Abstract
Background:
MET amplification and MET exon 14 alterations (METex14) in lung cancers impart sensitivity to MET kinase inhibitors. Fluorescence in situ hybridization (FISH), next-generation sequencing (NGS), and IHC, have been used to evaluate MET dependency. Here, we determined the association of MET IHC with METex14 mutations and MET amplification.
Methods:
We collected data from a tri-institutional cohort from LCMC2 (Lung Cancer Mutation Consortium). All patients had metastatic lung adenocarcinomas and no prior targeted therapies. MET IHC positivity was defined by H-score ≥ 200 using SP44 antibody and MET amplification by copy number fold change ≥ 1.8x using NGS or MET/CEP7 ratio > 2.2 using FISH.
Results:
We tested tissue from 181 patients for MET IHC, MET amplification, and METex14 mutations. Overall, 71/181 (39%) were MET IHC positive, 3/181 (2%) were MET amplified, and 2/181 (1%) harbored MET exon 14 mutations. Of MET-amplified cases, 2 cases were FISH positive with MET/CEP7 of 3.1 and 3.3, 1 case was NGS positive with fold change 4.4x, and 1/3 of the cases were MET IHC positive. Of IHC positive cases, 1/71 (1%) were MET amplified and 2/71 (3%) were METex14 mutated. Of MET IHC negative cases, 2/110 (2%) were MET amplified.
Conclusions:
In this study, nearly all MET IHC positive cases are negative for MET amplification or METex14 mutations. MET IHC can also miss patients with MET amplification. The limited number of MET amplified cases in this cohort makes it challenging to demonstrate an association between MET IHC and MET amplification. Nevertheless, IHC appears to be an inefficient screen for these genomic changes. MET amplification or METex14 mutations can best be detected by FISH and a multiplex NGS panel.
Keywords: MET exon 14, MET amplified, Lung cancer, Next-generation Sequencing, FISH, immunohistochemistry
Introduction
The hepatocyte growth factor receptor (MET) has been shown to be an oncogenic driver in lung cancers. MET pathway activation, either by MET amplification or a splice site alteration in exon 14 (METex14), facilitates lung cancer growth, survival, and metastasis1, 2. In lung cancers, both MET mutation and amplification are primary oncogenic drivers. MET amplification is also a mechanism of acquired resistance to EGFR- and ALK-targeted therapies.
MET pathway activation by MET amplification occurs by constitutive signaling through protein expression and kinase activation. De novo MET amplification occurs in 1% to 5% of lung cancers, depending on the assay and positivity cut-point used2. A global consensus regarding the appropriate cut-off for MET amplification based on gene copy number has yet to be reached3. One classification scheme by Camidge et al. proposed various categories of MET/CEP7 ratio as follows: low: ≥ 1.8 to ≤ 2.2; intermediate: > 2.2 to < 5; or high: ≥ 5 (although a later classification changed the intermediate cut-point to > 2.2 to < 4 and high cut-point to ≥ 4) and has been applied in clinical settings when treating patients with MET inhibitors3.
METex14 mutations produce a skipping alteration that prevents the MET receptor from being degraded, resulting in increased MET activity. METex14 mutations occur in 3% to 4% of patients with lung adenocarcinomas based on studies employing hybrid capture NGS4. These mutations impart sensitivity to MET tyrosine kinase inhibitors (TKI), including cabozatinib, crizotinib, tepotinib, and capmatinib2, 5.
Some have suggested that MET IHC can serve as a potential predictive marker for MET kinase inhibitor activity. However, studies that used IHC for MET-targeted therapies have been unsuccessful thus far6, 7. Moreover, there is growing evidence that MET IHC may not be a good screening test for MET amplification or METex14 mutation in lung cancer8.
The Lung Cancer Mutation Consortium 2 (LCMC2) was a multi-institutional effort established in 2010 to investigate the frequency of oncogenic drivers in lung adenocarcinoma9. Using a tri-institutional cohort derived from the LCMC experience (University of Colorado, Dana Farber Cancer Institute, Memorial Sloan Kettering), we set out to determine the association of MET IHC testing with MET amplification or METex14 mutation status in patients with metastatic lung adenocarcinomas.
