OBJECTIVES:
Loss-of-function mutations of BMPR1A cause juvenile polyposis syndrome (JPS), but large genomic deletions in BMPR1A are rare, reported in few families only, and data regarding the associated phenotype are limited.
METHODS:
We investigated clinical features and genomic data of 7 extended seemingly unrelated families with a genomic deletion of the entire coding region of BMPR1A. We defined mutation size, mutation prevalence, and tumor pathogenesis using whole-genome sequencing, targeted genotyping, and haplotype analysis.
RESULTS:
Patients with JPS from 7 families of Bukharin Jewish ancestry carried a deletion of 429 kb, encompassing the BMPR1A coding sequence and 8 downstream genes. Haplotype analysis and testing controls identified this as a common founder mutation occurring in 1/124 individuals of Bukharin origin. Tumor testing did not demonstrate loss of heterozygosity. Among carriers, JPS was almost fully penetrant, but clinical features varied widely, ranging from mild to very severe, including pan-enteric polyps, gastritis, and colorectal, esophageal, and testicular cancer, and carriers with phenotypes, which would not have raised suspicion of JPS.
DISCUSSION:
The phenotype in this large cohort was extremely variable, although all carriers shared the same variant and the same genetic background. New observations include a preponderance of adenomatous rather than juvenile polyps, possible association with testicular cancer, and unexpected upper gastrointestinal involvement.
INTRODUCTION
Juvenile polyposis syndrome (JPS, OMIM 174900) is a rare autosomal dominant disorder, affecting between 1 in 100,000 and 1 in 160,000 (1), characterized by hamartomatous polyps and increased risk of gastrointestinal (GI) cancer. JPS is diagnosed clinically when a person has any one of the following: (i) more than 5 juvenile polyps of the colon or rectum; (ii) juvenile polyps in other parts of the GI tract; or (iii) any number of juvenile polyps and one or more affected family members (National Comprehensive Cancer Network [NCCN] guidelines) (2). Up to 60% of individuals with clinically defined JPS are now found to exhibit mutations in SMAD4 or BMPR1A genes (3). JPS polyps are typically colonic with edematous, markedly inflamed lamina propria, with cystic dilation and smooth muscle proliferation. Although dysplastic polyps may appear with variable histology, one study reported “mixed polyposis syndrome” with polyps containing variable pathology of adenomatous, hyperplastic, and juvenile features caused by small base pair deletions in the BMPR1A gene (4); adenomas comprise less than 10% of JPS polyps (5).
JPS can involve the entire GI tract. Although colonic phenotype is similar between patients with SMAD4 and BMPR1A mutations, upper GI and gastric polyposis is much more common in SMAD4 mutation (1,6,7). As reported by Aretz et al. (6), SMAD4 mutation carriers had a significantly higher frequency of gastric polyposis than did patients with BMPR1A mutations (83% vs 8%, respectively). All cases of gastric cancer occurred in families with SMAD4 mutations (6,7).
Lifetime risk estimates of GI cancers, mostly colorectal cancer (CRC), are highly variable, ranging from 14% to 55% in different series (1,8,9). Although surveillance guidelines exist, the NCCN guidelines for surveillance recommend referral of patients with JPS to a specialized team due to the rarity of the syndrome and complexities of diagnosis and management (2).
The molecular alterations involved in polyp and tumor formation in JPS are attributed to defective BMP signaling, where aberrant BMP signaling disrupts stem cell self-renewal and differentiation, contributing to tumor formation (10). Loss of heterozygosity (LOH) was reported in half of BMPR1A-related polyps, compatible with BMPR1A acting as a tumor suppressor gene (11). However, BMPR1A LOH has not been documented yet in cancerous tumors.
Most pathogenic variants in BMPR1A are point mutations or small deletions. Large deletions of BMPR1A are rare, accounting for approximately 6% of cases, many of them are contiguous with PTEN (6,12–14). Contiguous gene deletions may lead to more pronounced manifestations; however, the rarity and variability of BMPR1A deletions not including PTEN have made genotype-phenotype relationships of large BMPR1A deletions difficult to assess (14).
We identified a deletion of the entire coding region of the BMPR1A gene among and investigated the clinical features in over 50 individuals from 7 unrelated families. This cohort enables expanding our knowledge about this rare predisposition syndrome.
METHODS
Patients were identified at 3 medical institutions in Israel. Clinical and pathology data were retrieved from medical records. This study was approved by the institutional review boards. The series of control patients genotyped for the deletion were Bukharin individuals referred for prenatal carrier screening or for genetic counseling due to other conditions.
