Abstract
Neurosteroids positively modulate GABA-A receptor (GABAAR) channel activity by binding to a transmembrane domain intersubunit site. Understanding the interactions in this site that determine neurosteroid binding and its effect is essential for the design of neurosteroid-based therapeutics. Using photo-affinity labeling and an ELIC-α1GABAAR chimera, we investigated the impact of mutations (Q242L, Q242W and W246L) within the intersubunit site on neurosteroid binding. These mutations, which abolish the thermal stabilizing effect of allopregnanolone on the chimera, reduce neither photolabeling within the intersubunit site nor competitive prevention of labeling by allopregnanolone. Instead, these mutations change the orientation of neurosteroid photolabeling. Molecular docking of allopregnanolone in WT and Q242W receptors confirms that the mutation favors re-orientation of allopregnanolone within the binding pocket. Collectively, the data indicate that mutations at Gln242 or Trp246 that eliminate neurosteroid effects do not eliminate neurosteroid binding within the intersubunit site, but significantly alter the preferred orientation of the neurosteroid within the site. The interactions formed by Gln242 and Trp246 within this pocket play a vital role in determining the orientation of the neurosteroid that may be necessary for its functional effect.
Keywords: neurosteroid, GABA receptor, photoaffmity labeling, mass spectrometry, ELIC, mutation
1. INTRODUCTION
Neurosteroids are endogenous modulators of GABA-A receptor (GABAAR) function. The positive allosteric effect of neurosteroids, such as allopregnanolone, on GABAAR channel activity underlies the therapeutic potential of these sterols as anesthetics, anxiolytics and anti-epileptics (1-3). Neurosteroids potentiate the GABAAR by direct interactions at specific sites within the transmembrane domains (TMDs) (4,5). Recent photo-affinity labeling and crystallographic studies have shown that neurosteroids bind to a specific site between adjacent subunits that mediates GABAAR potentiation (6-12). A next step beyond binding site identification is determination of the molecular interactions within the site that determine the affinity and orientation of ligand binding, and subsequent transduction of the binding signal. Mutagenesis studies that discriminate between these aspects of ligand action are particularly lacking for lipophilic small molecules such as neurosteroids that bind within receptor TMDs. A detailed understanding of the pharmacophore of neurosteroid binding and its relationship to allosteric modulation of the GABAAR is essential for the rational design of neurosteroid analogues that bind specific GABAAR isoforms or produce specific effects.
Previously, a chimera composed of the Gloeobacter violaceus ligand-gated ion channel (GLIC) extracellular domain (ECD) and the α1 subunit of the GABAAR TMD was crystallized with the neurosteroid, tetrahydro-deoxycorticosterone, bound to the α1 GABAAR TMD intersubunit site (8). This intersubunit binding site was also identified using a chimera composed of the Erwinia chrysanthemi ligand-gated ion channel (ELIC) ECD and α1 GABAAR TMD crystallized with the anesthetic neurosteroid analogue, alphaxalone (12). These studies revealed two key residues predicted to be important for neurosteroid binding. Gln242 in transmembrane helix 1 (TM1) of the α1 subunit forms a hydrogen bond with the neurosteroid C3α hydroxyl, and Trp246 also in TM1 of α1 forms a hydrophobic ring-stacking interaction with the neurosteroid ring structure (8,12). Previous functional studies have demonstrated that mutations of these residues (Gln242 and Trp246) abolish neurosteroid potentiation (4,8,9,12-15). Based on these findings, it was suggested that these mutations, particularly mutation of Gln242 to a bulky Trp, abolish potentiation by reducing or eliminating neurosteroid binding (8,13,16). There is, however, no experimental evidence to evaluate whether these mutations alter neurosteroid binding or affect transduction of the binding signal.
To probe the interactions that determine neurosteroid binding in this intersubunit site, we performed neurosteroid photo-affinity labeling of WT and mutant GABAAR TMDs and analyzed the photolabeled proteins with intact protein and middle-down mass spectrometry (MS). This approach has proven to be an effective method to characterize neurosteroid binding in the prokaryotic pentameric ligand-gated ion channel (pLGIC), GLIC (17), and the human α1β3GABAAR (7). Here, we extend this approach by testing the effect of mutations on the efficiency, stoichiometry and sites of neurosteroid photolabeling in order to evaluate whether these mutations reduce neurosteroid binding, change binding orientation or affect transduction of the binding signal. To enable the use of intact protein MS analysis of photolabeled GABAAR TMDs and the expression of multiple mutants, we expressed an xELIC-α1GABAAR chimera, which was recently shown to be a useful model protein of allosteric modulation of the GABAAR by propofol (18) and the neurosteroid anesthetic, alphaxalone (12). We find that mutations that disrupt putative interactions in the intersubunit binding site (Q242L, Q242W, W246L) and abolish neurosteroid effect (13,14), do not abrogate neurosteroid binding, but rather alter the orientation of binding within the intersubunit pocket. Similarly, neurosteroids photolabel ELIC-α1β3GABAAR heteromeric receptors in both orientations that are observed in photolabeled ELIC-α1GABAAR WT and Q242L/W chimeras. The result is consistent with the fact that the β3 subunit contains a Trp instead of the essential Gln in position 242 in the α1 subunit. Our data indicate that the binding of neurosteroids in the intersubunit pocket shows less specificity to structural perturbations than previously predicted (8,16), and indicate the importance of binding orientation that enables key sites of ligand-receptor contact in the mechanism of neurosteroid modulation of GABAARs.
2. MATERIALS AND METHODS
2.1. Construct design
The ELIC-α1GABAAR and ELIC-α1β3GABAAR constructs were designed by fusing the ECD of ELIC to the TMD of human GABAAR (12,18). The intracellular loop amino acids were substituted by the short linker -GVE- sequence. A deca-histidine tag and a maltose binding protein (MBP) were incorporated at the amino-terminal, and a tobacco etch virus protease cleavage site was inserted between MBP and ELIC-GABAAR (12).
2.2. Expression and purification
Mutagenesis was performed as described in previous reports (12,18) by oligonucleotide-directed mutagenesis using Phu polymerase (Thermo Fisher Scientific) verified by sequencing. ELIC-GABAAR and ELIC-α1β3GABAAR were expressed in OverExpress™ C43(D3) E. coli, derived from Rosetta™(DE3) cells under double selection with kanamycin and chloramphenicol for ELIC-GABAAR and triple selection with kanamycin, chloramphenicol and ampicillin for ELIC-α1β3GABAAR. respectively. A few isolated colonies were used to inoculate 100 mL of Luria-Bertani Broth (LB) (Sigma) media with the antibiotics and grown overnight at 37 °C in a shaker at 250 rpm. The overnight culture was then diluted 1:100 into 6 × 1 L of LB media with antibiotics and grown to an optical density of 0.7–0.8. All six liters were harvested (5000 rpm, 15 min, 4 °C) and resuspended in 2 L of LB media supplemented with 0.5 M sorbitol. The concentrated cells were equilibrated in a shaker at 15 °C and 250 rpm for 1 h before inducing expression with 0.2 mM isopropyl β-D-1-thiogalactopyranoside. The cells were harvested after 20 hours expression, re-suspended in Buffer A (20 mM Tris pH 7.5, 100 mM NaCl), cOmplete EDTA-free protease inhibitor cocktail (Roche Applied Science), and DNAase, lysed using an Avestin Emulsiflex C-5 at 15,000 psi, pelleted by ultracentrifugation, and homogenized. The fusion protein was extracted with 1% DDM. Solubilized protein was purified using amylose resin and eluted using Buffer A, 40 mM maltose and 0.02% DDM. His-MBP-(ELIC-GABAAR) was digested overnight with tobacco etch virus protease, cleaned up using a reverse Ni-NTA purification, and injected onto a Sephadex 200 Increase 10/300 column, which yielded pentameric protein in Buffer A + 0.02% DDM.
2.3. Thermal stability of the protein
Detergent-solubilized purified receptors were used in thermal stability experiments. Samples were aliquoted in thin-walled 0.5 ml microcentrifuge tubes and heated at 30 to 80 °C for 1 h. Samples were run on a Sephadex 200 Increase 10/300 column in 20 mM Tris, pH 7.5, 100 mM NaCl, 0.02% DDM. The UV absorbance intensity of the pentameric size exclusion chromatography (SEC) peak was monitored and the peak heights of heated relative to unheated (4 °C) control sample were determined. To assess whether allopregnanolone was able to thermally stabilize the detergent–receptor complex, purified protein was incubated in varying concentrations of allopregnanolone at 65 °C for 1 h before implementing SEC analysis as described above. It was apparent that 10 μM allopregnanolone was sufficient to increase thermal stability. Analysis of statistical significance comparing the UV absorbance intensity of the pentameric SEC peaks was determined using one-way ANOVA with post hoc Bonferroni correction (GraphPad Prism 6).
