Abstract
G protein-coupled receptor 137b (GPR137b) is an orphan seven-pass transmembrane receptor of unknown function. In mouse, Gpr137b is highly expressed in osteoclasts in vivo and is upregulated during in vitro differentiation. To elucidate the role that GPR137b plays in osteoclasts, we tested the effect of GPR137b deficiency on osteoclast maturation and resorbing activity. We used CRISPR/Cas9 gene editing in mouse-derived ER-Hoxb8 immortalized myeloid progenitors to generate GPR137b-deficient osteoclast precursors. Decreasing Gpr137b in these precursors led to increased osteoclast differentiation and bone resorption activity. To explore the role of GPR137b during skeletal development, we generated zebrafish deficient for the ortholog gpr137ba. Gpr137ba-deficient zebrafish are viable and fertile and do not display overt morphological defects as adults. However, analysis of osteoclast function in gpr137ba−/− mutants demonstrated increased bone resorption. Micro-computed tomography evaluation of vertebral bone mass and morphology demonstrated that gpr137ba-deficiency altered the angle of the neural arch, a skeletal site with high osteoclast activity. Vital staining of gpr137ba−/− fish with calcein and alizarin red indicated that bone formation in the mutants is also increased, suggesting high bone turnover. These results identify GPR137b as a conserved negative regulator of osteoclast activity essential for normal resorption and patterning of the skeleton. Further, these data suggest that coordination of osteoclast and osteoblast activity is a conserved process among vertebrates and may have similar regulation.
Keywords: GPR137b, bone, osteoclast, zebrafish
1. Introduction
The skeleton is a dynamic structure, continuously remodeled by coordinated bone formation and degradation in response to physiological signals or environmental factors. The balance of remodeling through bone formation by osteoblasts and bone resorption by osteoclasts is highly regulated and conditions that cause an imbalance in these activities can lead to pathological conditions such as osteoporosis or osteopetrosis. Identification of the factors that act to regulate remodeling would serve to aid in remediation of these conditions, however our knowledge of these agents remains incomplete.
Osteoblasts derive from mesenchymal progenitors, differentiate and produce bone matrix in response to a wide variety of growth factors [1]. Osteoblasts regulate osteoclast activity through the production of RANKL and a decoy receptor for RANKL, osteoprotegrin (OPG). Osteoclasts are multinucleated myeloid cells derived from differentiation and fusion of hematopoietic precursors in response to M-CSF (macrophage colony stimulating factor) and RANKL (Receptor Activator of Nuclear factor Kappa b Ligand) stimulation. Mature osteoclasts degrade bone matrix by secreting proteolytic enzymes and acid into a sealed zone known as resorbing lacuna [2]. Several lines of evidence suggest that osteoclasts can stimulate bone formation by osteoblasts, both by releasing growth factors from the bone matrix during bone resorption and by directly secreting factors that promote osteoblast activity [3]. However, how this functional coupling is regulated is uncertain.
G protein coupled receptors (GPCRs) are surface receptors which integrate signaling pathways in response to a diverse set of ligands, including ions, photons, hormones, and other peptides. Ligand binding triggers activation of G-proteins and intracellular signaling cascades to regulate transcription and specific cellular responses [4]. Several GPCRs have been identified to play a critical role in skeletal development and bone turnover [5, 6]. Gpr137b is reported to be highly expressed in mouse osteoclasts, while, in comparison, is minimally detected in osteoblasts [7]. GPR137b is a lysosomal seven-pass transmembrane orphan receptor, although its ligand and function are unknown [8, 9]. A number of lysosomal proteins are required for osteoclasts to acidify the resorption lacunae and to degrade the extracellular matrix [10]. Here, we demonstrate GPR137b is essential for regulation of osteoclast function. Further, we explored the role of this orphan receptor in bone homeostasis in vivo using the zebrafish, Danio rerio, by generating a mutant lacking the Gpr137b ortholog gpr137ba. We provide functional evidence that GPR137b is a conserved negative regulator of osteoclast activity important for skeletal homeostasis and form.
2. Methods
2.1. Zebrafish and mouse husbandry
Cas9-EGFP mice on a C57BL/6J background (Jackson Laboratories, stock number 026179) [11] and C57BL/6J mice (Jackson Laboratories, stock number 000664) were socially housed and maintained under specific pathogen–free conditions and provided with irradiated food ad libitum. Animal research was conducted with the approval of the Institutional Animal Care and Use Committee of Harvard Medical School and conformed to relevant guidelines and laws.
The zebrafish included in this study were housed and handled in accordance with the Boston Children’s Hospital Animal Research Care Committee. A complete description of the husbandry and environmental conditions in housing for the fish used in these experiments is available as a collection in https://www.protocols.io/view/zebrafishdanio-rerio-environmental-summary-aquati-mijc54n. CRISPR/Cas9 mutations were introduced into the Tuebingen wildtype background.
2.2. Phylogenetic analysis
To examine the phylogenetic relationship of gpr137b orthologues, cDNA sequences of Astyanax mexicamis (cave fish), Danio rerio (zebrafish), Gasterosteus aculeatus (stickleback), Lepisosteus oculatus (spotted gar), Mus musculas (mouse), Oryzias latipes (medaka) and Scleropages formosus (Asian arowana) were retrieved from NCBI [27] and ENSEMBL [28] databases. Sequences were aligned using MUSCLE [29] multiple sequence alignment tool. A phylogenetic tree was constructed via Tamura-Nei model in MEGA version 7.0.18 [30]. The reliability of the tree was assessed by 500 bootstrapping replicates; branches with nodes having bootstrap support less than 70 were collapsed.
2.3. Osteoclast differentiation and resorption assay
In vitro experiments were performed in α-MEM (Cellgro) containing 10% fetal calf serum (Hyclone), 100 U penicillin and 100 μg/ml streptomycin (Cellgro) at 37°C with 5% CO2. Mouse bone marrow cells derived from wildtype mice were plated on a tissue culture dish. After 24 hours, non-adherent cells were collected and seeded on plastic with 20 ng/mL M-CSF (R&D Systems) alone (for bone marrow derived macrophages, BMM) or in combination with 5 ng/ml RANKL (PeproTech). After 3 days, the cytokines were replenished daily.
To visualize osteoclasts, cells were fixed in 10% formalin and incubated with tartrate-resistant acid phosphatase (TRAP) staining solution (100μg/ml Naphtol AS-Mx phosphate, 1% N,N-Dimethylformamide, 600μg/ml Fast Red LB Violet salt in 0.1M acetate buffer) at 37°C for 10-15 minutes. Positively stained cells with 3 or more nuclei were counted as osteoclasts. The area of each multinucleated TRAP+ cell was measured using ImageJ software and hand-drawn contours. The average cell area was calculated from 3 wells per sample, with >200 cells measured per well.
For the resorption assay, 1×105 cells were seeded in Corning Osteo Assay plates (Corning). After 7-10 days of culture, plates were stained with von Kossa solution (2.5% silver nitrate in aqueous solution) for 30 minutes at room temperature and developed with pyrogallol aqueous solution (1%). Photographs were acquired with a Leica DM2000 microscope and the resorbed area was quantified using Image J [12].
2.4. Osteoblast Differentiation
For mRNA expression analysis, C57BL/6/J mouse bones were minced and cultured in α-MEM (Cellgro) containing 10% fetal calf serum (Hyclone), 100 U penicillin, 100 μg/ml streptomycin, non-essential amino acids (Cellgro) and 1μM beta-mercaptoethanol (Sigma) at 37°C with 5% CO2. After 5 days, adherent bone marrow stromal cells were re-plated for differentiation in the same media containing 50 ng/mL ascorbic acid (Sigma Aldrich) and 5 mM beta-glycerophosphate (Sigma) (OB media) and cultured for an additional 14 days. For immunofluorescence, calvarial osteoblasts from C57BL/6J mice were isolated from 2-4 day old pups by serial digestion with a mix of collagenase type I and II and differentiated for 21 days in OB media.
2.5. Cas9-HoxB8 cell lines
Cas9-EGFP expressing Hoxb8 cells have been described previously [13]. Briefly, bone marrow mononuclear cells from Cas9-EGFP expressing mice [11] were isolated and expanded in medium containing stem cell factor (SCF, 40 ng/mL, Peprotech) and IL-3 (20 ng/mL, Peprotech) for 48h, spin-infected with murine stem cell virus encoding ER–HoxB8 and cultured in the presence of SCF (20 ng/mL) and β-estradiol (500 nM, Sigma Aldrich) for two days prior to addition of G418 for selection. Non-adherent immortalized cells grew in ~3 weeks, as previously described [13].
