Supplemental Digital Content is available in the text
Keywords: Escherichia coli, Klebsiella pneumoniae, β-lactamase, Carbapenem resistance, New Delhi metallo-β-lactamase
Abstract
Background:
Extended-spectrum β-lactamase (ESBL)-producing Escherichia coli (E. coli) and Klebsiella pneumoniae (K. pneumoniae) are the important pathogens causing pneumonia. This study aimed to investigate the clinical characteristics and molecular epidemiology of ESBL-producing E. coli and K. pneumoniae causing pneumonia at a large teaching hospital in China.
Methods:
We collected patient's clinical data and ESBL-producing E. coli and K. pneumoniae strains causing pneumonia (from December 2015 to June 2016) at a hospital in Wuhan. The susceptibilities, multi-locus sequence typing, homologous analysis, ESBL genes by polymerase chain reaction and sequencing were determined.
Results:
A total of 59 ESBL-producing strains (31 E. coli and 28 K. pneumoniae) isolated from patients with pneumonia were analyzed. The majority of strains were isolated from patients were with hospital-acquired pneumonia (37/59, 62.7%), followed by community-acquired pneumonia (13/59, 22.0%), and ventilator-related pneumonia (9/59, 15.3%). The E. coli ST131 (9 isolates, 29.0%) and K. pneumoniae ST11 (5 isolates, 17.9%) were the predominant sub-types. The most prevalent ESBL gene was CTX-M-14, followed by SHV-77, CTX-M-3, SHV-11, and CTX-M-27. At least 33 (55.9%) of the ESBL-producing strains carried two or more ESBL genes. The ISEcp1 and IS26 were found upstream of all blaCTX-M (CTX-Ms) and of most blaSHV (SHVs) (57.6%), respectively. Moreover, three ESBL-producing K. pneumoniae ST11 strains which were resistant to carbapenems carried the blaNDM-1 and blaKPC-2, two of which also bearing blaOXA-48 were resistant to all antibiotics (including Tigecycline).
Conclusions:
Hospital-acquired pneumonia is more likely correlated with ESBL-producing E. coli and K. pneumoniae. ESBL-producing E. coli ST131 and multi-drug resistance ESBL-producing, as well as New Delhi metallo-β-lactamase-1 (NDM-1) and Klebsiella pneumoniae carbapenemases-2 (KPC-2) bearing K. pneumoniae ST11 are spreading in patients with pneumonia in hospital.
Introduction
Extended-spectrum β-lactamase (ESBL)-producing Enterobacteriaceae, especially Escherichia coli (E. coli) and Klebsiella pneumoniae (K. pneumoniae), which are the important pathogens causing pneumonia,[1–3] pose distinct clinical challenges.[4,5] The prevalence of ESBL-producing E. coli and K. pneumoniae is high and varied from different geographical areas in China and type of infections.[6–9] It was reported that 67.7% of E. coli and 27.5% of K. pneumoniae isolated from patients in Shanghai with blood stream infections were found to be ESBL producers[9]; however, the percentage was 58.4% in E. coli and 43.0% in K. pneumoniae reported by a multi-center epidemiological study from other regions of China.[10] Of note, the prevalence of ESBL production in E. coli and K. pneumonia was caused by clonal transmission of predominant sub-types such as E. coli ST131[11–14] or K. pneumonia ST11.[15–18] So continuous surveillance on the prevalence of ESBL-producing E. coli and K. pneumoniae in different areas is of great significance. Therefore, the aim of this study was to investigate the clinical and molecular epidemiology characteristics of ESBL-producing E. coli and K. pneumoniae causing pneumonia at a large teaching hospital with 5000 beds in Hubei province, China.
Methods
Ethical approval
The study was approved by Tongji Medical College Ethics Committee, Huazhong University of Science and Technology (2018-S356).
Clinical strains, anti-microbial susceptibility testing, and confirmation test of ESBL production
All ESBL-producing E. coli and K. pneumoniae strains causing pneumonia and the clinical data were collected from December 2015 to June 2016 at a teaching hospital in Hubei province. Community-acquired pneumonia (CAP), hospital-acquired pneumonia (HAP), and ventilator-related pneumonia (VAP) were defined according to the guidelines.[19,20] Samples were inoculated using blood agar medium, and cultured at 37°C for 18 to 24 h. BD Phoenix™100 Automated Microbiology System (BD Ltd., Franklin Lakes, NJ, USA) was used to identify strains and anti-microbial susceptibility. Confirmation of ESBL production was performed by using double-disc synergy test. The modified Hodge test was applied to confirm carbapenemases-producing strains. All results were interpreted based on the Clinical and Laboratory Standards Institute guidelines.[21] The disks used for confirmation test were obtained from Beijing Tiantan Biological Products Corporation (China). E. coli (ATCC25922) and K. pneumoniae (ATCC700603) were used as quality control strains.
Microbiological studies
Clonal relationships were analyzed by multi-locus sequence typing (MLST) and enterobacterial repetitive inter-genic consensus polymerase chain reaction (ERIC-PCR). Bacterial DNA was prepared according to the manufacturer instructions of bacteria DNA extraction kit (TIANamp Bacteria DNA kit, China). MLST was performed using the standard seven housekeeping loci for E. coli and K. pneumoniae according to protocols at http://bigsdb.pasteur.fr. The ERIC-PCR was performed as follows: PCR amplifications were performed in a volume of 25.0 μL of reaction mixtures containing 12.5 μL Go Taq (Dalian TaKaRa Corporation, China); 1.0 μL of 10 ρmol ERIC1R (5′-ATG TAA GCT CCT GGG GAT-3′) and 1.0 μL of 10 ρmol ERIC2 (5′-AAG TAA GTG ACT GGG GGT GAGC-3′)[22]; 1.0 μL of DNA template; 9.5 μL of PCR grade water for ERIC-PCR; the ERIC-PCR thermal cycler (Eastwin Scientific equipments limited, China) program for this method followed Wei et al[22] Amplified PCR products stained with ethidium bromide were separated by electrophoresis on 1.5% (w/v) agarose (1st base) at 100 V for 30 to 40 min, the molecular size of fragments generated by electrophoresis was determined by comparison to 2-kb DNA ladders (Dalian TaKaRa Corporation), and band patterns were captured under an ultraviolet illuminator. The GelCompar software package (version 7.6; Applied Maths, Bionumerics, Belgium) was used to compare the band patterns of aggregated data. The pattern analysis was calculated using the dice correlation coefficient at 1.0% band position tolerance and unweighted pair group method using arithmetic average.