2.4. Photolabeling and intact protein MS analysis
The syntheses of neurosteroid photolabeling reagents (KK123 and KK200) are detailed in previous reports (17,19). Purified ELIC-α1GABAAR was photolabeled and analyzed by intact protein MS as described previously (17,20). For the intact protein MS analysis, 100 μg ELIC-α1GABAAR was mixed with neurosteroid photolabeling reagent at 100 μM, or 100 μM added three times with UV irradiation after each addition. 100 μl samples were irradiated in a quartz cuvette with > 320 nm UV light, treated with 250 mM dithiothreitol (DTT), and then precipitated with chloroform/methanol/water. The precipitated protein was washed three times with equal volumes of water and methanol, centrifuged, and the protein pellet reconstituted in 3 μl of 90% formic acid followed by 100 μl of 4:4:1 chloroform/methanol/water. Reconstituted samples were then analyzed in an Orbitrap Elite mass spectrometer (Thermo Fisher Scientific) by direct injection at 3 μl/min using a Max Ion API source with a HESI-II probe. Full spectra of photolabeled ELIC-α1GABAAR were acquired on the linear trap quadrapole using spray voltage of 4 kV, capillary temperature of 320 °C, and in-source dissociation of 30 V. Deconvolution of intact ELIC-α1GABAAR spectra was performed using UniDec (21).
2.5. Tryptic middle-down MS analysis
25 μg of photolabeled ELIC-α1GABAAR or ELIC-α1β3GABAAR was reduced with 5 mM tris(2-carboxyethyl)phosphine for 1 hr, alkylated with 2.5 mM N-ethylmaleimide for 45 min, and quenched with 5 mM DTT for 15 min. Samples were then digested with 10 μg of trypsin for 7 days at 4 °C; extended digestion at low temperature was necessary to obtain maximal recovery of TMD peptides. Next, formic acid was added to 1%, followed directly by LC-MS analysis on an Orbitrap Elite mass spectrometer. 20 μl samples were injected into a home-packed PLRP-S (Agilent) column (10 cm × 75 μm, 300 A), separated with an 85 min gradient from 10 to 95% acetonitrile, and introduced to the mass spectrometer at 800 nl/min with a nanospray source. MS acquisition was set as an MS1 Orbitrap scan (resolution of 60,000) followed by top 20 MS2 Orbitrap scans (resolution of 15,000) using data dependent acquisition, 15 s dynamic exclusion, and exclusion of 1+ and 2+ precursors. Fragmentation was performed using CID and HCD with normalized collision energies of 35 and 30%, respectively. Analysis of data sets was performed using Xcalibur (Thermo Fisher Scientific) to manually search for TM1, TM2, TM3 or TM4 tryptic peptides with or without neurosteroid photolabel modifications. Photolabeling efficiency was estimated by generating extracted chromatograms of unlabeled and labeled peptides, determining the area under the curve, and calculating the abundance of labeled peptide/(unlabeled + labeled peptide). Competitive inhibition of labeling was performed by labeling ELIC-α1GABAAR with 10 μM KK200 in the presence of 100 μM allopregnanolone. Analysis of statistical significance comparing the photolabeling efficiency of KK200 for ELIC-ELIC-α1GABAAR WT, Q241W or W245L mutant was determined using Student’s unpaired t-test (GraphPad Prism 6). MS2 spectra of photolabeled TMD peptides were analyzed by manual assignment of fragment ions with and without photolabel modification. Fragment ions were accepted based on the presence of a monoisotopic mass within 10 ppm mass accuracy. In addition to manual analysis, PEAKS database searches were performed for data sets of ELIC-GABAAR photolabeled with KK123 or KK200. Search parameters were set for a precursor mass accuracy of 10 ppm, fragment ion accuracy of 0.1 Da, up to 3 missed cleavages on either end of the peptide, false discovery rate of 0.1%, and variable modifications of methionine oxidation, cysteine carbamidomethylation, and KK123 or KK200 on any amino acid.
2.6. Native MS analysis
For native MS experiments, purified proteins from co-expressed ELIC-α1GABAAR and ELIC-β3GABAAR were concentrated to 1 mg/ml, and the buffer exchanged to 200 mM ammonium acetate buffer, pH 7.5, with 0.02% DDM using Biospin6 columns (BioRad). The buffer-exchanged sample was analyzed by static nanospray on a Thermo QExactive EMR. Instrument conditions were spray voltage 1.2 kV, capillary temperature 150 °C, ion transfer optics set with the injection flatapole, inter-flatapole lens, bent flatapole, transfer multiple as 8, 7, 6, 4 V, respectively, AGC target 3 × 106, resolution 8,750, CID 200 V, and CE 100 V. Spectra of ELIC-α1β3GABAAR were deconvoluted using UniDec (21).
2.7. Docking Simulations
Docking templates were generated using the GLIC-α1GABAAR crystal structure (PDB ID 5OSA) embedded in a 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylcholine (POPC) bilayer (8). Models of both the WT and mutant structures were prepared in Modeller using 5OSA as the template. Hydrogens and charges for both structures were obtained from the H++ (http://biophysics.cs.vt.edu), the structures were then submitted to the PPM server (https://opm.phar.umich.edu/ppm_server) for orientation into a lipid membrane. The correctly oriented receptors were then submitted to the CHARMM-GUI Membrane Builder server (http://www.charmm-gui.org) to build the fully solvated, membrane bound systems. For both simulations, the top membrane leaflet consisted of 166 POPC, the bottom leaflet consisted of 175 POPC lipids and the ionic strength was set to 0.15M KCl. The NAMD input files produced by CHARMM-GUI use a seven-step process of gradually loosening constraints in the simulation prior to production runs. Using WT and mutant receptor structures, 10-20 ns molecular dynamics trajectories were obtained using the CHARMM36 force field and NAMD (22). The resulting 10 and 20 ns trajectory was then processed using the utility mdtraj (23), to extract snapshots of the receptor at every 1 or 2 ns of the time frame, respectively. Docking was performed using AutoDock Vina on each of the 10 snapshots in order to capture receptor flexibility (24). Allopregnanolone was prepared by converting the sdf file from PubChem into a PDB file using Open Babel (25), and Gasteiger charges and free torsion angles were determined by AutoDock Tools. Docking simulations were performed with allopregnanolone using the GLIC-α1GABAAR WT and the Q242W mutant, and docking grid boxes with dimensions of 25 × 25 × 25 Ångströms encompassing the intersubunit pocket. All simulations were performed with 1 Å grid spacing, using a genetic algorithm with 250 runs, and otherwise default parameters. Docking was limited to an energy range of 3 kcal from the best docking pose and limited to a total of twenty unique poses. These were then clustered geometrically using the program DIVCF (26). The resulting clusters were ranked by Vina score, cluster size, and visually analyzed for compatibility with the KK200 photolabeling results. The clusters comprised of poses which were markedly distant from the photolabeled site were omitted when clusters were categorized into those where the neurosteroid A-ring was pointing intracellular (D-ring towards Phe298) or extracellular (D-ring towards Tyr309).