For osteoclast differentiation, Hoxb8 cells were washed twice with PBS, plated at 1×105 cells/well in 96-well tissue culture plates or 2×106 cells/well in 6-well plates and differentiated in the presence of 10 ng/mL RANKL, 20 ng/mL M-CSF and 2.5 ng/mL IL-1β (R&D systems), in the absence of SCF and β-estradiol.
2.6. CRISPR/Cas9 mutagenesis.
2.6.1. Gpr127b deficient Hoxb8 cells:
Lentivirus encoding a guide RNA (gRNA) for Gpr137b (CGTGAAGCTCGGCCTCACCG) or EGFP control (GAAGTTCGAGGGCGACACCC) were generated by the Broad Institute Genetic Perturbation Platform (Cambridge, MA) using the pXPR_003 vector. Cas9-EGFP Hoxb8 cells described above were spin-infected with gRNA lentivirus (multiplicity of infection 10) and selected in puromycin for 1 week prior to OC differentiation and evaluation of gene expression.
2.6.2. Zebrafish:
The Cas9 expressing vector pCS2_nCas9n [14] was used for in vitro transcription of Cas9 mRNA using the mMESSAGE mMACHINE SP6 kit (Life Technologies). The target sequence specific for gpr137ba (ENSDARG00000078448) was identified using ZiFiT Targeter [15] and complimentary oligos (zb-gRNA FW 5′-AAACGTCCATCAGTCTGCTGTT-3′; RV 5′-TAGGAACAGCAGACTGATGGAC-3′) were cloned downstream of the T7 promoter into the guide RNA cloning vector as previously described [14]. The linearized gRNA vector was then transcribed in vitro (MEGAShortscript T7 kit, Life Technologies) to produce gRNA. An RNA mixture (2nl) composed of gRNA (25 ng/μl) and Cas9 mRNA (400 ng/μl) was microinjected into one-cell stage zebrafish embryos. To detect CRISPR/Cas9 induced mutations, a 743bp genomic region containing gpr137ba exon 3 was amplified followed by T7 endonuclease digestion. To establish mutant lines, injected fish were grown to adulthood and outcrossed to AB wildtype fish. The nature of mutation created by CRISPR/Cas9 in the F1 progeny was identified by sequencing of subcloned PCR products. Carriers were then outcrossed to establish mutant lines. Genomic DNA extracted as previously described [16] from a fin clip of adult zebrafish was used for PCR genotyping (gpr137ba wt-fw 5′-CTGCTGTTCCTGTCC -3′; gpr137ba 5nt-del mutant-fw 5′-CTGTACCTGCTGTTCCTGTCCATC -3′;gpr137ba-rv 5′-ACCTTGGCCTCCAGGTAGAT-3′). Wild type (212bp) and mutant (218bp) amplicons were obtained by annealing the primers at 60°C or 64°C for the wild type and mutant mix, respectively.
2.7. RNA isolation and real-time quantitative PCR (qPCR)
RNA was isolated from cells or fish scales using Trizol reagent (Life Technologies). The scales were disrupted with bullet blender (Nextadvance Navy, 3 min, speed 8). The homogenized solution was then extracted with chloroform/isopropanol and 500ng of RNA was used to generate cDNA (Affinity Script RT-PCR cDNA Synthesis Kit, Agilent Technology). Real-time quantitative PCR (qPCR) was performed in duplicate using 10ng of cDNA per reaction using Sybr green reagent (Life Technologies). Primers for Hprt were previously described [20]; primers for mouse Gpr137b or zebrafish gpr137ba, gpr137bb, osteocalcin and sp7 were designed using Primerblast [21] (Table 1). Data was normalized to Hprt (mouse) or tubulin (zebrafish) housekeeping genes.
Table 1 –
Quantitative RT-PCR primer list.
| Gene | Species | Forward | Reverse |
|---|---|---|---|
| Gpr137b | M.musculus | CTACCTGGCCTCACTTTTCATC | GCCCGAGAGGTGTAGAGCA |
| Hprt | M.musculus | GTTAAGCAGTACAGCCCCAAA | AGGGCATATCCAACAACAAACTT |
| sp7 | D.rerio | GACCCTCACTGGACTGCTTC | CGAATTTGTTGCAGGTCGCA |
| osteocalcin | D.rerio | TCTTCTGCTGCCTGATGACTG | CGAGCTGAAATGGAGTCAGGT |
| gpr137ba | D.rerio | TCTGTACTTCGCACAGGTCA | ACAGGAACAGCAGGTACAGC |
| gpr137bb | D.rerio | GCCTGTACAAAGTGGCCAAG | ATCACGGTCACCTGACACAC |
| tubulin | D.rerio | GTACGTGGGTGAGGGTATGG | ACACAGCAGGCAGCATTTCTA |
2.8. Western Blot
Membrane proteins were isolated following published protocols [23]. Briefly, cells were lysed in distilled H2O, snap frozen in liquid nitrogen and subjected to multiple cycles of ultracentrifugation. The final pellet was resuspended in 8M Urea and 10mM DTT. Protein concentration was measured with the Bradford assay (BioRad). Membrane protein enriched extracts were separated by SDS-PAGE and transferred to nitrocellulose membranes, which were then incubated over-night at 4°C with the primary antibodies rabbit anti-mouse GPR137b (Proteintech) and anti-LAMP-1 (clone, 1D4B, Biolegend). The binding of the primary antibody was detected with enhanced chemiluminescence (ECL) detection reagent (GE Healthcare Lifesciences).
2.9. Immunofluorescence
Cultured osteoclasts and osteoblasts were fixed in 4% PFA and permeabilized with 0.3% Triton X-100. After blocking with 10% normal goad serum diluted in 0.25% BSA, cultures were incubated with anti-mouse GPR137b (Proteintech) followed by goat anti-rabbit IgG AlexaFluor 555 (Thermofisher) in PBS/0.25%BSA buffer. Co-staining with phalloidin (Thermofisher) and DAPI (Thermofisher) was performed to assist with cell identification. After imaging, wells were stained for alkaline phosphatase or TRAP activity as previously described [24]. Images were acquired using an Olympus FSX100 (epifluorescence) and Leica DM 2000 (brightfield).
2.10. Whole mount staining of scales
Zebrafish scales were collected bilaterally from anterior to posterior along the mid-line of the trunk of gpr137ba−/− and wildtype siblings.
2.10.1. TRAP staining:
To visualize osteoclasts, scales were fixed in 10% formalin and incubated with TRAP staining solution (100μg/ml Naphtol AS-Mx phosphate, 1% N,N-Dimethylformamide, 600μg/ml Fast Red LB Violet salt in 0.1M acetate buffer) at 37°C for 10-15 minutes. The number of scales with TRAP staining was manually counted. Ten scales per fish at 8wpf and 10 fish per group (gpr137ba−/− and wild type siblings) were evaluated.
2.10.2. Von Kossa staining:
Scales were stained with von Kossa solution (2.5% silver nitrate in aqueous solution) for 30 minutes at room temperature. Quantification of the percentage of resorbed area per scale was performed by applying a threshold to the image in Image J [12]. Ten scales per fish at 6, 8 and 12 wpf) and 10 fish per group (gpr137ba−/− and wild type siblings) were evaluated.
2.11. Micro-computed tomography (Micro-CT)
8wpf fish were euthanized with an overdose of tricaine methane sulfonate (MS222, 200-300 mg/L) solution, fixed in 10% formalin for 24h and preserved in 70% ethanol solution. Caudal vertebrae were imaged by micro-CT with a voxel size of 6 micron, using an X-ray tube potential of 55 kVp, an X-ray intensity of 0.145 mA and an integration time of 600 ms (Scanco mCT35). Vertebral analysis was performed using custom software to determine neural arch area and angle, vertebral volume and bone mineral density, as described previously [22]. With the exception of the arch angle, all parameters were normalized to the length of the fish, as described previously [22].
2.12. In vivo bone formation assay
For the in vivo bone formation assay, fish were stained overnight in 100μg/ml calcein (Sigma-Aldrich). After 14 days, fish were stained overnight in alizarin red solution (100μg/ml alizarin red, 10μM HEPES, pH 7.0, Sigma-Aldrich). Fish were then anesthetized with Tricaine and their infraorbital bones 3 (IO3) [25] and scales were imaged under a fluorescence stereomicroscope. We started this experiment when fish were 4, 6 and 8wpf and ended at 6, 8 and 12 wpf, respectively.
