Abstract
Special AT-rich sequence binding protein 2 (Satb2) is a matrix attachment region (MAR) binding protein. Satb2 impacts skeletal development by regulating gene transcription required for osteogenic differentiation. Although its role as a high-order transcription factor is well supported, other roles for Satb2 in skeletal development remain unclear. In particular, the impact of dosage sensitivity (heterozygous mutations) and variance on phenotypic severity is still not well understood. To further investigate molecular and cellular mechanisms of Satb2-mediated skeletal defects, we used the CRISPR/Cas9 system to generate Satb2 mutations in MC3T3-E1 cells. Our data suggest that, in addition to its role in differentiation, Satb2 regulates progenitor proliferation. We also find that mutations in Satb2 cause chromatin defects including nuclear blebbing and donut-shaped nuclei. These defects may contribute to a slight increase in apoptosis in mutant cells, but apoptosis is insufficient to explain the proliferation defects. Satb2 expression exhibits population-level variation and is mostly highly expressed from late G1 to late G2. Based on these data, we hypothesize that Satb2 may regulate proliferation through two separate mechanisms. First, Satb2 may regulate the expression of genes necessary for cell cycle progression in pre-osteoblasts. Second, similar to other MAR-binding proteins, Satb2 may participate in DNA replication. We also hypothesize that variation in the severity or penetrance of Satb2-mediated proliferation defects is due to stochastic variation in Satb2 binding to DNA, which may be buffered in some genetic backgrounds. Further elucidation of the role of Satb2 in proliferation has potential impacts on our understanding of both skeletal defects and cancer.
Keywords: Satb2, MAR-binding protein, chromatin defects, osteoblast differentiation
1. Introduction
Bone formation in development and regeneration requires proliferative expansion of pre-osteoblasts and their subsequent differentiation. These two processes rely on the function and temporal coordination of multiple transcriptional regulators. In particular, Runx2 and Sp7/Osx are essential to osteogenesis as deletion of either impairs osteoblast differentiation and eliminates bone formation (Komori et al., 1997; Nakashima et al., 2002). Runx2 regulates the expression of Sp7/Osx as well as the bone matrix proteins Ocn, Bsp, and Opn (Komori, 2005). Runx2 also promotes the expression of Atf4, a regulator of terminal differentiation in osteoblasts (Yang et al., 2004).
The function of these transcription factors is augmented by Satb2 (Special AT-rich sequence binding protein 2), a protein that binds to nuclear matrix-attachment regions (MARs). It is thought that Satb2 organizes chromatin looping at MARs such that the expression of cell-type specific differentiation genes may be enhanced (Britanova et al., 2005). In mice, loss of Satb2 results in skeletal hypoplasia and reduced expression of osteogenic genes including Ocn and Bsp (Dobreva et al., 2006). In contrast, Satb2 over-expression was found to promote bone formation and increase Ocn and Bsp expression (Gong et al., 2014; Zhang et al., 2011). These results are consistent with skeletal malformations in humans with SATB2-associated syndrome (SAS), which include bowing tibias, low bone mineral density with high fracture rate, osteoporosis, cleft palate, micrognathia, and maxillary hypoplasia (Boone et al., 2016; Leoyklang et al., 2007; Zarate and Fish, 2017; Zarate et al., 2018a; Zarate et al., 2018b).
One role of Runx2 in pre-osteoblasts is to down-regulate the miR cluster 23a/27a/24–2. These miRs target the 3’ UTR of Satb2, and thus Runx2 alleviates the repression of Satb2 (Hassan et al., 2010). These and other data have placed Satb2 downstream of Runx2 in the osteogenic differentiation program, with mutations in Satb2 largely thought to impair the expression of key genes involved in osteoblast maturation (Dobreva et al., 2006; Gong et al., 2014; Zhang et al., 2011). Although the majority of investigations into Satb2 function in osteogenesis have focused on its role in gene regulation, loss of Satb2 has also been reported to cause apoptosis in neural crest progenitors of the jaw (Britanova et al., 2006; Fish et al., 2011).
To further explore molecular and cellular mechanisms underlying Satb2-mediated defects in osteogenesis, we used the CRISPR/Cas9 system to generate mutations in the Satb2 locus in MC3T3-E1 cells, which are clonal pre-osteoblasts that have been widely used as a model of osteoblast differentiation (Xiao et al., 1997). Several modified colonies with different combinations of Satb2 alleles producing a range of Satb2 expression levels were selected for experimental analysis. Consistent with previous reports, we observe altered osteogenic gene expression in Satb2 mutants. However, we also find that reduction in Satb2 expression, even that of true heterozygotes, results in reduced pre-osteoblast proliferation. Further, mutations in Satb2 cause nuclear aberrations including nuclear blebbing and donut-shaped nuclei.
Our data point to a novel role for Satb2 in mediating pre-osteoblast proliferation, indicating that Satb2 is involved in both proliferative expansion of pre-osteoblasts and their subsequent differentiation. Previous studies failing to observe proliferation defects in Satb2 mutants may reflect buffering of this process in some genetic backgrounds. The potential role of genetic background in regulating susceptibility to Satb2-mediated proliferation defects may help explain variation in penetrance and severity of skeletal defects in SAS patients. Participation in both proliferation and differentiation processes makes Satb2 an interesting target for bone regeneration and osteoporosis therapies (Guo et al., 2016; Zhang et al., 2011). Finally, our observation that mutations in Satb2 both reduce proliferation and cause nuclear aberrations has implications for understanding its dichotomous potential for both tumor suppression and oncogenesis (Chen and Costa, 2018; Chen et al., 2019).
2. Materials and Methods
Cell lines
Mouse osteoblast precursor cells (MC3T3-E1 Subclone 4) obtained from ATCC were grown in αMEM supplemented with 10% FBS and Penicillin/Streptomycin. Passage numbers higher than 20 were discarded.
Generation of mutant lines using CRISPR/Cas9
To generate indel mutations near the start codon of Satb2 (Fig. 1A), we used a previously validated guide RNA, Satb2-272, ATCGGAGCAGCATGGAGCGG (Shinmyo et al., 2016). The guide RNA was cloned into pSpCas9(BB)-2A-Puro (PX459) V2.0 (Addgene plasmid # 62988), as previously described (Ran et al., 2013). Cells were transfected using the NEON transformation system (Invitrogen) using one 1200V pulse and a 30 ms pulse width and otherwise following the manufacturer’s protocol. 48-hours after electroporation, 3 μg/mL Puromycin was added to the cells for 24 hours. Surviving cells were diluted to 2.5 cells/ml and seeded into 96-well plates at 0.5 cells per well to obtain single cell colonies. Single colonies were grown to confluency and then expanded.
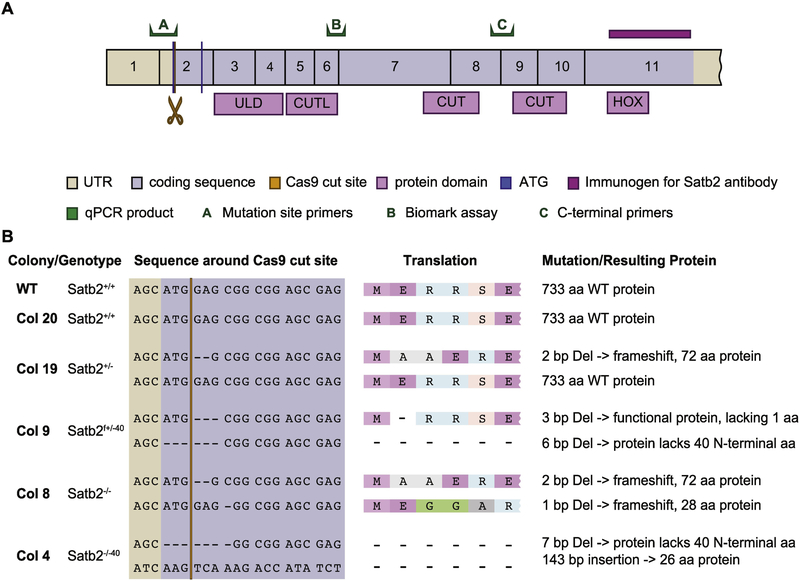
Figure 1: Overview of CRISPR strategy and outcomes.
A) Diagram of Satb2 protein with the location of the Cas9 cut site, qPCR primer sets, and the binding region of the antibody used in this study highlighted. B) Colony cell lines used in this study are described. The colony number, Satb2 genotype, cut site DNA sequence and translation, as well as protein product produced are listed.