3. RESULTS
3.1. Sites of neurosteroid photolabeling in the α1GABAAR TMD.
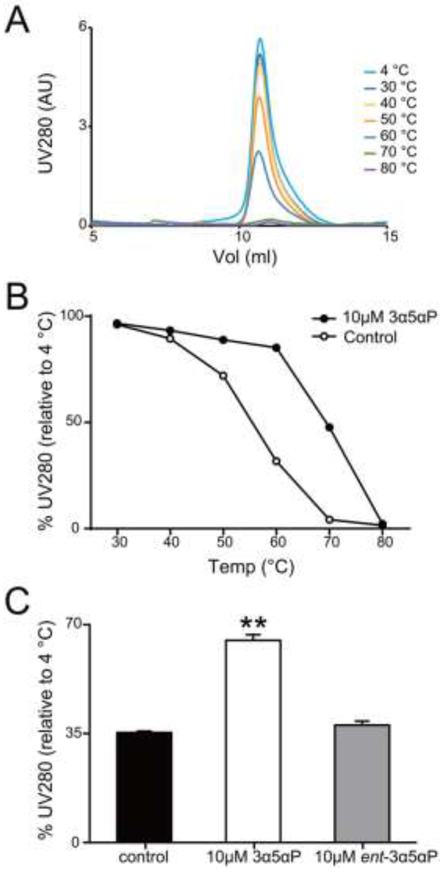
We previously showed that ELIC-α1GABAAR, a chimera consisting of an ELIC ECD and an α1GABAAR TMD, yields propylamine-activated currents (propylamine is an agonist for ELIC), which are potentiated by the neurosteroid anesthetic, alphaxalone (12). In this study, we photolabeled purified ELIC-α1GABAAR protein solubilized in n-dodccyl β-D-maltoside (DDM). We therefore sought to test the effect of neurosteroids on DDM-solubilized ELIC-α1GABAAR protein. Ligand or lipid binding can produce a thermal stabilizing effect on detergent-solubilized membrane protein oligomers assessed by size exclusion chromatography (27,28). A thermal stabilizing effect of neurosteroids was previously demonstrated in GLIC-α1GABAAR and β3GABAAR-α5GABAAR chimeras, and this effect was abolished by mutations that disrupt neurosteroid potentiation (8,9). We also found that the thermal stability of ELIC-α1GABAAR is significantly increased in the presence of allopregnanolone but not the enantiomer, ent-allopregnanolone (Fig. 1, B and C), recapitulating the enantioselectivity of neurosteroid potentiation of the GABAAR (2,5,29). Given the consistent correlation between neurosteroid thermal stabilization and potentiation of GABAAR-TMD chimeras, we subsequently utilized thermal stabilization as a reporter of neurosteroid effect in ELIC-α1GABAAR proteins. This assay has the advantage of using DDM-solubilized ELIC-α1GABAAR, which allows a more direct comparison of photolabeling data (neurosteroid binding) and thermal stabilization (effect).
FIGURE 1: Allopregnanolone thermally stabilizes the ELIC-α1GABAAR chimera.
(A) Gel filtration profiles of purified ELIC-α1GABAAR heated for 1h at a range of temperatures, showing the pentamer peak. (B) The percent intensity of the ELIC-α1GABAAR pentamer peak relative to 4 °C after heating at a range of temperatures in the absence or presence of 10 μM allopregnanolone (3α5αP). Data points are from single experiments. (C) The percent intensity of the ELIC-α1GABAAR pentamer peak relative to 4 °C after heating at 65 °C. The chimeric protein is thermally stabilized by 3α5αP but not ent-3α5αP at 10μM (n = 4, ± SEM). **P < 0.01 vs. control.
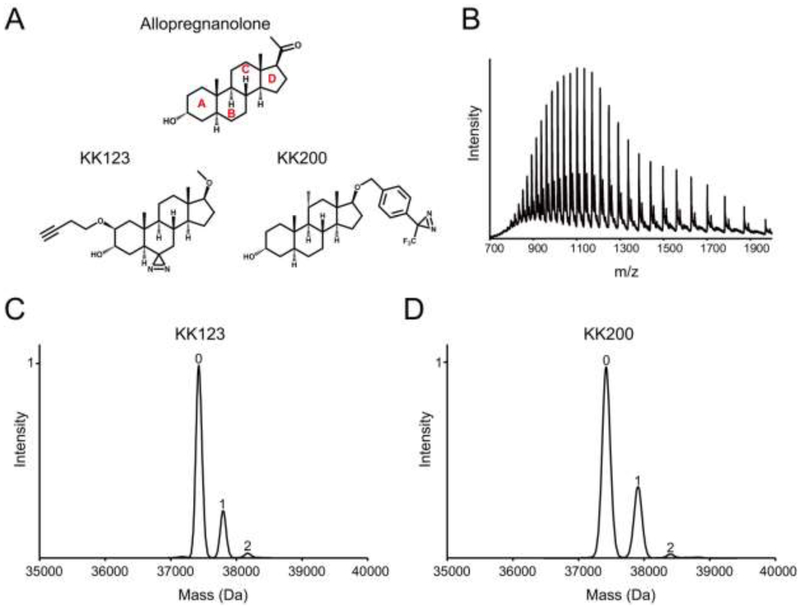
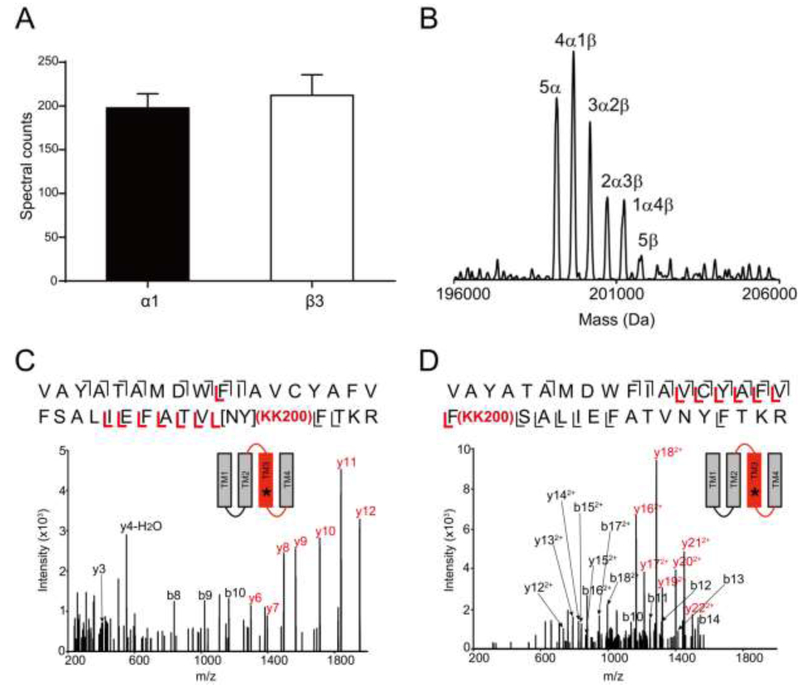
Next, we sought to determine whether our neurosteroid photolabeling reagents label the ELIC-α1GABAAR chimera with the same stoichiometry, sites and orientation as previously identified for the human α1β3GABAAR (7). Purified, DDM-solubilized ELIC-α1GABAAR was photolabeled with two established allopregnanolone-based photolabeling reagents, KK123 and KK200 (Fig. 2A), and the stoichiometry of labeling was determined using denatured intact protein MS. When ELIC-α1GABAAR was sequentially labeled three times with 100 μM KK123 or KK200 (total of 300 μM), we observed a minimum labeling stoichiometry of two (i.e. two neurosteroids per subunit) for both photolabeling reagents (Fig. 2, C and D). Although we cannot exclude the possibility that there are more than two photolabeled sites, the observed minimum stoichiometry is consistent with the two binding sites per subunit previously identified in GLIC and the human α1β3GABAAR (7,17).
FIGURE 2: Neurosteroids photolabel the ELIC-α1GABAAR subunit with a stoichiometry of two.
(A) Structures of allopregnanolone and the photolabeling reagents, KK123 and KK200. (B) Full spectrum of intact ELIC-α1GABAAR photolabeled with 100 μM KK123 three times. (C) and (D) Deconvoluted spectra of ELIC-α1GABAAR photolabeled with KK123 or KK200 at 100 μM three times. The number of photolabeling reagents covalently modifying each protein subunit is indicated by the number above the peak.