The area of total bone (calcein+alizarin red stains, TB) and pre-existent bone (calcein stain alone, CB) were measured with ImageJ software [26]. The percentage of new bone formed in the 14 days (the area stained with alizarin red only) was calculated with the formula: % new bone formed = 100 × (1-CB/TB).
2.13. Statistical Analysis
Comparisons between two groups were analyzed with unpaired Student’s t-test or Mann-Whitney test, for normal or nonparametric distributions, respectively. Analysis of variance (ANOVA) with Tukey’s multiple comparison test was used to compare 3 or more groups. The normality of the data was evaluated with the Shapiro-Wilk test, p-value of 0.05 or lower was considered significant. All statistical analyses were performed with GraphPad Prism software. Error bars represent standard deviation (SD) of biological replicates. All experiments were performed at least 3 times, except when otherwise indicated.
3. Results
3.1. GPR137b is expressed in mature mouse osteoclasts (OC).
A strategy for detecting regulators of osteoclast biology is to screen for genes differentially regulated among myeloid cell lineages. In broad expression analysis, Gpr137b came out as highly expressed in osteoclasts compared to other bone marrow myeloid cell types [7]. To further refine earlier evidence of expression differences between osteoclasts and other myeloid cells, we examined Gpr137b mRNA and protein in bone marrow-derived OC differentiated from myeloid precursors in the presence of RANKL using qPCR. Compared bone marrow macrophages (BMM) the expression of this receptor was highly abundant in mature osteoclasts (5 days after differentiation). Furthermore, Gpr137b expression increased during osteoclastogenesis, comparing pre-osteoclasts (pre-OC) formed after 2 days of RANKL exposure with mature osteoclasts exposed to RANKL for 7 days (Figure 1A). The higher expression in mature osteoclasts suggests a potential role for this receptor during late phases of differentiation or osteoclast function. To confirm GPR137b protein expression, lysosomal membrane preparations isolated from BMM and OC were probed for the presence of GPR137b, using LAMP-1 as loading control. GPR137b protein was detected in BMM, but was substantially increased in mature OC (Figure 1B). In contrast, Gpr137b expression was barely detectable in osteoblast cells (Figure 1C). Immunofluorescence of osteoclasts (Figure 1D) and osteoblasts (Figure 1E) differentiated in vitro confirmed the presence of GPR137b protein in osteoclasts but not in osteoblasts.
Figure 1 -. Gpr137b is expressed in mouse osteoclasts in vitro and in vivo.
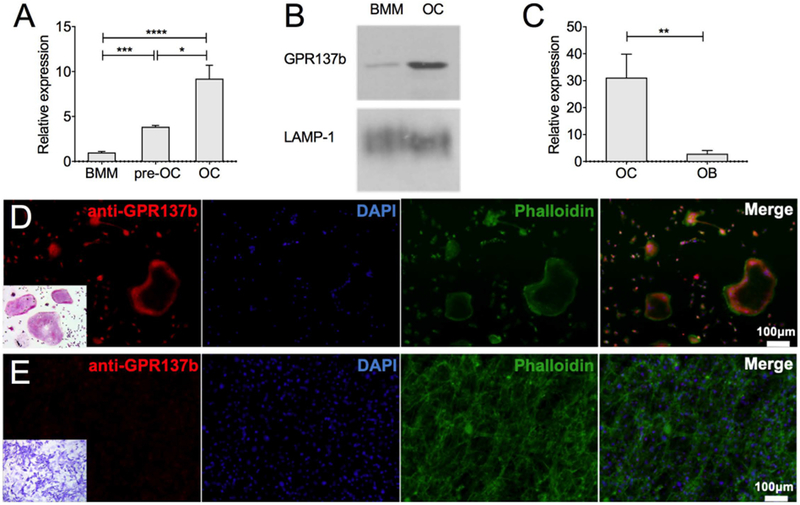
(A-E) Osteoclasts (OC) and bone marrow macrophages (BMM) were differentiated in vitro from bone marrow precursors with M-CSF alone (BMM) or in combination with RANKL (OC). Bone marrow stromal cell derived osteoblast (OB) cultures differentiated in the presence of ascorbic acid and beta-glycerophosphate for 14 days. (A) Gpr137b mRNA expression evaluated by qPCR in BMM, pre-OC and mature OC and (B) GPR137b protein detected by Western Blot in BMM and OC. Lysosomal-associated membrane protein 1 (LAMP-1) was used as loading control for the membrane protein-enriched extract. (C) Gpr137b mRNA expression in OB and OC by qPCR. (D-E) GPR137b expression in osteoclasts and osteoblasts in in vitro cultures. (D) Osteoclasts differentiated from bone marrow cells with M-CSF and RANKL in vitro stained with anti-GPR137b (red), phalloidin (green) and DAPI (blue); TRAP staining (inset) (E) osteoblasts differentiated in vitro from calvarial cells for 21 days and stained as in (D). Alkaline phosphatase staining performed as a control for OB (inset). Scale bar, 100 μm. Experiments were performed at least 3 times. Bars on graphs represent mean±SD, *p<0.05, ***p<0.001, ****p<0.0001 determined by ANOVA or unpaired Student’s t-test.
3.2. Loss-of-function of GPR137b promotes osteoclastogenesis and bone resorption.
In order to examine the role of GPR137b in osteoclast function, we used CRISPR/Cas9 to delete Gpr137b in Hoxb8-expressing immortalized mouse myeloid progenitors [31]. Hoxb8 cells can differentiate into osteoclasts when cultured in the presence of specific growth factors [13, 32, 33]. Bone marrow progenitors isolated from Cas9-expressing transgenic mice were used to generate Hoxb8 cells [13]. Cas9 expressing-Hoxb8 cells were then transduced with lentivirus expressing either a guide RNA (gRNA) complementary to a sequence located in exon 1 of the Gpr137b gene (Hoxb8::gRNAGpr137b) or a control gRNA targeting EGFP (Hoxb8::EGFP, Figures 2A, B). Cells were then differentiated in the presence of M-CSF, RANKL and IL-1 to generate osteoclasts. Compared to Hoxb8::EGFP, Hoxb8::gRNAGpr137b cells expressed substantially reduced amount of GPR137b protein (Figure 2C). Interestingly, TRAP staining demonstrated that Hoxb8::gRNAGpr137b precursors differentiated into more numerous and larger osteoclasts than control cells (Figures 2D–E). The population of cells in these preparations are heterogeneous, thus the clearest analysis of differentiation and function of osteoclasts is their ability to resorb matrix rather than expression of transcript levels of specific markers in the bulk population. Accordingly, we assessed resorption activity and found increased resorption by Hoxb8:gRNAGpr137b osteoclasts cultured on hydroxyapatite-covered wells (Figure 2F, G). These data indicate that in vitro reduction of GPR137b function enhances osteoclast differentiation and activity, suggesting that GPR137b acts as a repressor of osteoclastogenesis.
Figure 2 -. Gpr137b deficiency promotes OC differentiation and bone resorption in vitro.
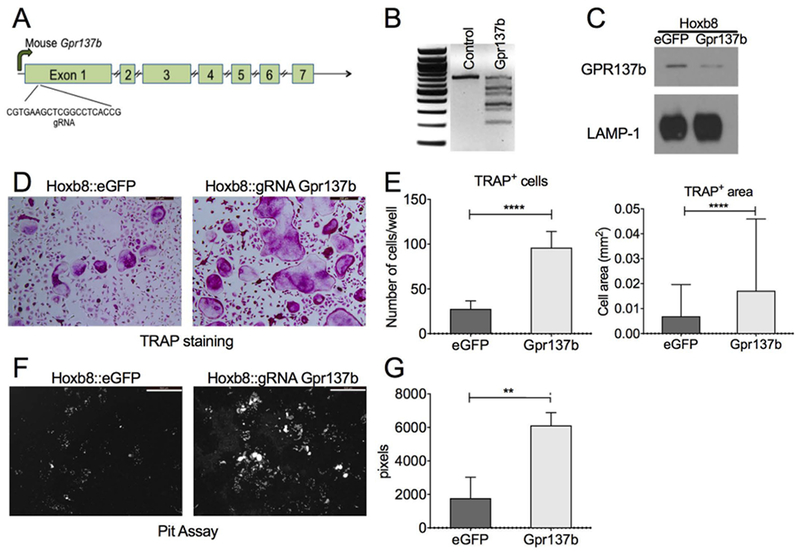
(A) Schematic of the Gpr137b gene indicating the gRNA target site on exon 1. (B-G) Differentiated Hoxb8 cells transduced with gRNA lentivirus. (B) Heteroduplex PCR followed by T7 Endonuclease I digestion detailing the presence of site-specific modification in the cells transduced with gRNA. (C) Western blot from membrane protein-enriched extracts showing GPR137b and LAMP-1 protein expression in control (eGFP) and transduced (Gpr137B−/−) cells. (D) Representative picture showing Hoxb8::eGFP and Hoxb8::gRNA gpr137b−/− cells differentiated on plastic and stained for TRAP activity. Scale bar 200μm. (E) Graph showing number of TRAP+ cells with 3 or more nuclei counted per cultured well and the average size of TRAP+ cells per well. (F-G) Hoxb8::eGFP and Hoxb8::gRNA gpr137b−/− were differentiated on osteo-assay plates. Cells were removed and the matrix was stained with von Kossa and pyrogallol to visualize the resorbtion pits (white spots). (F) Representative picture showing the matrix stained after 10 days of culture. Scale bar 500 μm. (G) Graph showing the quantification of resorbed area per well. Graphs represent mean±SD, **p<0.01, ****p<0.0001 determined by unpaired Student’s t-test or Mann Whitney test.