Genetic testing
Testing for the founder deletion was performed either by multigene panels, chromosomal microarray analysis, or directed polymerase chain reaction (PCR) analysis as detailed below. Whole-genome approach chromosomal microarray analysis was performed using single nucleotide polymorphism–based array platforms (Illumina, San Diego, CA). BROCA multigene panel was used for testing for genes known or suggested to harbor mutations leading to solid tumors (15). Sequencing was performed, and variants were evaluated as previously described (16,17). Genomic deletions and duplications were identified by analysis of BROCA sequence read depth (18). With the exception of the BMPR1A deletion, no potentially damaging variants were detected in any family in any of the 65 genes sequenced. Whole-genome sequencing: DNA (1 μg) from the proband of family 2 was sequenced to an average depth of 32x on a HiSeq X instrument. The fastq files were aligned to the hg19 reference gene using iSAAC v.01.15.02.08 (19), and copy number variations were called using Control-FREEC v6.4 (20). The breakpoints of BMPR1A copy number variations were identified by analysis of the split reads obtained by local realignment using Burrows-Wheeler Aligner-mem v0.7.12 (21) and visualization within integrative genomics viewer.
Directed PCR
Multiplex PCR was performed on whole blood genomic DNA using 2 primer pairs. One pair of primers that amplify a 541 bp inside the deletion area was used as the control amplicon of the wild-type allele, and second pair of primers flanking the deletion breakpoints, amplifying mutation-specific 359-bp fragment. The size of the deletion was determined by Sanger sequencing of the deletion junction in 2 affected individuals from 2 families.
Haplotype analysis
Six short tandem repeat (STR) markers that span a region of approximately 2.4 Mb flanking the deletion at 10q23.2 were used. The STR markers were used for PCR amplification of whole blood genomic DNA using fluorescently end-labeled primers and analyzed by capillary electrophoresis on a 3500 or ABI3130xl Genetic Analyzer using standard methods. We examined these STR markers in 7 individuals from 4 families and built the haplotype according to the mutation status of the family members. For 2 single affected individuals, it was not possible to reconstruct the haplotype with certainty, but we did see the allele sizes we expected, and haplotypes were built according to the hypothesis of a common mutation haplotype conservation.
LOH tumor testing was performed on 2 tumor samples (CRC from patient III-1 family 1 and testicular seminoma form patient III-5 family 2). The tumor area was identified by a pathologist on an hematoxylin & eosin slide. An appropriate region was selected from the corresponding unstained slide(s). Tumor cells consisted 60% of cells analyzed. DNA was extracted from formalin-fixed paraffin-embedded tumor and normal tissues using QIAamp DNA FFPE Tissue Kit (Qiagen, Hilden, Germany). The same STR markers used for haplotyping were used for haplotype comparison between normal and tumor tissue to check for LOH.
The standardized incidence ratios were computed as a ratio of observed to expected cancers. Person-years at risk were computed from date of birth up to December 31, 2015 (last date of follow-up) or the date of death for those who died of nonmalignant causes or the date of cancer diagnosis or total proctocolectomy. The expected number of malignancies was computed by applying the appropriate age and sex according to the tables of the Israeli cancer registry for CRC.
RESULTS
We report a genomic deletion of 429,426 bp (chr10:88,611,882-89,041,308 [hg19]), encompassing the entire coding region (exons 3-13) of BMPR1A, and the complete loci of 8 downstream genes. The deletion was heterozygous members from 7 families of Bukharin Jewish origin, who originate from a highly endogamous community in Central Asia for some 2,500 years. After the collapse of the former Soviet Union in 1991, most members immigrated to the United States and Israel. Genomic analysis among mutation carriers revealed a shared haplotype of at least 2.4 Mb flanking the BMPR1A region, reflecting a founder mutation. The deletion was present in 1 of 124 adult Bukharin Jewish controls.
Loss of heterozygosity
LOH was assessed in a colorectal tumor from patient 1-III-1 and in a testicular seminoma from patient 2-III-5. Both BMPR1A alleles were present in both tumors.