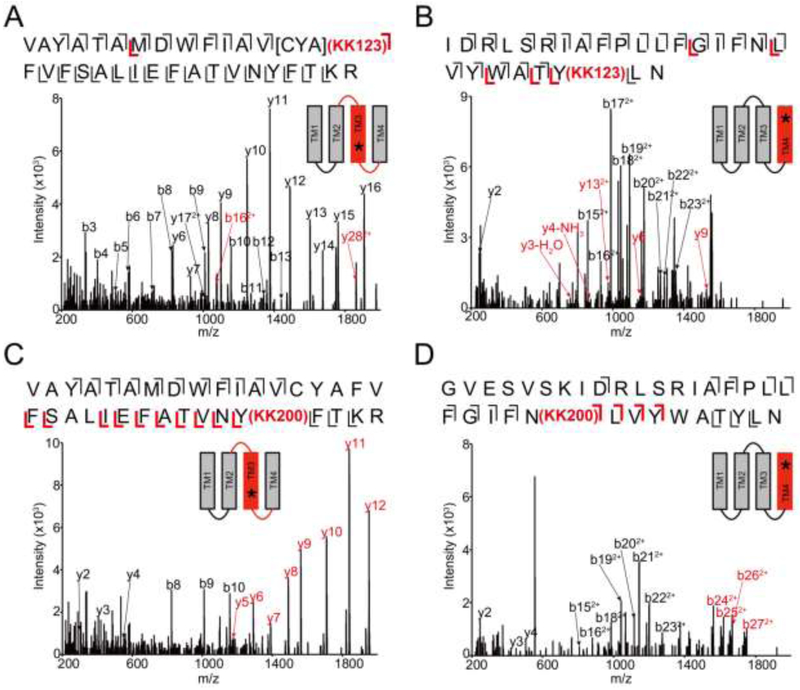
To identify the residues labeled by KK123 and KK200 in the ELIC-α1GABAAR TMD, we applied a middle-down MS analysis as previously described (17). Tryptic digests of the chimeric protein photolabeled with 100 μM KK123 showed labeling of [Cys293-Tyr294-Ala295] in TM3 and Tyr415 in TM4 with efficiencies of 7.8% for TM3 and 4.0% for TM4 (Fig. 3, A and B). 100 μM KK200 labeled Tyr309 in TM3 with an efficiency of 8.0%, and Asn408 in TM4 with an efficiency of 1.8% (Fig. 3, C and D). Fragmentation spectra of the corresponding unlabeled peptides and representative sequence coverage for the ELIC-α1GABAAR tryptic digest (99% and 94% by using HCD or CID fragmentation, respectively) are shown in supplementary data (Supplemental Fig. 1 and 2). For ease of comparison, we numbered amino acids within the ELIC-α1GABAAR TMD based on the full-length human α1GABAAR subunit. Examination of these photolabeled residues in the ELIC-α1GABAAR crystal structure shows that most of these residues delineate the same two neurosteroid binding sites identified in GLIC (17) and human α1β3GABAAR (7): an intersubunit (Tyr309) and intrasubunit site (Asn408 and Tyr415) (Fig. 4A). The intersubunit site is also consistent with the site identified in the GLIC-α1GABAAR, β3GABAAR-α5GABAAR and ELIC-α1GABAAR crystal structures (8,9,12). An exception to these established sites is the segment labeled by KK123 in TM3 (Cys293-Tyr294-Ala295) (Fig. 4A). While we cannot rule out the possibility that this represents a third distinct neurosteroid binding site, we interpret this labeled site with caution since it was not previously identified in GLIC or α1β3GABAAR, and since KK123 utilizes an aliphatic diazirine that preferentially labels nucleophilic residues (20,30) and may diffuse some distance from its binding site to label nucleophiles (e.g. Tyr294). In contrast, KK200 which contains a trifluoromethylphenyl-diazirine (TPD) group is expected to form a photo-reactive carbene that will rapidly label any residue in its proximity in the nanosecond timescale (17,30,31). As such, KK200 is our preferred reagent to precisely define neurosteroid binding modes (7,17). In the case of the intersubunit site in the α1GABAAR TMD, labeling of Tyr309 by KK200 defines a preferred orientation for the neurosteroid analogue where the C3α hydroxyl points away from the intracellular surface toward the center of the TMD, and likely forms a hydrogen bond with Gln242 (Fig. 4A), as demonstrated in the crystal structure of ELIC-α1GABAAR with alphaxalone (12).
FIGURE 3: Neurosteroids photolabel ELIC-α1GABAAR at an intersubunit and intrasubunit site.
(A) HCD fragmentation spectrum of the ELIC-α1GABAAR TM3 tryptic peptide photolabeled with 100 μM KK123. Black and red indicate fragment ions that do not contain or contain KK123, respectively. Schematic highlights in red identify the TMD being analyzed and the asterisk denotes the approximate location of KK123. (B) Same as (A) for TM4. (C) and (D) Same as (A) photolabeled with 100 μM KK200 for TM3 and TM4, respectively.
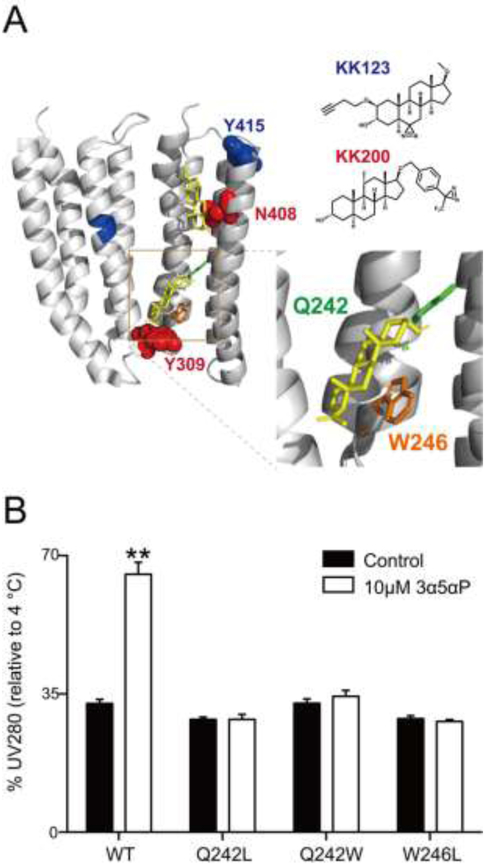
FIGURE 4: Mutations in ELIC-α1GABAAR ablate neurosteroid thermal stabilization.
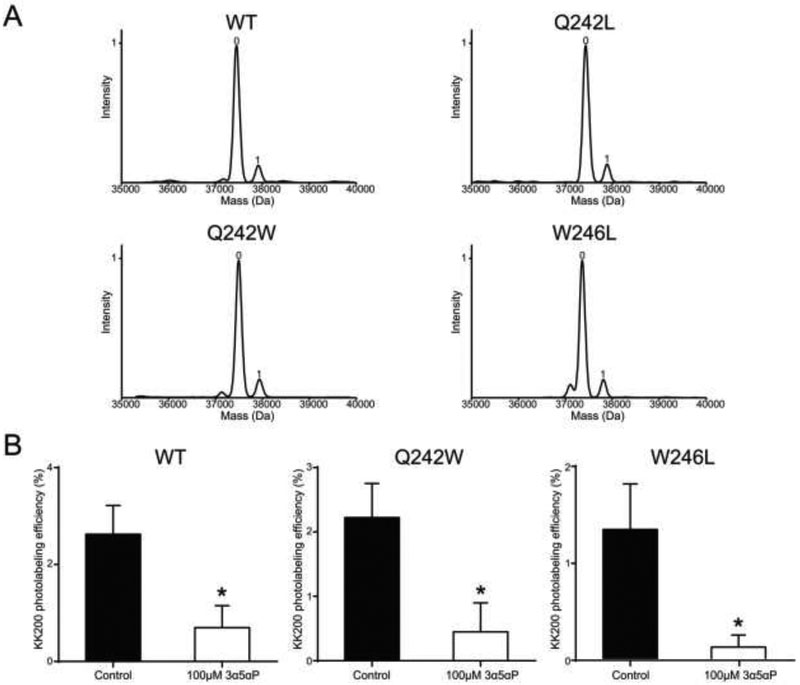
(A) Structure of the α1GABAAR-TMD highlighting the residues photolabeled by 100 μM KK123 (blue) or KK200 (red) in WT, and showing a representative docking pose for allopregnanolone (yellow) at both intersubunit and intrasubunit sites. Two adjacent subunits are shown and the channel pore is behind these subunits. A zoomed-in box of the intersubunit site shows the chains of residues where mutations were made (Q242-green, W246-orange). (B) The percent intensity of ELIC-α1GABAAR WT or mutant pentamer peaks relative to 4 °C after heating to a temperature that causes 70% peak decay (65 °C for WT; 52 °C for Q242L; 68 °C for Q242W; 60 °C for W246L). 10 μM allopregnanolone (3α5αP) thermally stabilizes WT but not the mutants (n = 3, ± SEM). **P < 0.01 vs. control.
