser T, Keller G, Kong CW, Li RA. Mechanism-based facilitated maturation of human pluripotent stem cell-derived cardiomyocytes. Circ Arrhythm Electrophysiol 2013;6:191–201. doi: 10.1161/CIRCEP.111.973420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Schaaf S, Shibamiya A, Mewe M, Eder A, Stöhr A, Hirt MN, Rau T, Zimmermann WH, Conradi L, Eschenhagen T, Hansen A. Human engineered heart tissue as a versatile tool in basic research and preclinical toxicology. PLoS One 2011;6:e26397. doi: 10.1371/journal.pone.0026397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Tulloch NL, Muskheli V, Razumova MV, Korte FS, Regnier M, Hauch KD, Pabon L, Reinecke H, Murry CE. Growth of engineered human myocardium with mechanical loading and vascular coculture. Circ Res 2011;109:47–59. doi: 10.1161/CIRCRESAHA.110.237206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kensah G, Lara A Roa, Dahlmann J, Zweigerdt R, Schwanke K, Hegermann J, Skvorc D, Gawol A, Azizian A, Wagner S, Maier LS, Krause A, Dräger G, Ochs M, Haverich A, Gruh I, Martin U. Murine and human pluripotent stem cell-derived cardiac bodies form contractile myocardial tissue in vitro. Eur Heart J 2013;34:1134–1146. doi: 10.1093/eurheartj/ehs349. [DOI] [PubMed] [Google Scholar]
- 163.Zhang D, Shadrin IY, Lam J, Xian HQ, Snodgrass HR, Bursac N. Tissue-engineered cardiac patch for advanced functional maturation of human ESC-derived cardiomyocytes. Biomaterials 2013;34:5813–5820. doi: 10.1016/j.biomaterials.2013.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Thavandiran N, Dubois N, Mikryukov A, Massé S, Beca B, Simmons CA, Deshpande VS, McGarry JP, Chen CS, Nanthakumar K, Keller GM, Radisic M, Zandstra PW. Design and formulation of functional pluripotent stem cell-derived cardiac microtissues. Proc Natl Acad Sci U S A 2013;110:E4698–E4707. doi: 10.1073/pnas.1311120110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Tiburcy M, Hudson JE, Balfanz P, Schlick SF, Meyer T, Liao ML Chang, Levent E, Raad F, Zeidler S, Wingender E, Riegler J, Wang M, Gold JD, Kehat I, Wettwer E, Ravens U, Dierickx P, van Laake L, Goumans MJ, Khadjeh S, Toischer K, Hasenfuss G, Couture LA, Unger A, Linke WA, Araki T, Neel B, Keller G, Gepstein L, Wu JC, Zimmermann WH. Defined engineered human myocardium with advanced maturation for applications in heart failure modeling and repair. Circulation 2017;135:1832–1847. doi: 10.1161/CIRCULATIONAHA.116.024145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Schwartz RE, Fleming HE, Khetani SR, Bhatia SN. Pluripotent stem cell-derived hepatocyte-like cells. Biotechnol Adv 2014;32:504–513. doi: 10.1016/j.biotechadv.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kiskinis E, Sandoe J, Williams LA, Boulting GL, Moccia R, Wainger BJ, Han S, Peng T, Thams S, Mikkilineni S, Mellin C, Merkle FT, Davis-Dusenbery BN, Ziller M, Oakley D, Ichida J, Di Costanzo S, Atwater N, Maeder ML, Goodwin MJ, Nemesh J, Handsaker RE, Paull D, Noggle S, McCarroll SA, Joung JK, Woolf CJ, Brown RH, Eggan K. Pathways disrupted in human ALS motor neurons identified through genetic correction of mutant SOD1. Cell Stem Cell 2014;14:781–795. doi: 10.1016/j.stem.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Peters DT, Henderson CA, Warren CR, Friesen M, Xia F, Becker CE, Musunuru K, Cowan CA. Asialoglycoprotein receptor 1 is a specific cell-surface marker for isolating hepatocytes derived from human pluripotent stem cells. Development 2016;143:1475–1481. doi: 10.1242/dev.132209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Musunuru K Personalized genomes and cardiovascular disease. Cold Spring Harb Perspect Med 2014;5:a014068. doi: 10.1101/cshperspect.a014068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Chen IY, Matsa E, Wu JC. Induced pluripotent stem cells: at the heart of cardiovascular precision medicine. Nat Rev Cardiol 2016;13:333–349. doi: 10.1038/nrcardio.