Analysis of mutant cell lines
DNA was extracted from single cell colonies using the ZR Duet DNA/RNA Miniprep Kit (Zymo). A 708 bp PCR product around the editing site of Satb2-272 was amplified using primers Satb2-seq-F ACTGGCCTGATCGTCTATCA and Satb2-seq-R GCCAGATCCTAGGTCTCTGT. PCR fragments were cloned into TOPO pCR4 (Invitrogen) and sequenced with M13 F+R primers using Sanger sequencing to determine the exact sequence of each allele.
Differentiation assay
An overview of the differentiation assay is shown in Figure 6A. Cells were plated in a 12-well plate at 5 × 104 cells per well. After 3 days, differentiation was induced (=T0) with Dulbecco’s modified Eagle Medium supplemented with 10% FBS, Penicillin/Streptomycin, 50 μg/ml beta-glycerophosphate, and 200 μM ascorbic acid. The media was changed every 2–3 days over the differentiation period (0–28 days). Mineralization was detected with 1% (w/v) Alizarin S pH 4.2 for 1 h and imaged on an Epson Perfection 4990 Photo scanner and an Olympus BX41 microscope. After imaging, Alizarin Red S was extracted from three separate differentiation assays using acetic acid following published protocols (Gregory et al., 2004) and the absorbance at 405 nm was measured on a SpectraMax M2 microplate reader.
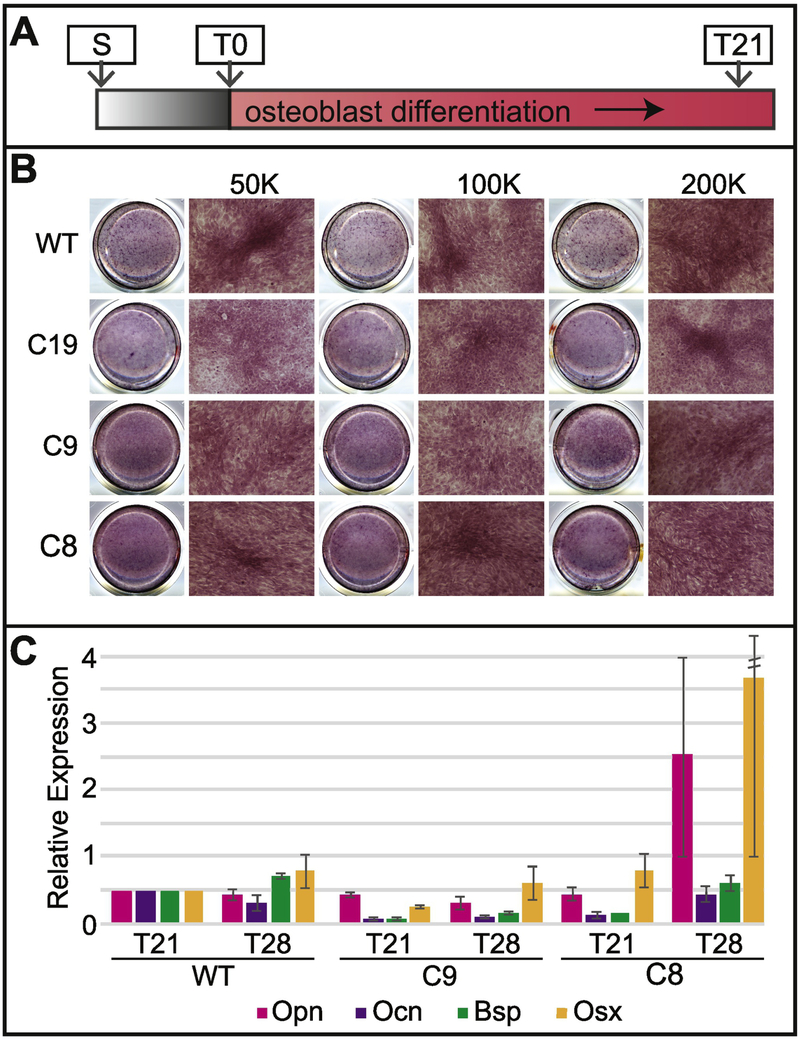
Figure 6: Mutations in Satb2 reduce, but don’t inhibit, osteogenic differentiation.
A) In vitro differentiation assay timeline. B) Overview scans of wells (left) and 10x magnification of Alizarin Red staining on WT, C19, C9, and C8 cells after 21 days (T21) in differentiation. Data are shown for seeding densities of 50,000 cells, 100,000 cells, and 200,000 cells. Note that clusters start to form when mutant cells are plated at higher density (compare asterisk in C19 at 200K to WT at 50K). C) Expression of Opn (pink bars), Ocn (purple bars), Bsp (green bars), and Osx (yellow bars) is shown relative to WT at T21. Data are the means of three experiments with standard deviation. Standard deviation for Osx in C8 at T28 is 5.01.
Immunohistochemistry
Cells were fixed in 4%PFA/PBS for 15 minutes, permeabilized with 0.1% Triton-X-100/PBS and then blocked in 5% FBS supplemented with 0.1% BSA for 1 h. For BrdU, cells were treated with an additional incubation in 2N HCl for 1 h at room temperature after the permeabilization. Primary antibodies were incubated overnight at 4°C in blocking solution at the following concentrations: mouse anti-Satb2 (SATBA4B10, SantaCruz) 1:300; mouse anti-alpha Tubulin (DM1A) 1:500; mouse anti-Lamin B1 (B-10, SantaCruz) 1:100; rabbit anti-Lamin A/C (Cell Signaling Technology) 1:100; rabbit anti-BrdU (Abcam) 1:50. Cells were imaged on a Nikon AR-1, a Leica Sp8, or a Zeiss Axiovert 200M microscope. Images were processed using the Fiji distribution of ImageJ and Adobe Photoshop CC 2018 (Schindelin et al., 2012).
Imaging flow cytometry
Cells for the flow analysis were plated at 4 × 105 four days before the experiment. After trypsinizing, cells were resuspended in media and counted using a hemocytometer. Cells were spun down, resuspended in PBS to 1 × 106, and incubated for 15 min at room temperature in the dark in 1x Annexin Binding Buffer (Invitrogen) supplemented with 2 μl Annexin V, Alexa Flour 488 conjugate (Invitrogen) and 0.5 μg/ml propidium iodide (Alfa Aesar). After a final centrifugation step, cells were resuspended in 100 μl 1x Annexin Binding Buffer and then analyzed immediately on an Amnis imaging flow cytometer with a 20x objective. Both Annexin V A488 and PI were excited using the 488 laser. Brightfield (channel 01 and 09, 435–480 nm and 570–595 respectively), Annexin V A488 (channel 02, 480–560 nm) and PI (channel 04, 595–642 nm) were measured and 10000 single cell events (see gating below) were counted and all events collected. Color compensation was achieved using single-stained samples that contained cells in which apoptosis was induced by incubating cells for 24–48h with 5 μM camptothecin. Focused cells were gated using the gradient root mean square of the bright field image, and bright field area and aspect ratio were used to gate single cells. Using fluorescence intensity of channel 02 (Annexin V A488) and channel 04 (PI), cells were divided into double negative (live) cells, Annexin V-positive cells (early apoptotic) and double positive cells (late apoptotic and necroptotic).
Satb2 over-expression
Satb2 was amplified from mouse cDNA using primers Satb2_cds_F CCAAATCGGAGCAGCATGG and Satb2_cds_R TTATCTCTGGTCGGTTTCGG. The PCR product was then cloned into the pCAGIG vector (gift from Connie Cepko; Addgene plasmid # 11159; http://n2t.net/addgene:11159; RRID:Addgene_11159) at the EcorRV cut site (Matsuda and Cepko, 2004). This vector contains a CMV enhancer, a CAG promoter, and an IRES-EGFP. Cloning was confirmed by sequencing. Expression was confirmed by qPCR and immunohistochemistry. Cells were transfected with Lipofectamine 3000 with a DNA concentration of 0.5μg/μl.
qPCR
RNA was collected with the illustra triplePrep kit (GE healthcare) or TRIzol (Invitrogen). RNA was DNase-treated with the TURBO DNA-Free kit (Invitrogen) and quantified using a Qubit 3 Fluorometer (Invitrogen). cDNA was generated with 400ng of RNA with iScript RT Supermix for RT-qPCR (BioRad). GAPDH and Satb2 primers were from (Gong et al., 2016), Satb2 cut (F: CCCCAGCCAGCCAAGTTTCA, R: GCCGGAGTCTGTTCACTACC), Runx-2 (F: ATTCGTCAACCATGGCCCAG, R: GAAACCCAGTTATGACTGCCC), Bsp/Ibsp (F: ATGGAGACGGCGATAGTTCC, R: ACACCCGAGAGTGTGGAAAG), Ocn/Bglap (F: TTCTGCTCACTCTGCTGACC, R: GCCGGAGTCTGTTCACTACC), Osx/Sp7 (F: GCCGGAGTCTGTTCACTACC, R: GCCGGAGTCTGTTCACTACC). Fold change was calculated using the delta-delta C(t) method (Livak and Schmittgen, 2001).