Methods
Patient recruitment, enrollment, and IRB approval
Data was collected from three of the institutions that participated in LCMC29. All sites obtained Institutional Review Board approval for this study. Patients undergoing further evaluation for the diagnosis or treatment of stage IV or recurrent lung adenocarcinomas were prospectively enrolled if they provided written informed consent, as previously described9. In addition, patients were eligible if they had no prior treatment with targeted therapy, a diagnosis of metastatic disease between May 2012 to January 2016, and adequate tissue for molecular analyses. All subjects enrolled were provided written informed consent. Epidemiologic and clinicopathologic data including age, sex, and cigarette smoking history were collected.
Pathology evaluation
Molecular testing and the diagnosis of lung adenocarcinoma was confirmed by pathologists at each institution. The diagnosis of lung adenocarcinoma was confirmed centrally with the pathology report and review of a hematoxylin- and eosin-stained histology slide or a scanned whole-slide image (Leica Biosystems Inc.). All testing was done in Clinical Laboratory Improvement Amendments (CLIA) laboratories.
IHC and FISH detection
MET amplification was determined by MET FISH (Roche/Ventana) and NGS. FISH assays were performed with laboratory-developed reagents as previously described10. Amplification by FISH was considered present when the MET/CEP7 ratio was > 2.2.
IHC for MET (clone SP44, Roche/Ventana) was independently validated at each site. MET IHC was defined as positive if the sample had an H-score ≥ 200, following a previously established method11. Pathologist training and interlaboratory proficiency testing were used for IHC scoring (Supplemental Methods).
Mutational analyses
Mutational analyses were performed using methods previously described9. Mutations included in these studies include AKT1, BRAF, EGFR, ERBB2, KRAS, MAP2K1, MET, NRAS, and PIK3CA. During the course of this study, many diagnostic laboratories converted from single gene testing to NGS methods. NGS technologies at each site are provided (Supplementary Table S2). They were independently validated for both wet-bench and bioinformatics components and were also centrally reviewed. MET exon 14 testing was performed at selected LCMC sites. MET amplification by NGS was considered to be amplified from NGS when copy number fold change (log2 ratio) is ≥ 1.8x assuming 50% tumor content. At least a 50X mean target coverage was needed for a sample to pass analysis.
Statistical Analysis
Mann-Whitney Test was used to compare categorical values. All reported p-values are for two-sided hypothesis tests conducted at the 0.05 level.
Results
One hundred eighty-one patients from three institutions had tissues tested for MET IHC, MET amplification, and METex14 mutation with FISH or NGS. The median age at diagnosis was 65 years, 57% (104/181) were women, and 71% (129/181) were current or former smokers (Table 1). The prevalence of MET amplification by FISH or NGS was 2% (3/181, 95% CI: 3.4 to 5.0%) (Figure 1). Two MET amplified cases were detected by FISH and 1 by NGS. MET IHC by H-score was negative in 2/3 of the patients (Table 2). MET IHC status was unchanged by MetMab scoring criteria. Two of these MET IHC negative patients also had a concurrent KRAS G12C mutation.
Table 1. Features of patients with metastatic lung adenocarcinomas with MET testing*.
The clinical characteristics of 181 patients with MET IHC and MET FISH or massively parallel sequencing testing are described.
