Abstract
Background.
Previous reports suggested that US methicillin-resistant Staphylococcus aureus (MRSA) strain epidemiology has changed since the rise of USA300 MRSA. We describe invasive MRSA trends by strain type.
Methods.
Data came from 5 Centers for Disease Control and Prevention Emerging Infections Program sites conducting population-based surveillance and collecting isolates for invasive MRSA (ie, from normally sterile body sites), 2005–2013. MRSA bloodstream infection (BSI) incidence per 100 000 population was stratified by strain type and epidemiologic classification of healthcare exposures. Invasive USA100 vs USA300 case characteristics from 2013 were compared through logistic regression.
Results.
From 2005 to 2013, USA100 incidence decreased most notably for hospital-onset (6.1 vs 0.9/100 000 persons, P < .0001) and healthcare-associated, community-onset (10.7 vs 4.9/100 000 persons, P < .0001) BSIs. USA300 incidence for hospital-onset BSIs also decreased (1.5 vs 0.6/100 000 persons, P < .0001). However, USA300 incidence did not significantly change for health-care-associated, community-onset (3.9 vs 3.3/100 000 persons, P = .05) or community-associated BSIs (2.5 vs 2.4/100 000 persons, P = .19). Invasive MRSA was less likely to be USA300 in patients who were older (adjusted odds ratio [aOR], 0.97 per year [95% confidence interval {CI}, .96–.98]), previously hospitalized (aOR, 0.36 [95% CI, .24–.54]), or had central lines (aOR, 0.44 [95% CI, .27–.74]), and associated with USA300 in people who inject drugs (aOR, 4.58 [95% CI, 1.16–17.95]).
Conclusions.
Most of the decline in MRSA BSIs was from decreases in USA100 BSI incidence. Prevention of USA300 MRSA BSIs in the community will be needed to further reduce burden from MRSA BSIs.
Keywords: MRSA, epidemiology, bloodstream infections, strains, infection control
Methicillin-resistant Staphylococcus aureus (MRSA) was once thought to be largely limited to healthcare settings but now also frequently causes infections in the community among persons lacking classic healthcare-related risk factors [1–4]. It is now well established that these community-associated MRSA infections are largely due to a clonal strain designated by pulsed-field gel electrophoresis (PFGE) typing as USA300 [1, 3]. This strain type is quite distinct from traditional healthcare strains, such as USA100 (the most common in the United States) [5].
However, shifts in the molecular epidemiology of MRSA may be occurring. Some reports have suggested that USA300 is also an increasingly frequent cause of MRSA infections acquired by hospitalized patients and that healthcare exposure is no longer a discriminator of USA100 vs USA300 infections [6–8]. These reports have been made in the broader context of estimates that the national burden of invasive MRSA infections in the United States has declined from approximately 111 000 invasive infections in 2005 to 72 000 in 2014 [9, 10].
Secular trends in the population incidence of MRSA bloodstream infections (BSIs) and epidemiologic characteristics of invasive MRSA infections by strain type were explored to shed light on reasons for declines in invasive MRSA incidence and to investigate whether USA300 continued to be associated with healthcare-related risk factors. We hypothesized that the findings might also provide insight into future directions for MRSA prevention.
METHODS
Data came from the Centers for Disease Control and Prevention’s (CDC) Emerging Infections Program (EIP) surveillance for invasive MRSA, an active laboratory- and population-based public health surveillance program that has been described previously [11]. Geographic diversity and representativeness are important qualities of the overall EIP surveillance network. The EIP MRSA surveillance program defines a case as isolation of MRSA from a normally sterile body site (eg, blood, bone, cerebrospinal fluid) from a resident of selected counties within the EIP state. Staff at each participating EIP site contact clinical laboratories routinely serving residents of their area for laboratory reports of invasive MRSA and record epidemiologic data obtained through medical record review on a standardized case report form. Based on healthcare exposures reported, cases are assigned to 1 of 3 epidemiologic classifications: hospital-onset; healthcare-associated, community-onset; or community-associated. Cases are considered hospital-onset if the initial invasive MRSA culture was obtained on the fourth day or later during a hospital admission. Cases are considered healthcare-associated, community-onset if the culture was obtained either from an outpatient or a patient during the first 3 days of a hospital admission, with at least 1 of the following risk factors: hospital admission, long-term care facility residence, dialysis, or surgery within the prior year; or central venous catheter within 2 days prior to the culture. Cases are considered community-associated if the culture was obtained either from an outpatient or a patient during the first 3 days of a hospital admission, without any of the above healthcare risk factors.
