Supplemental Digital Content is available in the text
Keywords: ATRX, DAXX, lung cancer, prognosis
Abstract
Molecular characterization of lung cancer specimens after radical surgery offers additional prognostic information and may help to guide adjuvant therapeutic procedures. The transcriptional regulators alpha thalassemia/mental retardation X-linked (ATRX) and death domain-associated protein (DAXX) have recently been described in different cancer entities as a useful prognostic biomarker. This study was initiated to explore their protein expression patterns and prognostic value in patients with operable lung cancer disease.
The protein abundance (in the following text also named protein expression) of ATRX and DAXX were analyzed by immunohistochemistry in 194 samples of squamous cell lung carcinoma (SQCLC), 111 samples of pulmonary adenocarcinoma (AC) and 40 samples of small cell lung cancer (SCLC). The protein levels of ATRX and DAXX were correlated with clinicopathological characteristics and patient outcome.
ATRX showed strong protein expression in 16.2% of AC, 11.9% of SQCLC, and 42.5% of SCLC. DAXX was highly expressed in 54.9% of AC, 76.2% of SQCLC, and 82.5% of SCLC. Immunostaining of both ATRX and DAXX were seen in 14.4% of AC, 11.3% of SQCLC, and 42.5% of SCLC. High protein expression of ATRX was a favorable prognostic marker for patients with AC (hazard ratio 0.38, P = .02). Sub-group analyses showed a significant correlation between ATRX and the clinical stage of SQCLC and SCLC. Histological grading and ATRX were also significantly associated in cases of SQCLC.
The presence of ATRX and DAXX are correlated with lung cancer histology. Strong ATRX protein expression is associated with a significantly longer overall survival in patients with AC.
1. Introduction
Lung cancer is the leading cause of cancer-related death worldwide. 14% of all newly diagnosed cancer cases in the US are attributed to lung cancer. Overall, lung cancer is responsible for more than 20% cancer deaths in men and women. The 5-year survival rate is about 16%.[1,2] By histology, lung cancer is divided into non-small cell lung cancer (NSCLC, 85% of all cases) and small-cell lung cancer (SCLC, 15% of all cases). The most predominant NSCLC subtypes are adenocarcinoma (AC) and squamous cell lung carcinoma (SQCLC).[3]
The therapy of choice for early-stage lung cancer is radical surgery followed by adjuvant chemotherapy.[4,5] So far, this decision is usually based on the initial Union for international cancer control (UICC) cancer stage.[4] While the TNM-classification is used to describe lung cancer stages, molecular characterization may be used as a complementary tool that offers additional prognostic information and may help to determine the best adjuvant therapeutic protocol.[1] Broad molecular characterization of lung cancer patients is even more important as lung cancer has been shown to be a disease with heterogeneous molecular biology.[6]
The transcriptional regulator proteins alpha thalassemia/mental retardation X-linked (ATRX) and death domain-associated protein (DAXX) are 2 subunits of a transcription/chromatin remodeling complex. These loss-of-expression (that can, eg, be detected by immunohistochemistry [IHC]) and thus loss-of-function of either of these genes are associated with the alternative lengthening of telomere (ALT) phenotype[7–10] and chromosome instability[7] in different cancer entities, eg, in neuroendocrine tumors where mutations of ATRX or DAXX lead to protein loss. Interaction of both ATRX and DAXX is required for the formation of the ATRX/DAXX complex. The complex acts as a chaperone for the histone H3.3 which is required for maintaining the heterochromatic state of telomeres after mitosis.[5,11,12] In mouse embryonic fibroblasts, ATRX is a binding partner of enhancer of zeste homolog 2 (EZH2) and loss of ATRX causes improper EZH2 targeting to ectopic intergenetic space. This results in decreased selectivity of EZH2-target trimethylation and upregulation of EZH2-target genes.[13] Furthermore, ATRX-deficient cells show higher sensitivity to DNA damaging agents like Cisplatin and 5-fluorouracile.[14,15]
ATRX and DAXX have recently been described in different cancer entities as useful prognostic biomarkers. Loss of ATRX or DAXX occurs in 40% of neuroendocrine pancreatic cancers.[7] Here the loss of function of ATRX/DAXX is a late event in tumorigenesis and a marker of poor prognosis, leading to a reversed DNA methylation.[16–18] ATRX is a known biomarker in gliomas and a hallmark of astrocytoma. In gliomas, loss of function of ATRX is associated with longer patient survival, especially in isocitrate dehydrogenase-wildtype glioblastoma patients.[19,20]
We, therefore, analyzed the ATRX and DAXX protein abundance in lung cancer patients, investigating 2 new potential markers for outcome of patients suffering from AC, SQCLC, and SCLC.