Identification of ESBL and carbapenemase genes and ESBL genetic environment
All ESBL producers were screened for the presence of plasmid carrying blaSHV, blaCTX-M, and blaTEM. Three ESBL-producing carbapenem-resistant strains were screened for the detection of carbapenemases: blaKPC, blaIMI, blaAIM, blaSME, blaGES, blaGIM, blaIMP, blaVIM, blaNDM, and blaOXA-48-like genes by PCR. The oligonucleotide primers specific for the ESBL and carbapenemase genes in the PCR assays were designed by Doyle and Wang.[7,23–25] We used previously described primers to investigate the surrounding regions of the blaSHV, blaCTX-M, and blaTEM genes.[7] All positive PCR products were sent to the Invitrogen Corporation (Shanghai, China) for sequencing. The nucleotide sequences were analyzed with the basic local alignment search tool online.
Statistical analysis
Percentages and frequencies were used to analyze categorical variables. A Chi-square test or Fisher exact test was used for categorical variables (invasive procedures). A P value <0.05 was considered to be statistically significant. Analyses were performed using SPSS 23.0 for MacIntosh (SPSS Inc., Chicago, IL, USA).
Results
Prevalence of E. coli and K. pneumoniae and patient characteristics
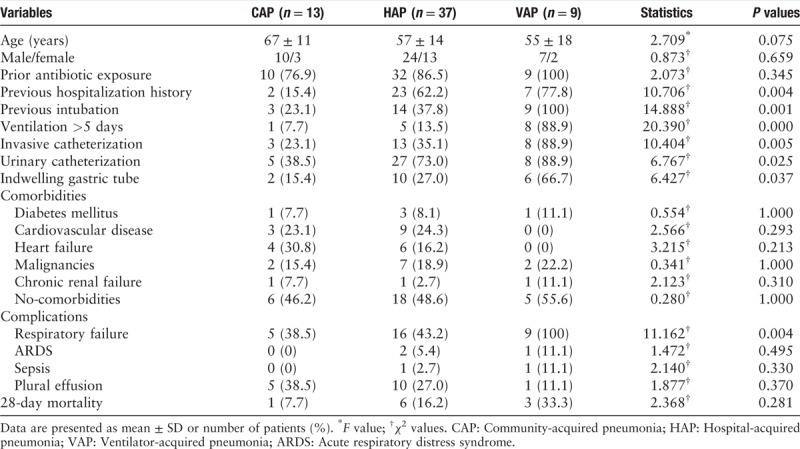
A total of 976 non-reduplicate E. coli and K. pneumoniae were collected from December 1st, 2015 to June 30th, 2016. Among these isolates, 28.7% (280/976) were confirmed as ESBL producers, and 59 strains (31 E. coli and 28 K. pneumoniae) isolated from non-reduplicate patients with pneumonia enrolled in this study. The flowchart of patient's enrollment is shown in Figure 1. Most of the strains (n = 54) were detected from sputum samples followed by blood samples (n = 5), and no strains were isolated from pleural effusion and alveolar lavage fluid. ESBL-producing E. coli and K. pneumoniae isolated from patients with pneumonia were mostly located in the neurosurgery department, followed by the cardiovascular department and the intensive care department. The mean age of the 59 adult patients was (59.0 ± 14.7) years and 35.6% (21/59) were elderly (>65years). Compared to patients with CAP, those with HAP and VAP had higher frequency of previous hospitalization history and invasive procedures, including invasive catheterization (χ2 = 10.404, P = 0.005), urinary catheterization (χ2 = 6.767, p = 0.025), indwelling gastric tube (χ2 = 6.427, P = 0.037) and intubation (χ2 = 14.888, P = 0.001), and the 28-day mortality in patients with HAP and VAP were also higher than those with CAP (χ2 = 2.368, P = 0.281). There were no statistical differences in the comorbidities, including diabetes mellitus, cardiovascular disease, heart failure, malignancies, and chronic renal failure among the three groups. Patients with HAP or VAP were more likely to have complications with respiratory failure, acute respiratory distress syndrome, sepsis, or plural effusion [Table 1].
Figure 1.
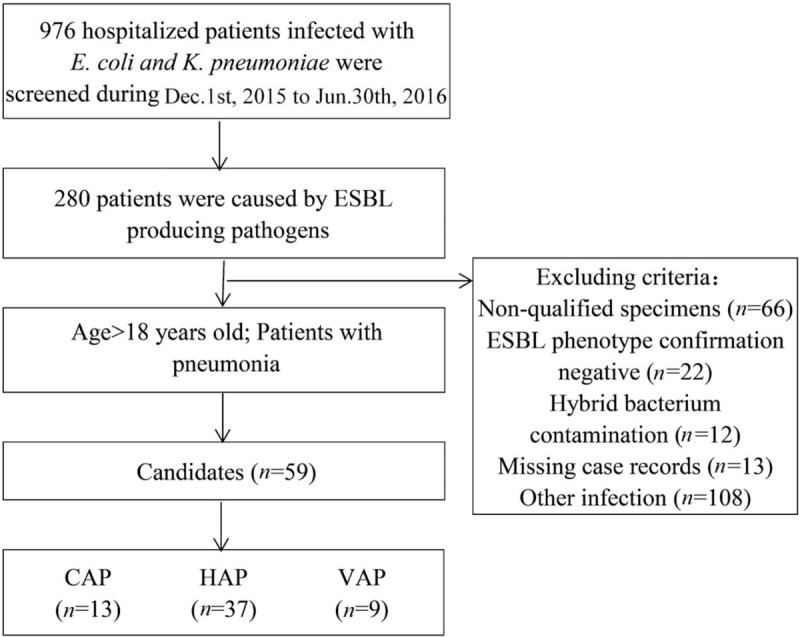
Flowchart of CAP, HAP, and VAP patients enrollment in this study. CAP: Community-acquired pneumonia; HAP: Hospital-acquired pneumonia; VAP: Ventilator-related pneumonia.