3.3. Generation of gpr137ba-deficient zebrafish.
Our analyses suggest an essential role for GPR137b in regulating osteoclast differentiation and activity. However, the complex interactions of intracellular coupling during bone homeostasis cannot be addressed in an in vitro setting. Zebrafish is emerging as a useful and cost-effective tool to study bone metabolism in vivo [34]. Many of the molecular pathways that regulate bone metabolism, including RANKL and M-CSF, are conserved across mammals and teleost fish species. In addition, like in mammals, zebrafish bone is cellular, containing osteoblasts, osteoclasts and osteocytes [35, 36]. Thus, we leveraged zebrafish to investigate the effects of loss of Gpr137b expression on bone development and remodeling.
The gpr137b gene is highly conserved across multiple species (Supplemental Table 1) [9]. All teleost fish share an ancestral whole genome duplication with lineage specific differentiation or loss of duplicated paralogues. Zebrafish retain two paralogs of gpr137b: gpr137ba and gpr137bb. Phylogenetic comparison among paralogues and orthologues showed that gpr137ba clustered closely to the mouse orthologue Gpr137b. Gpr137bb strongly differentiated (Supplemental Figure 1A, Supplemental Table 2). Analysis of the two paralogs during zebrafish development indicated that gpr137ba transcription increased in late juvenile stages coincident with the formation of the adult skeleton (Supplemental Figure 1B). In contrast, gpr137bb was mostly expressed at 4 days post-fertilization when ossification of the larval skeleton is just beginning (Supplemental Figure 1C). Thus, we focused on the zebrafish gpr137ba paralogue to examine the relevance of gpr137b function. We employed CRISPR/Cas9 targeted gene editing to generate a zebrafish mutant with predicted loss of gpr137ba function. Using gRNA designed against exon 3 of the gpr137ba gene, we generated and isolated a mutant line (mh107) harboring a 5bp deletion in the targeted region (Figure 3A, B). This allele is predicted to cause a frameshift mutation leading to a premature stop codon at amino acid 163 (Figure 3B, C). Assessment of mRNA for gpr137ba in bone showed substantially reduced expression in mutant fish compared with wild type siblings (Figure 3D), suggesting that stability of the mutant transcript was also impaired or, alternatively, the number of cells expressing gpr137ba in skeletal tissues was decreased.
Figure 3 -. Generation of mutant zebrafish deficient for gpr137ba.
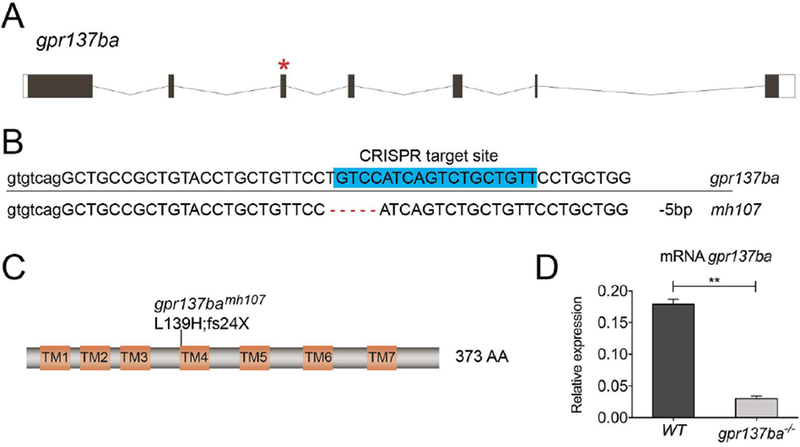
(A) Schematic of zebrafish gpr137ba indicating the gRNA target site on exon 3 (*). (B) Sequence of wild type and mutated gpr137ba gene showing deletion (mh107) leading to a frame shift and a predicted premature stop codon. (C) Cartoon of the predicted Gpr137ba protein showing the effect of the deletion (L139H;fs24X). (D) qPCR on cDNA from calvaria showing gpr137ba mRNA expression in wild type and gpr137ba−/− mutants. Graph represents mean±SD, n=3 where each sample represents a pool of 3 wildtype or 4 gpr137ba−/− fish, **p<0.01, unpaired Student’s t-test. TM= transmembrane domain.
3.4. Loss-of-function of gpr137ba in zebrafish increases bone resorption.
gpr137ba−/− zebrafish are viable and fertile and display no obvious adult phenotypes (Figure 4A). To examine skeletal remodeling in these fish, we took advantage of the fact that zebrafish scales are numerous independent skeletal units and are immediately accessible for study [37–39]. Staining scales for TRAP, we found that gpr137ba−/− zebrafish have a variable but similar percentage of TRAP+ scales as wild type siblings (Figure 4B, C). However, gpr137ba−/− fish have increased resorption activity on their scales, identified and quantified by the focal absence of von Kossa staining for mineralized matrix (Figure 4D, E). Thus, similar to mouse bone marrow derived osteoclasts, loss of gpr137ba in zebrafish appears to increase osteoclast activity.
Figure 4 -. Increased bone resorption in gpr137ba mutant zebrafish.
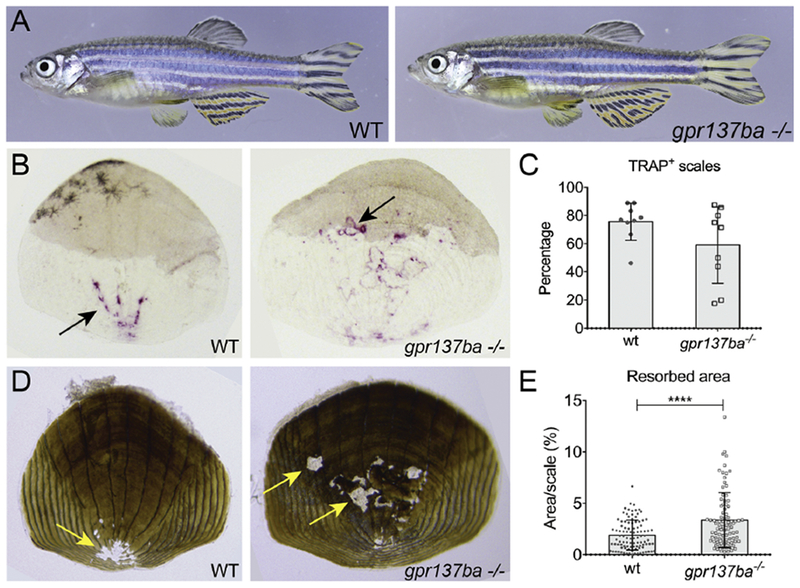
(A) Representative pictures of adult (12 wpf) gpr137ba−/− and wild type siblings. (B-E) Scales were collected from 8 weeks post fertilization. (B) Representative pictures of TRAP staining on scales indicated by black arrows. (C) Evaluation of the percentage of TRAP+ scales collected per individual gpr137ba−/− fish compared to wild type siblings (minimum 10 scales per fish were blindly evaluated). Graph represents mean±SD, n=10 fish. (D) Representative von Kossa staining of scales showing resorption pits (yellow arrow). (E) Quantification of percentage of resorbed area per individual scale collected from gpr137ba−/− and compared to wild type siblings. Graphs represent mean±SD, n=100 scales (10 fish) ****p<0.0001, Mann-Whitney test.