Single Cell qPCR
Cells were trypsinized and at T14 or later also treated with collagenase type 1 (Worthington) for 15–30 min at 37°C. Cells were loaded onto a C1 Single-Cell Preamp integrated fluidic circuit (IFC) with 17–25 μm capturing well size. Primers for pre-amplification were obtained from Fluidigm (Delta Gene Assays, see Supplementary Table 1). After single-cell capturing, the inlets of the IFC were microscopically checked for the presence of single cells. Empty inlets or inlets with two or more cells were recorded and excluded from the qPCR analysis. Lysis, reverse transcription and preamplification was performed according to protocol. Single cell qPCR was performed using the Fluidigm BioMark system on 96.96 Dynamic Array IFCs for Gene Expression using Delta Gene Assays and Sso Fast EvaGreen Supermix with Low ROX (Bio-Rad). Single cell qPCR was performed using the Fluidigm BioMark system on 96.96 Dynamic Array IFCs for Gene Expression using Delta Gene Assays and 2X SsoFast EvaGreen Supermix with low ROX (Bio-Rad). For data-processing, the Fluidigm Real-Time PCR Analysis Software v4.5.2 was used to eliminate failed assays. The default quality threshold cutoff (0.65) was used to identify potential artifacts. The baseline correction was to linear, and the Ct threshold detection method was set to Auto (Detectors). The Fluidigm Singular Analysis Toolset v3.6.2 was used in R to analyze single cell gene expression. The statistical computing software R was used to identify patterns in gene expression.
3. Results
3.1. Satb2 mutant lines
We selected several CRISPR/Cas9-modified cell lines with different types of mutations for analysis. Our selected colonies include two heterozygous lines and two homozygous mutant lines (Fig. 1). Of the heterozygous lines, one is a true heterozygote with one WT allele and one loss of function allele (colony 19), while the other heterozygote has an allele missing a single amino acid at the N-terminal end (with predicted WT function) and an allele with a N-terminal 6bp deletion that includes the ATG (colony 9). Of the two homozygous mutant lines, one is a complete loss of function mutant with two alleles carrying frameshift deletions (colony 8). The other has one allele with a 7bp deletion and one allele with a 143bp insertion, both removing the ATG (colony 4). We also included colony 20, a line a line that was subjected to our CRISPR/Cas9 protocol but was genotyped as WT after selection.
Satb2 mRNA and protein levels were evaluated in each of these lines by qPCR and Western blot, respectively. Using two sets of primers (A and C in Fig.1A), we find that no full-length mRNA is produced for any allele containing a mutation in the start codon (SupFig. 1A). Using primers directed against the C-terminal end of the gene, we find Satb2 mRNA is produced in all our cell lines. However, some of this mRNA (especially that of colony 8) is predicted to form non-functional (frame-shifted) protein or not be translated at all. Western blot confirmed colony 8 is a loss of function mutant (SupFig. 1B,C). Similarly, colony 19 produces roughly half the amount of Satb2 protein relative to WT, as expected for a true heterozygote. Interestingly, we found that the short N-terminal deletions found in colonies 9 and 4 produce a protein. Although the start codon is deleted in both affected alleles, another ATG exists 120bp downstream. Our data are consistent with this allele producing a Satb2 protein missing the first 40 amino acids. These 40 amino acids are not part of any previously described functional domains (Fig. 1A).
Immunostaining using an antibody targeted against the C-terminal end of Satb2 provides results consistent with our Western blot analyses (Fig. 2). Importantly, the immunostaining data confirm the production of protein lacking the N-terminal 40 amino acids (see C4 in Fig. 2). Also of note is that in colony 9, which produces 2 Satb2 proteins of different sizes, the shorter protein appears to be preferentially produced (SupFig. 1B,C). This preference for producing the shorter protein has been observed in 4 separate Western blots.
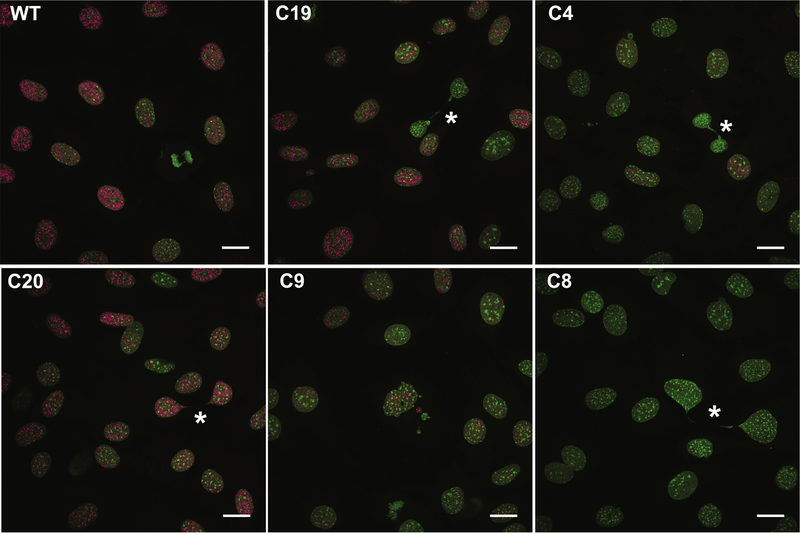
Figure 2: Mutations in Satb2 reduce protein levels.
Confocal maximum intensity projections showing representative Satb2 protein levels in undifferentiated (T0) cells of colonies used in this study. Note the presence of Satb2 protein in C4, indicating the production of a mutant protein, as well as the absence of protein in C8. DNA in green; Satb2 in pink. Asterisks highlight chromatin bridges. All scale bars represent 10 μm.
3.2. Satb2 and the cell cycle
When assessing Satb2 levels, we noticed that in all cell lines, Satb2 exhibits population variation (Fig. 2). In interphase cells, Satb2 ranges from intense to weak, although always associated with euchromatin. Satb2 is present in early mitosis, however it is no longer associated with chromatin, and is absent by late mitosis (Fig. 2 and 3A). As part of our experiments to investigate variation in osteogenic gene expression (described below), we performed single cell gene expression analyses. Interestingly, we found that in WT cells, Satb2 expression levels cluster among cell cycle regulators rather than with genes associated with osteogenic differentiation (SupFig. 2). Notably, the relative expression of a sub-set of cell cycle genes associated with late G1 and S phases separated the cells into two main clusters (SupFig. 2). In particular, the genes Cdc25a (G1-S), Cdc6 (S), Cdk2 (G1-S), and Cdk1 (S-M), along with Satb2, were highly expressed in one cluster relative to the other (see rectangle, SupFig. 2). In contrast, expression of genes associated with G1 such as Cdcn1, Cdcn2, Cdk4, and Cdk6, was more widely distributed among cells. These data suggest that Satb2 is cell cycle regulated and/or regulates cell cycle genes.