Table 1.
Demographic and clinical characteristics of 59 patients with pneumonia.
Susceptibility of E. coli and K. pneumoniae to anti-microbial agents
Among the 59 isolates, 31 E. coli and 28 K. pneumoniae strains were confirmed as ESBL producers. All ESBL-producing E. coli strains were multi-resistant to most of β-lactam antibiotics and fluoroquinolones, whereas they were sensitive to carbapenems, amikacin, and piperacillin-tazobactam. Unlike ESBL-producing E. coli, ESBL-producing K. pneumoniae strains showed poor susceptibility to piperacillin-tazobactam, but most strains showed higher sensitivity to carbapenems and amikacin. It is worth noting that there were three ESBL producing K. pneumoniae strains resistant to carbapenems, which were confirmed as carbapenemases producers by modified Hodge test [Supplementary Table 1].
MLST and ERIC-PCR
MLST experiments identified 21 unique sequence typings (STs) in 31 E. coli strains. The most prevalent ST was ST131 (n = 9, 29.0%) which was mainly located in the neurosurgery department (n = 4), followed by ST405 (n = 3, 9.7%) and the rest of 19 isolates belonged to different ST types [Figure 2]. E. coli ST131 isolates were predominately isolated from patients with HAP (n = 5), followed by VAP (n = 3), and the remaining one was isolated from a patient with CAP. The E. coli ST131 isolates showed lower susceptibility to β-lactam antibiotics and fluoroquinolones than non-ST131 isolates [Supplementary Table 1], furthermore, patients with pneumonia caused by E. coli ST131 isolates had higher proportion of respiratory failure and poor outcomes than those caused by non-ST131 isolates (data not shown). There were 20 STs identified among 28 K. pneumoniae strains, the most prevalent ST type was ST11 (n = 5, 17.9%), followed by ST23 (n = 4, 14.3%), ST37 (n = 2, 7.1%), and the remaining 17 strains corresponded to different ST types [Figure 3]. The K. pneumoniae ST11 isolates were detected from patients with VAP (n = 2) and HAP (n = 2), the remaining one was isolated from a patient with CAP. Among the five K. pneumoniae ST11, two located in the cardiovascular department were found to be the pan-drug resistance isolates; one K. pneumoniae ST11 strain was a multi-drug resistance (MDR) isolate, which was only susceptible to tigecycline; the remaining two K. pneumoniae ST11 strains were resistant to most antibiotics, but susceptible to carbapenems, tigecycline, and amikacin. We also discovered three new K. pneumoniae STs: ST2965, ST2966, and ST3003 [Figure 3]. Of note, the housekeeping gene (Phoe313) sequence in K. pneumoniae ST3003 was first reported. The ERIC-PCR results showed that 31 E. coli isolates were separated into 19 groups [Figure 2], and 28 K. pneumoniae isolates were separated into 14 groups [Figure 3]. There were several pulsotypes in clonal groups among E. coli ST131 isolates, and one predominant pulsotype included four isolates. The five K. pneumoniae ST11 isolates belonged to one single pulsotype.
Figure 2.
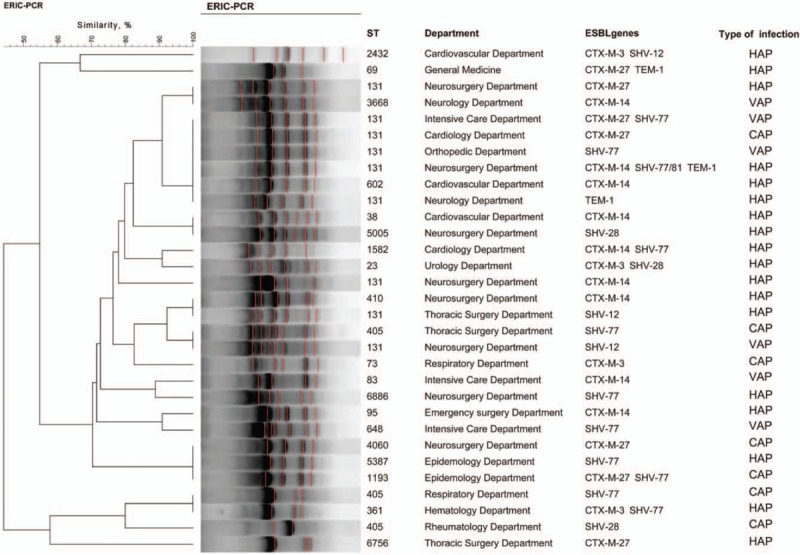
Cluster analysis of E. coli. isolates are grouped according to their Xal restriction patterns by BioNumerics version 7.6 software. CAP: Community-acquired pneumonia; HAP: Hospital-acquired pneumonia; VAP: Ventilator-related pneumonia.
Figure 3.
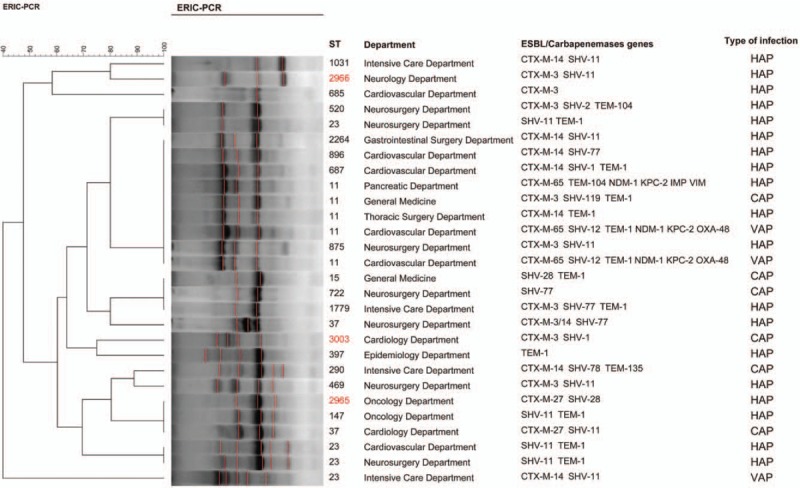
Cluster analysis of K. pneumoniae. Isolates are grouped according to their Xal restriction patterns by BioNumerics version 7.6 software. Three new K. pneumoniae ST numbers are marked in red. ST: Sequence typing. CAP: Community-acquired pneumonia; HAP: Hospital-acquired pneumonia; VAP: Ventilator-related pneumonia.
