Abstract
Deimination, a post-translational modification catalyzed by a family of enzymes called peptidylarginine deiminases (PADs), is the conversion of arginine into citrulline residues in a protein. Deimination has been associated with numerous physiological and pathological processes. Our aim was to study its implication in the homeostasis of human epidermis, where three PADs are expressed, namely PAD1, 2 and 3. Three-dimensional reconstructed human epidermis (RHEs) were treated for two days with increased concentrations (0–800 μM) of Cl-amidine, a specific PAD inhibitor. Cl-amidine treatments inhibited deimination in a dose-dependent manner and were not cytotoxic for keratinocytes. At 800 μM, Cl-amidine was shown to reduce deimination by half, alter keratinocyte differentiation, decrease the number of corneocyte layers, significantly increase the number of transitional cells, induce clustering of mitochondria and of heterogeneous vesicles in the cytoplasm of granular keratinocytes, and upregulate the expression of autophagy proteins, including LC3-II, sestrin-2 and p62/SQSTM1. LC3 and PADs were further shown to partially co-localize in the upper epidermis. These results demonstrated that Cl-amidine treatments slow down cornification and alter autophagy in the granular layer. They suggest that PAD1 and/or PAD3 play a role in the constitutive epidermal autophagy process that appears as an important step in cornification.
Keywords: deimination, citrullination, post-translational modification, skin, keratinocyte differentiation, autophagy, transitional cells
INTRODUCTION
Deimination (or citrullination) is the calcium-dependent post-translational conversion of arginine to citrulline residues in proteins. This modification alters the global charge of substrates and potentially induces changes in their conformation, interactions and thus functions. Deimination is catalyzed by a family of five enzymes called peptidylarginine deiminases (PADs; PAD1–4 and PAD6; Chavanas et al., 2004). Each PAD isotype possesses its own substrate specificity and displays a specific pattern of tissue expression (Nachat et al., 2005; Mechin et al., 2005, 2010; Moelants et al., 2012; Assohou-Luty et al., 2014; Cau et al., 2018). Although they have been associated with numerous physiological processes (such as regulation of gene expression, inflammation, immune responses and oogenesis) and implicated in various diseases (such as rheumatoid arthritis, multiple sclerosis, Alzheimer’s disease and cancers) (Méchin et al., 2007; Baka et al., 2012; Wang et al., 2013; Christophorou et al., 2014), the cellular functions of PADs are still poorly understood.
PAD1–3 isotypes are expressed in the epidermis where deiminated proteins are only detected in the cornified layer (Nachat et al., 2005). To date, five epidermal substrates of PADs have been certainly identified: keratins K1 and K10, filaggrin, filaggrin 2 and hornerin (Senshu et al., 1996; Tarcsa et al., 1996; Hsu et al., 2011; Hsu et al., 2017). However, they may be more numerous as suggested by mass spectrometry analysis (Winger et al., 2016). Keratins K1 and K10 are deiminated in the upper part of the cornified layer. The consequence of their deimination is suspected to induce relaxation of the corneocyte matrix. Filaggrin, a member of the S100-fused type protein (SFTP) family, facilitates the aggregation of keratin intermediate filaments during keratinocyte differentiation (Le Lamer et al., 2015). Filaggrin deimination disrupts its interaction with keratins and increases susceptibility to proteolysis by calpain 1, leading to the release of free amino acids and subsequent derivatives that contribute to the natural moisturizing factor (Harding and Scott, 1983; Kamata et al., 2009; Hsu et al., 2011). Lowering relative humidity drives filaggrin degradation through increase of deimination (Cau et al., 2G17). Deimination of filaggrin 2, another SFTP suspected to share the same fate as filaggrin, was also demonstrated to facilitate its proteolysis by calpain 1 (Hsu et al., 2G11). Finally, deimination of hornerin, a third SFTP that is incorporated into the cornified envelope, improves its crosslink by transglutaminases (Hsu et al., 2G17). Epidermal proteins are mainly deiminated by PAD1 and/or PAD3 in physiological conditions (Coudane et al., 2G11; Cau et al, 2017).
In order to further understand the physiological function of deimination in epidermal homeostasis, pan-PAD inhibitor Cl-amidine (Knuckley et al., 2G1G; Jones et al., 2G12) was added to the culture medium of three-dimensional reconstructed human epidermis (RHEs). Characterization of the epidermis revealed that Cl-amidine treatments disturbed cornification and acts on autophagy.
RESULTS
In vitro inhibition by Cl-amidine of recombinant PAD1, PAD2 and PAD3 activity
To assess the efficacy of Cl-amidine against the three epidermal PADs, a recombinant form of human filaggrin (FLG-His) was deiminated in vitro by the recombinant PAD1, PAD2 or PAD3 at 37°C, in the presence of increased concentrations of the inhibitor. After incubation, the reaction products were analyzed by western blotting with the AMC antibodies specific for deiminated proteins (Supplementary Figure S1 online). A clear dose-dependent decrease in filaggrin deimination was observed. Cl-amidine was more specific for PAD1 than for PAD2 and PAD3 but was nevertheless able to inhibit the activity of the three isotypes.
Expression and activity of PAD1 and PAD3 in RHEs
RHEs were produced using primary keratinocytes from normal human skin. In order to test for the expression and activity of PADs, RHEs were analyzed by indirect immunofluorescence and western blotting with the AMC antibodies (Supplementary Figure S2a and S2b). Deiminated proteins were detected only in the stratum corneum, as previously observed in human interfollicular epidermis (Senshu et al., 1995; 1996; Nachat et al., 2005). Protein deimination increased from day 6 and stabilized around day 10. The expression of PAD mRNAs was analyzed by RT-qPCR (Supplementary Figure S2c). PADI4 and PADI6 transcripts were not detected in RHEs, as in the epidermis. RHEs showed no PADI2 expression as well, an isotype known to be expressed in human epidermis (Ishigami et al., 2002; Chavanas et al., 2004; Nachat et al., 2005). PADI1 and PADI3 were clearly expressed in RHEs. The PADI1 mRNA amount increased from day 4 to day 10, and tended to reach the level observed in the epidermis. The PADI3 mRNA level did not significantly vary between day 4 and day 10. However, it was much higher than in the epidermis. PAD1 and PAD3 were also detected in RHEs by indirect immunofluorescence and presented a pattern of expression similar to that in the epidermis (Supplementary Figure S2d).
