Abstract
Zinc finger and SCAN domain containing 4 (Zscan4) is a gene that is specifically expressed during zygotic genome activation (ZGA) in mouse preimplantation embryos, and a reduction of Zscan4 transcripts leads to developmental failure. In mouse embryonic stem cells (ESCs), Zscan4 is expressed transiently in as little as 1–5% of the cell population. Zscan4 has also been shown to enhance the efficiency of mouse induced pluripotent stem cells (iPSCs) generation and their quality. Although ZSCAN4 plays important roles in murine embryos and stem cells, its expression and role in bovine embryos is unknown. This study examines ZSCAN4 transcripts in bovine embryos at various developmental stages and attempts to elucidate the functions of ZSCAN4 during bovine preimplantation development. ZSCAN4 transcripts were found to be upregulated at the 8- and 16-cell stages. We next attempted ZSCAN4 downregulation in bovine early embryos by RNA interference and evaluated developmental competency and transcripts levels of genes involved in ZGA and iPSCs generation. Although the bovine embryos injected with ZSCAN4-siRNA could develop to the 8-cell stage, very few were developing beyond the 16-cell stage. PIWIL2 expression was reduced in ZSCAN4 downregulated embryos. It is possible that ZSCAN4 downregulated embryos fail to regulate gene expression during ZGA. Our results indicate that ZSCAN4 is an important factor for the preimplantation development of bovine embryos.
Keywords: Bovine embryo, Early development, Gene expression, RNA interference, ZSCAN4
Procedures for in vitro production (IVP) of embryos such as in vitro fertilization (IVF) and somatic cell nuclear transfer (SCNT) have played an increasingly important role in domestic animal production. However, the efficiency remains low both for embryo development and offspring production after embryo transfer. Although the cause of such low efficiency is unknown, it may be due to the abnormal epigenetic status in bovine embryos obtained from IVF or SCNT procedures [1,2,3]. In mammalian embryos, zygotic genome activation (ZGA) is a critical event in preimplantation development [4]. ZGA occurs at the 2-cell stage in mouse embryos [5], at the 4- to 8-cell stage in human [6] and pig embryos [7], and at the 8- to 16-cell stage in bovine embryos [8, 9]. After fertilization, the developmental program controlled by maternally inherited transcripts in oocytes is replaced by a program controlled by embryonic transcripts [4, 10]. In case of SCNT, the epigenetic information of the donor nucleus needs to be reprogrammed in the early embryos reconstructed with donor cell and recipient oocyte. The epigenetic status in SCNT embryos is changed to an embryonic state by reprogramming of the donor nucleus [11]. Therefore, ZGA and reprogramming after fertilization or SCNT are essential for establishing the epigenetic status in IVP embryos. However, limited information is available regarding the mechanisms of ZGA and nuclear reprogramming in domestic animal embryos.
The Zinc finger and SCAN domain containing 4 (Zscan4) gene was originally identified as specifically expressed during ZGA in the late 2-cell stage of mouse preimplantation embryos [12]. Subsequently, Zscan4 knockdown by small interfering RNA (siRNA) presented a progression delay from the 2- to 4-cell stage and, which resulted in implantation failure [12]. Furthermore, in mouse embryonic stem cells (ESCs), the expression of Zscan4 is transient and reversible with infrequent transcriptional activation in only 1–5% of the cell population at a given time point [12, 13]. Additionally, Zscan4 also enhances the efficiency of mouse induced pluripotent stem cells (iPSCs) generation and their quality [14, 15]. Within that period of Zscan4 transcription, further biological events occur, including transient expression of other ZGA-specific genes [16, 17], rapid telomere extension [13], and blockage of global protein translation [18]. These data suggest that, in addition to ZGA in mouse early embryos, Zscan4 plays a role in reprogramming the somatic cell nucleus for iPSCs generation. However, the expression status and role of ZSCAN4 in preimplantation development of bovine embryos is unclear.
The objectives of this study were to investigate the expression status of the ZSCAN4 gene in bovine embryos at the preimplantation stage and to evaluate the role of ZSCAN4 during the early development of bovine embryos using RNA interference targeting ZSCAN4.