In addition to collecting epidemiologic data, EIP sites requested isolates from clinical laboratories for characterization. In all sites except one (New York, described below), clinical laboratories provided a convenience sample of isolates. All isolates submitted by clinical laboratories underwent molecular characterization at CDC as previously described, including antimicrobial susceptibility testing by broth microdilution, SCCmec typing, and detection of staphylococcal toxins [5, 12]. Molecular typing methodology of MRSA isolates obtained through the EIP has evolved over the years [10]. Initially (2005–2007), PFGE was used to categorize isolates into USA groups as described in [5]. During 2008–2011, an algorithm to infer the USA type based on other molecular characteristics was validated internally and implemented (https://www.cdc.gov/HAI/settings/lab/inferred-PFGE-algorithm.html). In 2012, that algorithm was updated to incorporate spa typing results (https://www.cdc.gov/HAI/settings/lab/CCalgorithm.html).
For this analysis, epidemiologic data and isolates obtained from 14 counties continuously performing surveillance in 5 of the 9 EIP sites with MRSA surveillance data (California, Georgia, Minnesota, New York, and Tennessee) from 2005 to 2013 were analyzed. Available resources only allowed these 5 sites to collect isolates during the entire 2005–2013 period. For all analyses, MRSA strain type was categorized as USA100, USA300, or other (“other” comprising all strains except USA100/USA300). Data analyses were performed using SAS software versions 9.3 and 9.4 (SAS Institute, Cary, North Carolina). Statistical significance was determined based on P < .05.
Two data analyses were conducted. The first calculated annual incidence of MRSA BSIs (cases identified from blood isolates per 100 000 population) stratified by epidemiologic class and strain type. The completeness of isolate collection from each clinical laboratory in the surveillance area was determined by comparing isolates submitted with reports of MRSA infections from the epidemiologic surveillance data. Because only a convenience sample of isolates from a population area was submitted, strain incidence calculations were performed through the following process. To determine the numerator (number of cases), strain type for cases in which no isolate was available for testing (~30%) was imputed based on the distribution of variables correlated with strain type. Demographics, underlying medical conditions, infection type, timing of culture collection relative to hospital admission, year of collection, and EIP site were selected as relevant variables through logistic regression with backward elimination. Imputation was performed for cases from the subset of clinical laboratories in each EIP site from which either >50% of MRSA blood isolates from cases were tested at CDC (California, Georgia, Minnesota, Tennessee) or a random sample of blood isolates was collected (New York). Cases from other laboratories (ie, those with <50% of blood isolates tested at CDC and without a systematic sampling scheme for isolates) were not included in the numerator for incidence calculation. Imputation was only performed for blood isolates (greatest proportion of isolates available). In addition, to calculate incidence of cases from this subset of laboratories, the population denominator was modified to reflect the population served by these laboratories based on the market share of MRSA cases in the entire catchment area served by those clinical laboratories, following methods used in a previous study to estimate population incidence of community-acquired pneumonia [13]. Multiple imputation with 25 imputation datasets was performed to account for imputation errors and the results were combined to make statistical inference. The SAS procedures PROC MI and MIANALYZE were used to perform analyses of the imputation datasets. Statistical significance of trends by strain type and epidemiologic classification was determined through negative binomial regression. The Supplementary Methods and Supplementary Tables 1 and 2 provide additional details about isolate collection and imputation methods.
Second, to provide further insight into the epidemiology of invasive MRSA infections by strain type, epidemiologic characteristics corresponding to all isolates characterized at CDC as either USA100 or USA300 from these 5 sites (ie, from all laboratories submitting isolates, not limited to those used for the incidence calculations above and without imputation) in 2013 were compared. The year 2013 is the most recent for which isolates are available and characterized from all 5 of these sites. Categorical variables were compared using a χ2 or Fisher exact test, as appropriate. Continuous variables were compared using a Wilcoxon rank-sum test. A logistic regression with random intercepts for EIP site was fitted to evaluate factors that were independently associated with higher likelihood of a MRSA isolate being USA100 (compared to USA300). This binary logistic model is distinct from the imputation model described earlier. In the model, variables with P < .25 in bivariate analysis were included as potential exposures, with outcome of strain type (USA100 or USA300). Backward elimination was performed for variable selection, retaining variables with P < .05 in the final model. In addition, separately the proportion of USA300 (and corresponding 95% confidence intervals [CIs]) was calculated for different types of clinical infections.
The EIP MRSA activity was determined to be a nonresearch public health surveillance activity by human subjects advisors at the National Center for Emerging and Zoonotic Infectious Diseases, CDC, and this analysis was conducted as a part of the approved objectives of the surveillance project. The project was either deemed a nonresearch activity in participating EIP sites or was approved by sites’ institutional review boards with a waiver of informed consent.