Clinical presentation
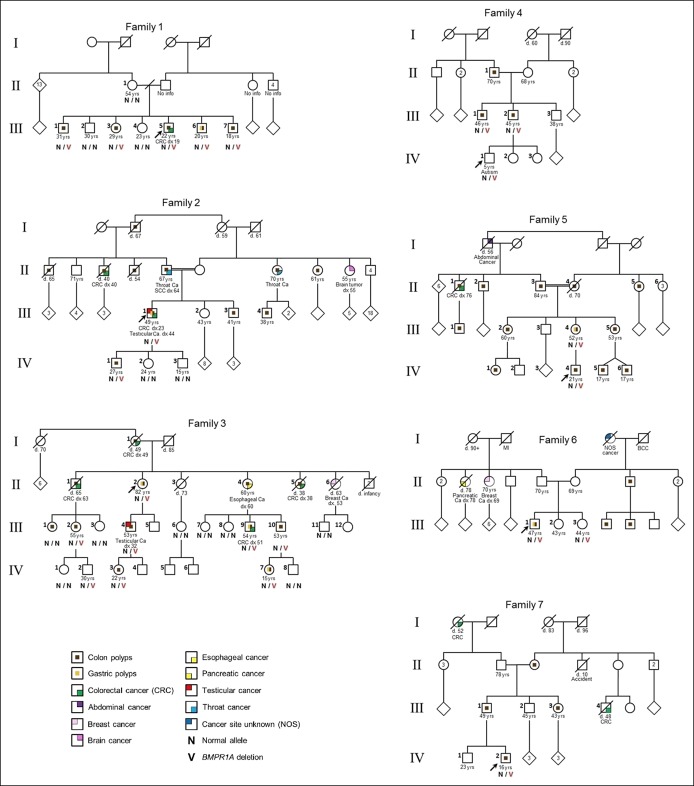
Clinical presentations among mutation carriers varied widely, both with and among families (Figure 1 and Table 1).
Figure 1.
Pedigrees of the 7 index individuals with the 429-kb deletion of BMPR1A germline mutation. The index persons are indicated by arrows (see main text and Table 1 for details). Circles represent females, and squares represent males. The age in years is at last contact, dx is age at cancer diagnosis, and d is age at death.
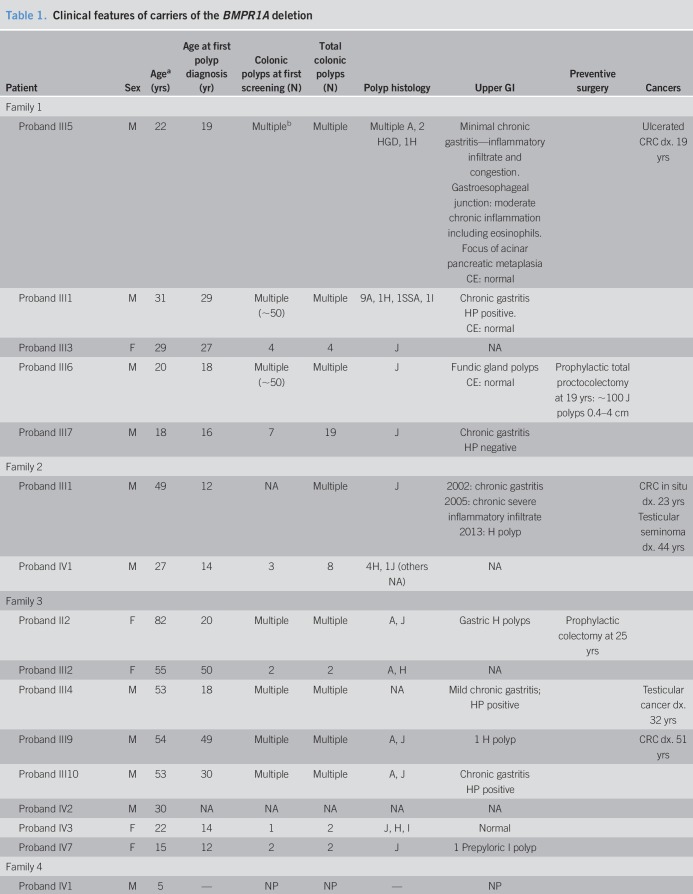
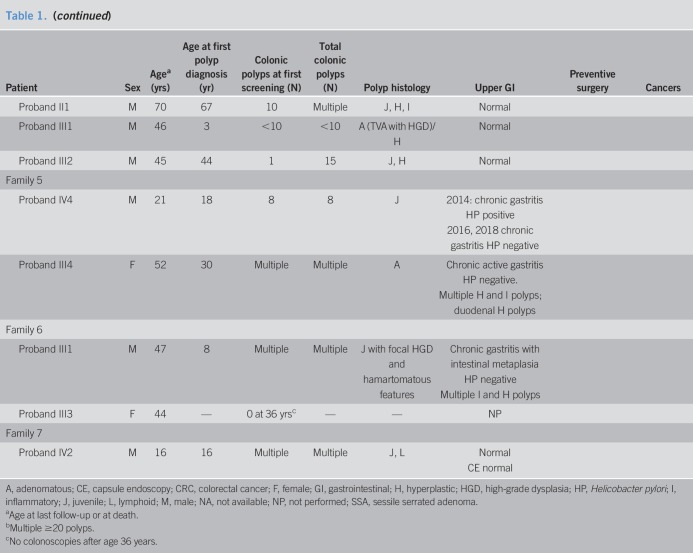
Table 1.