2. Material and methods
2.1. Tissue samples
Approval for using the patient material in this study was obtained from the Ethics Committee of the University Medical Center Goettingen (#1-2-08). Informed consent was obtained from all patients. All procedures were conducted in accordance with the declaration of Helsinki and institutional, state, and federal guidelines. Tissue samples were obtained from surgical resections at the Department of Thoracic Surgery of the University Medical Center Goettingen.
2.2. Immunohistochemical staining
Tissue samples were assembled in tissue microarrays before immunostaining. Immunohistochemical reactions were performed as described previously.[21] Briefly, 2-μm tissue sections were incubated in EnVision Flex Target Retrieval Solution, pH low (Dako/agilent, Sanata Clara CA) followed by incubation of primary antibodies against ATRX (Sigma, Sigma–Aldrich, St. Louis, MO 1:500) and DAXX (Sigma, 1:200) at room temperature for 20 minutes. Polymeric secondary antibodies coupled to horseradish peroxidase (EnVision Flex+, Dako) and DAB (Dako) were applied to visualize the sites of immunoprecipitations. Tissue samples were analyzed by light microscopy after counterstaining with Meyer's hematoxylin. Staining intensity was evaluated using a 3-tiered semiquantitative scoring system: 0 = negative; 1 = weakly positive; 2 = strongly positive.
2.3. Statistical analysis
Contingency table, Chi-square test, and Pearson coefficient were used for analyzing correlation between clinical and pathological features and ARTX or DAXX protein expression. Survival analysis was performed by using the Kaplan–Meier estimator and the log-rank test. P values <.05 were considered as significant. Microsoft Excel 2010 and GraphPad Prism (version 7.00 for Windows, GraphPad Software, La Jolla, CA, www.graphpad.com) was used for statistical analyses.
3. Results
3.1. Patient characteristics
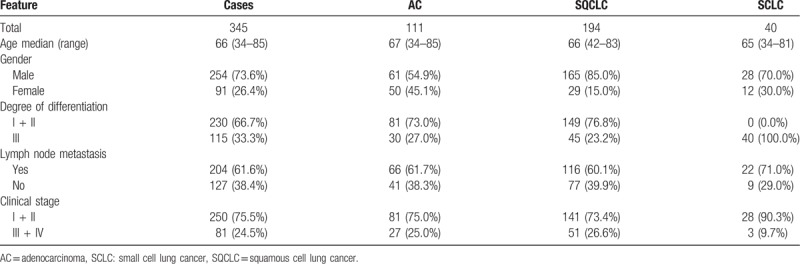
A total of 345 patients (254 male, 91 female) were included in this study. The median age was 66 years (range 34–85). Patients were diagnosed with either NSCLC (111 AC, 194 SQCLC) or SCLC (40 patients). All patients were treated with surgical tumor resection. According to UICC (7th edition) stage, 250 patients were classified as stage I or II and 81 patients as stage III or IV. R0 resection was achieved in 302 cases. Lymph node status was positive in 127 cases (for detailed patient characteristics refer to Table 1 and Supplementary Table 1).
Table 1.
Patient characteristics.