RESULTS
Incidence of MRSA BSIs by Strain Type and Epidemiologic Classification
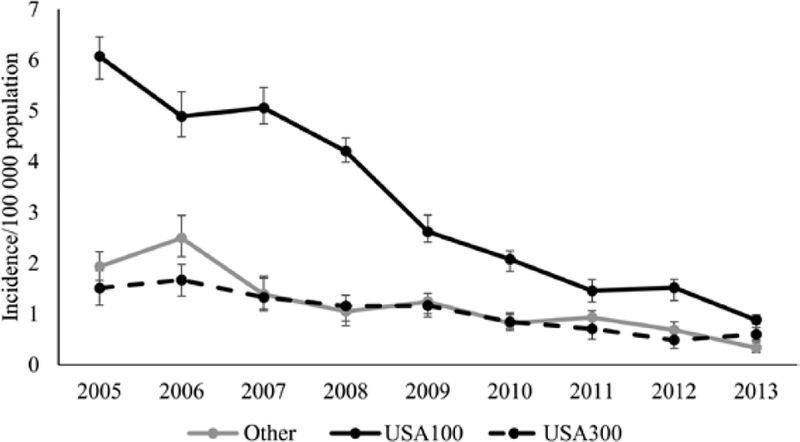
Incidence (per 100 000 population) of hospital-onset USA100 BSIs decreased (6.1 in 2005 to 0.9 in 2013) (Figure 1). The modeled annual decrease for that time period was 21% (95% CI, 17%–24%). Incidence of hospital-onset USA300 BSIs also decreased during that time (1.5 in 2005 vs 0.6 in 2013; modeled annual decrease: 13% [95% CI, 8%–18%]), as did incidence of hospital-onset MRSA BSIs from other strains (1.9 in 2005 vs 0.3 in 2013; modeled annual decrease: 17% [95% CI, 11%–22%]).
Figure 1.
Estimated incidence of hospital-onset methicillin-resistant Staphylococcus aureus bloodstream infections, by strain type, in 5 Emerging Infections Program sites, 2005–2013. Cases are considered hospital-onset if the initial culture was obtained on the fourth day or later during a hospital admission. Error bars represent the minimum and maximum incidence rates among imputation datasets.
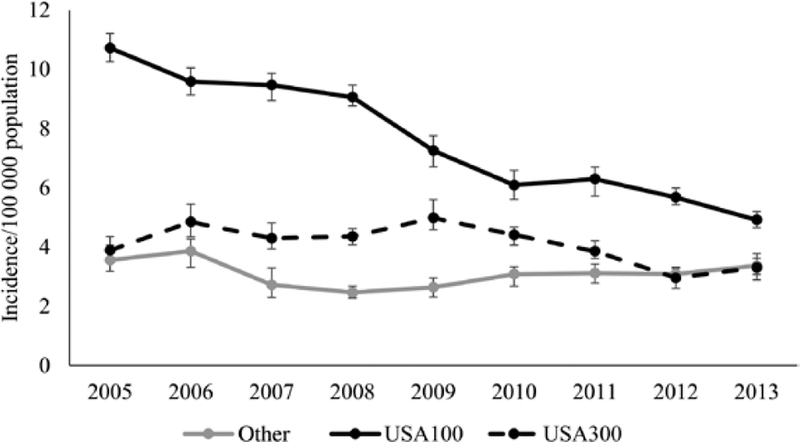
For healthcare-associated community-onset MRSA BSIs, incidence decreased for USA100 (10.7 in 2005 vs 4.9 in 2013) (Figure 2); the modeled annual decrease was 10% (95% CI, 8%–11%). Incidence did not change significantly for MRSA BSIs due to USA300 (3.9 in 2005 vs 3.3 in 2013; modeled annual decrease: 4% [95% CI, 0–7%]) or for other strains (3.6 in 2005 vs 3.4 in 2013; modeled annual decrease: 1% [95% CI, –3% to 5%]) (Figure 2).
Figure 2.
Estimated incidence of healthcare-associated, community-onset methicillin-resistant Staphylococcus aureus bloodstream infections, by strain type, in 5 Emerging Infections Program sites, 2005–2013. Cases are considered healthcare-associated, community-onset if the culture was obtained either from an outpatient or during the first 3 days of a hospital admission, from a patient with at least 1 of the following healthcare risk factors: hospital admission, long-term care facility residence, dialysis, or surgery within the prior year; or central venous catheter within 2 days prior to the culture. Error bars represent the minimum and maximum incidence rates among imputation datasets.