Dose-dependent inhibition of protein deimination in RHEs by Cl-amidine
In order to evaluate the efficacy of Cl-amidine to inhibit PAD activity in RHEs, various concentrations of the inhibitor, from 100 to 800 μM, were added in the culture medium at day 8. The RHEs were harvested 48 hours later and total proteins analyzed by western blotting with the AMC antibody. Cl-amidine treatment induced a decrease in the total quantity of deiminated proteins in a dose-dependent manner (Figure 1a). At 800 μM of Cl-amidine, the deimination rate was significantly reduced to a mean of 55± 30% (p = 0.014; n = 6; Figure 1a and b). Since the highest effect of Cl-amidine was obtained with a concentration of 800 μM, it was used for the rest of the study.
Figure 1. Dose-dependent inhibition of deimination in reconstructed human epidermis (RHEs) treated with Cl-amidine.
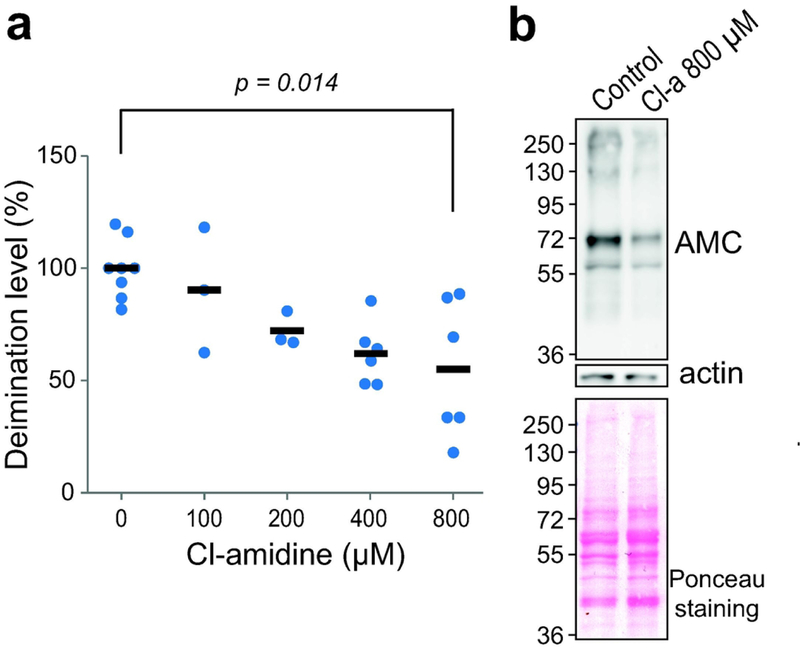
RHEs were treated with Cl-amidine (Cl-a) at different concentrations (0–800 μM) for 48 h and harvested on day 10 after air-liquid exposure. Total proteins were immunodetected with the AMC antibody. (a) Immunodetected deiminated proteins were quantified, normalized on Ponceau red staining and expressed relative to the value of the untreated (control) RHEs. Each sample value is represented by a dot and means are represented by dashes. (b) Representative blots of total protein extracts from RHEs produced without inhibitor (Control) or treated with 800 μM of Cl-amidine. Proteins were stained with Ponceau red or immunodetected with the indicated antibodies. Molecular mass markers are shown in kDa on the left.
Alteration of cornification in RHEs treated with Cl-amidine
The consequence of 800 μM Cl-amidine treatment was first analyzed using hematoxylin and eosin staining of RHEs (Figure 2a). Control and treated RHEs (n = 5) presented a similar morphology. Cl-amidine did not significantly decrease the cell viability (Figure 2b) and did not induce caspase-3 activation (data not shown). Moreover, the inhibitor did not affect keratinocyte proliferation, as evaluated using Ki67 immunodetection (21.13 ± 8.05 immunolabeled cells per RHE length in mm versus 16.92 ± 6.25 in controls; n = 3; p = 0.0719). To study the effect of Cl-amidine on RHEs at the ultrastructural level, transmission electron microscopy analysis was performed. In RHEs treated with Cl-amidine, we observed frequent unusual cells located between the granular and the cornified layers (Figures 2c and d). They were characterized by a marked cornified envelope, numerous heterogeneous vesicles and diffused large granules in the cytoplasm, and by a persistent nucleus in many cases. These characteristics are all the previously described hallmarks of transitional cells (Nix et al., 1965; Holbrook, 1994). A clear increase in the presence of these transitional cells was noted after Cl- amidine treatment (13 ± 1% in controls versus 88 ± 21%; p = 0.005; n = 4; Figure 2e). Accordingly, the number of corneocyte layers was shown to be significantly reduced (15.5 ± 4.3 in controls versus 12.2 ± 2.6; p = 0.05; n = 4; Figure 2f). Altogether, these results showed that treatment of RHEs with Cl-amidine during the last two days of culture disturbed the transformation of granular cells to corneocytes.
Figure 2. Increase of transitional cells and decreased number of corneocyte layers after Cl-amidine treatment.
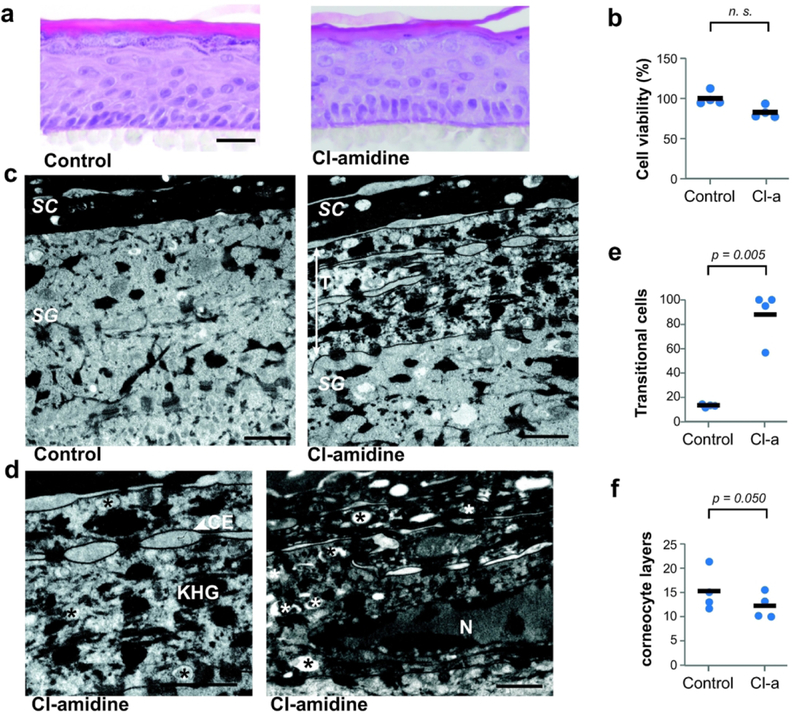
Reconstructed human epidermis (RHEs) were treated with 800 μM Cl-amidine (Cl-a) for 48 h (n = 4). (a) Treated and control RHEs were stained with hematoxylineosin. Scale bar = 25 μm. (b) The cell viability was evaluated using the MTT assay. (c and d) RHEs were analyzed by transmission electron microscopy. Scale bars = 1 μm. At the transition between stratum granulosum (SG) and stratum corneum (SC), transitional cells (T) are frequently observed in treated RHEs. They are characterized by a marked cornified envelope (CE), diffuse keratohyalin granules (KHG) and cytoplasmic vesicles (asterisks). Sometimes the nucleus (N) is observed. (e and f) The frequency of transitional cells and the number of corneocyte layers were quantified. Mean values for each RHE are represented by blue dots and means of all experiments by black dashes.