| All patients (n=181) | |
|---|---|
| Age at Diagnosis of Metastatic Disease | |
| median | 64 years |
| range | (18–90 years) |
| Sex | |
| female | 104 (57%) |
| male | 77 (43%) |
| Smoking history | |
| never smoker | 50 (28%) |
| former smoker | 118 (65%) |
| current smoker | 11 (6%) |
| unknown | 2 (1%) |
| MET IHC by H-Score | 181 (100%) |
| Positive | 71 (39%, 95% Cl: 32% to 47%) |
| Negative | 110 (61%, 95% Cl: 54% to 68%) |
| MET FISH | 85 (47%) |
| Positive | 2 (1%, 95% Cl: 0% to 4.2%) |
| Negative | 83 (46%, 95% Cl: 39% to 53%) |
| MET NGS | 181 (100%) |
| METex14 mutation | 2 (1%, 95% Cl: 0% to 4.2%) |
| MET amplification | 1 (1%, 95% Cl: 0% to 3.4%) |
| No MET mutation | 178 (98%, 95% Cl: 95% to 100%) |
Percentages may not total 100 because of rounding.
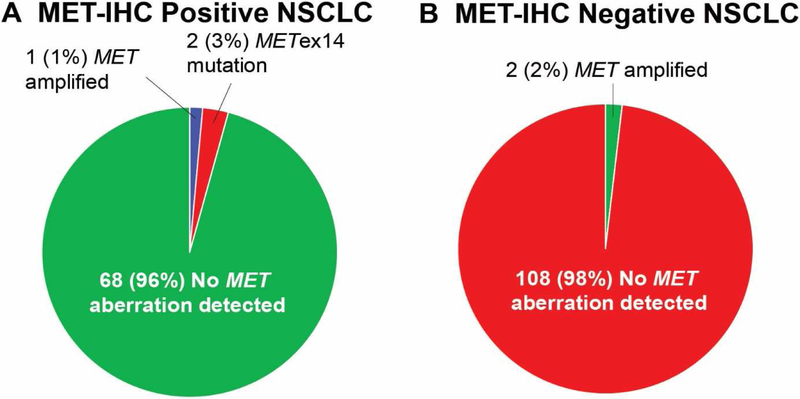
Figure 1. METex4 and MET amplification status in MET IHC positive and negative cases by H-score.
A) Of the MET IHC positive cases by H-score, MET amplification or METex14 was seen in 1% and 3% of cases, respectively. B) Of the MET IHC negative cases, only MET amplification was seen in 2% of cases.
Table 2. Patients with MET amplification or METex14 mutation.
Five patients with either MET amplification or METex14 mutation are identified. MET amplification was detected by FISH in 2/3 (67%) of the patients. Next-generation sequencing detected a concurrent KRAS G12C mutation in both. Amplification in case 3 was detected by next-generation sequencing. One out of three patients that were MET amplified were also MET IHC positive by H-score. METex14 was seen in two patients and both were MET IHC positive. These results were unchanged when using IHC status by MetMab scoring criteria.
| MET Amplification | MET mutation by NGS | Other Drivers | MET IHC by H-score | |
|---|---|---|---|---|
| Case 1 | Positive by FISH (MET/CEP7 3.1) | None | KRAS G12C | Negative |
| Case 2 | Positive by FISH (MET/CEP7 3.3) | None | KRAS G12C | Positive |
| Case 3 | MET amplified by NGS (Fold 4.4) | None | None | Negative |
| Case 4 | None | METex 14 | None | Positive |
| Case 5 | None | METex 14 | None | Positive |
METex14 was seen in 1% (2/181, 95% CI: 0% to 4.2%) (Table 1). The two patients with METex14 mutation were 73 and 83 years of age, female, and former smokers. MET IHC was positive in both of these cases (Table 2). Of note, neither case had concurrent MET amplification.
MET IHC was positive in 39% (71/181, 95% CI: 32 to 47%) (Table 1). Of the patients with MET IHC positive lung cancer, 1% (1/71) had MET amplification by FISH and 3% (2/71) had METex14 mutation by NGS. MET IHC was negative in 61% (110/181) of cases and, of these, 2% (2/110) were MET amplified (Figure 1B). Of the cases without amplification, MET IHC was positive in 39% (70/181). Two of these seventy MET IHC positive cases also had METex14 mutations.