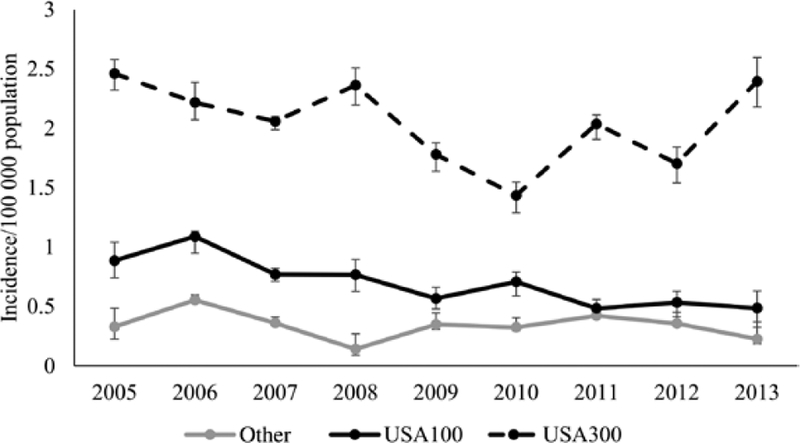
Community-associated MRSA BSI incidence decreased for USA100 (0.9 in 2005 vs 0.5 in 2013) (Figure 3); modeled annual decrease was 9% (95% CI, 3%–15%). In contrast, incidence did not significantly change for USA300 (2.5 in 2005 vs 2.4 in 2013; modeled annual decrease: 3% [95% CI, −1% to 7%]) or other strains (0.3 in 2005 vs 0.2 in 2013; modeled annual decrease: 3% [95% CI, −6% to 12%]).
Figure 3.
Estimated incidence of community-associated methicillin-resistant Staphylococcus aureus bloodstream infections, by strain type, in 5 Emerging Infections Program sites, 2005–2013. Cases are considered community-associated if the culture was obtained either from an outpatient or during the first 3 days of a hospital admission, from a patient without any of the following healthcare risk factors: hospital admission, long-term care facility residence, dialysis, or surgery within the prior year; or central venous catheter within 2 days prior to the culture. Error bars represent the minimum and maximum incidence rates among imputation datasets.
Overall, USA100 BSI incidence decreased from 17.7 to 6.3, and USA300 BSI incidence decreased from 7.9 to 6.3. The proportion that was USA300 increased from 25% to 38% overall.
Epidemiologic Descriptions of the Most Recent USA100 and USA300 Invasive MRSA Cases
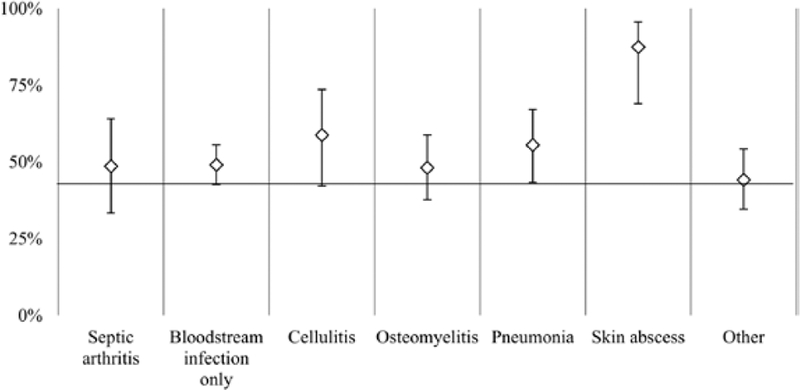
In 2013, isolates were available for 705 cases of invasive MRSA, of which 534 were characterized as either USA100 or USA300. Also represented in the collection were 81 USA500/Archaic/Iberian strain complex isolates (11%) and 32 USA800 (5%). USA200, USA400, USA600, USA700, USA1000, and USA1100 were also identified but each comprised ≤2% of the collection. Most invasive cases with skin abscess were USA300 (88% [95% CI, 69%–96%] of cases associated with skin abscess). The other most common infection syndromes associated with the cases were equally due to either USA100 or USA300 (Figure 4).
Figure 4.
Proportion of clinical infections associated with invasive methicillin-resistant Staphylococcus aureus cases due to USA300 (vs USA100) strain type, in 5 Emerging Infections Program sites, 2013. Cases with cellulitis, pneumonia, and skin abscess had an underlying invasive specimen source (eg, positive blood culture). The vertical error bars depict the corresponding 95% confidence intervals.
In bivariate analysis, invasive USA100 cases occurred more often in older patients, patients of white race, and patients with certain chronic medical comorbidities and prior healthcare exposures (Table 1). Invasive cases were more likely to be USA300 when occurring in patients with human immunodeficiency virus infection or injection drug use. Hospital-onset cases were not more likely to be caused by USA100 compared to USA300.
Table 1.