Alteration of keratinocyte differentiation in treated RHEs
We then analyzed late keratinocyte differentiation by RT-qPCR and western blotting (Figure 3a and 3b). At the mRNA level, the expression of filaggrin and transglutaminase-5 significantly decreased (0.5 fold, p = 0.0313 for FLG; 0.04 fold, p = 0.0313 for TG5; n = 3), the expression of transglutaminases 1 and 3 showed only a downward trend, and involucrin did not change. At the protein level, we observed a significant down-regulation of profilaggrin (0.79 ± 0.22 arbitrary units in controls versus 0.46 ± 0.10; p = 0.0030; n = 4) and filaggrin monomers (1.20 ± 0.36 arbitrary units in controls versus 0.84 ± 0.19; p = 0.0024; n = 4), but no changes in the expression of transglutaminases. We therefore looked at the transglutaminase activity using an in situ assay (Figure 3c). A significant decrease in the number of positive cell layers was noted (3.70 ± 0.96 in controls versus 1.65 ± 0.66;p < 0.0001; n = 2).
Figure 3. Effect of Cl-amidine treatments on keratinocyte differentiation.
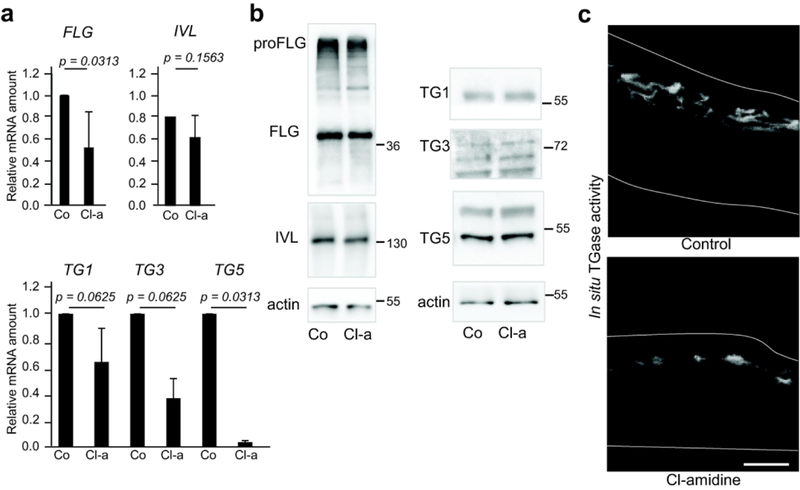
Reconstructed human epidermis (RHEs) were either untreated (Control or Co) or treated with 800 μM Cl-amidine (Cl-a) for 48 h (n= 2–4). RHEs were analyzed by (a) RT-qPCR, (b) western blotting and (c) an in situ transglutaminase activity assay. Molecular mass marker is indicated in kDa on the right. Scale bar = 25 μm. FLG, filaggrin; IVL, involucrin; TG, transglutaminase.
Effects of Cl-amidine on autophagy in RHEs
By electron microscopy analysis, accumulations of heterogeneous vesicles in the cytoplasm of the granular keratinocytes were often observed in RHEs treated with Cl-amidine for 48 hours, while this type of vesicles was sparse in the granular cells of control RHEs. Moreover, large vesicles were sometimes observed close to nuclei in the granular layer of treated RHEs (Figure 4a). These vesicles were suspected to be autophagy and nucleophagy related vesicles, which have previously been observed in the granular layer of the interfollicular normal epidermis (Akinduro et al., 2016). In addition, abnormal clustering of mitochondria was visible in the cytoplasm of granular keratinocytes (Figure 4b). This feature could also be related to autophagy, in particular macro-autophagy (Mizushima and Levine, 2010; Galluzi et al., 2017). Double membrane vesicular structures, also known as autophagosomes, are a hallmark of autophagy. Such transient and dynamic structures were not clearly observed after 48 hours of Cl-amidine treatment. However, double membrane vesicles with a size compatible with autophagosomes were observed (Figure 4c) in the cytoplasm of sub-granular and granular keratinocytes of RHEs treated at day 8 for shorter times (3, 6 and 24 hours). In order to back up the hypothesis of an amplified autophagy, RHEs were analyzed by western blotting with an antibody directed to microtubule-associated protein light chain 3 (LC3), the most widely used marker of autophagy. LC3 is known to be present in several forms in the cells, the conversion of the soluble LC3-I (~16 kDa) to the phosphatidylethanolamine bound LC3-II (~14 kDa) being associated with the formation of autophagosomes (Mizushima et al., 2010; Galluzi et al., 2017). In RHEs treated with Cl-amidine, both the amount of LC3-I + LC3-II and the amount of LC3-II alone increased (Figure 4d). Accordingly, an increase in the LC3 immunostaining intensity of treated RHEs was also observed (2.33 ± 1.27 arbitrary units in treated RHEs as compared to control values normalized to 1; p = 0.0020; n = 3), with some discrete punctate pattern (Figure 4e). The mRNA level of LC3B also increased (1.6 fold; p = 0.0313; n = 3; Figure 5a), suggesting a boost of autophagy. A similar tendency was shown for the mRNA amount of autophagy protein 5 (ATG5), a key protein for the formation of autophagic vesicles (Figure 5a).
Figure 4. Cl-amidine treatment alters autophagy in the granular cells.