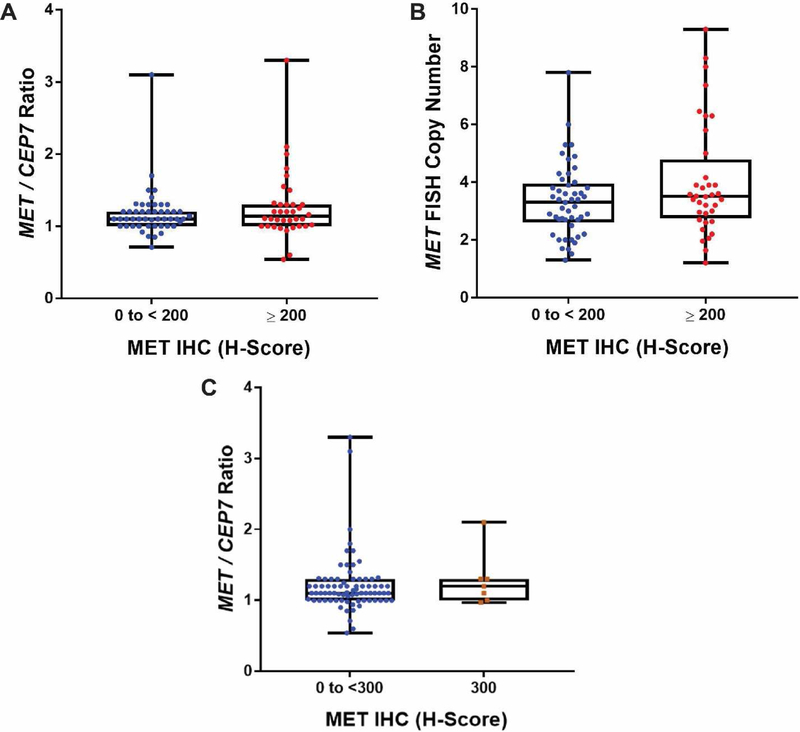
A total of 85 cases (47%) had both MET IHC and MET FISH performed. The median MET/CEP7 ratio of MET IHC negative cases (1.1, n = 49) was not different from that of MET IHC positive cases (1.14, n = 36; Mann-Whitney test, p = 0.57) (Figure 2). The median MET FISH copy number was also not different between MET IHC negative (3.3, n = 49) and that of positive cases (3.5, n = 36; Mann-Whitney test, p = 0.20) (Figure 2B). Using a higher cut-off to define IHC positive (H-score 300) did not select for MET amplified cases (Figure 2C).
Figure 2. Box plot representations of MET/CEP7 ratios and MET FISH copy numbers compared to MET IHC H-Score.
Boxes represent the interquartile range, which contains 50% of the values, whereas lines extend the entire range of values. A) Median MET/CEP7 ratios of MET H-scores from 0 to < 200 (1.1, n=49) and ≥ 200 (1.135, n=36) are not significantly different (Mann Whitney test: p=0.57). B) Median MET FISH copy number from 0 to < 200 (3.3, n=49) and ≥ 200 (3.5, n=36) are also not significantly different (Mann Whitney test: p=0.2). C) Median MET/CEP7 ratios continue to overlap when comparing MET H-scores from 0 to < 300 (1.1, n=78) and the max score (300) (1.2, n=7) (Mann Whitney test: p=0.58).
Discussion
Attempts to use MET IHC as a marker of MET dependency have largely been unsuccessful. Recently, MET IHC was shown to correlate poorly with MET/CEP7 ratio in all stages of sarcomatoid lung cancer studied8. In this tri-institutional cohort of patients with metastatic lung adenocarcinoma, more than 1/3 of cases were MET IHC positive, but only 2% were MET amplified. MET IHC also did not detect MET in 2 of the 3 MET amplified cases.