Selected Descriptive Characteristics of USA100 and USA300 Invasive Methicillin-resistant Staphylococcus aureus Cases, 5 Emerging Infections Program Sites, 2013
| Characteristic | USA100 (n = 266) |
USA300 (n = 268) |
PValue |
|---|---|---|---|
| Demographics | |||
| Age, y, median (IQR) | 65.5 (54–81) | 55 (40–65) | < .001 |
| Male sex | 154 (57.9) | 165 (61.6) | .39 |
| White race | 165 (62.0) | 125 (46.6) | .0004 |
| Medical comorbidities | |||
| Chronic liver disease | 23 (8.7) | 20 (7.5) | .62 |
| Chronic pulmonary disease | 54 (20.3) | 45 (16.8) | .30 |
| Chronic renal insufficiency | 101 (38.0) | 72 (26.9) | .006 |
| Chronic skin breakdown | 34 (12.8) | 36 (13.4) | .82 |
| Congestive heart failure | 77 (29.0) | 47 (17.5) | .002 |
| Smoker | 30 (11.3) | 78 (29.1) | < .0001 |
| Prior cerebrovascular accident | 46 (17.3) | 23 (8.6) | .003 |
| Decubitus ulcer | 38 (14.3) | 23 (8.6) | .04 |
| Dementia | 34 (12.8) | 20 (7.5) | .04 |
| Diabetes | 109 (41.0) | 109 (40.7) | .94 |
| Hemiplegia/paraplegia | 17 (6.4) | 13 (4.9) | .44 |
| HIV | 1 (0.4) | 18 (6.7) | < .0001 |
| Injection drug use | 3 (1.1) | 20 (7.5) | .0003 |
| Obesity | 48 (18.1) | 41 (15.3) | .39 |
| Peripheral vascular disease | 39 (14.7) | 25 (9.3) | .06 |
| Healthcare exposures | |||
| Hospital onset | 47 (17.7) | 41 (15.3) | .27 |
| Hospitalization in prior year | 191 (71.8) | 121 (45.2) | < .0001 |
| Surgery in prior year | 104 (39.1) | 55 (20.5) | < .0001 |
| Central venous catheter in place | 64 (24.1) | 35 (13.1) | .001 |
| Chronic dialysis patient | 55 (20.7) | 41 (15.3) | .11 |
| Long-term care facility residence in prior year | 63 (23.7) | 34 (12.7) | .001 |
Data are presented as no. (%) unless otherwise indicated.
Abbreviations: HIV, human immunodeficiency virus; IQR, interquartile range.
In multivariable analysis, only 4 characteristics were independently associated with differences in USA100 vs USA300 invasive MRSA infections (Table 2). Cases were more likely to be USA100 in patients who were older (adjusted odds ratio [aOR], 1.03 per year [95% CI, 1.02–1.04]), hospitalized in the prior year (aOR, 2.78 [95% CI, 1.87–4.16]), or with a recent central venous catheter (aOR, 2.25 [95% CI, 1.35–3.77]). Cases were less likely to be USA100 if associated with injection drug use (aOR, 0.22 [95% CI, .06–.86]).
Table 2.
Multivariate Analysis of Characteristics Associated With USA100 Invasive Methicillin-resistant Staphylococcus aureus Infection (Versus USA300), 2013
| Characteristic | Adjusted OR (95% Cl) for USA100 | PValue |
|---|---|---|
| Age (y) | 1.03 (1.02–1.04) | <0001 |
| Injection drug use | 0.22 (.06–.86) | .03 |
| Hospitalization in prior year | 2.78 (1.87–4.16) | <.0001 |
| Central venous catheter | 2.25 (1.35–3.77) | .002 |
Abbreviations: CI, confidence interval; OR, odds ratio.
DISCUSSION
In this surveillance area, overall USA100 BSI incidence declined by >60% during 2005–2013, including a >80% decline in hospital-onset USA100 BSI incidence. A decline in USA300 BSI incidence only occurred for hospital-onset cases (60% decrease); however, the incidence of USA300 BSIs did not increase in any epidemiologic category. USA100 infections were still more likely to be associated with some classic healthcare risk factors including central venous catheter use and prior hospitalization.
Several conclusions or observations can be made from these results. First, as other US investigators have reported [7, 8, 14–16], there is an overall increase in the proportion of overall MRSA BSIs due to USA300, including for hospital-onset BSIs. However, the change in proportion of BSIs due to USA300 can be attributed to greater declines in USA100 since, as noted above, invasive USA300 infections are not increasing in frequency on a population level in any setting.
Second, USA100 remains the strain that is most strongly associated with healthcare exposure. The majority of health-care-associated (ie, hospital-onset and healthcare-associated community-onset) MRSA BSIs were due to USA100. Notably, as described in an urban Chicago-area hospital [7], we did not find the hospital-onset epidemiologic category to be a significant predictor of USA100 vs USA300 MRSA. However, prior hospitalization and central venous catheter use were significant predictors of USA100 in our data.