3.2. Protein expression of ATRX and DAXX in lung cancer patients and correlation with clinicopathologic characteristics
The protein abundance (in the following text also named protein expression) of ATRX and DAXX was described as low (staining score 0 or 1, see material and methods) or high (staining score 2) and analyzed for each tumor entity (Fig. 1). The presence of both ATRX and DAXX was classified as low-low (both weak or no immunostaining), low-high (either 1 with strong immunostaining), or high-high (both with strong immunostaining). ATRX showed a strong immunostaining in 16.2% of AC, 11.9% in SQCLC and 42.5% in SCLC. DAXX was highly expressed in 55.0% of AC, 76.3% of SQCLC, and 82.5% of SCLC. Strong immunostaining of both ATRX and DAXX was seen in 14.5% of AC, 11.3% of SQCLC, and 42.5% of SCLC (Fig. 2 and Table 2). The simultaneous expression of ATRX and DAXX (expression pattern high-high) in SCLC was significantly (P < .001) more common in SCLC patients compared to NSCLC patients (AC and SQCLC). ATRX and DAXX immunostaining levels showed a weak but non-significant correlation (r = 0.39; P = .09; degrees of freedom [df] = 344). Subgroup analysis revealed that AC had the highest – albeit not significant – Pearson correlation coefficient (r = 0.47, P = .91, df = 110). Correlation coefficient of ATRX and DAXX immunostaining in SQCLC (r = 0.30, P = .032, df = 193) and SCLC (r = 0.32, P = .039, df = 39) was lower, but significant.
Figure 1.
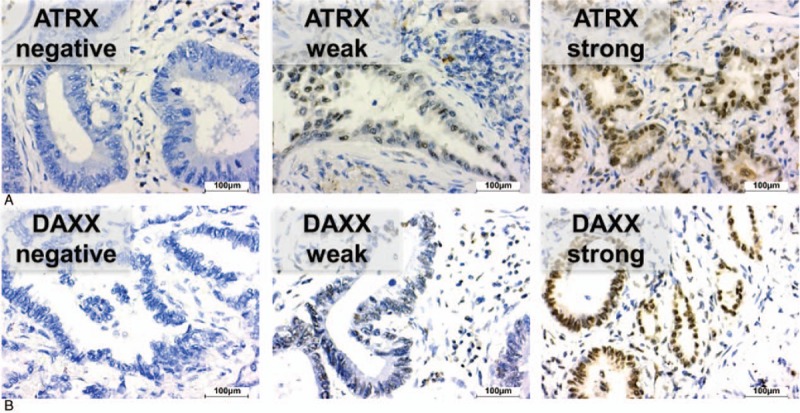
Representative immunohistochemical stainings with no (score 0), weak (score 1), and strong (score 2) immunostaining of ATRX (A) and DAXX (B) in pulmonary adenocarcinomas. Scale bar: 100 μm. ATRX = alpha thalassemia/mental retardation X-linked, DAXX = death domain-associated protein.
Figure 2.
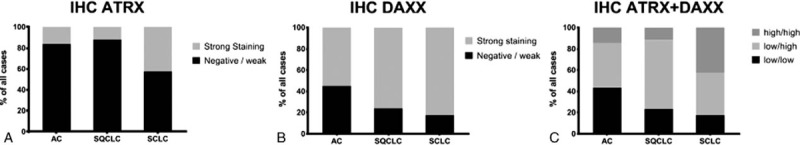
Protein expression of ATRX (A) and DAXX (B) sorted by entity. Combined protein expression of ATRX and DAXX was categorized as low/low, low/high and high/high (C). ATRX = alpha thalassemia/mental retardation X-linked, DAXX = death domain-associated protein.
Table 2.
Expression pattern of ATRX and DAXX.

Protein abundance of ATRX and DAXX is correlated with cancer entity and histological grading (P < .001). Nodal status, age, gender, or UICC stage showed no association with protein expression of either marker (Table 3). We next analyzed the influence of ATRX and DAXX for each cancer entity separately. ATRX protein expression correlated with UICC stage in both SQCLC (P = .03) and SCLC (P = .02). SQCLC samples classified as UICC stage III or IV showed high protein expression more often than tumors in UICC stage I and II (19.4% vs 8.5%). Histological grading in SQCLC was also significantly correlated with ATRX protein expression as strong immunostaining of ATRX was found more often in samples with a higher grading (26.7% vs 7.4%, P = .0005) (Tables 4–6).