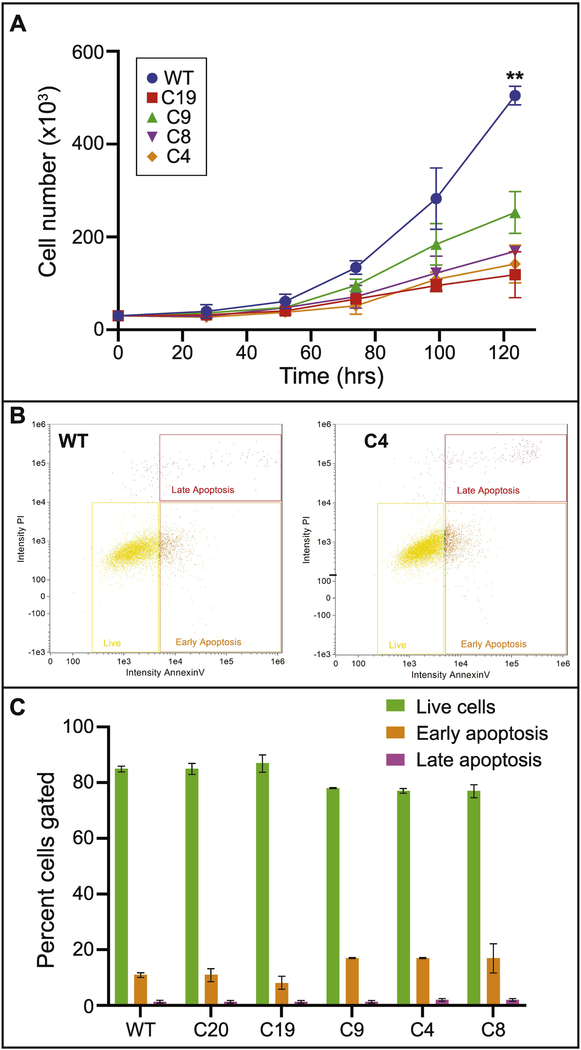
Figure 3: Mutations in Satb2 reduce pre-osteoblast proliferation rates.
A) Growth curves for undifferentiated (T0) cells. Data represent the means of three replicate experiments, with 95% confidence intervals shown. Asterisks indicate significance at p<0.0001. B) Representative flow cytometry detection of Annexin V (x-axis) and propidium iodide (y-axis) staining in WT (left) and C4 (right) cells. Yellow rectangles indicate live cells, orange boxes indicate early apoptotic cells, and red boxes indicate late apoptotic cells. C) Quantification of flow cytometry for live cells (green bars), early apoptotic cells (orange bars), and late apoptotic cells (magenta bars) for all colonies is shown. Data represent the means of two replicates with standard deviation.
To further investigate Satb2 variation during the cell cycle, we evaluated Satb2 levels in cells that had been treated with BrdU for 2 hours. We binned cells into 3 classes of Satb2 levels. Low to no Satb2 protein is observed in cells in anaphase through early G1 (see Fig. 2 and SupFig3). BrdU positive cells, indicating cells in S and G2 have high levels of Satb2, however, high Satb2 levels are also observed in BrdU negative cells (SupFig. 3). Taken together, these data suggest that Satb2 is most active from late G1 to late G2.
3.3. Mutations in Satb2 reduce pre-osteoblast proliferation
Previous work has reported a role for Satb2 in regulating osteogenic differentiation, but not found a role in osteoblast proliferation (Dobreva et al., 2006). We find that mutations in one or more Satb2 alleles result in reduced proliferative capacity of pre-osteoblasts (Fig. 3). All of the mutant cell lines proliferate significantly more slowly than WT (p<0.0001 for all colonies relative to WT). Notably, colony 4, which expresses only Satb2 protein with the 40aa N-terminal deletion, proliferates even more slowly than all of the other lines, including colony 8, which lacks all Satb2 protein (Fig. 3A). Additionally, Satb2 mutant cells have a greater than 50% reduction in mitotic cells compared to WT cell lines (Table 1).
Table 1: Quantification of nuclear morphology.
Fixed cells were stained with Hoechst to visualize nuclear morphology. Nuclei were scored for mitosis, apoptosis, large size, donut-like morphology, and nuclear blebbing. For each colony, an area of larger than 3 mm2 was counted. Images used for counting are available at http://dx.doi.org/10.17632/6yfs85wyy6.1.
| Nuclear phenotype | WT | C20 | C19 | C9 | C8 | C4 |
|---|---|---|---|---|---|---|
| Mitotic | 51 | 50 | 16 | 18 | 11 | 23 |
| Apoptotic | 2 | 8 | 4 | 14 | 9 | 10 |
| Chromatin bridge | 15 | 4 | 24 | 15 | 6 | 5 |
| Large nucleus | 1 | 6 | 3 | 3 | 8 | 7 |
| Donut nucleus | 1 | 1 | 2 | 5 | 2 | 0 |
| Nuclear blebbing | 4 | 1 | 18 | 34 | 9 | 30 |
Decreased proliferation may be also be caused by increased apoptosis. We observed apoptotic cells in our immunostaining assays, but quantification of fixed cells did not indicate a significant increase in numbers (Table 1). This may be due to loss of cells during fixation and staining and/or lack of appropriate markers. Therefore, we turned to imaging flow cytometry to quantify dead and dying cells using Annexin V to identify early apoptotic and propidium iodide to identify late apoptotic cells. There is some variation in early apoptotic cell numbers, but very little difference in late apoptotic cell numbers (Fig. 3B,C). We observe minor variation in these percentages between experiments, but the overall pattern of only subtle increases in apoptosis in our mutant cell lines is consistent. Notably, our loss of function mutant (colony 8) has similar numbers of apoptotic cells as WT (and colony 20), yet still proliferates much more slowly. Taken together with our quantifications on fixed cells, these data indicate that mutations in Satb2 mildly increase apoptosis, but that apoptosis is not sufficient to explain reductions in proliferation.
3.4. Mutations in Satb2 generate nuclear aberrations
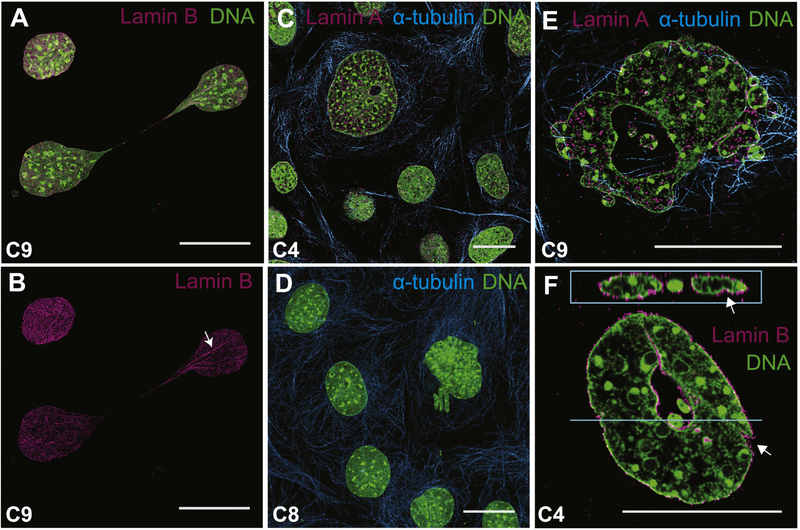
In our immunostaining assays to detect Satb2 levels, we noticed a number of nuclear aberrations in the mutant cell lines. We defined 4 types of nuclear aberration (Fig. 4): chromatin bridges (Fig. 4A,B), nuclei with diameters more than twice the normal size (large nuclei; Fig. 4C), nuclear blebbing or chromatin herniation (Fig. 4D–F), and donut-shaped nuclei (Fig. 4E,F). We quantified the abundance of these aberrations in fixed cells (Table 1). All 4 of these aberrations appear more frequently in all of our mutant cell lines relative to WT, suggesting that the aberrations are specific to mutations in Satb2. However, to address the possibility that these aberrations are a consequence of CRISPR/Cas9 treatment, we also quantified cells from colony 20, a WT cell line.
Figure 4: Mutations in Satb2 cause aberrant nuclear morphology.
Confocal images of Satb2 mutant osteoblasts showing representative nuclear aberrations. A,B) C9 cells with a chromatin bridge. Lamin B immunostaining highlights nuclear folds being pulled into bridge (arrow in B). C) Large cell with small hole shown relative to other normal-sized cells. D) Chromatin herniation from nucleus. E, F) Nuclear blebbing and donut-shaped nuclei in C9 (E) and C4 (F) cells. Note the presence of nuclear folds in F (arrows). DNA in green; alpha-tubulin in teal; lamin in pink (Lamin A in C and E; Lamin B in A, B, and F). Images in A, B, C, and D are maximum intensity projections. Images in E and F are single confocal sections. Panel F includes a z-section at the top. All scale bars represent 10 μm.
In both WT and colony 20 cells, the presence of nuclear aberrations was rare, and always more subtle than those observed in the mutant cell lines. For example, chromatin bridges, which were the most common aberrant phenotype seen in WT cells, are typically present as very thin string-like strands between nuclei (see Fig. 2B). In contrast, chromatin bridges in the mutant cell lines were often thick as shown in Figure 4B, where Lamin B immunostaining shows folds in DNA within the bridge. Similar folds in DNA can be observed in Figure 4F. Finally, while WT cells exhibit the occasional small nuclear bleb, we only observe the catastrophic blebbing shown in Figure 4E in the mutant cell lines. Large panel overviews of all 6 cell lines from which quantifications were made are available on Mendeley (http://dx.doi.org/10.17632/6yfs85wyy6.1).