Third, these results support a conclusion that healthcare infection control interventions contributed to reductions in invasive MRSA that have been seen in the United States. As outlined below, the finding that strain-specific trends differ substantially according to healthcare exposure categories provides evidence that activities in healthcare settings themselves are causing the MRSA trends. Indeed, some healthcare infection control activities have clearly contributed to substantial reductions in invasive MRSA. A well-known example is the large reductions that have been observed in central line–associated BSI burden resulting primarily from improving central line insertion practices [17–19]. Some have also attributed decreases in MRSA infections in Department of Veterans Affairs hospitals to a healthcare infection control bundle that included strategies aimed at reducing transmission of MRSA [20].
Alternative explanations that have been proposed for observed reductions in MRSA incidence are (1) increases in use of antibiotics effective against MRSA, or (2) a byproduct of natural variation in strain ecology [21–24]. The first explanation seems less likely to be the sole explanation for these reductions given existing data about epidemiology of antibiotic use. For example, trimethoprim-sulfamethoxazole use in outpatients has increased in the United States during this period [25, 26]. However, for community-associated MRSA BSIs, incidence decreased for USA100 only even though USA300 strains tend to have higher susceptibility to trimethoprim-sulfamethoxazole [5].
Evaluating the second explanation is more difficult, as there is no clear a priori expectation of what natural variation would cause. At a minimum though, we do not see evidence that USA300 is expanding its prevalence in the population, which is one of the specific explanations that has been proposed [24]. Researchers using other methods have also suggested that the historic expansion and dissemination of USA300 may have reached a plateau [27]. Therefore, USA300 expansion as a cause for the decline in USA100 is not a phenomenon supported by our data.
If it is true that healthcare infection prevention has been a dominant reason for these trends, the differences between USA100 vs USA300 trends within the same epidemiologic class are intriguing. It suggests that even within the same setting there are strain-specific differences in effectiveness or impact of healthcare prevention strategies. This possibility has been raised by other investigators [28, 29]. We suggest that an explanation for the lack of decrease in healthcare-associated, community-onset USA300 incidence is that current infection prevention practices are primarily only successful for preventing USA100 infections outside the hospital setting. One plausible specific mechanism by which this could occur would be that healthcare-associated, community-onset USA100 MRSA infections are more likely to have been acquired through transmission in healthcare settings, whereas patients with health-care-associated, community-onset USA300 MRSA infections may have acquired these strains in the community. This is plausible as USA100 strains typically have greater levels of antimicrobial resistance than USA300 strains [5] and therefore may be better able to survive when patients are frequently exposed to broad-spectrum antibiotics in healthcare settings [30]. An alternative mechanism could be differential effects of antimicrobial stewardship activities on changing prevalence of USA100 vs USA300. Future work to confirm whether the different trends are primarily due to differential transmission would have important implications for future prevention work. For example, although continued prevention of transmission of USA100 in hospitals is likely important to prevent a resurgence of these infections, the data also suggest that that additional strategies beyond simply intensifying reductions in transmission in hospitals are needed to substantially reduce the burden of health-care-associated USA300 infections.
The most significant limitation of this analysis is that, because of lack of availability of isolates, the trends reported here end in 2013 without subsequent information on current incidence of MRSA strain types. However, trends for MRSA BSI incidence in the overall EIP area have not greatly changed after 2013 (unpublished data). Other limitations of this work include isolates not being available for all cases, so the results are subject to the accuracy of the strain imputation and incidence estimation process; specifically, it is possible that true USA300 incidence has changed for community-associated and healthcare-associated, community-onset BSIs and we did not have sufficient precision to detect this. This surveillance area only represents a fraction of the United States. However as noted previously, existing published single-center reports about trends for MRSA strains outside of our surveillance area show consistent results [7, 16]. In addition, the epidemiologic trends (stratified by epidemiologic class but without strain information) in these 5 EIP sites match those of the additional 4 EIP sites from which epidemiologic surveillance data are available but not a consistent collection of isolates. Furthermore, we do not have information on trends for colonizing strains; some of our conclusions depend on an assumption that trends for invasive infections would mirror those of colonization, which we are unable to evaluate with these data. We also are unable to make inferences about relative contribution of different healthcare interventions on the historical declines in MRSA BSI incidence. A final limitation is the use of PFGE and spa typing as the predominant typing methods, which can cause misclassification of sublineages within clonal complex 8 [31, 32]. A strength of this analysis is the multisite and population-based nature of the data. Population-based data provide assurance that we measure actual increases or decreases in burden rather than an effect of shifts in where patients seek healthcare.
