To the editor:
Homeodomain-containing proteins are broadly involved in regulating development and cell fate decisions (Bürglin et al. 2016). The homeodomain-only protein (known as HOP or HOPX) is unique within this family of proteins due to its inability to bind DNA and is expressed abundantly in differentiated layers of the epidermis (Yang et al. 2010; Obarzanek-Fojt et al. 2011), suggesting a potential role for HOPX in epidermal differentiation. HOPX governs a balance between proliferation and differentiation in the heart (Chen et al. 2002), lungs (Yin et al. 2006), and muscle (Kee et al. 2007), but its role in epidermal homeostasis is controversial (Mariotto et al. 2016). In one study, experimental HOPX depletion in keratinocytes inhibited expression of filaggrin and transglutaminase-1, suggesting that it is required to promote differentiation (Obarzanek-Fojt et al. 2011). By contrast, a second study found that HOPX knockdown resulted in loricrin and involucrin upregulation, whereas its overexpression in immortalized HaCaT keratinocytes repressed loricrin and involucrin (Yang et al. 2010). This conversely suggested that HOPX inhibits differentiation.
The uncertainty to HOPX function has important implications because it has been identified as a key genetic determinant in epidermal differentiation (Lopez-Pajares et al. 2015) and psoriasis (Li et al. 2014). In this context, we sought to better define the role of HOPX in keratinocyte differentiation. We first aimed to place HOPX within the network of regulators that control epidermal development. To identify candidate regulatory enhancers near HOPX, we examined ENCODE data for histone 3, lysine 27 acetylation (H3K27Ac) and DNase-I hypersensitivity sites in neonatal human epidermal keratinocytes (DHS NHEK). A strong peak ~6 kb downstream of the 3’ end of HOPX showed coincident H3K27Ac and DHS enrichment (Figure 1a). We also examined chromatin immunoprecipitation-seq (ChIP-seq) data with a focus on transcription factors active in skin. This identified a binding site for the transcription factor ZNF750 overlapping with the H3K27Ac/DHS peak. ZNF750 is a p63 target that promotes differentiation in human epidermis (Sen et al. 2012).
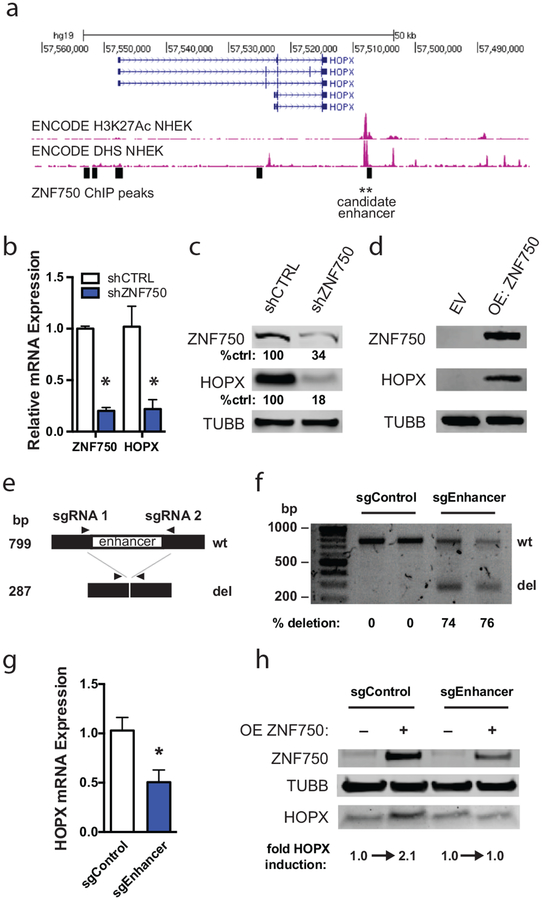
Figure 1. HOPX is induced by ZNF750 binding at a downstream enhancer.