3.2. The role of Gln242 and Trp246 on neurosteroid binding in the α1GABAAR TMD.
To assess the effect of neurosteroid binding to the intersubunit photolabeled site in ELIC-α1GABAAR, we generated mutations of two key residues within this binding pocket—Q242L, Q242W, and W246L (Fig. 4A)—and examined their impact on the thermal stabilizing effect of allopregnanolone. All mutants purified as stable monodisperse pentamers in DDM. All three mutants also reduced the thermal stabilizing effect of allopregnanolone in ELIC-α1GABAAR (Fig. 4B), consistent with the recently reported effect that these mutations reduce neurosteroid potentiation in this ELIC-α1GABAAR chimera as well as human GABAARs and the GLIC-α1GABAAR chimera (4,8,12,13).
The primary aim of this study is to test whether the loss of these functionally important interactions disrupt neurosteroid binding. To address this, we photolabeled each ELIC-α1GABAAR mutant with 100 μM KK200. Of note, the observed stoichiometry of labeling of WT at 100 μM is one per subunit (Fig. 5A). The probability of observing a doubly-labeled species is the product of the labeling efficiencies at each site. Thus, the observed singly-labeled peak includes subunits labeled at both the intersubunit and intrasubunit site (Fig. 5A, Table 1); a doubly-labeled species was not observed at this concentration (unlike labeling three times at 100 μM) because of the lower labeling efficiency. Analysis of the labeled mutants by intact protein MS showed labeling efficiencies identical to WT (11.3% for WT; 11.8% for Q242L; 11.7% for Q242W; and 11.5% for W246L) (Fig. 5A, Table 1), suggesting that neurosteroids still bind to mutant ELIC-α 1GABAARs. This finding contradicts the assertion that these mutations, particularly Q242W which may introduce steric hindrance to the intersubunit binding site, disrupt neurosteroid binding (8,16). To confirm that the endogenous neurosteroid, allopregnanolone, also binds to the intersubunit site of the Q242W or W246L mutant TMD, we tested whether 100 μM allopregnanolone competitively reduces the labeling efficiency of 10 μM KK200 in WT, Q242W and W246L mutants. Indeed, allopregnanolone reduced the photolabeling efficiency of KK200 for TM3 (i.e. the intersubunit site) to the same extent in WT and both mutants (Fig. 5B), suggesting that allopregnanolone binds to the intersubunit site in both WT and the mutants. Moreover, using the lower concentration of 10 μM, KK200 labeled TM3 in WT and the mutants with the following efficiencies (% ± SEM): 2.6 ± 0.6 for WT (n = 4); 2.2 ± 0.1 for Q242L (n = 2); 2.2 ± 0.5 for Q242W (n = 4); 1.4 ± 0.5 for W246L (n = 4). These efficiencies are statistically indistinguishable further suggesting that the mutations do not significantly alter neurosteroid binding affinity at the intersubunit site.
FIGURE 5: KK200 photolabels WT and mutant ELIC-α1GABAAR with similar efficiencies, and photolabeling is prevented by allopregnanolone.
(A) Deconvoluted spectra of the ELIC-α1GABAAR WT and mutants photolabeled with 100 μM KK200. The number of photolabeling reagents covalently modifying each protein subunit is indicated by the number above the peak. (B) Photolabeling efficiency of TM3 in ELIC-α1GABAAR WT, Q242W and W246L mutants by 10 μM KK200 in the absence or presence of 100 μM allopregnanolone (n = 4, ± SEM). *P < 0.05 vs. control.
TABLE 1:
Photolabeling efficiencies of KK200 in ELIC-α1GABAAR.
| ELIC-α1GABAAR | Intact protein MS | Middle-down MS | |
|---|---|---|---|
| TM3 | TM4 | ||
| WT | 11.3 | 8.0 | 1.8 |
| Q242L | 11.8 | 6.1 | 1.9 |
| Q242W | 11.7 | 5.2 | 1.7 |
| W246L | 11.5 | 3.7 | 1.2 |
Labeling efficiencies of KK200 (%) in intact protein MS and middle-down MS analyses for the ELIC-α1GABAAR WT and the mutated receptors.
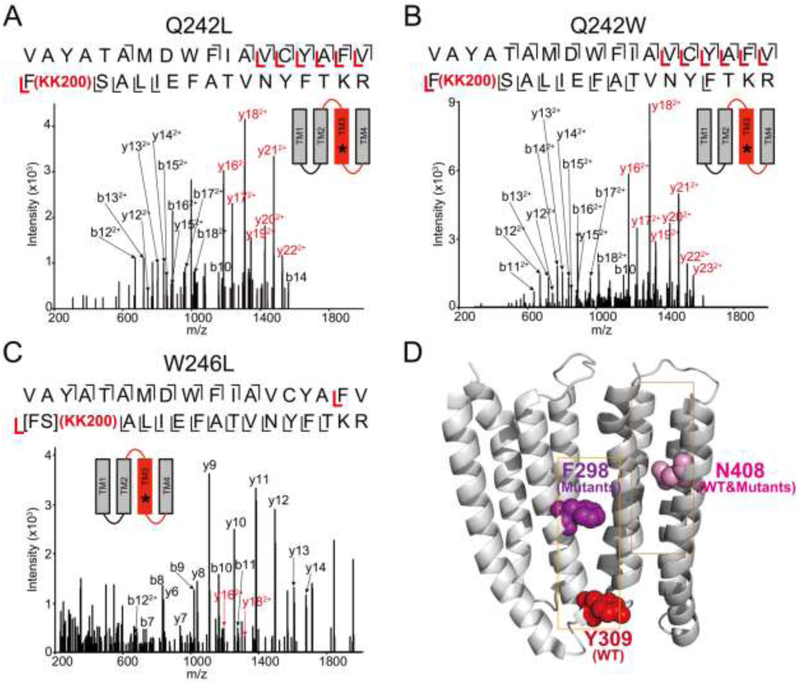
Next, we used a middle-down MS analysis to identify the residues labeled by 100 μM KK200 in each mutant. Just as in WT, KK200 labeled Asn408 (photolabeling efficiency: 1.8% for WT; 1.9% for Q242L; 1.7% for Q242W; 1.2% for W246L) in TM4 (i.e. the intrasubunit site) in all three mutants (Table 1). Surprisingly, unlike WT, where KK200 labeled Tyr309 in TM3 which forms the intracellular end of the intersubunit pocket (Fig. 4A), KK200 labeled Phe298 in the Q242L and Q242W mutants (photolabeling efficiency: 6.1% for Q242L; 5.2% for Q242W) (Fig. 6, A and B, Table 1). KK200 also labeled TM3 in the W246L mutant (photolabeling efficiency of 3.7%) (Fig. 6C, Table 1), and the labeled amino acid was localized to either Phe298 or Ser299 (Fig. 6C). Phe298 is located at the extracellular end of the intersubunit pocket (Fig. 6D). We previously showed that the equivalent amino acid in GLIC, Phe267, was also photolabeled by KK200 consistent with a neurosteroid binding orientation where the C3α hydroxyl points intracellular (17). Thus, KK200 photolabeling of Phe298 in all three mutants, instead of Tyr309, indicates that Q242 and W246 are important determinants of neurosteroid binding orientation. Fragmentation spectra of the corresponding unlabeled peptides in the mutated receptors are shown in supplementary data (Supplemental Fig. 3).
FIGURE 6: Mutations in ELIC-α1GABAAR alter the orientation of KK200 photolabeling.
(A) CID fragmentation spectrum of the ELIC-α1GABAAR TM3 tryptic peptide photolabeled with 100 μM KK200 in the Q242L mutant. Black and red indicate fragment ions that do not contain or contain KK200, respectively. Schematic highlights in red identify the TMD being analyzed and the asterisk denotes the approximate location of KK200. (B) Same as (A) in the Q242W mutant. (C) HCD fragmentation spectrum of the ELIC-α1 GABAAR TM3 tryptic peptide photolabeled with 100 μM KK200 in the W246L mutant. (D) Structure of the α1GABAAR-TMDs highlighting the residue, Tyr309 in the intersubunit site photolabeled by KK200 in WT in red. Shown in purple is Phe298 in the intersubunit site, which is photolabeled by KK200 in the mutants. Asn408 shown in pink is photolabeled by KK200 in both WT and mutants in the intrasubunit site.