Two cases of METex14-mutated lung cancer were also MET IHC positive. Other studies have also shown that lung cancers with METex14 mutations are often IHC positive12–14. These studies also show that MET IHC is a poor screening strategy for METex14, considering how frequently MET IHC is positive in lung cancers9, 13. In this study, 96% of MET IHC positive cases had no detectable MET amplification or METex14 mutation. Finally, no clear association was observed between IHC and MET amplification by FISH in this study. The low number of METex14 mutations or MET amplification by NGS makes it difficult to determine an association with MET IHC. The sensitivity and specificity of H-score for evaluating for a MET genomic aberration (either MET exon14 mutation or MET amplification by FISH or NGS) in this series were both 0.6.
This study was limited by the low number of MET amplified and mutated cases. The prevalence of MET amplification (2%) in our cohort was similar to earlier reports. However, this prevalence was lower than that reported in LCMC2 as a whole. This discrepancy is likely due to the use of different cut-offs for MET amplification in post-hoc analyses between our cohort and that in LCMC29. Furthermore, differences in the prevalence of MET amplification between cohorts may be limited by the precision of current methodologies. Recently, it was shown that there was a surprising amount of intratumoral heterogeneity of MET copy number gain and amplification as determined by FISH15. In addition, NGS could potentially miss some cases of MET amplification that would otherwise be called by FISH. How well MET amplification by NGS correlates with MET amplification by FISH is not well-understood and needs to be further explored15.
The prevalence of METex14 (1%) is also lower than that reported in the literature, which may be related to poor coverage of relevant target regions in METex14 with earlier versions of the NGS panels used in this study. Issues with METex14 coverage with older NGS panels has been described and newer generation NGS panels, which were implemented as the study progressed, provide better coverage of these regions16. Although the low number of MET amplification and METex14 mutation in this study makes it difficult to draw a strong conclusion about the diagnostic accuracy of MET IHC, the large number of false-positive MET IHC cases in this cohort suggests that MET IHC is a poor screen for MET amplification and METex14 mutation.
Multiple trials have used MET IHC as a predictive marker for MET-directed therapies, such as onartuzumab, but have largely been unsuccessful6, 7. In contrast, ongoing studies with MET tyrosine kinase inhibitors have seen more success with using high MET copy numbers (gene copy number >5) and MET/CEP7 ratios as predictive markers1, 3. Coupling the results of these trials with growing literature showing that MET IHC inadequately selects for MET amplification or METex14 mutations strongly challenges its use as a screen for MET dependency. Multiplex next-generation sequencing panels in use today detect actionable targets (like EGFR, ALK, ROS1, and BRAF) and nearly always assess MET copy number and MET exon 14 mutations. We recommend that tissue should be prioritized for NGS and FISH over IHC to test for actionable MET mutations or amplification in lung adenocarcinomas.
Supplementary Material
Supplementary Figure S1. Box plot representations of MET FISH copy number compared to MET IHC by MetMab score. Boxes represent the interquartile range, which contains 50% of the values, whereas lines extend the entire range of values. Positive and negative IHC by MetMab is represented by red and blue dots, respectively. There is no significant difference in MET FISH copy number among MetMab scores using one-way ANOVA by ranks (p=0.41). The sensitivity and specificity of MetMab scoring for detecting a MET genetic abnormality is 0.6 and 0.4, respectively.
Acknowledgements:
This research was supported in part by the National Cancer Institute of the National Institutes of Health (T32 CA009207, P30 CA008748) and the Lung Cancer Research Foundation
Disclosures:
Dr. Guo reports grants from National Cancer Institute of the National Institutes of Health (T32 CA009207, P30 CA008748), grants from Lung Cancer Research Foundation, during the conduct of the study.
Dr. Aisner reports personal fees from Bayer Oncology, personal fees from Genentech, personal fees from AbbVie, personal fees from Bristol Myers Squibb, outside the submitted work.
Dr. Bunn reports grants from Lung Cancer Mutation Consortium, during the conduct of the study.
Dr. Johnson reports grants from Novartis, grants from Canon Medical Systems (previously Toshiba), outside the submitted work; In addition, Dr. Johnson has a patent EGFR Mutation Testing with royalties paid to Dr. Johnson.