3.5. Satb2 over-expression reduces nuclear aberrations in mutant cells
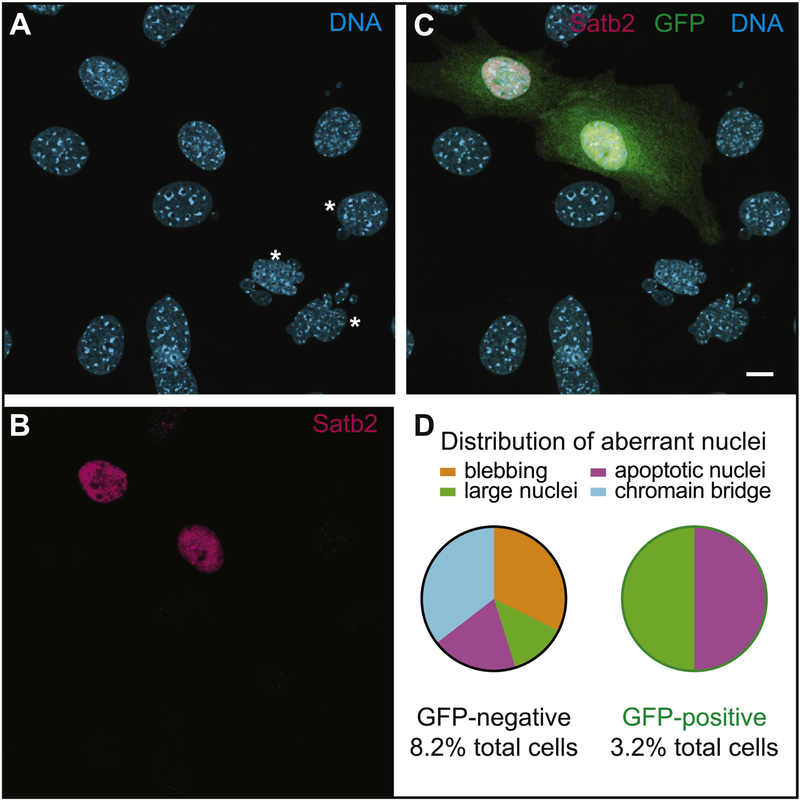
To further investigate the specificity of these defects, we cloned Satb2 into a mammalian expression vector with an IRES-GFP (Matsuda and Cepko, 2004), and transfected C4 cells. Transfected cells express both Satb2 and GFP (Fig. 5). We also confirmed over-expression by qPCR (SupFig. 1A). We did not find any effect on proliferation (data not shown). This may be due to incomplete transfection efficiency or due to the tendency of Satb2 over-expression to drive differentiation (Hu et al., 2015). Although we used a low DNA concentration for transfection, we cannot exclude this possibility. We did, however, find that GFP-positive C4 cells have a reduced incidence of abnormal nuclei, with 8.2% for GFP-negative and 3.2% for GFP-positive. Apoptosis (1.6% in both) and large nuclei (1.1% and 1.6%, respectively) occur at a similar percentage in the two populations, however, we did not observe any donut-shaped nuclei or excessive blebbing in GFP positive cells (Fig. 5D).
Figure 5: Over-expression of Satb2 in mutant cells reduces the frequency of abnormal nuclei.
Confocal images of C4 cells transfected with a Satb2-GFP over-expression plasmid. A) DNA stained with Hoechst. Asterisks highlight abnormal nuclei. B) Satb2 (magenta) is detected in nuclei of transfected cells. C) Overlay showing GFP-positive transfected cells. D) Pie-chart showing distribution of different types of nuclear aberrations observed in GFP-negative and GFP-positive C4 cells. Scale bar represents 10 μm.
3.6. Mutations in Satb2 reduce osteogenic differentiation
To investigate how Satb2 mutations in our cell lines affect osteogenic differentiation, we performed an in vitro differentiation assay (Fig. 6). Differentiation was evaluated by histology (alizarin red) and gene expression (qPCR). All of our mutant cell lines appear to differentiate less completely than WT cells (Fig. 6; SupFig4A). While all of the lines exhibit some mineralization, as noted by Alizarin red staining, none of the colonies form as many nodules of punctate staining as observed in WT cells (Fig. 6B). To further quantify mineralization, we extracted the Alizarin from the wells and measured its absorption at 405nm (SupFig4B). The trend is for all mutant lines to have less Alizarin, but only colony 19 is significantly less (p<0.05). We also found that the expression of bone matrix genes is reduced in all Satb2 mutant cell lines (Fig. 6C).
As reported above, our mutant cell lines have proliferation defects. Therefore, we tested whether differentiation potential was affected by cell density at the onset of differentiation. We plated cells at 3 different densities prior to differentiation, and also extended our differentiation protocol by an additional week (28 vs. 21 days). Increasing cell number at the onset of differentiation appears to improve osteogenic output as determined by the presence of mineralized nodules (Fig. 6B). Additionally, extending the differentiation time leads to increased expression of bone matrix proteins (Fig. 6C). Interestingly, colony 8, the loss of function line, differentiates better than colonies 9 and 19, two heterozygous lines. These data suggest that proliferation defects reducing cell density at the onset of differentiation contribute to reduced osteogenesis in cells with Satb2 mutations.
3.6. Satb2 mutations increase gene expression variance
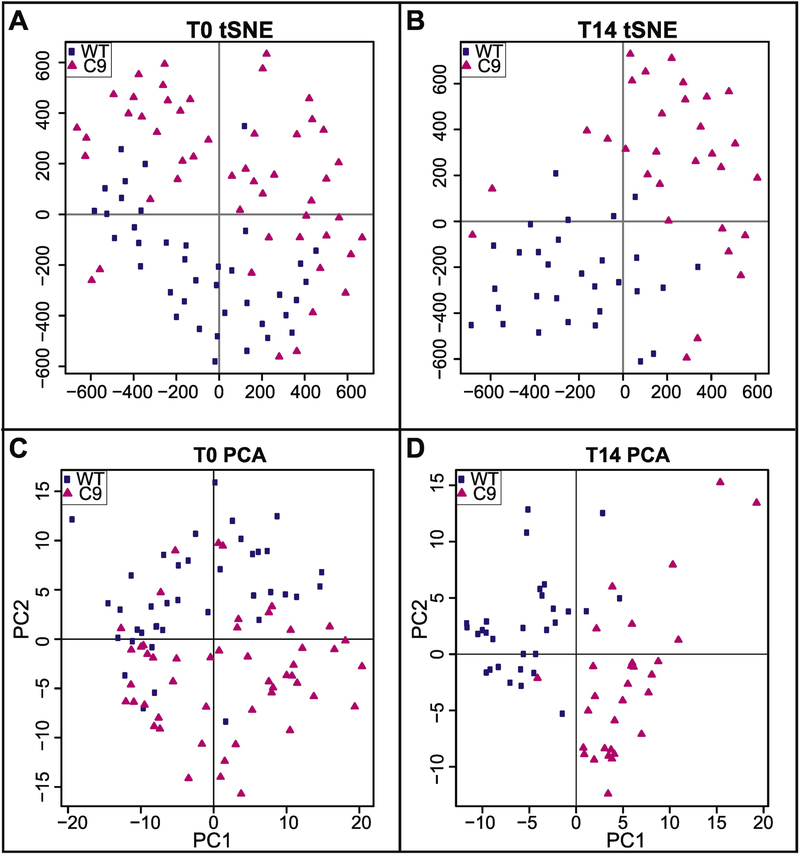
We used single cell gene expression analyses to evaluate how mutations in Satb2 affect osteogenic gene expression. Expression was quantified for 93 genes including 66 related to osteogenesis, 18 related to cell cycle progression, and 7 related to apoptosis (Supplementary Table 1). Down-regulation of Satb2 alters the expression profile of differentiating (T14) osteoblasts (Fig. 7 and SupFig. 5). Single cell expression profiles can be visualized using tSNE plots (Fig. 7A,B) which represent all the variation in the dataset and PCA plots (Fig. 7C,D) which represent 46% and 28% of the variation, respectively. As differentiation proceeds, gene expression becomes increasing distinct between WT and mutant cells. At T0, there is no significant difference between the means or variance the two populations, however by T14, gene expression shows more variance between individual mutant cells than is observed in WT cells (T0, F=1.11, n.s; T14, F=3.19, p=0.01).