β-Lactamase genes characterization
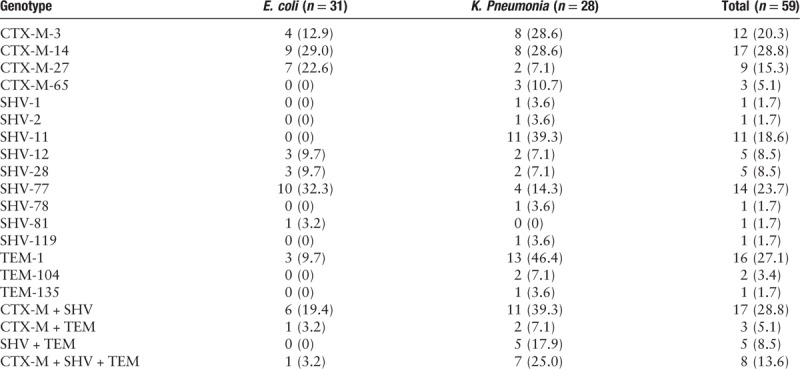
All 59 strains were confirmed to carry plasmid-mediated β-lactamase genes by PCR. Sequence analysis revealed that blaCTX-M, blaSHV, and blaTEM genes were present in 41, 40, and 19 isolates, respectively. A total of 55.9% isolates (33/59) harbored two or more ESBL genes. The main type of blaCTX-M was blaCTX-M-14 (17 isolates), followed by blaCTX-M-3 (12 isolates), blaCTX-M-27 (9 isolates), and blaCTX-M-65 (3 isolates). Group II, III, or V blaCTX-M were not detected. Sequencing results indicated that the most prevalent ESBL blaSHV was SHV-77 (n = 14, 23.7%), which mutated on the 637 base compared with non-ESBL enzymes SHV-77 (data not shown), followed by SHV-11 (n = 11, 18.6%), blaSHV-28 (n = 5), blaSHV-12 (n = 5), blaSHV-2 (n = 1), blaSHV-78 (n = 1), blaSHV-81 (n = 1), blaSHV-1 (n = 1), and blaSHV-119 (n = 1). Twenty-five (42.4%) isolates carried two ESBL genes, and eight (13.6%) isolates had three ESBL genes [Table 2]. Three ESBL producing K. pneumoniae ST 11 strains, which were resistant to carbapenems, were detected to carry blaNDM-1 (three isolates), blaKPC-2 (three isolates), blaOXA-48 (two isolates), blaIMP (one isolates), and blaVIM (one isolates) [Figure 3].
Table 2.
Genotypes of ESBL-producing E. coli and K. pneumoniae isolates, n (%).
Genetic environment of blaCTX-M and blaSHV
ISEcp1 was found upstream of the start codon of all CTX-Ms. The region between the end of ISEcp1 and the start codon of CTX-Ms was an identical 42 base pairs (bp) length sequence, the ISEcp1 was disrupted by IS1X2 768 bp from the 3′non-coding region of the ISEcp1 in all CTX-Ms bearing strains (data not shown). A non-coding region which belonged to truncated IS26 element provided the promoter to the blaSHV gene, the truncated IS26 element has also been found further from the start codon of blaSHV-77, blaSHV-12, blaSHV-28, and blaSHV-11 genes (data not shown).
Discussion
This study analyzed the clinical and molecular characteristics of ESBL-producing E. coli and K. pneumoniae strains isolated from patients with pneumonia at the large teaching hospital, and the results demonstrated that the prevalence of ESBL-producing E. coli and K. pneumoniae was lower than that reported in other regions of China; however, the worldwide spreading E. coli ST131 and K. pneumoniae ST11 were prevalent in patients with pneumonia in the hospital.
In this study, we found the prevalence of ESBL-producing E. coli and K. pneumoniae was 33.5% and 19.4%, respectively (average 28.7%), which was lower than that reported by Yu et al (35.7%) and Hawser et al (48.2%).[26,27] In accordance to other studies,[28,29] our results also showed that the prevalence rates of ESBL-producing E. coli and K. pneumoniae in HAP were higher than that in CAP, which suggested that HAP was more likely correlated with ESBL producers than CAP. Additionally, patients with HAP or VAP caused by ESBL producers had a higher 28-day mortality than those with CAP. High proportion of co-existence of ESBL genes, multidrug resistant and excessive complications had been put forward to poor outcomes in patients with HAP or VAP. However, more than one-fifths of the patients were CAP, which was much higher than that reported by other studies,[30,31] this indicated that there was a trend of ESBL-producing strains being community outbreak.
The most predominant ESBL genes was CTX-M-14, which was consistent with other reports in Asia-Pacific region.[32–34] For all blaCTX-M, the ISEcp1 insertion sequence had been detected. The close relationship between blaCTX-M and ISEcpl had been frequently reported,[35,36] these results demonstrated the important role of ISEcpl in the worldwide spread of blaCTX-M. In contrast to blaCTX-M, SHV-11 was the main blaSHV in our study, followed by SHV-77. It is worth noting that SHV-77 detected in our study was an encoding ESBL enzyme, which was different from the study in which it was reported as a non-ESBL enzyme.[37] Sequencing analysis demonstrated that it may be attributed to the mutation of the coding region of blaSHV-77. Furthermore, IS26 was detected upstream of most of blaSHV, the blaSHV genes associated with IS26 were regulated by the strong promoter, which was consistent with previous studies.[38,39] These findings suggested that IS26 could play an important role in expression of blaSHV and contribute to transmission of blaSHV.