Table 3.
ATRX and DAXX expression sorted by clinical features.
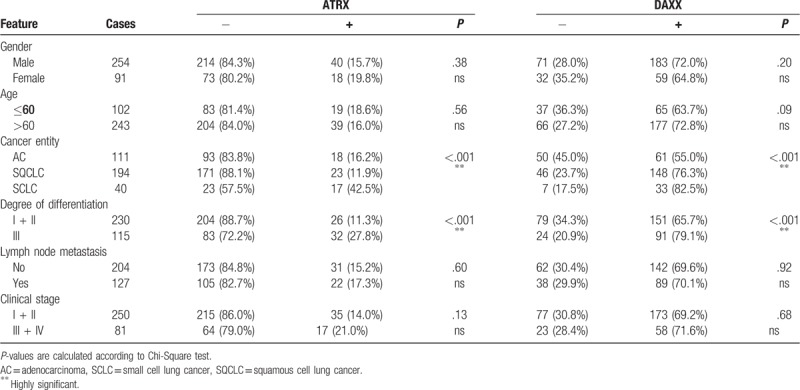
Table 4.
ATRX and DAXX expression in adenocarcinoma sorted by clinical features.
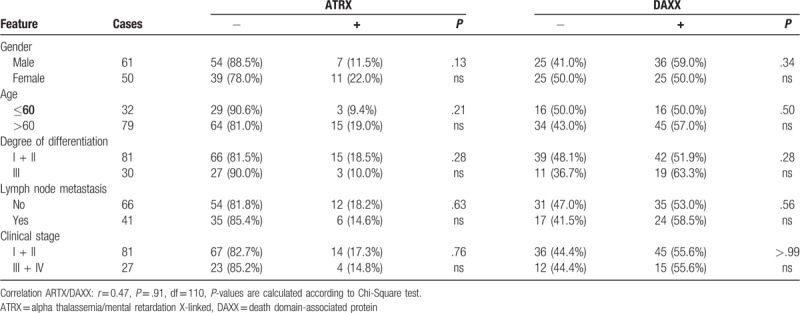
Table 6.
ATRX and DAXX expression in small cell lung carcinoma sorted by clinical features.
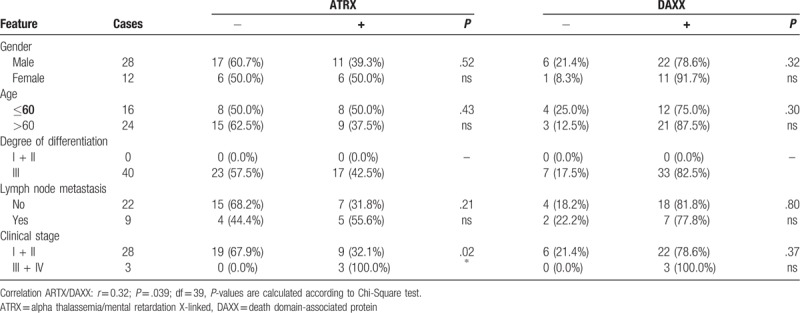
Table 5.
ATRX and DAXX expression in squamous cell lung carcinoma sorted by clinical features.

3.3. Impact of ARXT and DAXX protein expression on prognosis
AC patients with a high ATRX protein abundance showed a significantly better overall survival in Kaplan-Meyer-Estimation (median survival 102 vs 35 months, hazard ratio = 2.61, P = .022, Fig. 3A, top). On the other hand, AC patients with high DAXX protein expression showed a moderate, albeit not significant, worse overall survival (median survival 35 vs 44 months, P = .216, Fig. 3A, bottom). Taking both ATRX and DAXX into account, Kaplan-Meyer-analysis showed a significantly better outcome in the subgroup of AC patients with both high ATRX and DAXX levels (median survival low/low = 44 months, low/high = 34 months, high/high = 102 months, P = .032, Fig. 4A). By contrast, overall survival of patients with SQCLC and SCLC showed no significant correlation with ATRX or DAXX (Figs. 3B + C and 4B and C).