Figure 7: Reduction in Satb2 increases gene expression variance.
Upper panels show t-distributed Stochastic Neighbor Embedding (tSNE) plots of single cell gene expression in wild-type (WT; blue squares) and colony 9 (C9; pink triangles) cells at A) T0 (initial dimensions= 80, perplexity=15, iterations 4000, error was 0.528 at the 4000th iteration) and B) T14 (initial dimensions= 80, perplexity=10, iterations 3000, error was 0.524 at the 3000th iteration). Lower panels show plots of principal components 1 and 2 from Principal Components Analysis (PCA) on single cell gene expression in WT (blue squares) and C9 (pink triangles) cells at C) T0 and D) T14.
The increased variation of gene expression patterns in mutant cells can also be observed by evaluating the number of genes that correlate with Satb2 (SupFig. 5). At T14, the expression of 16 genes positively correlates with Satb2 in WT cells (correlation coefficient of 0.5 and above). In contrast, only 4 genes show correlated expression with Satb2 in T14 mutant cells. No genes in either cell population negatively correlate with Satb2. These data suggest that large-scale alterations to gene expression occur when Satb2 is down-regulated, and that Satb2 mutations lead to increased population variation in osteoblast gene expression.
4. Discussion
4.1. Cell biological mechanism of osteogenic defect
Although it is generally accepted that loss of Satb2 causes bone hypoplasia and low bone mineral density, there is some discrepancy among the reported molecular and cellular mechanisms mediating these phenotypes. In a study focusing on the developing jaw, loss of Satb2 was associated with progenitor cell death, and more specific effects of Satb2 on osteogenic differentiation (in the facial or axial skeleton) were not reported (Britanova et al., 2006). That study also found that Satb2+/− heterozygous mice have phenotypes of the craniofacial skeleton that are intermediate between WT and mutant. In a separate study, loss of Satb2 was associated with deficiencies in osteogenic gene expression (Dobreva et al., 2006). Notably, Dobreva and colleagues (2006) reported that Satb2+/− mice were phenotypically normal and used them as controls to evaluate gene expression defects in Satb2−/− mice. In addition to not finding evidence of apoptosis (even in homozygous mutants), Dobreva and colleagues (2006) also report that proliferation of osteoblasts was not affected, although these data were not shown. In general, the craniofacial phenotype shown by Dobreva and colleagues (2006) appears less severe than that shown by Britanova and colleagues (2006). Here, we find that mutations in Satb2 cause both proliferation and differentiation defects. We also find a dosage effect, where loss of even a single allele of Satb2 causes defects in osteogenesis. Thus, we hypothesize that Satb2 has two cell biological roles in osteogenesis: 1) regulation of pre-osteoblast proliferation and 2) subsequent osteoblast differentiation.
The different results reported in the previous studies described above may be related to differences in genetic background (although the background used in the Dobreva study was not specified). Our work, conducted in the C57BL/6 background (which is the same background used by Britanova and colleagues), indicates that Satb2 mutations increase apoptosis (although not to the extent they observed) in pre-osteoblasts. Additionally, we show that increasing cell density at the onset of proliferation increases differentiation potential. Taken together, these data indicate that proliferation defects in osteogenic progenitors contribute to deficits in bone mineral production, particularly in the C57BL/6 background. Disruption to osteogenic gene expression has been reported in all loss of Satb2 studies, suggesting it is a more penetrant phenotype. If the C57BL/6 background is susceptible to both proliferation and differentiation defects, it would also explain why the jaw phenotype reported by Britanova and colleagues (2006) is more severe. Further, this data raises the interesting possibility for modifiers with the potential to buffer the proliferation defects to be discovered.
4.2. Molecular mechanism of osteogenic defect
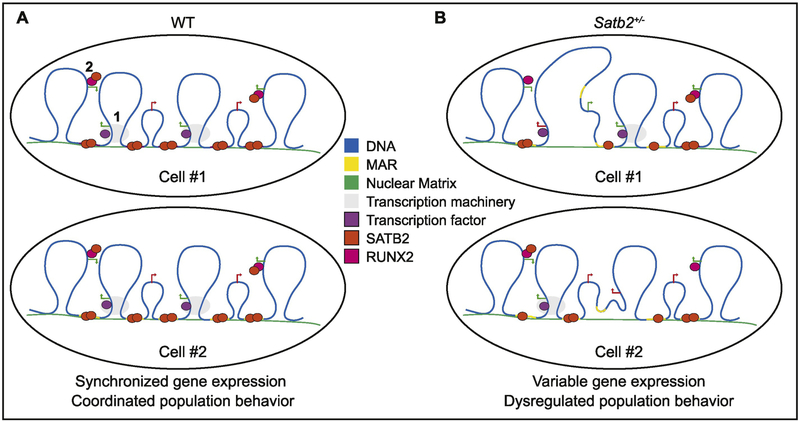
MAR-binding proteins have at least two distinct molecular roles, transcriptional regulation and DNA replication (Ottaviani et al., 2008b). MAR-binding proteins regulate transcription by defining the borders of chromatin loops anchored in the nuclear matrix (Fig. 8A). The anchoring sites form discrete territories of genome organization that serve as transcription hubs, where transcription factors, histone modifying enzymes, and other proteins aggregate to promote gene expression (Cai et al., 2003; Gyorgy et al., 2008; Kirillov et al., 1996; Ottaviani et al., 2008b). Mutations reducing MAR-binding proteins may limit their ability to properly modify chromatin and/or participate in protein-protein interactions mediating gene transcription thus disrupting the coordinated regulation of cell-type specific genes (Fig. 8B). For example, in Satb1 mutant mice, gene expression is dysregulated during differentiation of thymocytes and T cells (Alvarez et al., 2000). Similarly, we observe a significant shift in gene expression and increased variation in gene expression in Satb2 mutant cells. We do not observe increased variance in gene expression in proliferating osteoblasts (T0), but this may be an artifact of the genes used in our single-cell analysis, which are heavily biased towards osteogenic differentiation genes.
Figure 8: Model of Satb2-mediated variation in molecular and cellular outcomes.
Satb2 in known to regulate gene expression through high-order chromatin organization, including looping of chromatin to associate long-distance regulatory elements with promotors into “transcription factories” (1). Satb2 also associates with other transcription factors, including Runx2, to more directly regulate gene expression (2). A) In WT cells, Satb2-mediated gene regulation in a population is coordinated. B) When Satb2 is reduced, it is predicted to fail to bind to some of its DNA and/or protein partners randomly. Thus, cell to cell variation in gene expression will exist in a population, contributing to molecular and cellular variation in severity of phenotypes.
However, our finding that proliferation is affected in pre-osteoblasts indicates an additional role for Satb2. Satb2 may also mediate this effect through regulation of transcription of genes important for cell cycle progression. Notably, in a different study in MC3T3-E1 cells where Satb2 was over-expressed, genes associated with cell cycle and proliferation were more upregulated than genes associated with differentiation (Kim et al., 2012). However, the possibility that Satb2 is more directly involved in DNA replication by binding at MARs remains open, especially as Satb2 is highly expressed from late G1 through G2. If Satb2 is required for DNA replication (either directly through binding at MARs or indirectly through gene regulation), its reduction may cause replication stress, which is associated with DNA damage (Zeman and Cimprich, 2014). Further investigation into Satb2 involvement in DNA replication could elucidate mechanisms underlying DNA damage observed in Satb2 mutants.
4.3. N-terminal domain of Satb2
Several genetic modifications of the SATB2 locus, including deletions, point mutations, duplications, chromosomal translocations, and frameshift and splice site mutations have been implicated in causing SAS (Zarate and Fish, 2017). Most of these mutations have been hypothesized to mediate disease phenotypes through haploinsufficiency of SATB2. However, few mutations have been functionally tested. In the case of one nonsense mutation, the mutant SATB2 mRNA produces a truncated protein retaining the SATB2 dimerization domain, and lacking the DNA-binding motifs, thus suggesting a dominant negative effect (Leoyklang et al., 2007; Leoyklang et al., 2013). Some SATB2 activity remains in cells expressing this truncated protein, however, it remains unclear how these levels compare to SATB2 mutations resulting in loss of function. More generally, it is still not well understood how heterozygous loss of function mutations affect SATB2 mRNA and protein levels.