3.5. Loss-of-function of gpr137ba in zebrafish alters skeletal morphology.
In teleost fishes, like other vertebrates, remodeling is important in shaping form and function of the skeleton. In zebrafish and other small laboratory fishes, structures of the vertebrae are sensitive to changes in remodeling [17, 22, 41, 42]. Thus, we looked at the formation of vertebrae in order to address functional consequences of gpr137ba deficiency. During late zebrafish juvenile development (14-21 dpf), osteoclasts accumulate around forming vertebral arches [17]. We assessed changes in vertebral morphology of gpr137ba−/− mutants by micro-computed tomography (micro-CT) using previously described methods for zebrafish [22]. Radiographic analysis of comparably aged and sized adults focused on the first caudal vertebrae, C1 (Figure 5A, B). Measures of vertebral morphology in gpr137ba mutant fish, including tissue bone mineral density (BMD), and volume were comparable to wild type siblings (Figure 5C, D). Vertebral arch area, radius and length were also similar (data not shown). However, the angle that the neural arch forms with the vertebral body had significant higher amplitude in gpr137ba−/− mutants (Figure 5E). Similar results were obtained with caudal vertebrae 2 (data not shown). Previous analysis of csf1ra−/− mutant zebrafish, which have reduced number of osteoclasts, showed a similar but opposite modification (reduction) of arch angle [22]. This suggests that the angle between the arches and the vertebral bodies is sensitive to the alteration of osteoclast development and activity. This finding is consistent with the hypothesis that GPR137b acts as a repressor of osteoclast activity.
Figure 5 -. Analysis of caudal vertebrae reveals altered bone morphology in gpr137ba mutants.
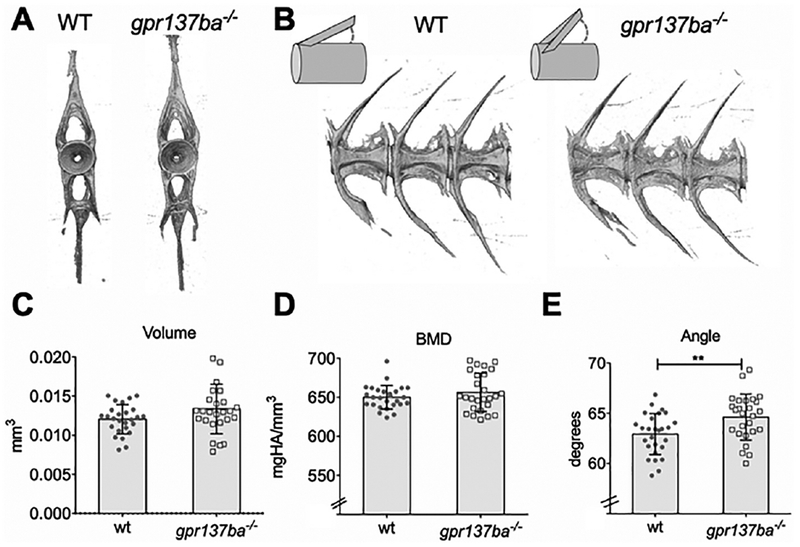
gpr137ba−/− and wild type zebrafish siblings were scanned by micro-CT (voxel size 6 microns). Representative images of (A) frontal and (B) lateral views of vertebrae from gpr137ba−/− and wildtype siblings. The first caudal vertebra was analyzed (C-E) Quantification of morphometry (C) volume, (D) bone mineral density (BMD) and (E) angle (n=26 wildtype, n=27 gpr137ba−/−). Graphs represent mean±SD, **p<0.01, unpaired Student’s t-test.
3.6. Gpr137ba-deficient zebrafish have high bone turnover.
During skeletal homeostasis the action of osteoclasts and osteoblasts is balanced to maintain bone shape and form. Through coupling events that are not well delineated, increased resorption typically results in a reciprocal increase in osteoblast activity. Given the lack of obvious teratology in the mutant, we hypothesized that the increased osteoclast activity in gpr137ba−/− zebrafish is balanced by increased bone formation, such that the effect of the mutation on overall skeletal morphology is minimal.
To examine osteoblast function in this mutant, we first examined the expression of hallmark osteoblast markers in adult scales of mutants and wild type siblings. Expression of osteocalcin and sp7 (osterix) were increased in gpr137b−/− scales compared to age and size matched wild type siblings suggesting increased osteoblast function (Supplemental Figure 2A, B). In order to measure bone formation rate in vivo, we used a dual alizarin red and calcein labeling methodology to determine bone accrual over time [43, 44]. We focused on bone formation in scales and the infraorbital bones, as both have a simple flat shape useful for determining changes in growth rate. The area of the new bone formed in a 14 day interval was measured for scales (Supplemental Figure 2C,D) and infraorbital bone 3 (Figure 6A, B). We find that the area of new bone formed between 8-10 wpf was significantly larger in gpr137ba−/− zebrafish at both of these sites, suggesting an increased rate of bone formation compared to wild type siblings.
Figure 6. Effect of gpr137ba−/− on skeletal homeostasis during zebrafish development.
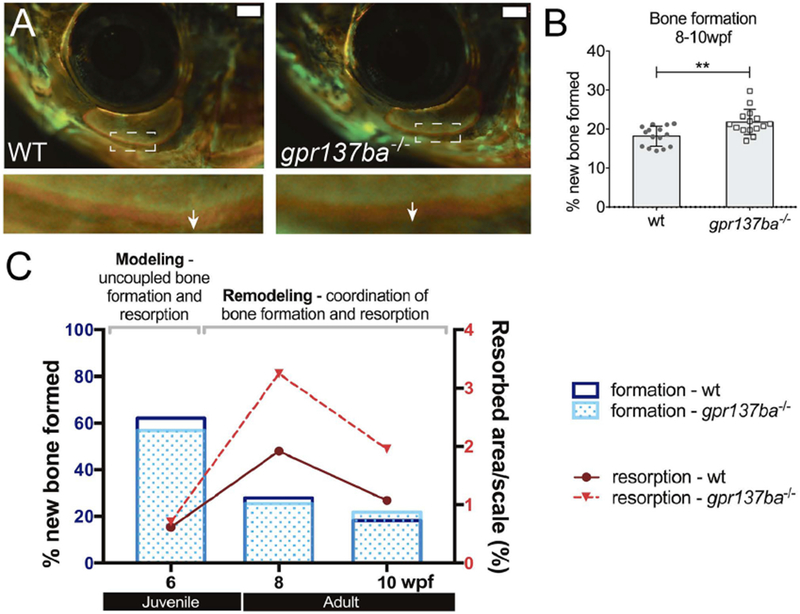
Bone formation in gpr137ba deficient fish as measured by differential fluorophore incorporation (A) Representative images showing the infraorbital bone 3 (IO3) on live fish. Inset shows the difference between red and green staining (white arrow) representing the new bone formed in 14 days. Scale bar: 1mm. (B) Quantification of new bone formed during 14 days in IO3. Graph represents mean ± SD, data points represent the average between left and right IO3, **p<0.01, unpaired Student’s t-test. (C) Model showing changes in bone homeostasis during development in wild type and gpr137ba−/− zebrafish. Juvenile fish show an initial phase of bone modeling characterized by uncoupled bone formation and bone resorption. At this age, bone formation largely supersedes bone resorption. When fish reach adulthood, the rate of bone formation gradually decreases while bone resorption increases. During this phase, bone formation and resorption are coupled in space and time. Detailed bone formation and resorption data during wild type and gpr137ba−/− zebrafish growth is shown in supplemental figure 3.
In addition, when we compared bone formation with bone resorption at specific time-points over a period of 6 weeks (6-12 wpf), we can clearly delineate an initial modelling phase (6 wpf) characterized by an extraordinarily high rate of bone formation and very low bone resorption. This initial modeling phase is followed by a remodeling phase in adulthood defined by a gradual decrease of bone formation accompanied by increased resorption (Figure 6C and Supplemental Figure 3). Overlapping gpr137ba−/− and wild type data, the mutant shows a significant increase in bone resorption at 8wpf that is maintained throughout time and is accompanied by a reactive and significant increase in bone formation at 10 wpf. Thus, we conclude that, in young adult fish, loss of gpr137ba results in increased bone remodeling.
4. Discussion
Osteoclasts are specialized cells of the hematopoietic lineage that resorb bone by secreting acid and proteolytic enzymes onto the mineralized surface of bone. These cells are key to the remodeling and shaping of the skeleton in all vertebrates. In this study, we describe the conserved function of an orphan G-protein coupled receptor, GPR137b, that is primarily expressed in osteoclasts and acts a negative regulator of osteoclast activity.