Clinical features of carriers of the BMPR1A deletion
Family 1
The proband (III-5) was diagnosed with CRC and multiple polyps at age 19 years. Tumor pathology showed adenocarcinoma with mucinous, tubular, and villotubular patterns with high cytological grade. Sampled polyps were first diagnosed as hyperplastic and adenomatous, but reclassified as JPS with dysplasia following genetic diagnosis. Four siblings also carry the deletion. Two siblings had multiple and 2 had few colonic polyps. The polyp pathology was JP in 3 siblings (as many as 100 polyps) and variable polyps in another with adenomatous, inflammatory, sessile serrated polyps seen. All had gastritis with one having fundic gland polyps. The proband's mother is not a carrier. No information is available about the father or his family members. Three carriers had capsule endoscopy without small bowel polyps.
Family 2
The proband (III-1) had multiple polyps at age 12 years, CRC at age 23 years, and testicular seminoma at 44 years. Gastroscopy showed chronic gastritis and a hyperplastic polyp. The proband's parents are first cousins, with relatives with polyposis, CRC, and other tumors throughout the family. Only his children were tested so far. The proband's son is a carrier, with few polyps, first diagnosed at age 14 years.
Family 3
The 75-year-old proband (II-2) had multiple polyps at the age of 20 years and underwent preventive colectomy at 25 years. Multigene testing detected the large deletion in BMPR1A. The proband's mother and 2 of her siblings died of CRC at ages 49, 38, and 63 years, respectively. Two of her sisters had had prophylactic colectomy in their 20s, but both succumbed to GI cancer, one had CRC and the other had esophageal cancer. The proband's siblings died by the time genetic diagnosis was possible, but most members from generation III and IV underwent targeted testing. All carriers had polyps. Number and age of polyp onset varied significantly; patient III-9 had multiple polyps and CRC, patient III-10 had multiple polyps and seminoma, and others (III-1 and III-2) had only few polyps by age 50 years. Upper GI findings included esophageal carcinoma, gastritis, gastric polyps, and a prepyloric polyp at the age 14 years (IV-7).
Family 4
The deletion was revealed incidentally in the proband (IV-1) when he underwent chromosomal analysis due to developmental delay. During genetic counseling, the father reported colonic polyps since childhood in his brother. The father, his brother, and their father carry the deletion. Endoscopy revealed multiple variable colonic polyps in all adult carriers.
Family 5
The proband (IV-4) had 8 juvenile polyps by age 18 years. His maternal grandparents are first cousins, with polyposis and CRC reported from both sides of the family. His mother (III-4) had multiple colonic adenomas by age 30 years and multiple gastric and duodenal polyps of inflammatory and hyperplastic histology. Both the proband and his mother had gastritis negative for Helicobacter pylori (HP).
Family 6
The proband (III-1) had multiple juvenile polyps from age 8 years. Upper endoscopy revealed multiple gastric hyperplastic and inflammatory polyps and HP-negative chronic gastritis with intestinal metaplasia and foveolar hyperplasia. The proband's sister, age 44 years, is a carrier and has not yet had colonoscopy. Multiple polyps were reported in a maternal uncle and cousins, but none had genetic testing.
Family 7
The proband (IV-2) had multiple juvenile polyps diagnosed at age 16 years. His father, paternal aunt, and grandmother report multiple colonic polyps. No polyps on endoscopy or small bowel capsule. Two other relatives had CRC at young ages (Figure 1). None of them had genetic testing yet.
DISCUSSION
Describing multiple individuals (up to 50 individuals with polyposis) harboring the same BMPR1A mutation allows the delineation of features associated with a large BMPR1A deletion and assessment of phenotypic variability in carriers of the same mutation.