Figure 3.
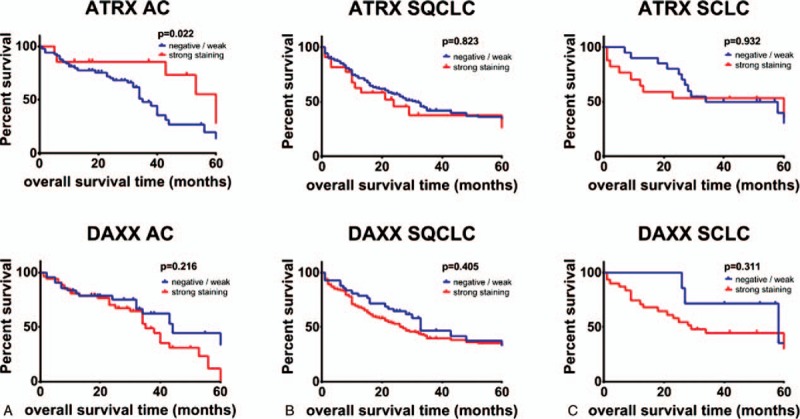
Kaplan–Meier analysis of OS in all patients with adenocarcinoma (A), SQCLC (B) and SCLC (C). Patients were grouped according to protein expression of ARXT (top) and DAXX (bottom). The P value is from a log-rank test. ATRX = alpha thalassemia/mental retardation X-linked, DAXX = death domain-associated protein, OS = overall survival, SCLC = small cell lung cancer, SQCLC = squamous cell lung carcinoma.
Figure 4.
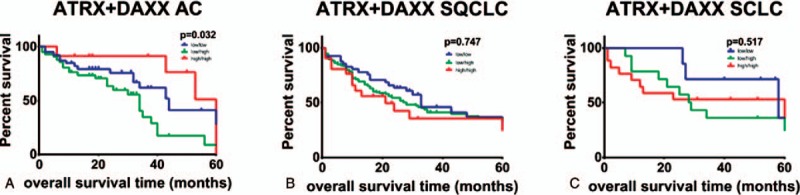
Kaplan–Meier analysis of OS in which all patients with adenocarcinoma (A), SQCLC (B) and SCLC (C) were grouped according to protein expression of ATRX + DAXX. The p value is from a log-rank test. ATRX = alpha thalassemia/mental retardation X-linked, DAXX = death domain-associated protein, OS = overall survival, SCLC = small cell lung cancer, SQCLC = squamous cell lung carcinoma.
4. Discussion
Surgery is the treatment of choice for early-stage lung cancer. According to the recommendations of the European Society of Medical Oncology an adjuvant chemotherapy regimen should be offered to patients suffering from UICC stage II and IIIA lung cancer. Data on UICC stage I is inconclusive. While adjuvant treatment leads to a benefit in patients with UICC stage IB and a tumor size >4 cm, patients with UICC stage IA cancer do not profit from therapy. According to the guidelines, the subgroup of UICC stage IB should be considered for adjuvant treatment.[4,22–24] A prognostic biomarker that would give additional information supporting a decision for or against adjuvant chemotherapy would be an important improvement.