MLST analysis demonstrated that E. coli ST131 was the most prevalent strain of ESBL-producing E. coli causing pneumonia in our hospital. E. coli ST131 strain originally had risen to prominence as early as 2003, receiving increasing attention due to its rapid global dissemination and frequent multi-drug-resistant phenotype.[40] Previous studies reported that E. coli ST131 strain was prevalent in patients with urinary tract infections[41] and blood stream infections[42]; however, there were few studies about the prevalence of E. coli ST131 in patients with pneumonia. Although Cha et al[43] reported that CTX-M-15-producing E. coli ST131 has emerged and disseminated among patients with HAP in South Korea, Thailand and the Philippines, limited information existed about its clinical impacts on patients with pneumonia from China. To our knowledge, this is the first study to identify E. coli clone ST131 strain prevalent in patients with pneumonia in the mainland of China. Consistent with previous study,[43] our study also demonstrated that the E. coli ST131 was the most prevalent strain causing HAP. However, more importantly, our study showed E. coli ST131 strains were higher anti-microbial resistant than non-ST131 strains, and patients with pneumonia caused by E. coli ST131 strains had poor prognosis owing to acute respiratory failure, these results indicated that the ST131 strains were more virulent than non-ST131 isolates. The high resistant and virulent ST131 strain may also be associated with the widespread nature of this strain, due to the more adaptive characteristic than non-ST131 in the hospital environment.[44] Additionally, previous studies indicated that E. coli ST131 which was from phylogenetic group B2 has a fitness advantage owing to their group B2 genomic backbone and this may attribute to the remarkable success prevalence of ST131 in hospital worldwide.[45–47] However, the mechanism about the genomic backbone contributing to the fitness advantage was still unclear, which deserved further exploration.
In contrast to E. coli, our study demonstrated that K. pneumoniae ST11 was the major type of ESBL-producing K. pneumoniae strains. It was reported that K. pneumoniae ST11, which was multi-drug resistant and had highly transmissible characteristics, was the predominant clone of KPC-producing K. pneumoniae in China.[48,49] In our study, most (60.0%) of the K. pneumoniae ST11 strains resistant to carbapenems carried blaNDM-1 and blaKPC-2, this was different from previous studies that blaKPC-2 was reported as the prevalent gene in China,[50–52] and blaNDM-1 or blaOXA-48 was the predominant gene type in other countries.[53–55] Previous studies reported that K. pneumoniae ST11 was the dominant strain causing UTI and bacteremia, but the CTX-M-24 and KPC-2 producing K. pneumoniae ST11 in patients with VAP was hospital outbreak and dissemination in other regions of China.[15,52,56] However, in our study, K. pneumoniae ST11 strains, which mainly carried CTX-M-65, were isolated from patients with HAP, VAP, or CAP.
Previous studies reported that dissemination of K. pneumoniae ST11 related to mobile genetic elements (MGEs).[57,58] MGEs including transposons, integrons, prophages, integrative and conjugative elements, and genomic islands, which carried anti-microbial resistance and adaptability associated genes, made K. pneumoniae ST11 formidable adaptive and caused drug-resistant infections in hospitals.[59,60] Our results indicated epidemic trends of multi-drug or pandrug resistant K. pneumoniae ST11 harboring blaNDM-1 and blaKPC-2 in patients with pneumonia in the hospital, which is a serious threat to the public health. Thus, continuous surveillance on the prevalence of K. pneumoniae ST 11 was of great significance to control the outbreak of the resistant strain.
In summary, we documented the emergence and dissemination of multidrug resistant E. coli ST131 and K. pneumoniae ST11 causing pneumonia at a Chinese teaching hospital, which is a major threat for hospitalized patients. Meanwhile, our study revealed that the blaCTX-M and blaSHV genes were mobilized by ISEcpl and IS26. These results underline the paramount significance of the consequent surveillance of MDR ESBL-producing E. coli and K. pneumoniae strains in hospital.
Acknowledgements
The author would like to thank Dr. Sean James Dickinson for English language editing, and the team of curators of the Institut Pasteur MLST and whole genome MLST databases for curating the data and making them publicly available at http://bigsdb.pasteur.fr.
Funding
This work was supported by a grant from the National Natural Science Foundation of China (No. 81500005).
Conflicts of interest
None.
Supplementary Material
Footnotes
How to cite this article: Liu J, Du SX, Zhang JN, Liu SH, Zhou YY, Wang XR. Spreading of extended-spectrum β-lactamase-producing Escherichia coli ST131 and Klebsiella pneumoniae ST11 in patients with pneumonia: a molecular epidemiological study. Chin Med J 2019;00:00–00. doi: 10.1097/CM9.0000000000000368
Jing Liu and Shuai-Xian Du contributed equally to this study.
The abstract of this manuscript was presented at the Annual Conference of Respiratory Diseases of Chinese Medical Association – 2018 (Nineteenth National Academic Conference on Respiratory Diseases).