Clinical features were variable, with polyp burden differing by both age at diagnosis and histology. Several carriers had adenomatous and sessile serrated polyps, rather than classic juvenile polyps, allowing conclusive diagnosis of JPS only after genetic testing (3). CRC was diagnosed in 6/27 (22.0%) carriers; 3 carriers had undergone prophylactic colectomy. The youngest patient with CRC, at 19 years, had a severe phenotype, with multiple polyps, including high-grade dysplastic adenomatous polyps. The mean age at CRC diagnosis was 42.1 years (±14.0 years, range 19–63 years), with standardized incidence ratio 1.16 for males and 1.43 for females, comparable with previous reports (6). We were surprised by the finding of upper GI involvement, with polyps found in 7/18 patients (39%), including 2 at ages 12 and 18 years. One patient died of esophageal cancer. Gastritis was noted in 9/18 carriers whose stomachs were biopsied, with 3 of the 7 of the available HP test results being negative, and 1 patient was initially tested positive, then tested negative twice, but still had gastritis. The clinical significance in terms of gastric cancer risk and screening recommendations is unclear because cancer in JPS has been reported primarily among SMAD4 carriers (1,5). Testicular tumors were reported in 2/17 male carriers (12%) from 2 different families. This could be incidental, but should be further explored, especially since sex-cord tumors have been reported in Peutz-Jeghers syndrome.
Phenotypic variability for JPS has been reported in the past; however, this is the largest cohort describing a single mutation with a similar genetic background, showing full penetrance but intra- and inter-familial nonallelic variability. The variability in cancer occurrence and age at onset could stem from differences in surveillance, but variation applies also for age and burden of polyps, with polyps detected in 5 children younger than 15 years. Possible explanations include modifying effects of other genes, differences in environmental exposures, and chance. Our finding of nonjuvenile polyposis both of the stomach and the colon may be important in understanding the molecular pathway of polyp progression, as well as a clinical challenge in diagnosing carriers with JPS with predominance of polyps with nonjuvenile features.
Large gene deletions in cancer predisposition syndromes may be associated with more severe or earlier phenotypic onset, such as MSH2-associated Lynch syndrome (22) and neurofibromatosis type 1 (23). For BMPR1A deletions, it has been suggested that the contiguous deletion involving PTEN is responsible for a more sever phenotype (14). The phenotypic variability shown in here is not indicative for a more severe phenotype associated with a BMPR1A deletion.
Both BMPR1A alleles were present in the tumors tested. Blatter et al. (11) reported loss of BMPR1A wild-type allele in 5 of 9 juvenile polyps, but no loss in adenomas (11). Although other mechanisms of loss of function were not excluded, the presence of both BMPR1A alleles in the tumors suggests that, as with SMAD4, complete somatic loss of BMPR1A may not be required for BMPR1A-associated cancer.
In summary, among patients with genomic deletion of the BMPR1A gene, JPS was almost fully penetrant, but age at onset, polyp burden and histology, and cancer occurrence were highly variable, with unexpected upper GI involvement. These observations contribute to the clinical delineation of JPS. As the mutational spectrum of BMPR1A grows with increasingly widespread genetic testing, these observations can inform surveillance measures for carriers.
CONFLICTS OF INTEREST
Guarantor of the article: Yael Goldberg and S. Lieberman, PhD.
Specific author contributions: Every author of the article has made substantial contributions to the planning of the work that led to the manuscript. M.S., D.K., E.H., S.G., A.T., J.H., S.C., M.M., R.B., L.H.K., E.G., and I.K. substantially contributed to acquisition of data. L.B.S. and T.P. obtained funding and contributed to critical revision of the manuscript. S.L., R.B., T.W., Z.L., M.-C.K., E.L.-L., and Y.G. substantially contributed to study concept and design, acquisition of data, analysis and interpretation of data, drafting of the manuscript, and critical revision of the manuscript.
Financial support: Grant NIH R35 CA197458.
Potential competing interests: All authors declare that they have no conflicts of interest.
REFERENCES
- 1.Latchford AR, Neale K, Phillips RK, et al. Juvenile polyposis syndrome: A study of genotype, phenotype, and long-term outcome. Dis Colon Rectum 2012;55:1038–43. [DOI] [PubMed] [Google Scholar]
- 2.https://www.nccn.org/professionals/physician_gls/default.aspx. Accessed on February 1, 2019.