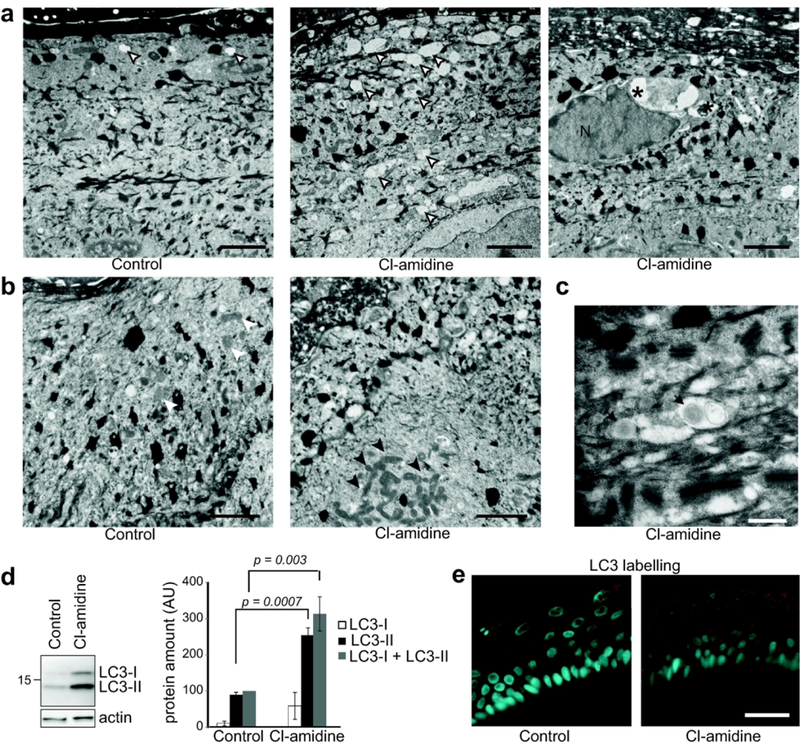
Reconstructed human epidermis (RHEs) samples were either untreated (Control) or treated with 800 μM Cl-amidine. (a-c) RHEs (n = 4) were analyzed by transmission electron microscopy. In the treated RHEs, accumulation of vesicles (a) and clustering of mitochondria (b; arrow heads) are observed in granular keratinocytes. Vesicles are indicated by open arrow heads when they are diffuse in the cytoplasm, and by asterisks when they are near to the nucleus (N). Double membrane vesicles (black arrows) are observed in the subgranular keratinocytes (c). Scale bars = 1 μm (in a and b), 500 nm (in c). (d) Total proteins were immunodetected with anti-LC3 and anti-actin antibodies. Immunodetected LC3-I, LC3-II and (LC3-I + LC3-II) were quantified, normalized to actin and expressed as percentage relatively to the control total LC3. (e) Representative images of endogenous LC3 indirect immunofluorescence detection (n = 3). Sometimes punctate labelling could be observed, as illustrated. Scale bar = 25 μm.
Figure 5. Induction of ATF4/SESN2 pathway after Cl-amidine treatment.
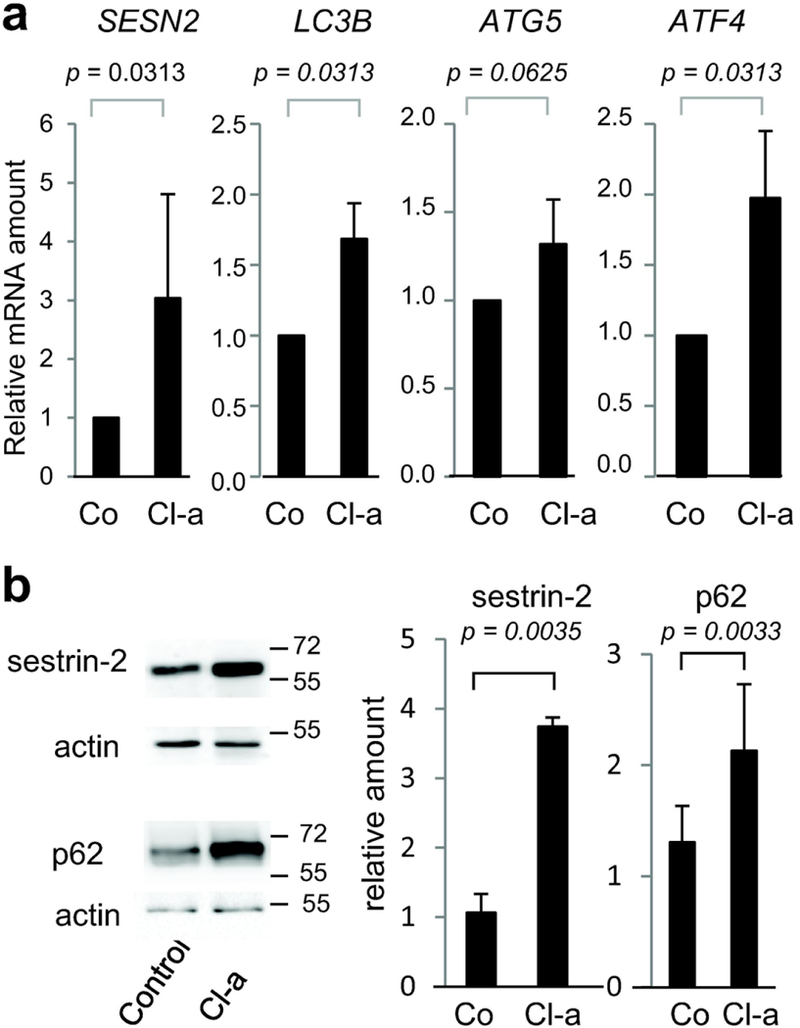
Reconstructed human epidermis (n = 4) were untreated (Co) or treated with 800 μM of Cl-amidine (Cl-a) for 48 h. (a) Expression of SESN2, MAP1LC3B, ATG5, and ATF4 genes was analyzed by RT-qPCR. (b) Nonidet P40 soluble proteins were immunodetected with anti-SESN2 and anti-actin antibodies (upper left). Total protein extracts were immunodetected with anti-p62/SQSTM1 and anti-actin antibodies (lower left). Molecular mass marker is indicated in kDa on the right. Immunodetected sestrin-2 and p62 were quantified, normalized to actin and expressed relatively to the control (right).
Since the PAD4 inhibitor YW3–56 has been previously found to regulate autophagy in the human osteosarcoma U2OS cell line through upregulation of the activation transcription factor 4 (ATF4), which in turn upregulates the expression of sestrin-2 (SESN2) (Wang et al., 2012, 2015), we tested whether a similar pathway could also be affected in Cl-amidine treated RHEs. The treatment significantly increased ATF4 and SESN2 expression at the mRNA level (Figure 5a). A clear increase in expression of SESN2 was also observed at the protein level (Figure 5b). Furthermore, the expression of p62 autophagy receptor (or sequestosome-1, SQSTM1), that functions as a bridge between polyubiquitinated cargos and autophagosomes (Puissant et al., 2012), was increased (2.13 ± 0.60 versus 1.31 ± 0.33 for controls; n = 4; p = 0.0033; Figure 5b).