References
- 1.Fritsche TR, Sader HS, Stilwell MG, Dowzicky MJ, Jones RN. Antimicrobial activity of tigecycline tested against organisms causing community-acquired respiratory tract infection and nosocomial pneumonia. Diagn Microbiol Infect Dis 2005; 52:187–193. doi: 10.1016/j.diagmicrobio.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 2.Wang Y, Zhang R, Li W, Feng Y, Leng T. Serious antimicrobial resistance status of pathogens causing hospital-acquired lower respiratory tract infections in North China. J Int Med Res 2009; 37:899–907. doi: 10.1177/147323000903700336. [DOI] [PubMed] [Google Scholar]
- 3.Song JH, Chung DR. Respiratory infections due to drug-resistant bacteria. Infect Dis Clin North Am 2010; 24:639–653. doi: 10.1016/j.idc.2010.04.007. [DOI] [PubMed] [Google Scholar]
- 4.Jean SS, Hsueh PR. High burden of antimicrobial resistance in Asia. Int J Antimicrob Agents 2011; 37:291–295. doi: 10.1016/j.ijantimicag.2011.01.009. [DOI] [PubMed] [Google Scholar]
- 5.Dhillon RH, Clark J. ESBLs: a clear and present danger? Crit Care Res Pract 2012; 2012:625170.doi: 10.1155/2012/625170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.An S, Chen J, Wang Z, Wang X, Yan X, Li J, et al. Predominant characteristics of CTX-M-producing Klebsiella pneumoniae isolates from patients with lower respiratory tract infection in multiple medical centers in China. FEMS Microbiol Lett 2012; 332:137–145. doi: 10.1111/j.1574-6968.2012.02586.x. [DOI] [PubMed] [Google Scholar]
- 7.Wang XR, Chen JC, Kang Y, Jiang N, An SC, Gao ZC. Prevalence and characterization of plasmid-mediated blaESBL with their genetic environment in Escherichia coli and Klebsiella pneumoniae in patients with pneumonia. Chin Med J 2012; 125:894–900. doi: 10.3760/cma.j.issn.0366-6999.2012.05.029. [PubMed] [Google Scholar]
- 8.Wang Y, Zhang R, Liu W. Distribution and drug resistance of pathogenic bacteria in ventilator-associated pneumonia at a local hospital of North-eastern China. Infect Drug Resist 2018; 11:2249–2255. doi: 10.2147/IDR.S172598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quan J, Zhao D, Liu L, Chen Y, Zhou J, Jiang Y, et al. High prevalence of ESBL-producing Escherichia coli and Klebsiella pneumoniae in community-onset bloodstream infections in China. J Antimicrob Chemother 2017; 72:273–280. doi: 10.1093/jac/dkw372. [DOI] [PubMed] [Google Scholar]
- 10.Liu XJ, Lyu Y, Li Y, Xue F, Liu J. Trends in antimicrobial resistance against Enterobacteriaceae strains isolated from blood: a 10-year epidemiological study in mainland China (2004-2014). Chin Med J 2017; 130:2050–2055. doi: 10.4103/0366-6999.213407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rogers BA, Ingram PR, Runnegar N, Pitman MC, Freeman JT, Athan E, et al. Community-onset Escherichia coli infection resistant to expanded-spectrum cephalosporins in low-prevalence countries. Antimicrob Agents Chemother 2014; 58:2126–2134. doi: 10.1128/AAC.02052-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang S, Zhao SY, Xiao SZ, Gu FF, Liu QZ, Tang J, et al. Antimicrobial resistance and molecular epidemiology of Escherichia coli causing bloodstream infections in three hospitals in Shanghai, China. PLoS One 2016; 11:e147740.doi: 10.1371/journal.pone.0147740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li B, Lu Y, Lan F, He Q, Li C, Cao Y. Prevalence and characteristics of ST131 clone among unselected clinical Escherichia coli in a Chinese university hospital. Antimicrob Resist Infect Control 2017; 6:118.doi: 10.1186/s13756-017-0274-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen X, Zou Q, Zhang W, Wang R, Yu F, Chen Y. Clinical features and microbiological characteristics of hospital- and community-onset Escherichia coli bloodstream infection. J Med Microbiol 2019; 68:178–187. doi: 10.1099/jmm.0.000904. [DOI] [PubMed] [Google Scholar]
- 15.Hu L, Liu Y, Deng L, Zhong Q, Hang Y, Wang Z, et al. Outbreak by ventilator-associated ST11 K. pneumoniae with co-production of CTX-M-24 and KPC-2 in a SICU of a tertiary teaching hospital in central China. Front Microbiol 2016; 7:1190.doi: 10.3389/fmicb.2016.01190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiao SZ, Wang S, Wu WM, Zhao SY, Gu FF, Ni YX, et al. The resistance phenotype and molecular epidemiology of Klebsiella pneumoniae in bloodstream infections in Shanghai, China, 2012-2015. Front Microbiol 2017; 8:250.doi: 10.3389/fmicb.2017.00250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu J, Yu J, Chen F, Yu J, Simner P, Tamma P, et al. Emergence and establishment of KPC-2-producing ST11 Klebsiella pneumoniae in a general hospital in Shanghai, China. Eur J Clin Microbiol Infect Dis 2018; 37:293–299. doi: 10.1007/s10096-017-3131-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu L, Feng Y, Tang G, Lin J, Huang W, Qiao F, et al. Carbapenem-resistant isolates of the Klebsiella pneumoniae complex in Western China: the common ST11 and the surprising hospital-specific types. Clin Infect Dis 2018; 67:S263–S265. doi: 10.1093/cid/ciy662. [DOI] [PubMed] [Google Scholar]
- 19.Torres A, Niederman MS, Chastre J, Ewig S, Fernandez-Vandellos P, Hanberger H, et al. International ERS/ESICM/ESCMID/ALAT guidelines for the management of hospital-acquired pneumonia and ventilator-associated pneumonia: Guidelines for the management of hospital-acquired pneumonia (HAP)/ventilator-associated pneumonia (VAP) of the European Respiratory Society (ERS), European Society of Intensive Care Medicine (ESICM), European Society of Clinical Microbiology and Infectious Diseases (ESCMID) and Asociacion Latinoamericana del Torax (ALAT). Eur Respir J 2017; 50:1700582.doi: 10.1183/13993003.00582-2017. [DOI] [PubMed] [Google Scholar]