(A) Chromatin immunoprecipitation-sequencing tracks around the HOPX genomic locus. ZNF750 binding sites are represented as black rectangles. Histone 3, lysine 27 acetylation = H3K27Ac. DNase hypersensitivity site = DHS. (B) Quantitative RT-PCR of control and ZNF750-depleted (shZNF750) keratinocytes differentiated for 6 days. n=2, error bars=SEM. * p<0.05. (C) Western blot of day 5 differentiated keratinocytes following short hairpin RNA-induced depletion of ZNF750 (shZNF750). Quantitation of band intensity performed using Li-Cor software. Band intensities were normalized to loading control and shCTRL band intensity. (D) Western blot of keratinocytes transduced to overexpress ZNF750 (OE ZNF750). Keratinocytes were infected with empty vector (EV) or ZNF750-HA. (E) Scheme of CRISPR/Cas9-mediated enhancer deletion and detection by PCR assay. (F) PCR assay to detect HOPX enhancer deletion. Cas9 was introduced into keratinocytes along with single guide RNAs flanking the candidate enhancer (sgEnhancer) or scramble control guides (sgControl). Quantitation of band intensity was performed using Bio-Rad Image Lab™ Software analysis. Band intensities were normalized to amplicon length to determine relative deletion efficiency. (G) Quantitative RT-PCR of HOPX expression in control and enhancer-deleted pooled differentiated keratinocytes. n=3, error bars=SEM. * p<0.05. (H) Western blot of differentiating sgControl and sgEnhancer keratinocytes transduced to overexpress ZNF750. Quantification was performed using LiCor software.
To evaluate if ZNF750 controls HOPX expression through binding of this enhancer, we first depleted ZNF750 using short hairpin RNA (shZNF750) and evaluated HOPX expression (Figure 1b–c). We used primary neonatal keratinocytes isolated from discarded surgical specimens with written, informed patient consent, under approval of a UCSD Institutional Review Board protocol. HOPX upregulation was strongly inhibited by ZNF750 depletion, suggesting that ZNF750 is required for HOPX induction. We then overexpressed ZNF750 in keratinocytes grown in progenitor conditions. Overexpression of ZNF750 (OE) stimulated strong HOPX expression (Figure 1d). Viewed together, these experiments showed that ZNF750 is both necessary and sufficient for HOPX induction.
Next, we used CRISPR/Cas9-mediated genomic editing to delete the candidate enhancer to determine if it was required for ZNF750 to control HOPX expression. Two short guide RNAs (sgRNAs) were designed to flank the ZNF750 genomic binding site (denoted as sgEnhancer) and infected into Cas9-expressing keratinocytes. PCR of genomic DNA was used to assess targeted deletion (schematized in Figure 1e). With this approach, the enhancer was deleted in ~75% of pooled keratinocytes (Figure 1f). We attempted multiple times to isolate individual sgEnhancer clones but could not propagate primary clones long enough to perform downstream experiments. Therefore, we compared pools of sgEnhancer keratinocytes against those infected with control sgRNAs (sgControl). Assessing HOPX mRNA induction, we found that HOPX mRNA induction was inhibited by ~50% in sgEnhancer keratinocytes (Figure 1g), consistent with this enhancer having a role to induce HOPX expression. To further test whether ZNF750 binding was specifically involved with this enhancer element, we overexpressed ZNF750 in differentiating sgControl vs. sgEnhancer keratinocytes. ZNF750 overexpression induced HOPX protein levels in sgControl cells, but did not lead to higher expression in the sgEnhancer cells (Figure 1h). Together, these experiments show that HOPX is a downstream activation target of the transcription factor ZNF750, which acts, at least in part, through binding an enhancer downstream of HOPX.