Dr. Kwiatkowski reports other from AADi, other from Revolution Medicines, other from Genentech, personal fees from Novartis, personal fees from AstraZeneca, outside the submitted work.
Dr. Sholl reports personal fees from LOXO Oncology, personal fees from Foghorn Therapeutics, personal fees from AstraZeneca, personal fees from Bristol Myers Squibb, grants from Roche, outside the submitted work.
Dr. Drilon reports grants from National Cancer Institute of the National Institutes of Health (P30 CA008748), grants from Lung Cancer Research Foundation, during the conduct of the study; personal fees from Medscape, personal fees from OncLive, personal fees from PeerVoice, personal fees from Physicians Education Resources, personal fees from Tyra Biosciences, personal fees from Targeted Oncology, personal fees from MORE Health, personal fees from Research to Practice, personal fees from Foundation Medicine, personal fees from Peerview, personal fees from AstraZeneca, personal fees from Genentech/Roche, personal fees from Bayer, personal fees from Ignyta, personal fees from Loxo, personal fees from TP Therapeutics, personal fees from Pfizer, personal fees from Blueprint Medicines, personal fees from Takeda, personal fees from Helsinn Therapeutics, personal fees from BeiGene, personal fees from Hengrui Therapeutics, personal fees from Exelixis, personal fees from Bayer, personal fees from Wolters Kluwer, grants from PharmaMar, outside the submitted work.
Dr. Kris reports grants from National Cancer Institute of the National Institutes of Health (T32 CA009207, P30 CA008748), grants from Lung Cancer Research Foundation, during the conduct of the study; personal fees and non-financial support from AstraZeneca, personal fees and non-financial support from Pfizer, personal fees and non-financial support from Regeneron, personal fees from WebMD, personal fees from OncLive, personal fees from Physicians Education Resources, personal fees from Prime Oncology, personal fees from Intellisphere, personal fees from Creative Educational Concepts, personal fees from Peerview, personal fees from i3 Health, personal fees from Paradigm Medical Communications, personal fees from AXIS, personal fees from Carvive Systems, personal fees from Research to Practice, grants and non-financial support from Genentech/Roche, grants from PUMA Biotechnology, grants from IBM, outside the submitted work.
Other authors have nothing to disclose.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wu YL, Zhang L, Kim DW, et al. Phase Ib/II Study of Capmatinib (INC280) Plus Gefitinib After Failure of Epidermal Growth Factor Receptor (EGFR) Inhibitor Therapy in Patients With EGFR-Mutated, MET Factor-Dysregulated Non-Small-Cell Lung Cancer. J Clin Oncol 2018:JCO2018777326. [DOI] [PubMed] [Google Scholar]
- 2.Drilon A, Cappuzzo F, Ou SI, et al. Targeting MET in Lung Cancer: Will Expectations Finally Be MET? J Thorac Oncol 2017;12:15–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Camidge DR, Otterson GA, Clark JW, et al. Crizotinib in patients (pts) with MET-amplified non-small cell lung cancer (NSCLC): Updated safety and efficacy findings from a phase 1 trial. . Journal of Clinical Oncology 2018:Abstr 9062. [Google Scholar]
- 4.Reungwetwattana T, Liang Y, Zhu V, et al. The race to target MET exon 14 skipping alterations in non-small cell lung cancer: The Why, the How, the Who, the Unknown, and the Inevitable. Lung Cancer 2017;103:27–37. [DOI] [PubMed] [Google Scholar]
- 5.Felip E, Horn L, Patel JD, et al. Tepotinib in patients with advanced non-small cell lung cancer (NSCLC) harboring MET exon 14-skipping mutations: Phase II trial. Journal of Clinical Oncology 2018;36:9016–9016. [Google Scholar]