- 20.Cao B, Huang Y, She DY, Cheng QJ, Fan H, Tian XL, et al. Diagnosis and treatment of community-acquired pneumonia in adults: 2016 clinical practice guidelines by the Chinese Thoracic Society, Chinese Medical Association. Clin Respir J 2018; 12:1320–1360. doi: 10.1111/crj.12674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clinical and Laboratory Standards Institute, Wayne Pa. Clinical and Laboratory Standards Institute: Performance Standards for Antimicrobial Susceptibility Testing. CLSI Supplement M100-S28. 2018. [Google Scholar]
- 22.Wei G, Pan L, Du H, Chen J, Zhao L. ERIC-PCR fingerprinting-based community DNA hybridization to pinpoint genome-specific fragments as molecular markers to identify and track populations common to healthy human guts. J Microbiol Methods 2004; 59:91–108. doi: 10.1016/j.mimet.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 23.Poirel L, Walsh TR, Cuvillier V, Nordmann P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis 2011; 70:119–123. doi: 10.1016/jdiagmicrobio.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 24.Doyle D, Peirano G, Lascols C, Lloyd T, Church DL, Pitout JD. Laboratory detection of Enterobacteriaceae that produce carbapenemases. J Clin Microbiol 2012; 50:3877–3880. doi: 10.1128/JCM.02117-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mlynarcik P, Roderova M, Kolar M. Primer evaluation for PCR and its Application for detection of carbapenemases in enterobacteriaceae. Jundishapur J Microbiol 2016; 9:e29314.doi: 10.5812/jjm.29314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu Y, Zhou W, Chen Y, Ding Y, Ma Y. Epidemiological and antibiotic resistant study on extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae in Zhejiang Province. Chin Med J 2002; 115:1479–1482. doi: 10.3760/cma.j.issn.0366-6999.2002.10.110. [PubMed] [Google Scholar]
- 27.Hawser SP, Bouchillon SK, Hoban DJ, Badal RE, Hsueh PR, Paterson DL. Emergence of high levels of extended-spectrum-beta-lactamase-producing gram-negative bacilli in the Asia-Pacific region: data from the study for monitoring antimicrobial resistance trends (SMART) program, 2007. Antimicrob Agents Chemother 2009; 53:3280–3284. doi: 10.1128/AAC.00426-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones RN. Microbial etiologies of hospital-acquired bacterial pneumonia and ventilator-associated bacterial pneumonia. Clin Infect Dis 2010; 51 Suppl 1:S81–S87. doi: 10.1086/653053. [DOI] [PubMed] [Google Scholar]
- 29.Kalil AC, Metersky ML, Klompas M, Muscedere J, Sweeney DA, Palmer LB, et al. Executive summary: management of adults with hospital-acquired and ventilator-associated pneumonia: 2016 clinical practice guidelines by the Infectious Diseases Society of America and the American Thoracic Society. Clin Infect Dis 2016; 63:575–582. doi: 10.1093/cid/ciw504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park HK, Song JU, Um SW, Koh WJ, Suh GY, Chung MP, et al. Clinical characteristics of health care-associated pneumonia in a Korean teaching hospital. Respir Med 2010; 104:1729–1735. doi: 10.1016/j.rmed.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 31.Shindo Y, Ito R, Kobayashi D, Ando M, Ichikawa M, Shiraki A, et al. Risk factors for drug-resistant pathogens in community-acquired and healthcare-associated pneumonia. Am J Respir Crit Care Med 2013; 188:985–995. doi: 10.1164/rccm.201301-0079OC. [DOI] [PubMed] [Google Scholar]
- 32.Shu JC, Chia JH, Kuo AJ, Su LH, Wu TL. A 7-year surveillance for ESBL-producing Escherichia coli and Klebsiella pneumoniae at a university hospital in Taiwan: the increase of CTX-M-15 in the ICU. Epidemiol Infect 2010; 138:253–263. doi: 10.1017/S0950268809990409. [DOI] [PubMed] [Google Scholar]
- 33.Ho PL, Chow KH, Lai EL, Lau EH, Cheng VC. Extended-spectrum-beta-lactamase-positive Escherichia coli mainly adds to, rather than replaces, extended-spectrum beta-lactamase-negative E. coli in causing bacteraemia in Hong Kong, 2000-10. J Antimicrob Chemother 2012; 67:778–780. doi: 10.1093/jac/dkr502. [DOI] [PubMed] [Google Scholar]
- 34.Sheng WH, Badal RE, Hsueh PR. Distribution of extended-spectrum beta-lactamases, AmpC beta-lactamases, and carbapenemases among Enterobacteriaceae isolates causing intra-abdominal infections in the Asia-Pacific region: results of the study for monitoring antimicrobial resistance trends (SMART). Antimicrob Agents Chemother 2013; 57:2981–2988. doi: 10.1128/AAC.00971-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Diestra K, Juan C, Curiao T, Moya B, Miro E, Oteo J, et al. Characterization of plasmids encoding blaESBL and surrounding genes in Spanish clinical isolates of Escherichia coli and Klebsiella pneumoniae. J Antimicrob Chemother 2009; 63:60–66. doi: 10.1093/jac/dkn453. [DOI] [PubMed] [Google Scholar]
- 36.Ma L, Lin CJ, Chen JH, Fung CP, Chang FY, Lai YK, et al. Widespread dissemination of aminoglycoside resistance genes armA and rmtB in Klebsiella pneumoniae isolates in Taiwan producing CTX-M-type extended-spectrum beta-lactamases. Antimicrob Agents Chemother 2009; 53:104–111. doi: 10.1128/AAC.00852-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mendonca N, Ferreira E, Louro D, Canica M. Molecular epidemiology and antimicrobial susceptibility of extended- and broad-spectrum beta-lactamase-producing Klebsiella pneumoniae isolated in Portugal. Int J Antimicrob Agents 2009; 34:29–37. doi: 10.1016/j.ijantimicag.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 38.Hammond DS, Schooneveldt JM, Nimmo GR, Huygens F, Giffard PM. Bla(SHV) genes in Klebsiella pneumoniae: different allele distributions are associated with different promoters within individual isolates. Antimicrob Agents Chemother 2005; 49:256–263. doi: 10.1128/AAC.49.1.256-263.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Turner MS, Andersson P, Bell JM, Turnidge JD, Harris T, Giffard PM. Plasmid-borne blaSHV genes in Klebsiella pneumoniae are associated with strong promoters. J Antimicrob Chemother 2009; 64:960–964. doi: 10.1093/jac/dkp338. [DOI] [PubMed] [Google Scholar]
- 40.Chen CM, Yu WL, Huang M, Liu JJ, Chen IC, Chen HF, et al. Characterization of IS26-composite transposons and multidrug resistance in conjugative plasmids from Enterobacter cloacae. Microbiol Immunol 2015; 59:516–525. doi: 10.1111/1348-0421.12289. [DOI] [PubMed] [Google Scholar]