- 3.Schreibman IR, Baker M, Amos C, et al. The hamartomatous polyposis syndromes: A clinical and molecular review. Am J Gastroenterol 2005;100:476–90. [DOI] [PubMed] [Google Scholar]
- 4.Cheah PY, Wong YH, Chau YP, et al. Germline bone morphogenesis protein receptor 1A mutation causes colorectal tumorigenesis in hereditary mixed polyposis syndrome. Am J Gastroenterol 2009;104:3027–33. [DOI] [PubMed] [Google Scholar]
- 5.Shaco-Levy R, Jasperson KW, Martin K, et al. Morphologic characterization of hamartomatous gastrointestinal polyps in Cowden syndrome, Peutz-Jeghers syndrome, and juvenile polyposis syndrome. Hum Pathol 2016;49:39–48. [DOI] [PubMed] [Google Scholar]
- 6.Aretz S, Stienen D, Uhlhaas S, et al. High proportion of large genomic deletions and a genotype phenotype update in 80 unrelated families with juvenile polyposis syndrome. J Med Genet 2007;44:702–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aytac E, Sulu B, Heald B, et al. Genotype-defined cancer risk in juvenile polyposis syndrome. Br J Surg 2015;102:114–8. [DOI] [PubMed] [Google Scholar]
- 8.Howe JR, Mitros FA, Summers RW. The risk of gastrointestinal carcinoma in familial juvenile polyposis. Ann Surg Oncol 1998;5:751–6. [DOI] [PubMed] [Google Scholar]
- 9.Brosens LA, van Hattem A, Hylind LM, et al. Risk of colorectal cancer in juvenile polyposis. Gut 2007;56:965–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wakefield LM, Hill CS. Beyond TGFβ: Roles of other TGFβ superfamily members in cancer. Nat Rev Cancer 2013;13:328–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blatter RH, Plasilova M, Wenzel F, et al. Somatic alterations in juvenile polyps from BMPR1A and SMAD4 mutation carriers. Genes Chromosomes Cancer 2015;54:575–82. [DOI] [PubMed] [Google Scholar]
- 12.van Hattem WA, Brosens LA, de Leng WW, et al. Large genomic deletions of SMAD4, BMPR1A and PTEN in juvenile polyposis. Gut 2008;57:623–7. [DOI] [PubMed] [Google Scholar]
- 13.Calva-Cerqueira D, Chinnathambi S, Pechman B, et al. The rate of germline mutations and large deletions of SMAD4 and BMPR1A in juvenile polyposis. Clin Genet 2009;75:79–85. [DOI] [PubMed] [Google Scholar]
- 14.Salviati L, Patricelli M, Guariso G, et al. Deletion of PTEN and BMPR1A on chromosome 10q23 is not always associated with juvenile polyposis of infancy. Am J Hum Genet 2006;79:593–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.http://tests.labmed.washington.edu/BROCA#BROCA_Gene_List. Accessed on January 1, 2017.
- 16.Shirts BH, Casadei S, Jacobson AL, et al. Improving performance of multigene panels for genomic analysis of cancer predisposition. Genet Med 2016;18:974–81. [DOI] [PubMed] [Google Scholar]
- 17.Nord AS, Lee M, King MC, et al. Accurate and exact CNV identification from targeted high-throughput sequence data. BMC Genomics 2011;12:184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.http://products.invitrogen.com/ivgn/product/4475346. Accessed on February 1, 2019.
- 19.Raczy C, Petrovski R, Saunders CT, et al. Isaac: Ultra-fast whole-genome secondary analysis on Illumina sequencing platforms. Bioinformatics 2013;29:2041–3. [DOI] [PubMed] [Google Scholar]
- 20.Boeva V, Popova T, Bleakley K, et al. Control-FREEC: A tool for assessing copy number and allelic content using next-generation sequencing data. Bioinformatics 2012;28:423–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pérez-Cabornero L, Infante Sanz M, Velasco Sampedro E, et al. Frequency of rearrangements in Lynch syndrome cases associated with MSH2: Characterization of a new deletion involving both EPCAM and the 5′ part of MSH2. Cancer Prev Res 2011;4:1556–62. [DOI] [PubMed] [Google Scholar]
- 23.Kehrer-Sawatzki H, Mautner VF, Cooper DN. Emerging genotype-phenotype relationships in patients with large NF1 deletions. Hum Genet 2017;136:349–76. [DOI] [PMC free article] [PubMed] [Google Scholar]