More than 1100 GPCR genes have been identified in the mouse genome and about 600 in the zebrafish. Many GPCR are highly conserved across species [45, 46]. Several GPCR family members expressed by osteoblasts are important skeletal regulators, including PTHR1, the receptor for parathyroid hormone (PTH) and PTH related peptide (PTHrP) [47]. Recent work has shown that GPR4, a receptor for protons, regulates RANKL expression in osteoblasts, indirectly promoting osteoclastogenesis [48]. The calcitonin receptor (CTR) is the only GPCR a role in osteoclast regulation extensively studied to date [5, 6, 49], however other GPCR have been implicated in osteoclast differentiation and/or function. Activation of GPR40, GPR41, GPR43 and GPR120 by lipid metabolites, or GPR55 by cannabinoids, has inhibitory effects on osteoclastogenesis [50–53] and LGR4 (GPR48) was shown to be an alternative, inhibitory receptor for RANKL [54]. We suggest that GPR137b is an important GPCR that is essential to restrain osteoclast function. The mechanism by which this, or other GPRs in this class, function in this capacity has yet to be defined.
Zebrafish is emerging as a valuable model to study the genetic and developmental aspects regulating bone dynamics and remodeling. Although quite distantly related to humans, zebrafish have become a reliable model for bone diseases such as osteogenesis imperfecta [34]. Additionally, fish have been used to study the effect of drug treatments such as glucocorticoids and bisphosphonates in models of osteoporosis [55–57]. Thus, zebrafish are predictive of bone phenotypes in mammalian bone and useful models for a growing set of human skeletal dysplasias. This is not surprising due to the fact that the skeleton is an ancestral trait shared by vertebrate lineages. Given the ease of genetic analysis and experimental accessibility, we chose to study the in vivo function of GPR137b on skeletogenesis using loss-of-function genetic models in the zebrafish.
Osteoclast–dependent bone remodeling in zebrafish and other teleost fish is abundant during formation of the vertebral arches during early juvenile stages (3-4 weeks post fertilization). As a demonstration of the role of remodeling in fish skeletal development, decreased osteoclast function in either zebrafish or medaka results in bone accumulation leading to deformity of the arches [17, 58]. Similarly, we have found that arch development is one of the more sensitive and precise measures of increased or reduced remodeling in the zebrafish [22]. The increased angle of the neural arches in gpr137ba mutants is consistent with these findings indicating an increase in remodeling activity. In contrast, csf1ra−/− zebrafish mutants which are deficient in osteoclasts exhibit decreased angle formation [22, 59]. We speculate that the subtleness of overt morphological defects of gpr137ba−/− mutants is due to compensation by osteoblasts. This hypothesis is supported by the increased expression of markers for bone formation in the scales and increased bone formation observed in gpr137ba mutant fish. While we cannot completely exclude a direct effect of gpr137ba-deficiency on osteoblast function, this is less likely as bone formation in juveniles with low bone resorption is similar to wild type. In mammals, osteoclast and osteoblast activity are coordinated by crosstalk mechanisms that include growth factors released from the matrix during osteoclast resorption (e.g. TGF-β and IGF-1) and factors secreted by osteoclasts, referred to as clastokines [3]. Whether coupling exists in fishes is unclear, though recent work in medaka suggests that it may [60].
Our data suggests that coupling mechanisms linking bone resorption and bone formation likely exist in zebrafish, as they do in mammals. Therefore, zebrafish may be a useful model to interrogate and identify signals that regulate the communication between bone cells. Our work points to a potential shift in regulation of coupling through development (Figure 6C). While early juvenile development is accompanied by increased growth and low resorption, this balance flips within a short period of time as growth of the skeleton slows. It is only during this period that we observe changes in osteoclast activity due to loss of gpr137ba. The lack of an early difference in the osteoclast activity may be simply due to efficient bone formation rate, wherein any resorption is being quickly accommodated. However, this shift may point to specific changes in coupling between these two skeletal cell types in development. Although this remains an open question, the timing of this change with fundamentally different stages of skeletal growth (development vs homeostasis) would lend support to specific differences in the regulation of coupling between osteoblast and osteoclasts.
Through analysis of gpr137ba-deficient zebrafish, we demonstrate the functional relevance of GPR137b for bone remodeling. Our results indicate that, similar to in vitro analyses of mouse osteoclasts, the absence of this receptor enhances osteoclastogenesis and resorption activity, leading to heightened bone resorption. We therefore propose that GPR137b is a conserved negative regulator of osteoclastogenesis in vertebrates, although the specific ligand(s) and signaling pathway(s) triggered are not known. Our data extends the class of conserved and essential inhibitory signals for osteoclast function [61–65]. Given advances in small molecule development to simulate or inhibit GPCR receptors [66]. GPR137b represents a potential target for drug treatment of osteoporosis or osteopetrosis.
Supplementary Material
Highlights.
Gpr137b is a seven-pass transmembrane receptor expressed by osteoclasts
Decreasing Gpr137b in osteoclast precursors led to increased osteoclast differentiation and bone resorption activity in vitro.
Loss-of-function gpr137ba zebrafish mutants demonstrated increased bone resorption
We propose that GPR137b is a conserved negative regulator of osteoclastogenesis in vertebrates
5. Acknowledgments
This work was supported by grants from US National Institutes of Health, grants R01 AR062590 (JFC), U01 DE024434 (MPH), R01 AR065538 and P30 AR070253 (PAN), and a T32 training grant (T32 AI007512). Additional support was provided by research cores funded by P30 AR066261, the Rheumatology Research Foundation Investigator Award (PYL) and K Supplement Award (JFC). JC-L would like to thank Dr. Matthew Warman for his support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict-of-interest Statement:
The authors declare that they have no conflict of interest
6. References
- [1].Marie PJ, Cohen-Solal M, The Expanding Life and Functions of Osteogenic Cells: From Simple Bone-Making Cells to Multifunctional Cells and Beyond, J Bone Miner Res 33(2) (2018) 199–210. doi: 10.1002/jbmr.3356. [DOI] [PubMed] [Google Scholar]
- [2].Crockett JC, Rogers MJ, Coxon FP, Hocking LJ, Helfrich MH, Bone remodelling at a glance, J Cell Sci 124(Pt 7) (2011) 991–8. doi: 10.1242/jcs.063032. [DOI] [PubMed] [Google Scholar]
- [3].Charles JF, Aliprantis AO, Osteoclasts: more than ‘bone eaters’, Trends Mol Med 20(8) (2014) 449–59. doi: 10.1016/j.molmed.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Katritch V, Cherezov V, Stevens RC, Structure-function of the G protein-coupled receptor superfamily, Annu Rev Pharmacol Toxicol 53 (2013) 531–56. doi: 10.1146/annurev-pharmtox-032112-135923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Wu M, Deng L, Zhu G, Li YP, G Protein and its signaling pathway in bone development and disease, Front Biosci (Landmark Ed) 15 (2010) 957–85. [DOI] [PubMed] [Google Scholar]
- [6].Keinan D, Yang S, Cohen RE, Yuan X, Liu T, Li YP, Role of regulator of G protein signaling proteins in bone, Front Biosci (Landmark Ed) 19 (2014) 634–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, Hodge CL, Haase J, Janes J, Huss JW 3rd, Su AI, BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources, Genome Biol 10(11) (2009) R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Spangenberg C, Winterpacht A, Zabel BU, Lobbert RW, Cloning and characterization of a novel gene (TM7SF1) encoding a putative seven-pass transmembrane protein that is upregulated during kidney development, Genomics 48(2) (1998) 178–85. doi: 10.1006/geno.1997.5170. [DOI] [PubMed] [Google Scholar]