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Liang P, Lan F, Lee AS, Gong T, Sanchez-Freire V, Wang Y, Diecke S, Sallam K, Knowles JW, Wang PJ, Nguyen PK, Bers DM, Robbins RC, Wu JC. Drug screening using a library of human induced pluripotent stem cell-derived cardiomyocytes reveals disease-specific patterns of cardiotoxicity. Circulation 2013;127:1677–1691. doi: 10.1161/CIRCULATIONAHA.113.001883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Braam SR, Tertoolen L, Casini S, Matsa E, Lu HR, Teisman A, Passier R, Denning C, Gallacher DJ, Towart R, Mummery CL. Repolarization reserve determines drug responses in human pluripotent stem cell derived cardiomyocytes. Stem Cell Res 2013;10:48–56. doi: 10.1016/j.scr.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 173.Stillitano F, Hansen J, Kong CW, Karakikes I, Funck-Brentano C, Geng L, Scott S, Reynier S, Wu M, Valogne Y, Desseaux C, Salem JE, Jeziorowska D, Zahr N, Li R, Iyengar R, Hajjar RJ, Hulot JS. Modeling susceptibility to drug-induced long QT with a panel of subject-specific induced pluripotent stem cells. Elife 2017;6:e19406. doi: 10.7554/eLife.19406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Burridge PW, Li YF, Matsa E, Wu H, Ong SG, Sharma A, Holmström A, Chang AC, Coronado MJ, Ebert AD, Knowles JW, Telli ML, Witteles RM, Blau HM, Bernstein D, Altman RB, Wu JC. Human induced pluripotent stem cell-derived cardiomyocytes recapitulate the predilection of breast cancer patients to doxorubicin-induced cardiotoxicity. Nat Med 2016;22:547–556. doi: 10.1038/nm.4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Sharma A, Burridge PW, McKeithan WL, Serrano R, Shukla P, Sayed N, Churko JM, Kitani T, Wu H, Holmströ A, Matsa E, Zhang Y, Kumar A, Fan AC, del Álamo JC, Wu SM, Moslehi JJ, Mercola M, Wu JC. High-throughput screening of tyrosine kinase inhibitor cardiotoxicity with human induced pluripotent stem cells. Sci Transl Med 2017;9:eaaf2584. doi: 10.1126/scitranslmed.aaf2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Matsa E, Burridge PW, Yu KH, Ahrens JH, Termglinchan V, Wu H, Liu C, Shukla P, Sayed N, Churko JM, Shao N, Woo NA, Chao AS, Gold JD, Karakikes I, Snyder MP, Wu JC. Transcriptome profiling of patient-specific human iPSC-cardiomyocytes predicts individual drug safety and efficacy responses in vitro. Cell Stem Cell 2016;19:311–325. doi: 10.1016/j.stem.2016.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Sharma A, Marceau C, Hamaguchi R, Burridge PW, Rajarajan K, Churko JM, Wu H, Sallam KI, Matsa E, Sturzu AC, Che Y, Ebert A, Diecke S, Liang P, Red-Horse K, Carette JE, Wu SM, Wu JC. Human induced pluripotent stem cell-derived cardiomyocytes as an in vitro model for coxsackievirus B3-induced myocarditis and antiviral drug screening platform. Circ Res 2014;115:556–566. doi: 10.1161/CIRCRESAHA.115.303810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Campbell KA, Terzic A, Nelson TJ. Induced pluripotent stem cells for cardiovascular disease: from product-focused disease modeling to process-focused disease discovery. Regen Med 2015;10:773–783. doi: 10.2217/rme.15.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Green RC, Berg JS, Grody WW, Kalia SS, Korf BR, Martin CL, McGuire AL, Nussbaum RL, O’Daniel JM, Ormond KE, Rehm HL, Watson MS, Williams MS, Biesecker LG; American College of Medical Genetics and Genomics. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med 2013;15:565–574. doi: 10.1038/gim.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Schulz WL, Tormey CA, Torres R. Computational approach to annotating variants of unknown significance in clinical next generation sequencing. Lab Med 2015;46:285–289. doi: 10.1309/LMWZH57BRWOPR5RQ. [DOI] [PubMed] [Google Scholar]