Here we show that mutations in Satb2 result in decreased mRNA and protein levels. Additionally, several observations from our work suggest that mutations may generate proteins with dominant negative effects. First, colony 4 (generating only protein with the N-terminal deletion) proliferates more slowly than colony 8 (complete loss of function). Colony 8 appears to have fewer nuclear aberrations than either colony carrying the N-terminal deletion. Finally, colony 8 differentiates better than the colonies carrying the N-terminal deletion. We used the Phyre2 software to predict the effect of this deletion on Satb2 structure (Kelley et al., 2015). Although we did not see any obvious alterations to Satb2 protein folding, we did find that the N-terminal domain is highly disordered (SupFig. 6). Disorder domains are important for protein dissociation (Umezawa et al., 2016). Chromatin architecture is dynamic and genomic anchors need to be altered throughout the cell cycle to adapt to changing genomic function (Heng et al., 2004; Ottaviani et al., 2008a). Additionally, chromatin remodeling complexes dissociate from chromatin during mitosis (Ma et al., 2015). Mutations in the Satb2 N-terminal region may therefore hinder its ability to dissociate from chromatin, disrupting its function and/or contribute to DNA damage.
4.5. Variation in disease phenotypes
Individuals with SAS exhibit a broad spectrum of skeletal abnormalities including tibial bowing, osteomalacia, osteopenia or osteoporosis, and they have a high risk for fractures (Zarate et al., 2018b). The incidence and severity of these skeletal anomalies varies among SAS patients, with some individuals having little to no skeletal problems (Zarate et al., 2018a). Age at presentation also reflects variability of severity with skeletal morbidity reported as early as infancy. Current available data suggests that for those individuals that have undergone dedicated evaluation for bone density, it is common to have lower age/gender adjusted BMD scores regardless of the type of genetic modification in SATB2. Overall, the current data from SAS patients is consistent with variation in penetrance of skeletal anomalies that is not associated with mutation type. Stochastic variation in Satb2 function upon its reduction in dosage may be one mechanism underlying variation in disease severity (Fig. 8). We hypothesize that genetic background also plays a role, with more severe phenotypes occurring in individuals that are more susceptible to pre-osteoblast proliferation defects.
Anti-resorptive drugs (e.g. bisphosphonates) which increase bone density and strength by inhibiting osteoclast activity, have been the main therapeutic option for individuals with childhood osteopenia in SAS (Zarate et al., 2018b). The results presented here showing a role for Satb2 in pre-osteoblast proliferation suggest that the low BMD in SAS may be a consequence of a decline in the number and vitality of osteoblasts. Therefore, drugs that directly target osteoblasts such as teriparatide, an anabolic agent that increases osteoblast formation and activates bone-lining cells (quiescent osteoblasts) to form new bone, could provide a biologically sound alternative treatment for osteopenia in SAS.
4.6. Implications for cancer
We observed several types of nuclear aberrations, including nuclear blebbing and chromatin herniation, that are also characteristic of cancer cells. A previous study in MC3T3-E1 cells found that Satb2 levels increase in response to oxidative stress. When Satb2 was reduced via siRNA in cells under oxidative stress, apoptosis increased (Wei et al., 2012). Similarly, Satb1 was recently found to participate in DNA repair associated with oxidative damage (Kaur et al., 2016). Reductions in Satb2 have also been reported in cancers associated with deficiencies in DNA mismatch repair proteins (Ma et al., 2018). Together, these data suggest that down-regulation of Satb2 may be involved in cancer through compromised DNA repair mechanisms. Paradoxically, over-expression of Satb2 is also associated with cancer in different cell types (Chen and Costa, 2018; Ma et al., 2018). Our finding that Satb2 is involved with proliferation and DNA damage may explain this dichotomy. Cell-type specific gene regulation may impact how alterations to Satb2 disrupt normal cell biology in cancer progression.
5. Conclusion
Previous work has described Satb2 is a high-order transcription factor regulating genes required for osteogenic differentiation. Our data suggest that, in addition to its role in differentiation, Satb2 regulates progenitor proliferation. Satb2 may promote pre-osteoblast proliferation through regulation of the expression of genes associated with cell cycle progression and/or it may have a role during DNA replication through its association with MARs. The involvement of Satb2 in separate molecular processes during osteogenesis, one of which may be buffered in certain genetic backgrounds, may help explain variation in disease phenotypes (Merkuri and Fish, 2019). We have also found that mutations in Satb2 cause chromatin defects. Further investigation of the mechanism underlying these defects has potential impacts on both skeletal defects and cancer.
Supplementary Material
Highlights.
Satb2 expression exhibits population-level variation in pre-osteoblast cells.
Mutations in Satb2 reduce pre-osteoblast proliferation
Reduction in Satb2 causes nuclear blebbing and other DNA aberrations.
Our data point to a novel role for Satb2 in proliferating osteoblasts.
Acknowledgements
This work was funded by NIH NIDCR R15DE026611. Generous technical help was provided by Jack Lepine (UML), Susanne Pechold (UMMS) and Kahraman Tanriverdi (UMMS). We thank the SATB2 Foundation and SAS families for sharing their stories and providing inspiration to solve mysteries underlying this disease.
Footnotes
Conflict of Interest
The authors declare that they have no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alvarez JD, Yasui DH, Niida H, Joh T, Loh DY, Kohwi-Shigematsu T. 2000. The MAR-binding protein SATB1 orchestrates temporal and spatial expression of multiple genes during T-cell development. Genes Dev 14: 521–535. [PMC free article] [PubMed] [Google Scholar]
- Boone PM, Chan YM, Hunter JV, Pottkotter LE, Davino NA, Yang Y, Beuten J, Bacino CA. 2016. Increased bone turnover, osteoporosis, progressive tibial bowing, fractures, and scoliosis in a patient with a final-exon SATB2 frameshift mutation. Am J Med Genet A 170: 3028–3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britanova O, Akopov S, Lukyanov S, Gruss P, Tarabykin V. 2005. Novel transcription factor Satb2 interacts with matrix attachment region DNA elements in a tissue-specific manner and demonstrates cell-type-dependent expression in the developing mouse CNS. Eur J Neurosci 21: 658–668. [DOI] [PubMed] [Google Scholar]
- Britanova O, Depew MJ, Schwark M, Thomas BL, Miletich I, Sharpe P, Tarabykin V. 2006. Satb2 haploinsufficiency phenocopies 2q32-q33 deletions, whereas loss suggests a fundamental role in the coordination of jaw development. Am J Hum Genet 79: 668–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S, Han HJ, Kohwi-Shigematsu T. 2003. Tissue-specific nuclear architecture and gene expression regulated by SATB1. Nat Genet 34: 42–51. [DOI] [PubMed] [Google Scholar]
- Chen QY, Costa M. 2018. Oncogenic and tumor suppressive roles of special AT-rich sequence-binding protein. J Carcinog 17: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen QY, Des Marais T, Costa M. 2019. Deregulation of SATB2 in carcinogenesis with emphasis on miRNA-mediated control. Carcinogenesis 40: 393–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobreva G, Chahrour M, Dautzenberg M, Chirivella L, Kanzler B, Farinas I, Karsenty G, Grosschedl R. 2006. SATB2 is a multifunctional determinant of craniofacial patterning and osteoblast differentiation. Cell 125: 971–986. [DOI] [PubMed] [Google Scholar]
- Fish JL, Villmoare B, Kobernick K, Compagnucci C, Britanova O, Tarabykin V, Depew MJ. 2011. Satb2, modularity, and the evolvability of the vertebrate jaw. Evol Dev 13: 549–564. [DOI] [PubMed] [Google Scholar]