This article provides insight into why invasive MRSA incidence in the United States has declined and offers specific hypotheses that can be tested in future studies. Most importantly, it illustrates that although substantial progress has been made in preventing invasive MRSA infections, only USA100 MRSA incidence has significantly decreased outside of hospitals. Strategies to prevent USA300 infections in the community among patients with and without healthcare exposure are particularly needed to ensure further substantial reductions in the burden of MRSA infections in the United States.
Supplementary Material
Acknowledgments.
The authors thank the Centers for Disease Control and Prevention (CDC) staff members Shirley Zhang for database assistance; Scott Fridkin (current affiliation: Emory University), Brandi Limbago, and John Jernigan for critical feedback and discussion of results; the CDC’s Emerging Infections Program (EIP) office for administrative support; and the following EIP site staff for data collection and project oversight at sites: Lauren Pasutti, Brittany Martin, Cindy Amezcua, Gretchen Rothrock, Arthur Reingold, Elizabeth Partridge, Maria Rosales (California EIP); Randy Van Dolson, Sasha Harb, Stepy Thomas, Monica M. Farley, Wendy Baughman, Amy Tunali, Janine Ladson, Jessica Reno, Betsy Stein, and Lewis Perry (Georgia EIP); Kathryn Como-Sabetti, Lindsey Lesher, and Jessica Nerby (Minnesota EIP); Anita Gellert, Christina Felsen (New York EIP); and Gail Hughett, Terri McMinn, Brenda Barnes, Karen Leib, and Katie Dyer (Tennessee EIP).
Financial support. This work was supported by a cooperative agreement through the CDC EIP (grant numbers U50CK000201 [California], U50CK00196 [Georgia], U50CK000204 [Minnesota], U50CK000199 [New York], and U50CK000198 [Tennessee]).
Footnotes
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Publisher's Disclaimer: Disclaimer. The findings and conclusions in this article are those of the authors and do not necessarily represent the official position of the CDC.
Potential conflicts of interest. W. S. has received personal fees from Pfizer, Merck, Dynavax, SutroVax, Shionogi, and Seqirus. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Fridkin SK, Hageman JC, Morrison M, et al. Methicillin-resistant Staphylococcus aureus disease in three communities. N Engl J Med 2005; 352:1436–44. [DOI] [PubMed] [Google Scholar]
- 2.Herold BC, Immergluck LC, Maranan MC, et al. Community-acquired methicillin-resistant Staphylococcus aureus in children with no identified predisposing risk. JAMA 1998; 279:593–8. [DOI] [PubMed] [Google Scholar]
- 3.Moran GJ, Krishnadasan A, Gorwitz RJ, et al. Methicillin-resistant S. aureus infections among patients in the emergency department. N Engl J Med 2006; 355:666–74. [DOI] [PubMed] [Google Scholar]
- 4.Naimi TS, LeDell KH, Como-Sabetti K, et al. Comparison of community- and health care-associated methicillin-resistant Staphylococcus aureus infection. JAMA 2003; 290:2976–84. [DOI] [PubMed] [Google Scholar]
- 5.Limbago B, Fosheim GE, Schoonover V, et al. Characterization of methicillin-resistant Staphylococcus aureus isolates collected in 2005 and 2006 from patients with invasive disease: a population-based analysis. J Clin Microbiol 2009; 47:1344–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seybold U, Kourbatova EV, Johnson JG, et al. Emergence of community-associated methicillin-resistant Staphylococcus aureus USA300 genotype as a major cause of health care-associated blood stream infections. Clin Infect Dis 2006; 42:647–56. [DOI] [PubMed] [Google Scholar]
- 7.Rhee Y, Aroutcheva A, Hota B, Weinstein RA, Popovich KJ. Evolving Epidemiology of Staphylococcus aureus bacteremia. Infect Control Hosp Epidemiol 2015; 36:1417–22. [DOI] [PubMed] [Google Scholar]
- 8.David MZ, Daum RS, Bayer AS, et al. Staphylococcus aureus bacteremia at 5 US academic medical centers, 2008–2011: significant geographic variation in community-onset infections. Clin Infect Dis 2014; 59:798–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Centers for Disease Control and Prevention. Active Bacterial Core Surveillance report, Emerging Infections Program Network, methicillin-resistant Staphylococcus aureus, 2005. Available at: https://www.cdc.gov/abcs/reports-findings/survre-ports/mrsa05.html. Accessed 21 November 2018
- 10.Centers for Disease Control and Prevention. Active Bacterial Core Surveillance report, Emerging Infections Program Network, methicillin-resistant Staphylococcus aureus, 2014. Available at: https://www.cdc.gov/abcs/reports-findings/survre-ports/mrsa14.html. Accessed 21 November 2018 [Google Scholar]