- 181.Manrai AK, Funke BH, Rehm HL, Olesen MS, Maron BA, Szolovits P, Margulies DM, Loscalzo J, Kohane IS. Genetic misdiagnoses and the potential for health disparities. N Engl J Med 2016;375:655–665. doi: 10.1056/NEJMsa1507092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Nelson TJ, Terzic A. Induced pluripotent stem cells: an emerging theranostics platform. Clin Pharmacol Ther 2011;89:648–650. doi: 10.1038/clpt.2010.304. [DOI] [PubMed] [Google Scholar]
- 183.Beers J, Linask KL, Chen JA, Siniscalchi LI, Lin Y, Zheng W, Rao M, Chen G. A cost-effective and efficient reprogramming platform for large-scale production of integration-free human induced pluripotent stem cells in chemically defined culture. Sci Rep 2015;5:11319. doi: 10.1038/srep11319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Paull D, Sevilla A, Zhou H, Hahn AK, Kim H, Napolitano C, Tsankov A, Shang L, Krumholz K, Jagadeesan P, Woodard CM, Sun B, Vilboux T, Zimmer M, Forero E, Moroziewicz DN, Martinez H, Malicdan MC, Weiss KA, Vensand LB, Dusenberry CR, Polus H, Sy KT, Kahler DJ, Gahl WA, Solomon SL, Chang S, Meissner A, Eggan K, Noggle SA. Automated, high-throughput derivation, characterization and differentiation of induced pluripotent stem cells. Nat Methods 2015;12:885–892. doi: 10.1038/nmeth.3507. [DOI] [PubMed] [Google Scholar]
- 185.Terstegge S, Laufenberg I, Pochert J, Schenk S, Itskovitz-Eldor J, Endl E, Brüstle O. Automated maintenance of embryonic stem cell cultures. Biotechnol Bioeng 2007;96:195–201. doi: 10.1002/bit.21061. [DOI] [PubMed] [Google Scholar]
- 186.Thomas RJ, Anderson D, Chandra A, Smith NM, Young LE, Williams D, Denning C. Automated, scalable culture of human embryonic stem cells in feeder-free conditions. Biotechnol Bioeng 2009;102:1636–1644. doi: 10.1002/bit.22187. [DOI] [PubMed] [Google Scholar]
- 187.Conway MK, Gerger MJ, Balay EE, O’Connell R, Hanson S, Daily NJ, Wakatsuki T. Scalable 96-well plate based iPSC culture and production using a robotic liquid handling system. J Vis Exp 2015;99:e52755. doi: 10.3791/52755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Amit M, Laevsky I, Miropolsky Y, Shariki K, Peri M, Itskovitz-Eldor J. Dynamic suspension culture for scalable expansion of undifferentiated human pluripotent stem cells. Nat Protoc 2011;6:572–579. doi: 10.1038/nprot.2011.325. [DOI] [PubMed] [Google Scholar]
- 189.Zweigerdt R, Olmer R, Singh H, Haverich A, Martin U. Scalable expansion of human pluripotent stem cells in suspension culture. Nat Protoc 2011;6:689–700. doi: 10.1038/nprot.2011.318. [DOI] [PubMed] [Google Scholar]
- 190.Hemmi N, Tohyama S, Nakajima K, Kanazawa H, Suzuki T, Hattori F, Seki T, Kishino Y, Hirano A, Okada M, Tabei R, Ohno R, Fujita C, Haruna T, Yuasa S, Sano M, Fujita J, Fukuda K. A massive suspension culture system with metabolic purification for human pluripotent stem cell-derived cardiomyocytes. Stem Cells Transl Med 2014;3:1473–1483. doi: 10.5966/sctm.2014-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Kempf H, Kropp C, Olmer R, Martin U, Zweigerdt R. Cardiac differentiation of human pluripotent stem cells in scalable suspension culture. Nat Protoc 2015;10:1345–1361. doi: 10.1038/nprot.2015.089. [DOI] [PubMed] [Google Scholar]
- 192.Fonoudi H, Ansari H, Abbasalizadeh S, Larijani MR, Kiani S, Hashemizadeh S, Zarchi AS, Bosman A, Blue GM, Pahlavan S, Perry M, Orr Y, Mayorchak Y, Vandenberg J, Talkhabi M, Winlaw DS, Harvey RP, Aghdami N, Baharvand H. A universal and robust integrated platform for the scalable production of human cardiomyocytes from pluripotent stem cells. Stem Cells Transl Med 2015;4:1482–1494. doi: 10.5966/sctm.2014-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Chen VC, Ye J, Shukla P, Hua G, Chen D, Lin Z, Liu JC, Chai J, Gold J, Wu J, Hsu D, Couture LA. Development of a scalable suspension culture for cardiac differentiation from human pluripotent stem cells. Stem Cell Res 2015;15:365–375. doi: 10.1016/j.scr.