- Gong Y, Lu J, Yu X, Yu Y. 2016. Expression of Sp7 in Satb2-induced osteogenic differentiation of mouse bone marrow stromal cells is regulated by microRNA-27a. Mol Cell Biochem 417: 7–16. [DOI] [PubMed] [Google Scholar]
- Gong Y, Qian Y, Yang F, Wang H, Yu Y. 2014. Lentiviral-mediated expression of SATB2 promotes osteogenic differentiation of bone marrow stromal cells in vitro and in vivo. Eur J Oral Sci 122: 190–197. [DOI] [PubMed] [Google Scholar]
- Gregory CA, Gunn WG, Peister A, Prockop DJ. 2004. An Alizarin red-based assay of mineralization by adherent cells in culture: comparison with cetylpyridinium chloride extraction. Anal Biochem 329: 77–84. [DOI] [PubMed] [Google Scholar]
- Guo Y, Dong SS, Chen XF, Jing YA, Yang M, Yan H, Shen H, Chen XD, Tan LJ, Tian Q, Deng HW, Yang TL. 2016. Integrating Epigenomic Elements and GWASs Identifies BDNF Gene Affecting Bone Mineral Density and Osteoporotic Fracture Risk. Sci Rep 6: 30558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyorgy AB, Szemes M, de Juan Romero C, Tarabykin V, Agoston DV. 2008. SATB2 interacts with chromatin-remodeling molecules in differentiating cortical neurons. Eur J Neurosci 27: 865–873. [DOI] [PubMed] [Google Scholar]
- Hassan MQ, Gordon JA, Beloti MM, Croce CM, van Wijnen AJ, Stein JL, Stein GS, Lian JB. 2010. A network connecting Runx2, SATB2, and the miR-23a~27a~24–2 cluster regulates the osteoblast differentiation program. Proc Natl Acad Sci U S A 107: 19879–19884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng HH, Goetze S, Ye CJ, Liu G, Stevens JB, Bremer SW, Wykes SM, Bode J, Krawetz SA. 2004. Chromatin loops are selectively anchored using scaffold/matrix-attachment regions. J Cell Sci 117: 999–1008. [DOI] [PubMed] [Google Scholar]
- Hu N, Feng C, Jiang Y, Miao Q, Liu H. 2015. Regulative Effect of Mir-205 on Osteogenic Differentiation of Bone Mesenchymal Stem Cells (BMSCs): Possible Role of SATB2/Runx2 and ERK/MAPK Pathway. Int J Mol Sci 16: 10491–10506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur S, Coulombe Y, Ramdzan ZM, Leduy L, Masson JY, Nepveu A. 2016. Special AT-rich Sequence-binding Protein 1 (SATB1) Functions as an Accessory Factor in Base Excision Repair. J Biol Chem 291: 22769–22780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10: 845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim IS, Jeong SJ, Kim SH, Jung JH, Park YG, Kim SH. 2012. Special AT-rich sequence-binding protein 2 and its related genes play key roles in the differentiation of MC3T3-E1 osteoblast like cells. Biochem Biophys Res Commun 417: 697–703. [DOI] [PubMed] [Google Scholar]
- Kirillov A, Kistler B, Mostoslavsky R, Cedar H, Wirth T, Bergman Y. 1996. A role for nuclear NF-kappaB in B-cell-specific demethylation of the Igkappa locus. Nat Genet 13: 435–441. [DOI] [PubMed] [Google Scholar]
- Komori T 2005. Regulation of skeletal development by the Runx family of transcription factors. J Cell Biochem 95: 445–453. [DOI] [PubMed] [Google Scholar]
- Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, Sato M, Okamoto R, Kitamura Y, Yoshiki S, Kishimoto T. 1997. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 89: 755–764. [DOI] [PubMed] [Google Scholar]
- Leoyklang P, Suphapeetiporn K, Siriwan P, Desudchit T, Chaowanapanja P, Gahl WA, Shotelersuk V. 2007. Heterozygous nonsense mutation SATB2 associated with cleft palate, osteoporosis, and cognitive defects. Hum Mutat 28: 732–738. [DOI] [PubMed] [Google Scholar]
- Leoyklang P, Suphapeetiporn K, Srichomthong C, Tongkobpetch S, Fietze S, Dorward H, Cullinane AR, Gahl WA, Huizing M, Shotelersuk V. 2013. Disorders with similar clinical phenotypes reveal underlying genetic interaction: SATB2 acts as an activator of the UPF3B gene. Hum Genet 132: 1383–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Ma C, Olevian DC, Lowenthal BM, Jayachandran P, Kozak MM, Chang DT, Pai RK. 2018. Loss of SATB2 Expression in Colorectal Carcinoma Is Associated With DNA Mismatch Repair Protein Deficiency and BRAF Mutation. Am J Surg Pathol 42: 1409–1417. [DOI] [PubMed] [Google Scholar]
- Ma Y, Kanakousaki K, Buttitta L. 2015. How the cell cycle impacts chromatin architecture and influences cell fate. Front Genet 6: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda T, Cepko CL. 2004. Electroporation and RNA interference in the rodent retina in vivo and in vitro. Proc Natl Acad Sci U S A 101: 16–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkuri F, Fish JL. 2019. Developmental processes regulate craniofacial variation in disease and evolution. Genesis 57: e23249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K, Zhou X, Kunkel G, Zhang Z, Deng JM, Behringer RR, de Crombrugghe B. 2002. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell 108: 17–29. [DOI] [PubMed] [Google Scholar]
- Ottaviani D, Lever E, Mitter R, Jones T, Forshew T, Christova R, Tomazou EM, Rakyan VK, Krawetz SA, Platts AE, Segarane B, Beck S, Sheer D. 2008a. Reconfiguration of genomic anchors upon transcriptional activation of the human major histocompatibility complex. Genome Res 18: 1778–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottaviani D, Lever E, Takousis P, Sheer D. 2008b. Anchoring the genome. Genome Biol 9: 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. 2013. Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8: 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9: 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinmyo Y, Tanaka S, Tsunoda S, Hosomichi K, Tajima A, Kawasaki H. 2016. CRISPR/Cas9-mediated gene knockout in the mouse brain using in utero electroporation. Sci Rep 6: 20611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa K, Ohnuki J, Higo J, Takano M. 2016. Intrinsic disorder accelerates dissociation rather than association. Proteins 84: 1124–1133. [DOI] [PubMed] [Google Scholar]
- Wei JD, Lin YL, Tsai CH, Shieh HS, Lin PI, Ho WP, Chen RM. 2012. SATB2 participates in regulation of menadione-induced apoptotic insults to osteoblasts. J Orthop Res 30: 1058–1066. [DOI] [PubMed] [Google Scholar]
- Xiao G, Cui Y, Ducy P, Karsenty G, Franceschi RT. 1997. Ascorbic acid-dependent activation of the osteocalcin promoter in MC3T3-E1 preosteoblasts: requirement for collagen matrix synthesis and the presence of an intact OSE2 sequence. Mol Endocrinol 11: 1103–1113. [DOI] [PubMed] [Google Scholar]
- Yang X, Matsuda K, Bialek P, Jacquot S, Masuoka HC, Schinke T, Li L, Brancorsini S, Sassone-Corsi P, Townes TM, Hanauer A, Karsenty G. 2004. ATF4 is a substrate of RSK2 and an essential regulator of osteoblast biology; implication for Coffin-Lowry Syndrome. Cell 117: 387–398. [DOI] [PubMed] [Google Scholar]
- Zarate YA, Fish JL. 2017. SATB2-associated syndrome: Mechanisms, phenotype, and practical recommendations. Am J Med Genet A 173: 327–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarate YA, Smith-Hicks CL, Greene C, Abbott MA, Siu VM, Calhoun A, Pandya A, Li C, Sellars EA, Kaylor J, Bosanko K, Kalsner L, Basinger A, Slavotinek AM, Perry H, Saenz M, Szybowska M, Wilson LC, Kumar A, Brain C, Balasubramanian M, Dubbs H, Ortiz-Gonzalez XR, Zackai E, Stein Q, Powell CM, Schrier Vergano S, Britt A, Sun A, Smith W, Bebin EM, Picker J, Kirby A, Pinz H, Bombei H, Mahida S, Cohen JS, Fatemi A, Vernon HJ, McClellan R, Fleming LR, Knyszek B, Steinraths M, Velasco Gonzalez C, Beck AE, Golden-Grant KL, Egense A, Parikh A, Raimondi C, Angle B, Allen W, Schott S, Algrabli A, Robin NH, Ray JW, Everman DB, Gambello MJ, Chung WK. 2018a. Natural history and genotype-phenotype correlations in 72 individuals with SATB2-associated syndrome. Am J Med Genet A 176: 925–935. [DOI] [PubMed] [Google Scholar]
- Zarate YA, Steinraths M, Matthews A, Smith WE, Sun A, Wilson LC, Brain C, Allgove J, Jacobs B, Fish JL, Powell CM, Wasserman WW, van Karnebeek CD, Wakeling EL, Ma NS. 2018b. Bone health and SATB2-associated syndrome. Clin Genet 93: 588–594. [DOI] [PubMed] [Google Scholar]
- Zeman MK, Cimprich KA. 2014. Causes and consequences of replication stress. Nat Cell Biol 16: 2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Tu Q, Grosschedl R, Kim MS, Griffin T, Drissi H, Yang P, Chen J. 2011. Roles of SATB2 in osteogenic differentiation and bone regeneration. Tissue Eng Part A 17: 1767–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.