These results indicated that HOPX lies within a genetic pathway known to activate keratinocyte differentiation. To further substantiate this result, we assessed the genome-wide transcriptional impact of depleting HOPX. Prior in vitro studies of HOPX used transfection of short interfering RNAs, which caused either incomplete and transient protein depletion (Obarzanek-Fojt et al. 2011) and/or were partially performed in immortalized HaCaT cells (Yang et al. 2010). To generate durable HOPX depletion in a primary cell and tissue context, we used stable short hairpin knockdown (shHOPX) in primary neonatal keratinocytes and regenerated organotypic epidermal tissue. Stable knockdown depleted HOPX mRNA by 98.4% +/− 0.1% (SEM, n=3) compared to the control (shCTRL), and reduced HOPX to undetectable levels on Western blot (Figure 2a).
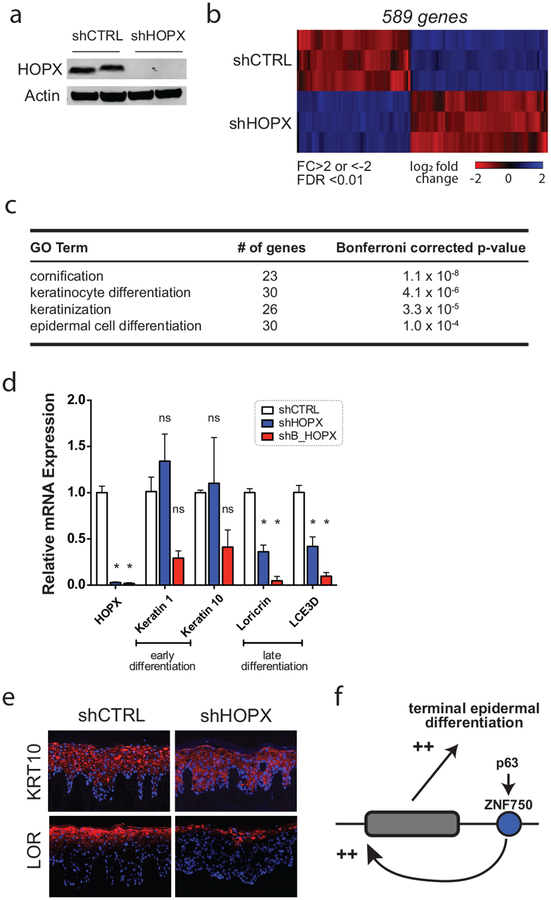
Figure 2. Depletion of HOPX inhibits late epidermal differentiation.
(A) Western blot following short hairpin RNA-induced depletion of HOPX (shHOPX). shCTRL = control. (B) RNA-sequencing of HOPX-depleted keratinocytes. Heatmap representing the differentially-expressed genes (DEGs) associated with HOPX depletion. DEG threshold was assigned as fold change (FC) >2 or <−2, calculated false discovery rate (FDR)<0.01. (C) Gene Ontology (GO) enrichment analysis of DEGs altered by HOPX depletion using PANTHER classification. (D) Quantitative RT-PCR of differentiation-associated transcripts with HOPX depletion using two independent short hairpin RNAs against HOPX. N=3, error bars=SEM. * p<0.05. ns = not significant. (E) Immunofluorescence staining of control and HOPX-depleted regenerated epithelial tissue. (F) Model of genetic p63-ZNF750-HOPX relationship in terminal keratinocyte differentiation.
We performed whole transcriptome sequencing of shCTRL and shHOPX keratinocytes differentiated for 6 days in vitro (data available at NCBI GEO #GSE125152). Using a threshold of 2-fold expression change and an ANOVA/false discovery rate <0.01, HOPX depletion resulted in 589 differentially expressed genes (DEGs; Figure 2b). Gene Ontology analysis using PANTHER classification (Mi et al. 2013) showed strong enrichment for genes involved with cornification and keratinocyte differentiation (Figure 2c). To confirm these results, we validated expression of key differentiation-related transcripts using quantitative RT-PCR and a second unrelated HOPX-targeting hairpin RNA (shB_HOPX; Figure 2d). HOPX depletion did not significantly affect early differentiation transcripts keratins 1 and 10, but repressed late epidermal differentiation genes loricrin and late cornified envelope 3D. To evaluate this phenotype in a three-dimensional tissue context, we regenerated human epidermal tissue with shCTRL and shHOPX keratinocytes. Concordant with qRT-PCR results, protein expression of keratin 10 (KRT10) was unchanged, whereas expression of the terminal differentiation protein, loricrin (LOR), was diminished with HOPX knockdown (Figure 2e). Taken together, these data confirm that HOPX is a positive regulator of epidermal late differentiation.