- 6.Spigel DR, Edelman MJ, O’Byrne K, et al. Results From the Phase III Randomized Trial of Onartuzumab Plus Erlotinib Versus Erlotinib in Previously Treated Stage IIIB or IV Non-Small-Cell Lung Cancer: METLung. J Clin Oncol 2017;35:412–420. [DOI] [PubMed] [Google Scholar]
- 7.Neal JW, Dahlberg SE, Wakelee HA, et al. Erlotinib, cabozantinib, or erlotinib plus cabozantinib as second-line or third-line treatment of patients with EGFR wild-type advanced non-small-cell lung cancer (ECOG-ACRIN 1512): a randomised, controlled, open-label, multicentre, phase 2 trial. Lancet Oncol 2016;17:1661–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mignard X, Ruppert AM, Antoine M, et al. c-MET Overexpression as a Poor Predictor of MET Amplifications or Exon 14 Mutations in Lung Sarcomatoid Carcinomas. J Thorac Oncol 2018;13:1962–1967. [DOI] [PubMed] [Google Scholar]
- 9.Aisner DL, Sholl LM, Berry LD, et al. The Impact of Smoking and TP53 Mutations in Lung Adenocarcinoma Patients with Targetable Mutations-The Lung Cancer Mutation Consortium (LCMC2). Clin Cancer Res 2018;24:1038–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sholl LM, Aisner DL, Varella-Garcia M, et al. Multi-institutional Oncogenic Driver Mutation Analysis in Lung Adenocarcinoma: The Lung Cancer Mutation Consortium Experience. J Thorac Oncol 2015;10:768–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koeppen H, Yu W, Zha J, et al. Biomarker analyses from a placebo-controlled phase II study evaluating erlotinib+/−onartuzumab in advanced non-small cell lung cancer: MET expression levels are predictive of patient benefit. Clin Cancer Res 2014;20:4488–4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Awad MM, Oxnard GR, Jackman DM, et al. MET Exon 14 Mutations in Non-Small-Cell Lung Cancer Are Associated With Advanced Age and Stage-Dependent MET Genomic Amplification and c-Met Overexpression. J Clin Oncol 2016;34:721–730. [DOI] [PubMed] [Google Scholar]
- 13.Lambros L, Uguen A. MET Immunohistochemistry Should Be Avoided in Selecting Non-small-cell Lung Cancers Requiring MET Exon 14 Skipping Mutation Analysis. Clin Lung Cancer 2018. [DOI] [PubMed] [Google Scholar]
- 14.Tong JH, Yeung SF, Chan AW, et al. MET Amplification and Exon 14 Splice Site Mutation Define Unique Molecular Subgroups of Non-Small Cell Lung Carcinoma with Poor Prognosis. Clin Cancer Res 2016;22:3048–3056. [DOI] [PubMed] [Google Scholar]
- 15.Lai GGY, Lim TH, Lim J, et al. Clonal MET Amplification as a Determinant of Tyrosine Kinase Inhibitor Resistance in Epidermal Growth Factor Receptor-Mutant Non-Small-Cell Lung Cancer. J Clin Oncol 2019;37:876–884. [DOI] [PubMed] [Google Scholar]
- 16.Poirot B, Doucet L, Benhenda S, et al. MET Exon 14 Alterations and New Resistance Mutations to Tyrosine Kinase Inhibitors: Risk of Inadequate Detection with Current Amplicon-Based NGS Panels. J Thorac Oncol 2017;12:1582–1587. [DOI] [PubMed] [Google Scholar]
- 17.Cheng DT, Mitchell TN, Zehir A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J Mol Diagn 2015;17:251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sholl LM, Do K, Shivdasani P, et al. Institutional implementation of clinical tumor profiling on an unselected cancer population. JCI insight 2016;1:e87062. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1. Box plot representations of MET FISH copy number compared to MET IHC by MetMab score. Boxes represent the interquartile range, which contains 50% of the values, whereas lines extend the entire range of values. Positive and negative IHC by MetMab is represented by red and blue dots, respectively. There is no significant difference in MET FISH copy number among MetMab scores using one-way ANOVA by ranks (p=0.41). The sensitivity and specificity of MetMab scoring for detecting a MET genetic abnormality is 0.6 and 0.4, respectively.