Partial co-localization of PADs and LC3 in the upper reconstructed epidermis
To further consolidate the possibility that PADs are involved in the autophagic process during cornification, cross-sections of RHEs were analyzed by double-labelling indirect immunofluorescence with antibodies against PADs (either PAD1 or PAD3) and LC3. Observation with an SP8 confocal microscope revealed partial co-localization of endogenous LC3 with both PADs at the transition between the granular and the cornified layers (Supplementary Figures S3 and S4). Quantitative analysis using the Manders’ colocalization coefficients from 0.1471 to 0.2236 demonstrated this partial overlap (Supplementary Table S1).
DISCUSSION
Autophagy is a process conserved from yeast to humans, by which eukaryote cells degrade their own components within lysosome-related organelles, and recycle their molecular constituents (Galuzzi et al., 2017). It allows the cell to survive when energy or nutrient sources are deficient, but also to adapt to various stresses, e.g., autophagy clears abnormal macromolecules and organelles induced by redox conditions or senescence. Autophagy anomalies have been described in various human diseases including cancers, autoimmune diseases and neurodegenerative diseases, making autophagy a new therapeutic target (for reviews see Mizushima and Komatsu, 2011; Jiang and Mizushima 2014; Li et al., 2016; Dikic and Elazar, 2018). Early indications that autophagy plays a role during keratinocyte differentiation date back to the 1970s: numerous vesicles containing protein material have been observed in the cytoplasm of transitional cells in the epithelium of the rumen of cattle just before the disappearance of granular keratinocyte organelles and nuclei (Lavker and Maltosy, 1970). Then, using keratinocytes grown in monolayer, the induction of autophagy markers (LC3-II, ATG5/ATG12, etc.) has been demonstrated during differentiation or after UV stress (Aymard et al., 2011; Zhao et al., 2013). Experiments with organotypic cultures have reinforced the link between autophagy and keratinocyte differentiation (Chick et al., 2014). Autophagy has then been shown to be constitutively active in the upper mouse epidermis (Rossiter et al., 2013). More recently, Akinduro et al. (2016) have demonstrated that i) the establishment of the epidermal barrier during fetal mouse development coincides with induction of autophagy markers and ii) nucleophagy (a selective autophagy directed against nuclei) is constitutively active in the granular keratinocytes of mouse and human skin. Inversely, the keratinocytes of mice deficient for autophagy (Atg7−/−) show a delay in differentiation (Yoshihara et al., 2015), and autophagy seemed required for keratinocyte differentiation in organotypic human skin (Monteleon et al., 2018). Furthermore, the AKT1/mTORC1/mTORC2 signaling loop involved in autophagy regulation also appear to be involved in skin barrier maintenance through control of filaggrin expression/processing (Naeem et al., 2017).
PADs have already been associated with autophagy in other cell types. For example, inhibition of PAD4-mediated deimination regulates autophagic flux in cancer cells (Wang et al., 2012, 2015); the induction of autophagy in monocytes and fibroblasts with rapamycin leads to PAD4 activation and to deimination of proteins (Sorice et al., 2016); PADs are activated during neutrophil extracellular trap production and the associated autophagy (Valesini et al., 2015).
In this report, we demonstrated that treatment of RHEs with Cl-amidine pan-PAD inhibitor impedes cornification as shown by a reduced number of corneocyte layers and an increase in transitional cell number. Accumulation of cytosolic heterogeneous vesicles (some being close to the nucleus), presence of double membrane vesicles and clustering of mitochondria, associated to the increased immunodetected amount of LC3, sestrin-2 and p62 demonstrated clear perturbations of autophagy. Finally, the partial co-localization of PADs with LC3 indicates a link between deimination and constitutive autophagy in granular keratinocytes. These data confirm the involvement of autophagy in the last step of keratinocyte terminal differentiation and strongly suggests that deimination is necessary for the control of this catabolic mechanism. However, it is possible that cornification is reduced by Cl-amidine independently of its effect on autophagy, through down-regulation of filaggrin for example. Indeed, absence of filaggrin is deleterious for keratinocyte differentiation (Pendaries et al, 2014).
Reduction of cornification in RHEs with Cl-amidine also allowed us to clearly observe transitional cells, a cellular state barely detectable in normal human epidermis, and features characteristic of the cornification-related autophagic process. Correspondingly, the deimination of proteins in the stratum corneum is diminished in the epidermis of psoriasis patients (Ishida-Yamamoto et al., 2000), where the expression or location of most of the autophagy markers is deregulated (Akinduro et al., 2016). Enhanced number of transitional cells and accumulation of vesicles into the granular keratinocytes have already been reported in the epidermis of some lamellar ichthyosis patients (Kolde et al., 1985; Haftek et al., 1996). It seems therefore essential to decipher the dynamic autophagic process in skin physiological and pathological conditions. Additional in vivo studies are necessary to detail the effect of PADs, and know whether PAD modifiers could be relevant to skin pathology. Cl-amidine has already been shown to be effective to treat mice with models of human diseases, e.g. colitis (Chumanevich et al., 2011) and collagen induced arthritis (Willis et al, 2011).
An alternative, but not mutually exclusive, interpretation of our data is that autophagy increased in keratinocytes as a response to a cellular stress. However, to the best of our knowledge, Cl-amidine cytotoxicity for normal cells has not been reported in the literature, and we have shown here that Cl-amidine treatment of RHEs neither changed keratinocyte viability nor induced caspase-3 activation. In addition, we did not observed any ultrastructural changes in the basal and first suprabasal cells of treated RHEs. We nevertheless could not exclude the involvement of an aggrephagy process. Indeed, autophagy is involved in response to endoplasmic reticulum stress due to unfolded protein accumulation (review in Pitale et al., 2017 and Galluzzi et al., 2017). Since Cl-amidine is an irreversible inhibitor that makes a covalent bound in the active site of PADs, elimination of the PAD-inhibitor complex (misfolded or aggregated) might be required, through p62 involvement (Puissant et al., 2012).
In conclusion, in human epidermis, deimination and PADs are now clearly related to the keratinocyte differentiation and to the autophagic process occurring at the transitional switch from granular to cornified layers.
MATERIALS AND METHODS
Chemicals and antibodies
Cl-amidine was synthetized as described previously (Knuckley et al., 2010) and stored at −80°C until used. The antibodies used in this study have been previously described (Cau et al., 2017; Nachat et al., 2005; Guerrin et al., 2003) or are described in the Supplementary Table S2.