In summary, these results demonstrate that HOPX functions downstream of ZNF750, a well-defined transcription factor in the skin that is activated by p63 (Sen et al. 2012). In this context, we propose that HOPX functions within a p63-ZNF750-HOPX pathway to upregulate key proteins required for terminal epidermal differentiation (Figure 2f), providing clarity to previous studies showing conflicting results. Future studies will aim to decipher the molecular pathways and mechanisms operating downstream of HOPX that engage the late differentiation program.
Supplementary Material
Acknowledgements
Research reported in this publication was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health under award K08AR067853 to B.K.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
The authors declare no conflicts of interest.
Data Availability Statement
Datasets related to this article can be found at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE125152, hosted at NCBI Gene Expression Omnibus under accession #GSE125152.
References
- Bürglin TR and Affolter M. Homeodomain proteins: an update. Chromosoma. 2016;125:497–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Kook H, Milewski R, Gitler AD, Lu MM, Li J, et al. Hop is an unusual homeobox gene that modulates cardiac development. Cell. 2002;110::713–723. [DOI] [PubMed] [Google Scholar]
- Kee HJ, Kim J-R, Nam K-I, Park HY, Shin S, Kim JC, et al. Enhancer of polycomb1, a novel homeodomain only protein-binding partner, induces skeletal muscle differentiation. J Biol Chem. 2007; 282:7700–7709. [DOI] [PubMed] [Google Scholar]
- Li B, Tsoi LC, Swindell WR, Gudjonsson JE, Tejasvi T, Johnston A, et al. Transcriptome analysis of psoriasis in a large case-control sample: RNA-seq provides insights into disease mechanisms. J Invest Dermatol. 2014; 134:1828–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Pajares V, Qu K, Zhang J, Webster DE, Barajas BC, Siprashvili Z, et al. A LncRNA-MAF:MAFB Transcription Factor Network Regulates Epidermal Differentiation. Dev Cell. 2015; 32:693–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariotto A, Pavlova O, Park H-S, Huber M, Hohl D. HOPX: The Unusual Homeodomain-Containing Protein. J Invest Dermatol. 2016; 136:905–911. [DOI] [PubMed] [Google Scholar]
- Obarzanek-Fojt M, Favre B, Kypriotou M, Ryser S, Huber M, Hohl D. Homeodomain-only protein HOP is a novel modulator of late differentiation in keratinocytes. Eur J Cell Biol. 2011; 90:279–290. [DOI] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Casagrande JT, Thomas PD. Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc 2013; 8:1551–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen GL, Boxer LD, Webster DE, Bussat RT, Qu K, Zarnegar BJ, et al. ZNF750 Is a p63 Target Gene that Induces KLF4 to Drive Terminal Epidermal Differentiation. Dev Cell. 2012;22:669–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J-M, Sim SM, Kim H-Y, Park GT. Expression of the homeobox gene, HOPX, is modulated by cell differentiation in human keratinocytes and is involved in the expression of differentiation markers. Eur J Cell Biol. 2010; 89:537–546. [DOI] [PubMed] [Google Scholar]
- Yin Z, Gonzales L, Kolla V, Rath N, Zhang Y, Lu MM, et al. Hop functions downstream of Nkx2.1 and GATA6 to mediate HDAC-dependent negative regulation of pulmonary gene expression. Am J Physiol Lung Cell Mol Physiol. 2006; 291: L191–L199. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.