Of note, the efficiencies of labeling in W246L at both TM3 and TM4 were lower than WT. Since the overall efficiency of labeling of W246L determined by intact protein MS was the same as WT, we surmise that the efficiency of neurosteroid labeling of both TM3 and TM4 in W246L is similar to WT. Neurosteroid labeling likely renders the W246L mutant less suitable for our middle-down tryptic digest, perhaps because the photolabeled protein is more prone to aggregation. This may lead to an artifactually low labeling efficiency at the peptide level, whereas the intact protein analysis which utilizes a precipitation and formic acid re-solubilization approach is not susceptible to such aggregation, and should accurately report the proportion of unphotolabeled and photolabeled protein species.
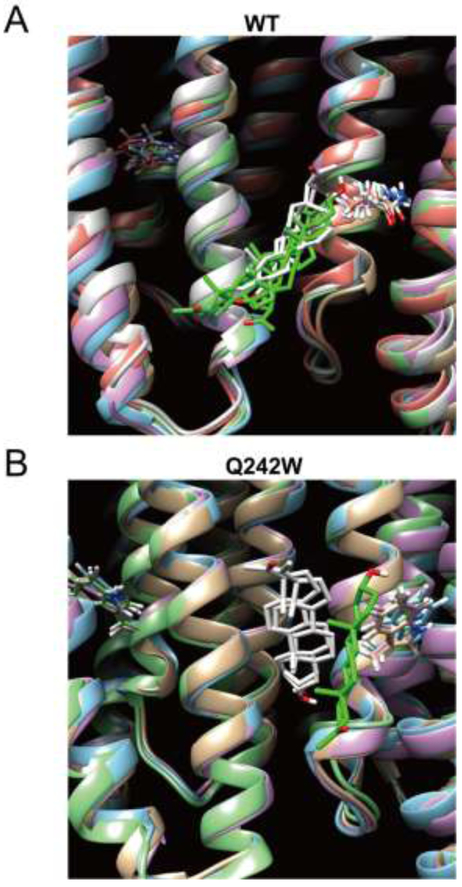
It is possible that the changes in photolabeling observed for KK200 are not reflective of binding for the endogenous neurosteroid, allopregnanolone. To address this possibility, we examined computational docking of allopregnanolone to the WT and Q242W α1GABAAR TMD. We utilized the GLIC-α1GABAAR crystal structure as a template of the α1GABAAR TMD, generated the Q242W mutation, and performed 10 ns of molecular dynamics (MD) simulations for WT and Q242W mutant receptors embedded in a POPC bilayer. Ten snapshots of each receptor were extracted at 1 ns intervals and used for docking simulations of allopregnanolone within the intersubunit pocket using Autodock Vina. Docking results of allopregnanolone to WT from each snapshot were then clustered together and yielded multiple clusters with the majority of poses similar to the reported crystal structure (8,9,12), oriented with the neurosteroid A-ring pointing extracellular and the C3α hydroxyl proximal to Gln242 (Fig. 7A, Table 2, Supplemental Table 1). In contrast, docking of allopregnanolone to Q242W resulted in different binding orientations where the largest number of poses was oriented with the A-ring pointing intracellular (Fig. 7B, Table 2, Supplemental Table 1). The respective binding modes are consistent with KK200 labeling of Tyr309 in WT and Phe298 in the Q242W mutant. In addition, the range of docking scores between WT and Q242W were not significantly different, consistent with our photolabeling results, which suggest that neurosteroid binding affinity is minimally affected by the Q242W mutation (Table 2). To test the reproducibility of these results over a longer time scale, we extended the MD simulation to 20 ns, and extracted an additional ten snapshots every 2 ns. Docking results using these snapshots as templates were comparable to those from the first analysis with respect to the preferred orientation of allopregnanolone (Table 2, Supplemental Table 1). C-α RMSD values were measured over the duration of 20 ns MD simulation showing that the receptor backbone structures have reached an equilibrium within the simulation times (Supplemental Fig. 4).
FIGURE 7: Allopregnanolone docking poses within the intersubunit pocket in the α1GABAAR TMD of WT and the Q242W mutant.
(A) Representative poses from each cluster for allopregnanolone docked within the intersubunit site in WT. The receptor snapshot corresponding to each pose is shown along with the Gln242 side chain for each subunit. Green indicates clusters where the A ring points extracellular, and white indicates clusters where the A-ring points intracellular. (B) Same as (A) for the Q242W mutant.
TABLE 2:
Docking results of allopregnanolone in the α1GABAAR TMD.
| Receptor | Total # docked poses |
% poses with A ring pointing extracellular |
% poses with A ring pointing intracellular |
Vina score ranges | |||
|---|---|---|---|---|---|---|---|
| 10 ns | 20 ns | 10 ns | 20 ns | 10 ns | 20 ns | ||
| WT | 192 | 52.6 | 59.9 | 35.9 | 21.3 | −7.8 — −5.6 | −6.7 — −5.5 |
| Q242W | 192 | 16.6 | 18.9 | 46.4 | 52.6 | −6.1 — −5.1 | −7.5 — −5.5 |
Results of 10 and 20 ns docking simulations for allopregnanolone in α1GABAAR TMD in WT and the Q242W mutant. Docking poses within the intersubunit pocket were compiled and parsed into two general orientations: A ring pointing extracellular or intracellular.
In summary, the Q242L, Q242W and W246L mutations in ELIC-α1GABAAR do not significantly alter the binding affinity of KK200 at the intersubunit site, but rather the orientation of the neurosteroid within this pocket.
3.3. The role of the β3GABAAR TMD in neurosteroid binding orientation.
We also co-expressed ELIC-α1GABAAR with an ELIC-β3GABAAR chimera in an effort to examine heteromeric receptors and the role of the β3GABAAR TMD on neurosteroid binding in the intersubunit pocket. Co-expression of ELIC-α1GABAAR with ELIC-β3GABAAR subunits yielded monodisperse pentamers in DDM. Label-free MS quantitation using spectral counting of peptides in tryptic digests demonstrated equal expression of α1 and β3 subunits (Fig. 8A) with close to full sequence coverage for both subunits (99% for ELIC-α1 and 100% for ELIC-β3 by using HCD fragmentation; 95% for ELIC-α1 and 96% for ELIC-β3 by CID) (Supplemental Fig. 5 and 6). We then utilized native MS analysis of the pentamer chimera to directly examine the combinations of α1 and β3 subunits formed in E. coli. Remarkably, this analysis demonstrated the presence of all six possible combinations of α1 and β3 subunits, although α1-containing pentamers were more abundant in the native MS spectrum (Fig. 8B). Given the equal expression of α1 and β3 subunits determined by label-free quantitation, we favor the conclusion that ELIC-α1GABAAR and ELIC-β3GABAAR randomly co-assemble as pentamers. β3-containing pentamers show lower abundance by native MS possibly because they are less efficiently de-solvated or more prone to dissociate in the gas phase. In support of this, expression of ELIC-β3GABAAR alone resulted in protein that mostly aggregated when solubilized in DDM.
FIGURE 8: KK200 photolabels the intersubunit site in ELIC-α1β3GABAAR in both orientations.
(A) Spectral counts of ELIC-α1GABAAR and ELIC-β3GABAAR TMD tryptic peptides in ELIC-α1β3GABAAR chimeric protein (n = 8, ± SEM). (B) Deconvoluted native spectrum of ELIC-α1β3GABAAR pentamer; labeled peaks represent combinations of ELIC-α1GABAAR and ELIC-β3GABAAR subunits within the pentamer. The mass of each peak (5α, 199,156 Da; 4α1β, 199,672 Da; 3α2β, 200,176 Da; 2α3β, 200,688 Da; 1α4β, 201,216 Da; 5β, 201,768 Da) likely represents ELIC-α1β3GABAAR pentamers with bound adducts or lipids. (C) and (D) HCD and CID fragmentation spectra of the ELIC-α1 subunit TM3 tryptic peptides in ELIC-α1β3GABAAR photolabeled with 100 μM KK200, respectively.