Production of RHEs
Primary normal human keratinocytes were isolated from abdominal skin samples, from five females (24–48 years old) without any history of skin diseases undergoing plastic surgery, obtained from Genoskin (Toulouse, France), following written informed consent of the donors and as agreed by the French Ministry of Research (#AC-2017–2897). Five keratinocyte banks were thus constituted. RHEs were produced in a humidified atmosphere with 5% CO2 at 37°C, as previously described (Frankart et al., 2012; Pendaries et al., 2014).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
RT-qPCR was performed as previously described (Méchin et al., 2010; Cau et al., 2017). The primers used have been described previously (Méchin et al, 2010) or are listed in the Supplementary Table S3.
Transmission electron microscopy analysis
RHEs were processed as described previously (Reynier et al., 2016) and observed with an HT7700 electron microscope (Hitachi, Tokyo, Japan). Quantifications were performed for RHEs produced from 4 different donors (at least 1 RHE per donor). The numbers of corneocyte layers were quantified on 3 independent areas for each epidermis (2500x magnification images). The frequency of transitional cells was estimated using an average of 30 pictures covering the area of transition between the stratum granulosum and the stratum corneum (2500x magnification images) per RHE and corresponds to the number of images presenting transitional cells in relation to the total number of analyzed pictures. Autophagosomes were searched on 10,000x magnification images obtained from RHEs (controls and treated for 3, 6 8 and 24 hours) produced from 2 donors.
In situ transglutaminase activity
In situ transglutaminase activity assays were performed as previously described (Cau et al., 2017) on cryosections of untreated and treated RHEs. The number of cell layers that depicted transglutaminase activity was evaluated in duplicate on 6 different areas of each of 10 images acquired per condition.
SP8 confocal microscopy and co-localization analysis
Sections of fixed RHEs were incubated with rabbit antibodies specific for either PAD1 or PAD3 and with the monoclonal antibody anti-LC3 (Supplementary Table S3). After incubation with a mix of the corresponding secondary antibodies, slides were mounted in Mowiol. Consecutive images were captured using a 63x objective with a Leica SP8 confocal microscope (Leica Microsystems, Nanterre, France) using Acquisition of Z-stacks mode with a depth of 50 nm (voxel of 50 nm3). To quantify co-localization, Manders coefficients, that are proportional to the amount of fluorescence of the colocalizing voxels in each color channel, were quantified using Imaris software (Bitplane, Zurich, Switzerland) on a single selected focal plan image. To point out the co-localizations, RGB profilers (comparison of the emission spectra of the fluorochromes along a line) were produced from the same images, using the Fiji plugin of ImageJ package (Schindelin et al., 2012).
Statistical analysis
Data is presented as mean ± standard deviation. Statistical differences were determined with Student’s t-tests when normality has been demonstrated using the Shapiro-Wilk normality test, or when not, with the non-parametric Wilcoxon-test. Differences were considered significant when the p-value (p) was less than 0.05.
Supplementary Material
ACKNOWLEDGEMENTS
We are indebted to Dr Pascal Descargues (Genoskin, Toulouse, France) for the human skin samples, Prof. Yves Poumay (University of Namur, Belgium) and Dr Valérie Pendaries-Rahoul (UDEAR) for their useful advice on RHE production, and to all members of the electron microscopy (Toulouse University), histopathology and cellular imaging (Toulouse Rio Imagerie, INSERM U1043) facilities. We thank Candide Alioli, Carole Pons and Géraldine Gasc for their helpful assistance. This work was supported in part by NIH grant R35 GM118112 (PRT) and by grants from CNRS, Toulouse University, and INSERM. We also thank the funding by the French Societies for Dermatology (SFD) and for Dermatological Research (SRD). Laura Cau was supported by the French Ministry of Research and Technology, and gratefully acknowledges the SILAB–Jean PAUFIQUE Corporate Foundation for its support. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Abbreviations:
- LC3
microtubule-associated protein light chain 3
- PAD
peptidylarginine deiminase
- RHE
reconstructed human epidermis
- SFTP
S100-fused type protein
Footnotes
DATA AVAILABILITY
All data are available on request to the corresponding author.
CONFLICT OF INTEREST
PRT declares the following competing financial interest(s): he was a founder, consultant, and chair of the scientific advisory board of Padlock Therapeutics which was acquired by Bristol Myers Squibb in 2016 and is entitled to payments if certain milestones are met. He is a consultant for Celgene and Disarm Therapeutics. The other authors state no conflict of interest.
REFERENCES
- Akinduro O, Sully K, Patel A, Robinson DJ, Chikh A, McPhail G, et al. Constitutive Autophagy and Nucleophagy during Epidermal Differentiation. J Invest Dermatol 2016; 136:1460–70. [DOI] [PubMed] [Google Scholar]
- Assohou-Luty C, Raijmakers R, Benckhuijsen WE, Stammen-Vogelzangs J, de Ru A, van Veelen PA, et al. The human peptidylarginine deiminases type 2 and type 4 have distinct substrate specificities. Biochim Biophys Acta BBA - Proteins Proteomics 2014; 1844:829–36. [DOI] [PubMed] [Google Scholar]
- Aymard E, Barruche V, Naves T, Bordes S, Closs B, Verdier M, et al. Autophagy in human keratinocytes: an early step of the differentiation? Exp Dermatol 2011; 20:263–8. [DOI] [PubMed] [Google Scholar]
- Baka Z, György B, Geher P, Buzas EI, Falus A, Nagy G. Citrullination under physiological and pathological conditions. Joint Bone Spine 2012; 79:431–6. [DOI] [PubMed] [Google Scholar]
- Cau L, Pendaries V, Lhuillier E, Thompson P, Serre G, Takahara H, et al. Lowering relative humidity level increases epidermal protein deimination and drives human filaggrin breakdown. J Dermatol Sci 2017; 86:106–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cau L, Mechin M-C, Simon M. Peptidylarginine deiminases and deiminated proteins at the epidermal barrier. Exp Dermatol 2018; in press. [DOI] [PubMed] [Google Scholar]