- [9].Gao J, Xia L, Lu M, Zhang B, Chen Y, Xu R, Wang L, TM7SF1 (GPR137B): a novel lysosome integral membrane protein, Mol Biol Rep 39(9) (2012) 8883–9. doi: 10.1007/s11033-012-1755-0. [DOI] [PubMed] [Google Scholar]
- [10].Lacombe J, Karsenty G, Ferron M, Regulation of lysosome biogenesis and functions in osteoclasts, Cell Cycle 12(17) (2013) 2744–52. doi: 10.4161/cc.25825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Platt RJ, Chen S, Zhou Y, Yim MJ, Swiech L, Kempton HR, Dahlman JE, Parnas O, Eisenhaure TM, Jovanovic M, Graham DB, Jhunjhunwala S, Heidenreich M, Xavier RJ, Langer R, Anderson DG, Hacohen N, Regev A, Feng G, Sharp PA, Zhang F, CRISPR-Cas9 knockin mice for genome editing and cancer modeling, Cell 159(2) (2014) 440–55. doi: 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Schneider CA, Rasband WS, Eliceiri KW, NIH Image to ImageJ: 25 years of image analysis, Nat Methods 9(7) (2012) 671–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Lee PY, Sykes DB, Ameri S, Kalaitzidis D, Charles JF, Nelson-Maney N, Wei K, Cunin P, Morris A, Cardona AE, Root DE, Scadden DT, Nigrovic PA, The metabolic regulator mTORC1 controls terminal myeloid differentiation, Sci Immunol 2(11) (2017). doi: 10.1126/sciimmunol.aam6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Jao LE, Wente SR, Chen W, Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system, Proc Natl Acad Sci U S A 110(34) (2013) 13904–9. doi: 10.1073/pnas.1308335110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh JR, Joung JK, Efficient genome editing in zebrafish using a CRISPR-Cas system, Nat Biotechnol 31(3) (2013) 227–9. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Truett GE, Heeger P, Mynatt RL, Truett AA, Walker JA, Warman ML, Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT), Biotechniques 29(1) (2000) 52, 54. [DOI] [PubMed] [Google Scholar]
- [17].Chatani M, Takano Y, Kudo A, Osteoclasts in bone modeling, as revealed by in vivo imaging, are essential for organogenesis in fish, Dev Biol 360(1) (2011) 96–109. doi: 10.1016/j.ydbio.2011.09.013. [DOI] [PubMed] [Google Scholar]
- [18].Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, Campbell DS, Parant JM, Yost HJ, Kanki JP, Chien CB, The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs, Dev Dyn 236(11) (2007) 3088–99. doi: 10.1002/dvdy.21343. [DOI] [PubMed] [Google Scholar]
- [19].O’Gorman S, Dagenais NA, Qian M, Marchuk Y, Protamine-Cre recombinase transgenes efficiently recombine target sequences in the male germ line of mice, but not in embryonic stem cells, Proc Natl Acad Sci U S A 94(26) (1997) 14602–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Urso K, Charles JF, Shull GE, Aliprantis AO, Balestrieri B, Anion Exchanger 2 Regulates Dectin-1-Dependent Phagocytosis and Killing of Candida albicans, PLoS One 11(7) (2016) e0158893. doi: 10.1371/journal.pone.0158893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL, Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction, BMC Bioinformatics 13 (2012) 134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Charles JF, Sury M, Tsang K, Urso K, Henke K, Huang Y, Russell R, Duryea J, Harris MP, Utility of quantitative micro-computed tomographic analysis in zebrafish to define gene function during skeletogenesis, Bone 101 (2017) 162–171. doi: 10.1016/j.bone.2017.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Lai X, Reproducible method to enrich membrane proteins with high purity and high yield for an LC-MS/MS approach in quantitative membrane proteomics, Electrophoresis 34(6) (2013) 809–17. doi: 10.1002/elps.201200503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Charles JF, Hsu LY, Niemi EC, Weiss A, Aliprantis AO, Nakamura MC, Inflammatory arthritis increases mouse osteoclast precursors with myeloid suppressor function, J Clin Invest 122(12) (2012) 4592–605. doi: 10.1172/JCI60920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Chang C, Franz-Odendaal TA, The zebrafish infraorbital bones: a descriptive study, Zebrafish 11(1) (2014) 50–6. doi: 10.1089/zeb.2013.0907. [DOI] [PubMed] [Google Scholar]
- [26].de Vrieze E, Moren M, Metz JR, Flik G, Lie KK, Arachidonic acid enhances turnover of the dermal skeleton: studies on zebrafish scales, PLoS One 9(2) (2014) e89347. doi: 10.1371/journal.pone.0089347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].O’Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, Rajput B, Robbertse B, Smith-White B, Ako-Adjei D, Astashyn A, Badretdin A, Bao Y, Blinkova O, Brover V, Chetvernin V, Choi J, Cox E, Ermolaeva O, Farrell CM, Goldfarb T, Gupta T, Haft D, Hatcher E, Hlavina W, Joardar VS, Kodali VK, Li W, Maglott D, Masterson P, McGarvey KM, Murphy MR, O’Neill K, Pujar S, Rangwala SH, Rausch D, Riddick LD, Schoch C, Shkeda A, Storz SS, Sun H, Thibaud-Nissen F, Tolstoy I, Tully RE, Vatsan AR, Wallin C, Webb D, Wu W, Landrum MJ, Kimchi A, Tatusova T, DiCuccio M, Kitts P, Murphy TD, Pruitt KD, Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation, Nucleic Acids Res 44(D1) (2016) D733–45. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Yates A, Akanni W, Amode MR, Barrell D, Billis K, Carvalho-Silva D, Cummins C, Clapham P, Fitzgerald S, Gil L, Giron CG, Gordon L, Hourlier T, Hunt SE, Janacek SH, Johnson N, Juettemann T, Keenan S, Lavidas I, Martin FJ, Maurel T, McLaren W, Murphy DN, Nag R, Nuhn M, Parker A, Patricio M, Pignatelli M, Rahtz M, Riat HS, Sheppard D, Taylor K, Thormann A, Vullo A, Wilder SP, Zadissa A, Birney E, Harrow J, Muffato M, Perry E, Ruffier M, Spudich G, Trevanion SJ, Cunningham F, Aken BL, Zerbino DR, Flicek P, Ensembl 2016, Nucleic Acids Res 44(D1) (2016) D710–6. doi: 10.1093/nar/gkv1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Edgar RC, MUSCLE: multiple sequence alignment with high accuracy and high throughput, Nucleic Acids Res 32(5) (2004) 1792–7. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Kumar S, Stecher G, Tamura K, MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets, Mol Biol Evol 33(7) (2016) 1870–4. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Wang GG, Calvo KR, Pasillas MP, Sykes DB, Hacker H, Kamps MP, Quantitative production of macrophages or neutrophils ex vivo using conditional Hoxb8, Nat Methods 3(4) (2006) 287–93. doi: 10.1038/nmeth865. [DOI] [PubMed] [Google Scholar]
- [32].Di Ceglie I, van den Akker GG, Ascone G, Ten Harkel B, Hacker H, van de Loo FA, Koenders MI, van der Kraan PM, de Vries TJ, Vogl T, Roth J, van Lent PL, Genetic modification of ER-Hoxb8 osteoclast precursors using CRISPR/Cas9 as a novel way to allow studies on osteoclast biology, J Leukoc Biol 101(4) (2017) 957–966. doi: 10.1189/jlb.1AB0416-180RR. [DOI] [PubMed] [Google Scholar]
- [33].Zach F, Mueller A, Gessner A, Production and Functional Characterization of Murine Osteoclasts Differentiated from ER-Hoxb8-Immortalized Myeloid Progenitor Cells, PLoS One 10(11) (2015) e0142211. doi: 10.1371/journal.pone.0142211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Mackay EW, Apschner A, Schulte-Merker S, A bone to pick with zebrafish, Bonekey Rep 2 (2013) 445. doi: 10.1038/bonekey.2013.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Witten PE, Huysseune A, A comparative view on mechanisms and functions of skeletal remodelling in teleost fish, with special emphasis on osteoclasts and their function, Biol Rev Camb Philos Soc 84(2) (2009) 315–46. doi: 10.1111/j.1469-185X.2009.00077.x. [DOI] [PubMed] [Google Scholar]
- [36].Apschner A, Schulte-Merker S, Witten PE, Not all bones are created equal - using zebrafish and other teleost species in osteogenesis research, Methods Cell Biol 105 (2011) 239–55. doi: 10.1016/B978-0-12-381320-6.00010-2. [DOI] [PubMed] [Google Scholar]