In this study, we investigated the protein expression of ATRX and DAXX in lung cancer and its impact on the outcome of lung cancer patients. ATRX was present in less than 20% of NSCLC cases while SCLC showed an expression in 42.5% of cases. DAXX protein expression was more common and was found in over 50% in AC and in more than 70% in SQCLC and SCLC. We observed a weak, but significant correlation between DAXX and ATRX in SQCLC and SCLC. This is not surprising, as both proteins interact to transport H3.3 to telomeric and pericentromeric chromatin. H3.3 inhibits homologous recombination repair and DNA-repair mechanisms.[25]
The protein expression pattern of ATRX and DAXX was correlated with lung cancer histology with SCLC showing the largest percentage of samples with a high protein abundance of ATRX and DAXX. We further observed a correlation between ATRX and DAXX and histological grading in SQCLC. Finally, we identified an association between UICC stage and ATRX in the sub-group of SQCLC and SCLC. It is noteworthy that we did not detect any association between DAXX and UICC stage – especially as one would expect less DAXX in advanced lung cancer because DAXX is known to prevent hypoxia-induced lung cancer cell metastasis.[26] Such an association between DAXX and UICC stage III/IV was previously described in esophageal cancer.[27] We could also not confirm the previously reported association between age and ATRX in patients suffering from Glioma[14,28] in our cohort of lung cancer patients. However, the association between age and ATRX loss was described for patients with Glioma and a mean age of 35.7 years.[28] Our cohort of lung cancer patients showed a median age of 66 years and therefore a later onset of the cancer disease.
Patients suffering from neuroendocrine pancreatic tumors show a shorter overall survival if they have a loss of function mutation (and therefore loss-of-expression) of ATRX or DAXX. A similar poor prognosis associated with loss of function for ATRX was also recently reported for uterine leiomyosarcomas.[18,29] In line with these findings, a subgroup analysis showed a worse outcome in AC patients with loss of ATRX protein expression. Loss-of function of ATRX or DAXX leads to ALT phenotype, a survival mechanism in cancer cells. At the same time, loss of these regulators reverses DNA methylation patterns making the cancer cell more susceptible for treatment with alkylating agents, which may explain why gliomas with loss of ARTX or DAXX are more susceptible to this treatment and why affected patients have a better prognosis.[30]
ALT sensitizes cancer cells also for treatment with methyltransferase and deacetylase inhibitors. The latter is already in use for hematological malignancies. So far, they have not shown results for solid tumors and are only used in combination therapies.[4,10,17] However, in our sub-group of AC patients with weak or no ATRX protein expression and a significantly shorter overall survival, a possible benefit of methyltransferase and deacetylase inhibitors might be a consideration for future research.
Subgroup analysis of SQCLC and SCLC patients showed no difference in patients’ outcome. A small study showed an association in SCLC patients between poor overall survival and ATRX mutations detected in plasma cell-free DNA.[31] We did not detect a correlation between ATRX and overall survival; however, this might be explained by the different approach (immunohistochemical analysis of protein expression in lung cancer tissue vs mutational analysis of ATRX gene in plasma cell-free DNA) and the small case number (n = 40) of our SCLC cohort.
In summary, analysis of ATRX and DAXX status in lung cancer patients adds relevant prognostic information and should be considered for patient stratification and therapeutic decisions.
Acknowledgments
The authors declare that they have no conflict of interest. We thank Jennifer Appelhans for her technical support. HB and OE are supported by the Deutsche Krebshilfe Foundation (grant: 70112551). HB is supported by the University medical center Göttingen and the Else-Kröner-Fresenius-Foundation. JB is supported by the “Göttinger Kolleg für Translationale Medizin” (funded by the Ministry for Science and Culture of Lower Saxony). SY is supported by the Chinese Scholarship Council.
Author contributions
Conceptualization: Sha Yao, Omar Elakad, Anna-Maria Lois, Jana Brünies, Marc Hinterthaner, Bernd C. Danner, Alexander Emmert, Hanibal Bohnenberger.
Data curation: Judith Buentzel, Sha Yao, Marc Hinterthaner, Hanibal Bohnenberger.
Formal analysis: Judith Buentzel, Hanibal Bohnenberger.
Funding acquisition: Judith Buentzel, Hanibal Bohnenberger.
Investigation: Judith Buentzel, Sha Yao, Omar Elakad, Marc Hinterthaner, Bernd C. Danner, Philipp Ströbel, Alexander Emmert, Hanibal Bohnenberger.
Methodology: Sha Yao, Omar Elakad, Anna-Maria Lois, Marc Hinterthaner, Bernd C. Danner, Philipp Ströbel, Alexander Emmert, Hanibal Bohnenberger.
Project administration: Jana Brünies, Philipp Ströbel, Alexander Emmert, Hanibal Bohnenberger.