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Jha R, Wu Q, Singh M, Preininger MK, Han P, Ding G, Cho HC, Jo H, Maher KO, Wagner MB, Xu C. Simulated microgravity and 3D culture enhance induction, viability, proliferation and differentiation of cardiac progenitors from human pluripotent stem cells. Sci Rep 2016;6:30956. doi: 10.1038/srep30956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Braam SR, Passier R, Mummery CL. Cardiomyocytes from human pluripotent stem cells in regenerative medicine and drug discovery. Trends Pharmacol Sci 2009;30:536–545. doi: 10.1016/j.tips.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 196.Gwathmey JK, Tsaioun K, Hajjar RJ. Cardionomics: a new integrative approach for screening cardiotoxicity of drug candidates. Expert Opin Drug Metab Toxicol 2009;5:647–660. doi: 10.1517/17425250902932915. [DOI] [PubMed] [Google Scholar]
- 197.Navarrete EG, Liang P, Lan F, Sanchez-Freire V, Simmons C, Gong T, Sharma A, Burridge PW, Patlolla B, Lee AS, Wu H, Beygui RE, Wu SM, Robbins RC, Bers DM, Wu JC. Screening drug-induced arrhythmia [corrected] using human induced pluripotent stem cell-derived cardiomyocytes and low-impedance microelectrode arrays. Circulation 2013;128(suppl 1):S3–13. doi: 10.1161/CIRCULATIONAHA.112.000570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Meyer T, Leisgen C, Gonser B, Günther E. QT-screen: high-throughput cardiac safety pharmacology by extracellular electrophysiology on primary cardiac myocytes. Assay Drug Dev Technol 2004;2:507–514. doi: 10.1089/adt.2004.2.507. [DOI] [PubMed] [Google Scholar]
- 199.Swanton C, Nicke B, Schuett M, Eklund AC, Ng C, Li Q, Hardcastle T, Lee A, Roy R, East P, Kschischo M, Endesfelder D, Wylie P, Kim SN, Chen JG, Howell M, Ried T, Habermann JK, Auer G, Brenton JD, Szallasi Z, Downward J. Chromosomal instability determines taxane response. Proc Natl Acad Sci U S A 2009;106:8671–8676. doi: 10.1073/pnas.0811835106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Nerbonne JM. Studying cardiac arrhythmias in the mouse: a reasonable model for probing mechanisms? Trends Cardiovasc Med 2004;14:83–93. doi: 10.1016/j.tcm.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 201.Shanks N, Greek R, Greek J. Are animal models predictive for humans? Philos Ethics Humanit Med 2009;4:2. doi: 10.1186/1747-5341-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Lacerda AE, Kuryshev YA, Chen Y, Renganathan M, Eng H, Danthi SJ, Kramer JW, Yang T, Brown AM. Alfuzosin delays cardiac repolarization by a novel mechanism. J Pharmacol Exp Ther 2008;324:427–433. doi: 10.1124/jpet.107.128405. [DOI] [PubMed] [Google Scholar]
- 203.Sirenko O, Cromwell EF, Crittenden C, Wignall JA, Wright FA, Rusyn I. Assessment of beating parameters in human induced pluripotent stem cells enables quantitative in vitro screening for cardiotoxicity. Toxicol Appl Pharmacol 2013;273:500–507. doi: 10.1016/j.taap.2013.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Pointon A, Harmer AR, Dale IL, Abi-Gerges N, Bowes J, Pollard C, Garside H. Assessment of cardiomyocyte contraction in human-induced pluripotent stem cell-derived cardiomyocytes. Toxicol Sci 2015;144:227–237. doi: 10.1093/toxsci/kfu312. [DOI] [PubMed] [Google Scholar]
- 205.Blinova K, Stohlman J, Vicente J, Chan D, Johannesen L, Hortigon-Vinagre MP, Zamora V, Smith G, Crumb WJ, Pang L, Lyn-Cook B, Ross J, Brock M, Chvatal S, Millard D, Galeotti L, Stockbridge N, Strauss DG. Comprehensive translational assessment of human-induced pluripotent stem cell derived cardiomyocytes for evaluating drug-induced arrhyth-mias. Toxicol Sci 2017;155:234–247. doi: 10.1093/toxsci/kfw200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Reynolds JG, Geretti E, Hendriks BS, Lee H, Leonard SC, Klinz SG, Noble CO, Lücker PB, Zandstra PW, Drummond DC, Olivier KJ Jr, Nielsen UB, Niyikiza C, Agresta SV, Wickham TJ. HER2-targeted liposomal doxorubicin displays enhanced anti-tumorigenic effects without associated cardiotoxicity. Toxicol Appl Pharmacol 2012;262:1–10. doi: 10.1016/j.taap.2012.04.008. [DOI] [PubMed] [Google Scholar]