- Chavanas S, Méchin M-C, Takahara H, Kawada A, Nachat R, Serre G, et al. Comparative analysis of the mouse and human peptidylarginine deiminase gene clusters reveals highly conserved non-coding segments and a new human gene, PADI6. Gene 2004; 330:19–27. [DOI] [PubMed] [Google Scholar]
- Chick A, Sanzà P, Raimondi C, Akinduro O, Warnes G, Chiorino G, et al. iASPP is a novel autophagy inhibitor in keratinocytes. J Cell Sci 2014; 127:3079–93. [DOI] [PubMed] [Google Scholar]
- Christophorou MA, Castelo-Branco G, Halley-Stott RP, Oliveira CS, Loos R, Radzisheuskaya A, et al. Citrullination regulates pluripotency and histone H1 binding to chromatin. Nature 2014; 507:104–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chumanevich AA, Causey CP, Knuckley BA, Jones JE, Poudyal D, Chumanevich AP, et al. Suppression of colitis in mice by Cl-amidine: a novel peptidylarginine deiminase inhibitor. Am J Physiol Gastrointest Liver Physiol 2011; 300: G929–G938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coudane F, Mechin M-C, Huchenq A, Henry J, Nachat R, Ishigami A, et al. Deimination and expression of peptidylarginine deiminases during cutaneous wound healing in mice. Eur J Dermatol 2011; 21:376–84. [DOI] [PubMed] [Google Scholar]
- Dikic I, Elazar Z. Mechanism and medical implications of mammalian autophagy. Nature Rev Mol Cell Biol 2018; 19:349–364. [DOI] [PubMed] [Google Scholar]
- Frankart A, Malaisse J, De Vuyst E, Minner F, de Rouvroit CL, Poumay Y. Epidermal morphogenesis during progressive in vitro 3D reconstruction at the air-liquid interface. Exp Dermatol 2012; 21:871–5. [DOI] [PubMed] [Google Scholar]
- Galluzzi L, Baehrecke EH, Ballabio A, Boya P, Bravo-San Pedro JM, Cecconi F et al. Molecular definitions of autophagy and related processes. EMBO J 2017; 36:1811–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrin M, Ishigami A, Mechin M-C, Nachat R, Valmary S, Sebbag M, et al. cDNA cloning, gene organization and expression analysis of human peptidylarginine deiminase type I. Biochem J 2003; 370:167–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haftek M, Cambazard F, Dhouailly D, Réano A, Simon M, Lachaux A et al. A longitudinal study of a harlequin infant presenting clinically as non-bullous congenital ichthyosiform erythroderma. Br J Dermatol 1996; 135:448–53. [PubMed] [Google Scholar]
- Harding CR, Scott IR Histidine-rich proteins (filaggrins): Structural and functional heterogeneity during epidermal differentiation. J Mol Biol 1983; 170:651–73. [DOI] [PubMed] [Google Scholar]
- Holbrook KA. Ultrastructure of the epidermis In Leigh IM, Lane EB, Watt FM, Ed, The Keratinocyte Handbook, Cambridge University Press; 1994; pp:3–39. [Google Scholar]
- Hsu C-Y, Henry J, Raymond A-A, Mechin M-C, Pendaries V, Nassar D, et al. Deimination of Human Filaggrin-2 Promotes Its Proteolysis by Calpain 1. J Biol Chem 2011; 286:23222–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu C-Y, Gasc G, Raymond A-A, Burlet-Schiltz O, Takahara H, Serre G, et al. Deimination of Human Hornerin Enhances its Processing by Calpain-1 and its Cross-Linking by Transglutaminases. J Invest Dermatol 2017; 137:422–9. [DOI] [PubMed] [Google Scholar]
- Ishida-Yamamoto A, Senshu T, Takahashi H, Akiyama K, Nomura K, Iizuka H. Decreased deiminated keratin K1 in psoriatic hyperproliferative epidermis. J Invest Dermatol 2000; 114:701–5. [DOI] [PubMed] [Google Scholar]
- Ishigami A, Ohsawa T, Asaga H, Akiyama K, Kuramoto M, Maruyama N. Human peptidylarginine deiminase type II: molecular cloning, gene organization, and expression in human skin. Arch Biochem Biophys 2002; 407:25–31. [DOI] [PubMed] [Google Scholar]
- Jiang P, Mizushima N. Autophagy and human diseases. Cell Res 2014; 24:69–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JE, Slack JL, Fang P, Zhang X, Subramanian V, Causey CP, et al. Synthesis and screening of a haloacetamidine containing library to identifiy PAD4 selective inhibitors. ACS Chem Biol 2012; 7:160–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamata Y, Taniguchi A, Yamamoto M, Nomura J, Ishihara K, Takahara H, et al. Neutral Cysteine Protease Bleomycin Hydrolase Is Essential for the Breakdown of Deiminated Filaggrin into Amino Acids. J Biol Chem 2009; 284:12829–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knuckley B, Causey CP, Jones JE, Bhatia M, Dreyton CJ, Osborne TC, et al. Substrate Specificity and Kinetic Studies of PADs 1, 3, and 4 Identify Potent and Selective Inhibitors of Protein Arginine Deiminase 3. Biochem (Mosc) 2010; 49:4852–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolde G, Happle R, Traupe H. Autosomal-Dominant Lamellar Ichthyosis: Ultrastructural Characteristics of a New Type of Congenital Ichthyosis. Arch Dermatol res 1985; 278:1–5. [DOI] [PubMed] [Google Scholar]
- Lavker RM, Matoltsy AG Formation of horny cells: the fate of cell organelles and differentiation products in ruminal epithelium. J Cell Biol 1970; 44:501–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Lamer M, Pellerin L, Reynier M, Cau L, Pendaries V, Leprince C, et al. Defects of corneocyte structural proteins and epidermal barrier in atopic dermatitis. Biol Chem 2015; 396:1163–79. [DOI] [PubMed] [Google Scholar]
- Li L, Chen X, Gu H. The signaling involving in autophagy machinery in keratinocytes and therapeutic approaches for skin diseases. Oncotarget 2016; 7:50682–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonsdale-Eccles JD, Teller DC, Dale BA. Characterization of a phosphorylated form of the intermediate filament-aggregating protein filaggrin. Biochemistry (Mosc) 1982; 21: 5940–8. [DOI] [PubMed] [Google Scholar]
- Méchin M-C, Enji M, Nachat R, Chavanas S, Charveron M, Ishida-Yamamoto A, et al. , The peptidylarginine deiminases expressed in human epidermis differ in their substrate specificities and subcellular locations. Cell Mol Life Sci 2005; 62:1984–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méchin M-C, Sebbag M, Arnaud J, Nachat R, Foulquier C, Adoue V, et al. Update on peptidylarginine deiminases and deimination in skin physiology and severe human diseases. Int J Cosmet Sci 2007; 29:147–68. [DOI] [PubMed] [Google Scholar]