Resources: Jana Brünies, Bernd C. Danner.
Software: Judith Buentzel, Anna-Maria Lois, Julia König, Hanibal Bohnenberger.
Supervision: Marc Hinterthaner, Philipp Ströbel, Alexander Emmert, Hanibal Bohnenberger.
Validation: Sha Yao, Hanibal Bohnenberger.
Visualization: Sha Yao, Hanibal Bohnenberger.
Writing – original draft: Judith Buentzel, Hanibal Bohnenberger.
Writing – review and editing: Judith Buentzel, Anna-Maria Lois, Jana Brünies, Julia König, Philipp Ströbel, Hanibal Bohnenberger.
Judith Buentzel orcid: 0000-0002-9531-7910.
Supplementary Material
Footnotes
Abbreviations: AC = pulmonary adenocarcinoma, ALT = alternative lengthening of telomere, ATRX = alpha thalassemia/mental retardation X-linked, DAXX = death domain-associated protein, df = degrees of freedom, EZH2 = enhancer of zeste homolog 2, NSCLC = non-small cell lung cancer, SCLC = small cell lung cancer, SQCLC = squamous cell lung carcinoma, UICC = union for international cancer control.
JB and SY share first authorship.
We acknowledge support by the German Research Foundation and the Open Access Publication Funds of the Göttingen University.
The authors have no conflicts of interest to disclose.
Supplemental Digital Content is available for this article.
References
- [1].Testa U, Castelli G, Pelosi E. Lung cancers: molecular characterization, clonal heterogeneity and evolution, and cancer stem cells. Cancers 2018;10:248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018;68:7–30. [DOI] [PubMed] [Google Scholar]
- [3].Bui KT, Cooper WA, Kao S, et al. Targeted molecular treatments in non-small cell lung cancer: a clinical guide for oncologists. J Clin Med 2018;7:192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Postmus PE, Kerr KM, Oudkerk M, et al. Early and locally advanced non-small-cell lung cancer (NSCLC): ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol Off J Eur Soc Med Oncol 2017;28:iv1–21. [DOI] [PubMed] [Google Scholar]
- [5].Rosen JE, Keshava HB, Yao X, et al. The natural history of operable non-small cell lung cancer in the national cancer database. Ann Thorac Surg 2016;101:1850–5. [DOI] [PubMed] [Google Scholar]
- [6].Levy MA, Lovly CM, Pao W. Translating genomic information into clinical medicine: lung cancer as a paradigm. Genome Res 2012;22:2101–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Heaphy CM, de Wilde RF, Jiao Y, et al. Altered telomeres in tumors with ATRX and DAXX mutations. Science 2011;333:425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Schwartzentruber J, Korshunov A, Liu X-Y, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 2012;482:226–31. [DOI] [PubMed] [Google Scholar]
- [9].Hechtman JF, Klimstra DS, Nanjangud G, et al. Performance of DAXX immunohistochemistry as a screen for DAXX mutations in pancreatic neuroendocrine tumors. Pancreas 2019;48:396–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].de Wilde RF, Heaphy CM, Maitra A, et al. Loss of ATRX or DAXX expression and concomitant acquisition of the alternative lengthening of telomeres phenotype are late events in a small subset of MEN-1 syndrome pancreatic neuroendocrine tumors. Mod Pathol Off J U S Can Acad Pathol Inc 2012;25:1033–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Dyer MA, Qadeer ZA, Valle-Garcia D, et al. ATRX and DAXX: Mechanisms and Mutations. Cold Spring Harb Perspect Med 2017;7: [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Di Domenico A, Wiedmer T, Marinoni I, et al. Genetic and epigenetic drivers of neuroendocrine tumours (NET). Endocr Relat Cancer 2017;24:R315–34. [DOI] [PubMed] [Google Scholar]