- 11.Dantes R, Mu Y, Belflower R, et al. National burden of invasive methicillin-resistant Staphylococcus aureus infections, United States, 2011. JAMA Intern Med 2013; 173:1970–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McDougal LK, Steward CD, Kilgore GE, Chaitram JM, McAllister SK, Tenover FC. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J Clin Micro 2003; 41:5113–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jain S, Self WH, Wunderink RG, et al. Community-acquired pneumonia requiring hospitalization among U.S. adults. N Engl J Med 2015; 373:415–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diekema DJ, Richter SS, Hellmann KP, et al. Continued emergence of USA300 methicillin-resistant Staphylococcus aureus in the United States: results from a nationwide surveillance study. Infect Control Hosp Epidemiol 2014; 35:285–92. [DOI] [PubMed] [Google Scholar]
- 15.David MZ, Cadilla A, Boyle-Vavra S, Daum RS. Replacement of HA-MRSA by CA-MRSA infections at an academic medical center in the midwestern United States, 2004–5 to 2008. PLoS One 2014; 9:e92760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanjilal S, Abdul Sater MR, Thayer M, et al. Trends in antibiotic susceptibility in Staphylococcus aureus in Boston, Massachusetts, 2000 to 2014. J Clin Microbiol 2017; 56:e01160–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wise ME, Scott RD 2nd, Baggs JM, et al. National estimates of central line-associated bloodstream infections in critical care patients. Infect Control Hosp Epidemiol 2013; 34:547–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weiner LM, Webb AK, Limbago B, et al. Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2011–2014. Infect Control Hosp Epidemiol 2016; 37: 1288–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burton DC, Edwards JR, Horan TC, Jernigan JA, Fridkin SK. Methicillin-resistant Staphylococcus aureus central line-associated bloodstream infections in US intensive care units, 1997–2007. JAMA 2009; 301:727–36. [DOI] [PubMed] [Google Scholar]
- 20.Jain R, Kralovic SM, Evans ME, et al. Veterans Affairs initiative to prevent methicillin-resistant Staphylococcus aureus infections. N Engl J Med 2011; 364:1419–30. [DOI] [PubMed] [Google Scholar]
- 21.Perencevich EN, Diekema DJ. Decline in invasive MRSA infection: where to go from here? JAMA 2010; 304:687–9. [DOI] [PubMed] [Google Scholar]
- 22.Jones M, Teng J, Huttner B, et al. Effects of initial anti-methicillin-resistant Staphylococcus aureus (MRSA) antibiotics on nosocomial MRSA acquisition and clinical cultures. In: IDWeek, San Diego, CA, 7–11 October 2015. [Google Scholar]
- 23.Jones M, Huttner B, Stevens V, et al. Lower methicillin-resistant Staphylococcus aureus (MRSA) acquisition and nosocomial cultures while on anti-MRSA antibiotics? In: IDWeek, San Diego, CA, 7–11 October 2015. [Google Scholar]
- 24.Kallen AJ, Mu Y, Bulens S, et al. Health care-associated invasive MRSA infections, 2005–2008. JAMA 2010; 304:641–8. [DOI] [PubMed] [Google Scholar]
- 25.Suda KJ, Hicks LA, Roberts RM, Hunkler RJ, Taylor TH. Trends and seasonal variation in outpatient antibiotic prescription rates in the United States, 2006 to 2010. Antimicrob Agents Chemother 2014:58:2763–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.US Food and Drug Administration, Center for Drug Evaluation and Research, Office of Surveillance and Epidemiology. Drug use review (systemic antibacterial drug products). Available at: https://www.fda.gov/downloads/Drugs/DrugSafety/InformationbyDrugClass/UCM319435.pdf. Accessed 21 November 2018 [Google Scholar]
- 27.Planet PJ. Life after USA300: the rise and fall of a superbug. J Infect Dis 2017; 215:S71–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cassone M, McNamara SE, Perri MB, Zervos M, Mody L. Impact of intervention measures on MRSA clonal type and carriage site prevalence. mBio 2016; 7:e00218–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Senn L, Clerc O, Zaneti G, et al. The stealthy superbug: the role of asymptomatic enteric carriage in maintaining a long-term hospital outbreak of ST228 methicillin-resistant Staphylococcus aureus. mBio 2016; 7:e02039–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Magill SS, Edwards JR, Beldavs ZG, et al. Prevalence of antimicrobial use in US acute care hospitals, May-September 2011. JAMA 2014; 312:1438–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frisch MB, Castillo-Ramirez S, Petit RA III, et al. Invasive methicillin-resistant Staphylococcus aureus USA500 strains from the U.S. Emerging Infections Program constitute three geographically distinct lineages. Mbio 2018; 3:e00571–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bowers JR, Driebe EM, Albrecht V, et al. Improved subtyping of Staphylococcus aureus clonal complex 8 strains based on whole-genome phylogenetic analysis. mSphere 2018; 3:e00464–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.