- Méchin M-C, Coudane F, Adoue V, Arnaud J, Duplan Charveron H, et al. Deimination is regulated at multiple levels including auto-deimination of peptidylarginine deiminases. Cell Mol Life Sci 2010; 67:1491–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat Cell Biol 2010; 12:823–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Levine B. Methods in Mammalian Autophagy Research. Cell 2010; 140:313–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell 2011; 147:728–41. [DOI] [PubMed] [Google Scholar]
- Moelants EAV, Mortier A, Van Damme J, Proost P, Loos T. Peptidylarginine deiminases: physiological function, interaction with chemokines and role in pathology. Drug Discov Today Technol 2012; 9: e261–80. [DOI] [PubMed] [Google Scholar]
- Monteleon CL, Agnihotri T, Dahal A, Liu M, Rebecca VW, Beatty GL, et al. Lysosomes Support the Degradation, Signaling, and Mitochondrial Metabolism Necessary for Human Epidermal Differentiation. J Invest Dermatol 2018; 138:1945–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachat R, Méchin M-C, Takahara H, Chavanas S, Charveron M, Serre G, et al. Peptidylarginine Deiminase Isoforms 1–3 Are Expressed in the Epidermis and Involved in the Deimination of K1 and Filaggrin. J Invest Dermatol 2005; 124:384–93. [DOI] [PubMed] [Google Scholar]
- Naeem AS, Tommasi C, Cole C, Brown SJ, Zhu Y, Way B, et al. A mechanistic target of rapamycin complex ½ (mTORC1)/V-Akt murine thymoma viral oncogene homolog 1 (AKT1)/cathepsin H axis controls filaggrin expression and processing in skin, a novel mechanism for skin barrier disruption in patients with atopic dermatitis. J Allergy Clin Immunol 2017; 139:1228–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nix TE, Nordquist RE, Everett MA. Ultrastructural changes induced by ultraviolet light in human epidermis: Granular and transitional cell layers. J Ultrastruct Res 1965; 12: 547–73. [DOI] [PubMed] [Google Scholar]
- Pendaries V, Malaisse J, Pellerin L, Le Lamer M, Nachat R, Kezic S, et al. Knockdown of Filaggrin in a Three-Dimensional Reconstructed Human Epidermis Impairs Keratinocyte Differentiation. J Invest Dermatol 2014; 134:2938–46. [DOI] [PubMed] [Google Scholar]
- Pitale PM, Gorbatyuk O, Gorbatyuk M. Neurodegeneration: Keeping ATF4 on a Tight Leash. Front Cell Neurosci 2017;11:410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puissant A, Fenouille N, Auberger P, When autophagy meets cancer through p62/SQSTM1 Am J Cancer Res 2012; 2:397–413. [PMC free article] [PubMed] [Google Scholar]
- Reynier M, Allart S, Gaspard E, Moga A, Goudounéche D, Serre G, et al. Rab11a Is Essential for Lamellar Body Biogenesis in the Human Epidermis. J Invest Dermatol 2016; 136:1199–209. [DOI] [PubMed] [Google Scholar]
- Rossiter H, König U, Barresi C, Buchberger M, Ghannadan M, Zhang CF, et al. Epidermal keratinocytes form a functional skin barrier in the absence of Atg7 dependent autophagy. J Dermatol Sci 2013; 71:67–75. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods 2012; 9:676–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senshu T, Akiyama K, Kan S, Asaga H, Ishigami A, Manabe M. Detection of Deiminated Proteins in Rat Skin: Probing with a Monospecific Antibody After Modification of Citrulline Residues. J Invest Dermatol 1995; 105:163–9. [DOI] [PubMed] [Google Scholar]
- Senshu T, Kan S, Ogawa H, Manabe M, Asaga H. Preferential Deimination of Keratin K1 and Filaggrin during the Terminal Differentiation of Human Epidermis. Biochem Biophys Res Commun 1996; 225:712–9. [DOI] [PubMed] [Google Scholar]
- Sorice M, Iannuccelli C, Manganelli V, Capozzi A, Alessandri C, Lococo E, et al. Autophagy generates citrullinated peptides in human synoviocytes: a possible trigger for anti-citrullinated peptide antibodies. Rheumatology (Oxford) 2016; 55:1374–85. [DOI] [PubMed] [Google Scholar]
- Tarcsa E, Marekov LN, Mei G, Melino G, Lee S-C, Steinert PM. Protein Unfolding by Peptidylarginine Deiminase SUBSTRATE SPECIFICITY AND STRUCTURAL RELATIONSHIPS OF THE NATURAL SUBSTRATES TRICHOHYALIN AND FILAGGRIN. J Biol Chem 1996; 271:30709–16. [DOI] [PubMed] [Google Scholar]
- Valesini G, Gerardi MC, Iannuccelli C, Pacucci VA, Pendolino M, Shoenfeld Y. Citrullination and autoimmunity. Autoimmun Rev 2015; 14:490–7. [DOI] [PubMed] [Google Scholar]
- Wang Y, Li P, Wang S, Hu J, Chen XA, Wu J, et al. Anticancer peptidylarginine deiminase inhibitors regulate autophagy flux and the mammalian target of rapamycin complex 1 activity. J Biol Chem 2012; 287:25941–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Wang Y. Peptidylarginine deiminases in citrullination, gene regulation, health and pathogenesis. Biochim Biophys Acta BBA - Gene Regul Mech 2013; 1829:1126–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Chen XA, Hu J, Jiang JK, Li Y, Chan-Salis KY, et al. ATF4 Gene Network Mediates Cellular Response to the Anticancer PAD Inhibitor YW3–56 in Triple-Negative Breast Cancer Cells. Mol Cancer Ther 2015; 14:877–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis VC, Gizinski AM, Banda NK, Causey CP, Knuckley B, Cordova KN, et al. N-α-benzoyl-N5-(2-chloro-1-iminoethyl)-L-ornithine amide, a protein arginine deiminase inhibitor, reduces the severity of murine collagen-induced arthritis. J Immunol 2011; 186:4396–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winget JM, Finlay D, Mills KJ, Huggins T, Bascom C, Isfort RJ, et al. Quantitative proteomic analysis of stratum corneum dysfunction in adult chronic atopic dermatitis. J Invest Dermatol 2016; 136:1732–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihara N, Ueno T, Takagi A, Oliva Trejo JA, Haruna K, Suga Y, et al. The significant role of autophagy in the granular layer in normal skin differentiation and hair growth. Arch Dermatol Res 2015; 307:159–69. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Zhang CF, Rossiter H, Eckhart L, König U, Karner S, et al. Autophagy is induced by UVA and promotes removal of oxidized phospholipids and protein aggregates in epidermal keratinocytes. J Invest Dermatol 2013; 133:1629–37. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