- [13].Sarma K, Cifuentes-Rojas C, Ergun A, et al. ATRX directs binding of PRC2 to Xist RNA and Polycomb targets. Cell 2014;159:869–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Haase S, Garcia-Fabiani MB, Carney S, et al. Mutant ATRX: uncovering a new therapeutic target for glioma. Expert Opin Ther Targets 2018;22:599–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Conte D, Huh M, Goodall E, et al. Loss of Atrx sensitizes cells to DNA damaging agents through p53-mediated death pathways. PloS One 2012;7:e52167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Singhi AD, Liu T-C, Roncaioli JL, et al. Alternative lengthening of telomeres and loss of DAXX/ATRX expression predicts metastatic disease and poor survival in patients with pancreatic neuroendocrine tumors. Clin Cancer Res Off J Am Assoc Cancer Res 2017;23:600–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Schmitt AM, Marinoni I, Blank A, et al. New genetics and genomic data on pancreatic neuroendocrine tumors: implications for diagnosis, treatment, and targeted therapies. Endocr Pathol 2016;27:200–4. [DOI] [PubMed] [Google Scholar]
- [18].Marinoni I, Kurrer AS, Vassella E, et al. Loss of DAXX and ATRX are associated with chromosome instability and reduced survival of patients with pancreatic neuroendocrine tumors. Gastroenterology 2014;146:453–60. [DOI] [PubMed] [Google Scholar]
- [19].Cai J, Chen J, Zhang W, et al. Loss of ATRX, associated with DNA methylation pattern of chromosome end, impacted biological behaviors of astrocytic tumors. Oncotarget 2015;6:18105–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Pekmezci M, Rice T, Molinaro AM, et al. Adult infiltrating gliomas with WHO 2016 integrated diagnosis: additional prognostic roles of ATRX and TERT. Acta Neuropathol (Berl) 2017;133:1001–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Bohnenberger H, Kaderali L, Ströbel P, et al. Comparative proteomics reveals a diagnostic signature for pulmonary head-and-neck cancer metastasis. EMBO Mol Med 2018;10:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Artal Cortés Á, Calera Urquizu L, Hernando Cubero J. Adjuvant chemotherapy in non-small cell lung cancer: state-of-the-art. Transl Lung Cancer Res 2015;4:191–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Salazar MC, Rosen JE, Wang Z, et al. Association of delayed adjuvant chemotherapy with survival after lung cancer surgery. JAMA Oncol 2017;3:610–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Strauss GM, Herndon JE, Maddaus MA, et al. Adjuvant paclitaxel plus carboplatin compared with observation in stage IB non-small-cell lung cancer: CALGB 9633 with the Cancer and Leukemia Group B, Radiation Therapy Oncology Group, and North Central Cancer Treatment Group Study Groups. J Clin Oncol Off J Am Soc Clin Oncol 2008;26:5043–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Fan H-C, Chen C-M, Chi C-S, et al. Targeting telomerase and ATRX/DAXX inducing tumor senescence and apoptosis in the malignant glioma. Int J Mol Sci 2019;20:1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Lin C-W, Wang L-K, Wang S-P, et al. Daxx inhibits hypoxia-induced lung cancer cell metastasis by suppressing the HIF-1(/HDAC1/Slug axis. Nat Commun 2016;7:13867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Ko TY, Kim JI, Park ES, et al. The clinical implications of death domain-associated protein (DAXX) expression. Korean J Thorac Cardiovasc Surg 2018;51:187–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Wiestler B, Capper D, Holland-Letz T, et al. ATRX loss refines the classification of anaplastic gliomas and identifies a subgroup of IDH mutant astrocytic tumors with better prognosis. Acta Neuropathol (Berl) 2013;126:443–51. [DOI] [PubMed] [Google Scholar]
- [29].Tsuyoshi H, Yoshida Y. Molecular biomarkers for uterine leiomyosarcoma and endometrial stromal sarcoma. Cancer Sci 2018;109:1743–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Nandakumar P, Mansouri A, Das S. The role of ATRX in glioma biology. Front Oncol 2017;29:236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Du M, Thompson J, Fisher H, et al. Genomic alterations of plasma cell-free DNAs in small cell lung cancer and their clinical relevance. Lung Cancer Amst Neth 2018;120:113–21. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