The fact that α1 and β3 subunits randomly co-assemble limits the utility of this ELIC-α3GABAAR chimera as a model of the human α1β3 heteromeric GABAAR, which is thought to predominantly form specific isoform combinations. Nevertheless, we hypothesized that, since the β3GABAAR TMD contains a Trp in the position equivalent to Gln242 in α1, KK200 may photolabel the intersubunit pocket in both orientations due to the presence of α1+/α1− and α1+/β3− interfaces. Indeed, KK200 labeled both Tyr309 (Fig. 8C) and Phe298 (Fig. 8D) in TM3 of ELIC-α1GABAAR. Fragmentation spectra of the corresponding unlabeled peptides are shown in supplementary (Supplemental Fig. 7). We were unable to detect KK200-photolabeled TMD peptides of ELIC-β3GABAAR to assess labeling orientation in β3+/α1− or β3+/β3− interfaces. The reason for this is unclear, although it is likely because ELIC-3GABAAR peptides are more prone to aggregate in the presence of neurosteroid or when photolabeled. However, we previously showed that KK200 labels human α1β3GABAAR in the intersubunit pocket at the β3+/α1− interface with an orientation where the neurosteroid C3α hydroxyl points extracellular adjacent to Gln242 (7). Moreover, we have also shown that neurosteroids photolabel the intersubunit pocket in the β3 homopentamer (6). Collectively, these data suggest that neurosteroids bind to the intersubunit pocket formed by all α1 and β3 interface combinations, but that the presence of Gln242 at the extracellular end of this pocket drives a preferred binding orientation that is essential for the functional effect of neurosteroids.
4. DISCUSSION
Photolabeling in human α1β3GABAAR and crystallographic studies in ELIC-α1GABAAR, GLIC-α1GABAAR, and β3GABAAR-α5GABAAR chimeras have identified an intersubunit neurosteroid binding site that is critical for potentiation of agonist-elicited currents (7-9,12). Within this site, two key residues, Gln242 and Trp246, are shown to form hydrogen bond and hydrophobic ring-stacking interactions, respectively, with the neurosteroid (8,9,12), and mutations of these residues abolish the neurosteroid potentiating effect (4,8,12-14). It has also been suggested that the mutation Q242W sterically prevents neurosteroid binding (8,13,16); however, there is no experimental evidence examining the impact of these mutations on neurosteroid binding. Understanding the role of these side chains and the effect of mutations within the intersubunit pocket on neurosteroid binding is essential for rational drug design, which uses binding measures as a readout for neurosteroid effect. We were able to experimentally examine neurosteroid binding in the intersubunit site and the effect of mutations by using an ELIC-GABAAR chimera, and a neurosteroid photolabeling approach that combines a TPD-based labeling reagent and MS analysis. The results demonstrate that mutations of Gln242 and Trp246 do not reduce neurosteroid binding but rather alter the orientation and position of binding within this site.
The central finding of this study is that mutations of Gln242 and Trp246 in the intersubunit pocket of the α1GABAAR TMD, which disrupt neurosteroid effect, do not significantly reduce neurosteroid binding. This is evidenced by the fact that KK200 photolabels WT and mutant ELIC-α1GABAAR at the intersubunit site with the same efficiencies at both 10 μM and 100 μM, and that allopregnanolone competitively inhibits labeling in WT and mutants to the same extent. While we recognize that photolabeling efficiencies do not equate with binding affinities, the above data suggest that relative affinities of binding are not markedly perturbed by the mutations. Neurosteroids also bind to the intersubunit pocket at interfaces consisting of all four possible combinations of α1 and β3 isoforms (6,7). Collectively, these findings are consistent with molecular modeling studies including our docking simulations, which show that the binding energies of neurosteroids are very similar within the intersubunit pocket of WT and Q242W receptors or interfaces consisting of α1, β3, or γ2 subunits (32-34), and that hydrophobic interactions are the primary contributor to binding (9). The intersubunit neurosteroid binding site in the GABAAR, therefore, appears to be a hydrophobic “pocket” where binding affinity is not significantly affected by amino acid perturbations or variations that exist in other subunit isoforms. In fact, this intersubunit pocket is likely occupied by other lipid molecules including cholesterol, which also photolabels this site in GLIC (35), and phospholipids, which are resolved in this site in the GLIC crystal structure (36).
If mutations of Gln242 and Trp246 do not abolish the neurosteroid binding, how do they abolish neurosteroid effect? Our results confirm that these amino acid side chains form key interactions with the neurosteroid as evidenced by the marked change in KK200 photolabeling orientation within the intersubunit pocket of Gln242 and Trp246 mutants. Multiple studies support the presence of a hydrogen bond between the C3α hydroxyl and Gln242, and modeling studies have suggested that this interaction mediates transduction of the neurosteroid potentiating effect (32,33). The W246L mutant which alters neurosteroid binding orientation would preclude this hydrogen bond interaction that appears to be essential for neurosteroid effect. Alternatively, a simple hydrogen bond may not be the sole interaction necessary for neurosteroid effect, but rather a specific binding orientation that may result in other interactions. Indeed, neurosteroid analogues lacking a C3α hydroxyl or equivalent hydrogen donor have been shown to potentiate the GABAAR (7,13). The role of binding orientation also raises the interesting hypothesis that non-functional neurosteroid analogues (e.g. enantiomers, 3β-neurosteroids) do not potentiate the GABAAR because they bind in the intersubunit pocket with an altered orientation. Neurosteroid analogues that bind to the intersubunit site in different orientations may be competitive antagonists.
It also bears noting that the amino acids photolabeled by KK200 (Tyr309 in WT, and Phe298 in Q242L and Q242W) provide only a single anchor point for binding site identification. While we cannot be certain of the exact location of the neurosteroid backbone, photolabeling, crystallographic and modeling studies support the presence of a conserved steroid binding pocket containing Trp246 (7-9,12,32,33). Thus, for the Q242L and Q242W mutants, it is likely that the neurosteroid orientation has flipped such that the steroid backbone continues to interact with Trp246 and the TPD group points extracellular to label Phe298. The same flipped binding orientation was identified in GLIC where KK200 photolabeled the equivalent residue, Phe267, and multiple neurosteroid reagents were used to map binding to the intersubunit pocket (17).
Our photolabeling results in ELIC-α1GABAAR clearly identify two sites per subunit, an intersubunit and intrasubunit site. This is consistent with our findings in GLIC (17) and human α1β3GABAAR (7) showing the presence of a functionally important intrasubunit site. In contrast, crystal structures of neurosteroid with GLIC-α1GABAAR (8) and ELIC-α1GABAAR (12) only demonstrate binding to the intersubunit site. The reason for this discrepancy is unclear, but may be because neurosteroids bind to the intrasubunit site with lower affinity or because proteins with neurosteroid bound to this site fail to crystallize. Future studies examining the pharmacophore of neurosteroid binding to the intrasubunit site should be performed in native GABAARs since the extracellular domain may contribute to the structure of this site.
In summary, using an ELIC-GABAAR chimera as a model of the GABAAR TMD, we determined that mutations in the intersubunit binding pocket that abolish neurosteroid effect (Q242L, Q242W, W246L) do not significantly reduce neurosteroid binding. This result suggests that the intersubunit pocket does not require specific interactions to accommodate a neurosteroid, but rather a hydrophobic space with a volume that is not very stringent. Our data also suggest that interactions formed by Gln242 and Trp246 within this pocket determine a binding orientation where the C3α hydroxyl points into the membrane. Altering neurosteroid orientation may prevent a hydrogen bond interaction with Gln242 or potentially other interactions that are essential for the transduction of the neurosteroid effect. Identification of these interactions within other neurosteroid sites of action such as the intrasubunit binding site may provide an avenue for designing site-specific and isoform-specific modulators of GABAAR activity.
Supplementary Material
HIGHLIGHTS.
Neurosteroids photolabel ELIC-α1GABAAR at an intersubunit and intrasubunit site.
Mutations in the intersubunit site do not reduce neurosteroid photolabeling.
Mutations in the intersubunit site alter the orientation of neurosteroid binding.
Neurosteroids photolabel ELIC-α1β3GABAAR in multiple orientations.
ACKNOWLEDGEMENTS
We are grateful to Michael Gross and the Washington University Biomedical Mass Spectrometry Resource for use of their Thermo QExactive EMR. The authors thank Dr. Joe Henry Steinbach for insightful advice and valuable suggestions.