- [37].Kitamura K, Takahira K, Inari M, Satoh Y, Hayakawa K, Tabuchi Y, Ogai K, Nishiuchi T, Kondo T, Mikuni-Takagaki Y, Chen W, Hattori A, Suzuki N, Zebrafish scales respond differently to in vitro dynamic and static acceleration: analysis of interaction between osteoblasts and osteoclasts, Comp Biochem Physiol A Mol Integr Physiol 166(1) (2013) 74–80. doi: 10.1016/j.cbpa.2013.04.023. [DOI] [PubMed] [Google Scholar]
- [38].Pasqualetti S, Banfi G, Mariotti M, The zebrafish scale as model to study the bone mineralization process, J Mol Histol 43(5) (2012) 589–95. doi: 10.1007/s10735-012-9425-z. [DOI] [PubMed] [Google Scholar]
- [39].Carnovali M, Luzi L, Banfi G, Mariotti M, Chronic hyperglycemia affects bone metabolism in adult zebrafish scale model, Endocrine 54(3) (2016) 808–817. doi: 10.1007/s12020-016-1106-3. [DOI] [PubMed] [Google Scholar]
- [40].To TT, Witten PE, Renn J, Bhattacharya D, Huysseune A, Winkler C, Rankl-induced osteoclastogenesis leads to loss of mineralization in a medaka osteoporosis model, Development 139(1) (2012) 141–50. doi: 10.1242/dev.071035. [DOI] [PubMed] [Google Scholar]
- [41].Cheah FS, Winkler C, Jabs EW, Chong SS, Tgfbeta3 regulation of chondrogenesis and osteogenesis in zebrafish is mediated through formation and survival of a subpopulation of the cranial neural crest, Mech Dev 127(7-8) (2010) 329–44. doi: 10.1016/j.mod.2010.04.003. [DOI] [PubMed] [Google Scholar]
- [42].Yu T, Graf M, Renn J, Schartl M, Larionova D, Huysseune A, Witten PE, Winkler C, A vertebrate-specific and essential role for osterix in osteogenesis revealed by gene knockout in the teleost medaka, Development 144(2) (2017) 265–271. doi: 10.1242/dev.139550. [DOI] [PubMed] [Google Scholar]
- [43].Kimmel CB, DeLaurier A, Ullmann B, Dowd J, McFadden M, Modes of developmental outgrowth and shaping of a craniofacial bone in zebrafish, PLoS One 5(3) (2010) e9475. doi: 10.1371/journal.pone.0009475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Jeradi S, Hammerschmidt M, Retinoic acid-induced premature osteoblast-to-preosteocyte transitioning has multiple effects on calvarial development, Development 143(7) (2016) 1205–16. doi: 10.1242/dev.129189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Fredriksson R, Schioth HB, The repertoire of G-protein-coupled receptors in fully sequenced genomes, Mol Pharmacol 67(5) (2005) 1414–25. doi: 10.1124/mol.104.009001. [DOI] [PubMed] [Google Scholar]
- [46].Hanlon CD, Andrew DJ, Outside-in signaling--a brief review of GPCR signaling with a focus on the Drosophila GPCR family, J Cell Sci 128(19) (2015) 3533–42. doi: 10.1242/jcs.175158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Cheloha RW, Gellman SH, Vilardaga JP, Gardella TJ, PTH receptor-1 signalling-mechanistic insights and therapeutic prospects, Nat Rev Endocrinol 11(12) (2015) 712–24. doi: 10.1038/nrendo.2015.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Okito A, Nakahama K, Akiyama M, Ono T, Morita I, Involvement of the G-protein-coupled receptor 4 in RANKL expression by osteoblasts in an acidic environment, Biochem Biophys Res Commun 458(2) (2015) 435–40. doi: 10.1016/j.bbrc.2015.01.142. [DOI] [PubMed] [Google Scholar]
- [49].Masi L, Brandi ML, Calcitonin and calcitonin receptors, Clin Cases Miner Bone Metab 4(2) (2007) 117–22. [PMC free article] [PubMed] [Google Scholar]
- [50].Wauquier F, Philippe C, Leotoing L, Mercier S, Davicco MJ, Lebecque P, Guicheux J, Pilet P, Miot-Noirault E, Poitout V, Alquier T, Coxam V, Wittrant Y, The free fatty acid receptor G protein-coupled receptor 40 (GPR40) protects from bone loss through inhibition of osteoclast differentiation, J Biol Chem 288(9) (2013) 6542–51. doi: 10.1074/jbc.M112.429084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Kim HJ, Yoon HJ, Kim BK, Kang WY, Seong SJ, Lim MS, Kim SY, Yoon YR, G Protein-Coupled Receptor 120 Signaling Negatively Regulates Osteoclast Differentiation, Survival, and Function, J Cell Physiol 231(4) (2016) 844–51. doi: 10.1002/jcp.25133. [DOI] [PubMed] [Google Scholar]
- [52].Cornish J, MacGibbon A, Lin JM, Watson M, Callon KE, Tong PC, Dunford JE, van der Does Y, Williams GA, Grey AB, Naot D, Reid IR, Modulation of osteoclastogenesis by fatty acids, Endocrinology 149(11) (2008) 5688–95. doi: 10.1210/en.2008-0111. [DOI] [PubMed] [Google Scholar]
- [53].Whyte LS, Ryberg E, Sims NA, Ridge SA, Mackie K, Greasley PJ, Ross RA, Rogers MJ, The putative cannabinoid receptor GPR55 affects osteoclast function in vitro and bone mass in vivo, Proc Natl Acad Sci U S A 106(38) (2009) 16511–6. doi: 10.1073/pnas.0902743106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Luo J, Yang Z, Ma Y, Yue Z, Lin H, Qu G, Huang J, Dai W, Li C, Zheng C, Xu L, Chen H, Wang J, Li D, Siwko S, Penninger JM, Ning G, Xiao J, Liu M, LGR4 is a receptor for RANKL and negatively regulates osteoclast differentiation and bone resorption, Nat Med 22(5) (2016) 539–46. doi: 10.1038/nm.4076. [DOI] [PubMed] [Google Scholar]
- [55].Pasqualetti S, Congiu T, Banfi G, Mariotti M, Alendronate rescued osteoporotic phenotype in a model of glucocorticoid-induced osteoporosis in adult zebrafish scale, Int J Exp Pathol 96(1) (2015) 11–20. doi: 10.1111/iep.12106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].de Vrieze E, van Kessel MA, Peters HM, Spanings FA, Flik G, Metz JR, Prednisolone induces osteoporosis-like phenotype in regenerating zebrafish scales, Osteoporos Int 25(2) (2014) 567–78. doi: 10.1007/s00198-013-2441-3. [DOI] [PubMed] [Google Scholar]
- [57].Barrett R, Chappell C, Quick M, Fleming A, A rapid, high content, in vivo model of glucocorticoid-induced osteoporosis, Biotechnol J 1(6) (2006) 651–5. doi: 10.1002/biot.200600043. [DOI] [PubMed] [Google Scholar]
- [58].To TT, Witten PE, Huysseune A, Winkler C, An adult osteopetrosis model in medaka reveals the importance of osteoclast function for bone remodeling in teleost fish, Comp Biochem Physiol C Toxicol Pharmacol 178 (2015) 68–75. doi: 10.1016/j.cbpc.2015.08.007. [DOI] [PubMed] [Google Scholar]
- [59].Parichy DM, Ransom DG, Paw B, Zon LI, Johnson SL, An orthologue of the kit-related gene fms is required for development of neural crest-derived xanthophores and a subpopulation of adult melanocytes in the zebrafish, Danio rerio, Development 127(14) (2000) 3031–44. [DOI] [PubMed] [Google Scholar]
- [60].Yu T, Witten PE, Huysseune A, Buettner A, To TT, Winkler C, Live imaging of osteoclast inhibition by bisphosphonates in a medaka osteoporosis model, Dis Model Mech 9(2) (2016) 155–63. doi: 10.1242/dmm.019091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Yeon JT, Choi SW, Kim SH, Arginase 1 is a negative regulator of osteoclast differentiation, Amino Acids 48(2) (2016) 559–65. doi: 10.1007/s00726-015-2112-0. [DOI] [PubMed] [Google Scholar]
- [62].Linares GR, Brommage R, Powell DR, Xing W, Chen ST, Alshbool FZ, Lau KH, Wergedal JE, Mohan S, Claudin 18 is a novel negative regulator of bone resorption and osteoclast differentiation, J Bone Miner Res 27(7) (2012) 1553–65. doi: 10.1002/jbmr.1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Zhao B, Ivashkiv LB, Negative regulation of osteoclastogenesis and bone resorption by cytokines and transcriptional repressors, Arthritis Res Ther 13(4) (2011) 234. doi: 10.1186/ar3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Stiffel V, Amoui M, Sheng MH, Mohan S, Lau KH, EphA4 receptor is a novel negative regulator of osteoclast activity, J Bone Miner Res 29(4) (2014) 804–19. doi: 10.1002/jbmr.2084. [DOI] [PubMed] [Google Scholar]
- [65].Kim JY, Min JY, Baek JM, Ahn SJ, Jun HY, Yoon KH, Choi MK, Lee MS, Oh J, CTRP3 acts as a negative regulator of osteoclastogenesis through AMPK-c-Fos-NFATc1 signaling in vitro and RANKL-induced calvarial bone destruction in vivo, Bone 79 (2015)242–51. doi: 10.1016/j.bone.2015.06.011. [DOI] [PubMed] [Google Scholar]
- [66].Miao Y, McCammon JA, G-protein coupled receptors: advances in simulation and drug discovery, Curr Opin Struct Biol 41 (2016) 83–89. doi: 10.1016/j.sbi.2016.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