- 41.Ismail MD, Ali I, Hatt S, Salzman EA, Cronenwett AW, Marrs CF, et al. Association of Escherichia coli ST131 lineage with risk of urinary tract infection recurrence among young women. J Glob Antimicrob Resist 2018; 13:81–84. doi: 10.1016/j.jgar.2017.12.006. [DOI] [PubMed] [Google Scholar]
- 42.Chung HC, Lai CH, Lin JN, Huang CK, Liang SH, Chen WF, et al. Bacteremia caused by extended-spectrum beta-lactamase-producing Escherichia coli sequence type ST131 and non-ST131 clones: comparison of demographic data, clinical features, and mortality. Antimicrob Agents Chemother 2012; 56:618–622. doi: 10.1128/AAC.05753-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cha MK, Kang CI, Kim SH, Thamlikitkul V, So TM, Ha YE, et al. Emergence and dissemination of ST131 Escherichia coli isolates among patients with hospital-acquired pneumonia in Asian Countries. Microb Drug Resist 2017; 23:79–82. doi: 10.1089/mdr.2016.0009. [DOI] [PubMed] [Google Scholar]
- 44.Johnson JR, Johnston B, Clabots C, Kuskowski MA, Castanheira M. Escherichia coli sequence type ST131 as the major cause of serious multidrug-resistant E. coli infections in the United States. Clin Infect Dis 2010; 51:286–294. doi: 10.1086/653932. [DOI] [PubMed] [Google Scholar]
- 45.Johnson JR, Clermont O, Menard M, Kuskowski MA, Picard B, Denamur E. Experimental mouse lethality of Escherichia coli isolates, in relation to accessory traits, phylogenetic group, and ecological source. J Infect Dis 2006; 194:1141–1150. doi: 10.1086/507305. [DOI] [PubMed] [Google Scholar]
- 46.Russo TA, Johnson JR. Proposal for a new inclusive designation for extraintestinal pathogenic isolates of Escherichia coli: ExPEC. J Infect Dis 2000; 181:1753–1754. doi: 10.1086/315418. [DOI] [PubMed] [Google Scholar]
- 47.Shaik S, Ranjan A, Tiwari SK, Hussain A, Nandanwar N, Kumar N, et al. Comparative genomic analysis of globally dominant ST131 clone with other epidemiologically successful extraintestinal pathogenic Escherichia coli (ExPEC) lineages. mBio 2017; 8:e01596-17.doi: 10.1128/mBio.01596-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qi Y, Wei Z, Ji S, Du X, Shen P, Yu Y. ST11, the dominant clone of KPC-producing Klebsiella pneumoniae in China. J Antimicrob Chemother 2011; 66:307–312. doi: 10.1093/jac/dkq431. [DOI] [PubMed] [Google Scholar]
- 49.Gu D, Dong N, Zheng Z, Lin D, Huang M, Wang L, et al. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis 2018; 18:37–46. doi: 10.1016/S1473-3099(17)30489-9. [DOI] [PubMed] [Google Scholar]
- 50.Cheng L, Cao XL, Zhang ZF, Ning MZ, Xu XJ, Zhou W, et al. Clonal dissemination of KPC-2 producing Klebsiella pneumoniae ST11 clone with high prevalence of oqxAB and rmtB in a tertiary hospital in China: results from a 3-year period. Ann Clin Microbiol Antimicrob 2016; 15:1.doi: 10.1186/s12941-015-0109-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guo X, Cao Z, Dai Z, Li Y, He X, Hu X, et al. Antimicrobial susceptibility and molecular epidemiology of multidrug-resistant Klebsiella pneumoniae in Central China. Japanese J Infect Dis 2017; 70:229–234. doi: 10.7883/yoken.JJID.2016.049. [DOI] [PubMed] [Google Scholar]
- 52.Zhan L, Wang S, Guo Y, Jin Y, Duan J, Hao Z, et al. Outbreak by hypermucoviscous Klebsiella pneumoniae ST11 Isolates with carbapenem resistance in a tertiary hospital in China. Front Cell Infect Microbiol 2017; 7:182.doi: 10.3389/fcimb.2017.00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Voulgari E, Zarkotou O, Ranellou K, Karageorgopoulos DE, Vrioni G, Mamali V, et al. Outbreak of OXA-48 carbapenemase-producing Klebsiella pneumoniae in Greece involving an ST11 clone. J Antimicrob Chemother 2013; 68:84–88. doi: 10.1093/jac/dks356. [DOI] [PubMed] [Google Scholar]
- 54.Jaidane N, Bonnin RA, Mansour W, Girlich D, Creton E, Cotellon G, et al. Genomic insights into colistin-resistant Klebsiella pneumoniae from a Tunisian Teaching Hospital. Antimicrob Agents Chemother 2018; 62:e01601–e016017. doi: 10.1128/AAC.01601-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Solgi H, Badmasti F, Giske CG, Aghamohammad S, Shahcheraghi F. Molecular epidemiology of NDM-1- and OXA-48-producing Klebsiella pneumoniae in an Iranian hospital: clonal dissemination of ST11 and ST893. J Antimicrob Chemother 2018; 73:1517–1524. doi: 10.1093/jac/dky081. [DOI] [PubMed] [Google Scholar]
- 56.Ko KS, Lee JY, Baek JY, Suh JY, Lee MY, Choi JY, et al. Predominance of an ST11 extended-spectrum beta-lactamase-producing Klebsiella pneumoniae clone causing bacteraemia and urinary tract infections in Korea. J Med Microbiol 2010; 59:822–828. doi: 10.1099/jmm.0.018119-0. [DOI] [PubMed] [Google Scholar]
- 57.Adler A, Khabra E, Paikin S, Carmeli Y. Dissemination of the blaKPC gene by clonal spread and horizontal gene transfer: comparative study of incidence and molecular mechanisms. J Antimicrob Chemother 2016; 71:2143–2146. doi: 10.1093/jac/dkw106. [DOI] [PubMed] [Google Scholar]
- 58.Fu P, Tang Y, Li G, Yu L, Wang Y, Jiang X. Pandemic spread of blaKPC-2 among Klebsiella pneumoniae ST11 in China is associated with horizontal transfer mediated by IncFII-like plasmids. Int J Antimicrob Agents 2019; [Epub ahead of print]. doi: 10.1016/j.ijantimicag.2019.03.014. [DOI] [PubMed] [Google Scholar]
- 59.Lam M, Wick RR, Wyres KL, Gorrie CL, Judd LM, Jenney A, et al. Genetic diversity, mobilisation and spread of the yersiniabactin-encoding mobile element ICEKp in Klebsiella pneumoniae populations. Microb Genom 2018; 4:e000196.doi: 10.1099/mgen.0.000196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Couve-Deacon E, Jove T, Afouda P, Barraud O, Tilloy V, Scaon E, et al. Class 1 integrons in Acinetobacter baumannii: a weak expression of gene cassettes to counterbalance the lack of LexA-driven integrase repression. Int J Antimicrob Agents 2019; 53:491–499. doi: 10.1016/j.ijantimicag.2018.11.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