FUNDING
This work was funded by grants from the National Institutes of Health (NIH) including K08GM126336 for W.W.C, R01GM108799 for A.S.E. and D.F.C., R01GM108580 for G.A., and R01GM056257 for P.T.; from Juntendo University for Y.S.; and from the Taylor Family Institute for Innovative Psychiatric Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Belelli D, and Lambert JJ (2005) Neurosteroids: endogenous regulators of the GABA(A) receptor. Nat Rev Neurosci 6, 565–575 [DOI] [PubMed] [Google Scholar]
- 2.Akk G, Covey DF, Evers AS, Steinbach JH, Zorumski CF, and Mennerick S (2007) Mechanisms of neurosteroid interactions with GABA(A) receptors. Pharmacol Ther 116, 35–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reddy DS, and Estes WA (2016) Clinical Potential of Neurosteroids for CNS Disorders. Trends Pharmacol Sci 37, 543–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hosie AM, Wilkins ME, da Silva HM, and Smart TG (2006) Endogenous neurosteroids regulate GABAA receptors through two discrete transmembrane sites. Nature 444, 486–489 [DOI] [PubMed] [Google Scholar]
- 5.Wittmer LL, Hu Y, Kalkbrenner M, Evers AS, Zorumski CF, and Covey DF (1996) Enantioselectivity of steroid-induced gamma-aminobutyric acidA receptor modulation and anesthesia. Mol Pharmacol 50, 1581–1586 [PubMed] [Google Scholar]
- 6.Chen ZW, Manion B, Townsend RR, Reichert DE, Covey DF, Steinbach JH, Sieghart W, Fuchs K, and Evers AS (2012) Neurosteroid analog photolabeling of a site in the third transmembrane domain of the beta3 subunit of the GABA(A) receptor. Mol Pharmacol 82, 408–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen ZW, Bracamontes JR, Budelier MM, Germann AL, Shin DJ, Kathiresan K, Qian MX, Manion B, Cheng WWL, Reichert DE, Akk G, Covey DF, and Evers AS (2019) Multiple functional neurosteroid binding sites on GABAA receptors. PLoS Biol 17, e3000157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laverty D, Thomas P, Field M, Andersen OJ, Gold MG, Biggin PC, Gielen M, and Smart TG (2017) Crystal structures of a GABAA-receptor chimera reveal new endogenous neurosteroid-binding sites. Nat Struct Mol Biol 24, 977–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller PS, Scott S, Masiulis S, De Colibus L, Pardon E, Steyaert J, and Aricescu AR (2017) Structural basis for GABAA receptor potentiation by neurosteroids. Nat Struct Mol Biol 24, 986–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Puthenkalam R, Hieckel M, Simeone X, Suwattanasophon C, Feldbauer RV, Ecker GF, and Ernst M (2016) Structural Studies of GABAA Receptor Binding Sites: Which Experimental Structure Tells us What? Front Mol Neurosci 9, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sigel E, and Steinmann ME (2012) Structure, function, and modulation of GABA(A) receptors. J Biol Chem 287, 40224–40231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Q, Wells MM, Aijunan P, Tillman TS, Cohen AE, Xu Y, and Tang P (2018) Structural basis of neurosteroid anesthetic action on GABAA receptors. Nat Commun 9, 3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akk G, Li P, Bracamontes J, Reichert DE, Covey DF, and Steinbach JH (2008) Mutations of the GABA-A receptor alpha1 subunit M1 domain reveal unexpected complexity for modulation by neuroactive steroids. Mol Pharmacol 74, 614–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bracamontes JR, Li P, Akk G, and Steinbach JH (2012) A neurosteroid potentiation site can be moved among GABAA receptor subunits. J Physiol 590, 5739–5747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hosie AM, Clarke L, da Silva H, and Smart TG (2009) Conserved site for neurosteroid modulation of GABA A receptors. Neuropharmacology 56, 149–154 [DOI] [PubMed] [Google Scholar]
- 16.Hannan S, and Smart TG (2018) Cell surface expression of homomeric GABAA receptors depends on single residues in subunit transmembrane domains. J Biol Chem [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng WWL, Chen ZW, Bracamontes JR, Budelier MM, Krishnan K, Shin DJ, Wang C, Jiang X, Covey DF, Akk G, and Evers AS (2018) Mapping two neurosteroid-modulatory sites in the prototypic pentameric ligand-gated ion channel GLIC. J Biol Chem 293, 3013–3027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kinde MN, Bu W, Chen Q, Xu Y, Eckenhoff RG, and Tang P (2016) Common Anesthetic-binding Site for Inhibition of Pentameric Ligand-gated Ion Channels. Anesthesiology 124, 664–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang X, Shu HJ, Krishnan K, Qian M, Taylor AA, Covey DF, Zorumski CF, and Mennerick S (2016) A clickable neurosteroid photolabel reveals selective Golgi compartmentalization with preferential impact on proximal inhibition. Neuropharmacology 108, 193–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Budelier MM, Cheng WWL, Bergdoll L, Chen ZW, Janetka JW, Abramson J, Krishnan K, Mydock-McGrane L, Covey DF, Whitelegge JP, and Evers AS (2017) Photoaffinity labeling with cholesterol analogues precisely maps a cholesterol-binding site in voltage-dependent anion channel-1. J Biol Chem 292, 9294–9304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marty MT, Baldwin AJ, Marklund EG, Hochberg GK, Benesch JL, and Robinson CV (2015) Bayesian deconvolution of mass and ion mobility spectra: from binary interactions to polydisperse ensembles. Anal Chem 87, 4370–4376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee J, Cheng X, Swails JM, Yeom MS, Eastman PK, Lemkul JA, Wei S, Buckner J, Jeong JC, Qi Y, Jo S, Pande VS, Case DA, Brooks CL 3rd, MacKerell AD Jr., Klauda JB, and Im W (2016) CHARMM-GUI Input Generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM Simulations Using the CHARMM36 Additive Force Field. J Chem Theory Comput 12, 405–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McGibbon RT, Beauchamp KA, Harrigan MP, Klein C, Swails JM, Hernandez CX, Schwantes CR, Wang LP, Lane TJ, and Pande VS (2015) MDTraj: A Modern Open Library for the Analysis of Molecular Dynamics Trajectories. Biophys J 109, 1528–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trott O, and Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31, 455–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, and Hutchison GR (2011) Open Babel: An open chemical toolbox. J Cheminform 3, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meslamani JE, Andre F, and Petitjean M (2009) Assessing the Geometric Diversity of Cytochrome P450 Ligand Conformers by Hierarchical Clustering with a Stop Criterion. J Chem Inf Model 49, 330–337 [DOI] [PubMed] [Google Scholar]
- 27.Hattori M, Hibbs RE, and Gouaux E (2012) A fluorescence-detection size-exclusion chromatography-based thermostability assay for membrane protein precrystallization screening. Structure 20, 1293–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller PS, and Aricescu AR (2014) Crystal structure of a human GABAA receptor. Nature 512, 270–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Covey DF (2009) ent-Steroids: novel tools for studies of signaling pathways. Steroids 74, 577–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Das J (2011) Aliphatic diazirines as photoaffinity probes for proteins: recent developments. Chem Rev 111, 4405–4417 [DOI] [PubMed] [Google Scholar]
- 31.Brunner J (1993) New photolabeling and crosslinking methods. Annu Rev Biochem 62, 483–514 [DOI] [PubMed] [Google Scholar]
- 32.Alvarez LD, and Pecci A (2018) Structure and dynamics of neurosteroid binding to the alpha1beta2gamma2 GABAA receptor. J Steroid Biochem Mol Biol 182, 72–80 [DOI] [PubMed] [Google Scholar]
- 33.Alvarez LD, Pecci A, and Estrin DA (2018) In Search of GABAA Receptor s Neurosteroid Binding Sites. J Med Chem [DOI] [PubMed] [Google Scholar]
- 34.Alvarez LD, Veleiro AS, and Burton G (2015) Exploring the molecular basis of action of ring D aromatic steroidal antiestrogens. Proteins 83, 1297–1306 [DOI] [PubMed] [Google Scholar]
- 35.Budelier MM, Cheng WWL, Chen ZW, Bracamontes JR, Sugasawa Y, Krishnan K, Mydock-McGrane L, Covey DF, and Evers AS (2018) Common binding sites for cholesterol and neurosteroids on a pentameric ligand-gated ion channel. Biochim Biophys Acta Mol Cell Biol Lipids 1864, 128–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sauguet L, Poitevin F, Murail S, Van Renterghem C, Moraga-Cid G, Malherbe L, Thompson AW, Koehl P, Corringer PJ, Baaden M, and Delarue M (2013) Structural basis for ion permeation mechanism in pentameric ligand-gated ion channels. EMBO J 32, 728–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.