Materials and Methods
In this study, we conducted three experiments, as follows. In experiment 1, we measured the transcript abundance of the ZSCAN4 gene in bovine oocytes and preimplantation embryos (Figs. 1 and 2). In experiment 2, we evaluated the effect of siRNA injection on ZSCAN4 expression in bovine embryos (Fig. 3) and the effect of ZSCAN4 downregulation on the development of bovine embryos (Table 2 and Fig. 4). In experiment 3, we determined gene expressions in bovine embryos derived from ZSCAN4-siRNA injection (Figs. 5 and 6). Experiment 1 was performed in the University of California, Davis, and experiments 2 and 3 were performed in The United Graduate School of Agricultural Sciences, Iwate University. All experiments were approved by the Animal Ethics Committee in each university.
Chemicals
All chemicals and reagents were purchased from Sigma-Aldrich (St. Louis, MO, USA) unless otherwise stated.
Oocyte collection and in vitro maturation
Cow ovaries were collected at a local slaughterhouse and maintained at room temperature during transportation to the laboratory. Cumulus-oocyte complexes (COCs) were aspirated from follicles of 2–8 mm. In experiment 1, 50 bovine COCs were matured in 500 µL modified M199 medium (Sigma M2154) supplemented with 0.1 mM ALA-glutamine, 0.2 mM sodium pyruvate, 5 mg/ml gentamicin, 50 ng/ml EGF, 50 ng/ml oFSH (National Hormone and Peptide Program, St. Torrance, CA, USA), 3 µg/ml bLH (National Hormone and Peptide Program), 0.1 mM cysteamine, and 10% FBS (Hyclone; South Logan, UT, USA). In experiment 2 and 3, 10 bovine COCs were matured in a 100 µl drop of IVMD-101 medium (Research Institute for the Functional Peptides, Yamagata, Japan) [19]. In vitro maturation (IVM) was performed at 38.5°C in a humidified atmosphere containing 5% CO2 in air for 22–24 h.
In vitro fertilization and in vitro culture
After IVM, COCs were fertilized in SOF-IVF medium containing 107.7 mM NaCl, 7.16 mM KCl, 1.19 mM KH2PO4, 0.49 mM MgCl2, 1.17 mM CaCl2, 5.3 mM sodium lactate, 25.07 mM NaHCO3, 0.20 mM sodium pyruvate, 0.5 mM fructose, 1X non-essential fatty acid, 5 μg/ml gentamicin, 10 μg/ml heparin, and 6 mg/ml BSA (experiment 1) or IVF-100 medium (Research Institute for the Functional Peptides, Yamagata, Japan; experiments 2 and 3) [19]. Cryopreserved semen was thawed, and sperms were washed twice by centrifugation (at 1800 rpm for 5 min) in IVF medium. Sperm were resuspended in the IVF medium at a final concentration of 1.0 × 106/ml (experiment 1) or 5.0 × 106/ml (experiment 2 and 3). Between 15 and 25 COCs were placed into each sperm suspension drop. COCs and sperm were incubated for 6 h (experiments 2 and 3) or 18 h (experiment 1) at 38.5°C in a humidified atmosphere containing 5% CO2 in air.
In experiment 1, embryos were cultured in KSOMaa Evolve Bovine media (ZEBV-100; Zenith Biotech, Guilford, CT, USA) supplemented with 4 mg/ml BSA at 38.5°C in a humidified atmosphere of 5% CO2, 5% O2 and 90% N2. On day 3 (IVF = Day 0), 5% FBS (Gemini Bio Products, West Sacramento, CA, USA) was supplemented into the culture drops. In a group of embryos, 50 μg/ml α-amanitin, which is an inhibitor of RNA synthesis, was added in the in vitro culture (IVC) medium, 4- and 8-cell embryos were collected at 28 and 40 h after IVC, respectively. In experiments 2 and 3, following microinjection of siRNA, embryos were cultured in modified TALP (mTALP) medium [20], with 0.1% BSA (fraction V) at 39°C in 5% CO2, 5% O2, and 90% N2. On Day 2, embryos were transferred to mTALP supplemented with 3% new-born calf serum (Invitrogen, Carlsbad, CA, USA) and subsequently cultured at 38.5°C in 5% CO2, 5% O2, and 90% N2 until Day 7. Rates of embryo development were assessed on Day 2 (2-cell ≤), Day 3 (8-cell ≤), Day 4 (16-cell ≤, 32-cell ≤), Day 5 (morula ≤) and Day 7 (blastocyst).
Design of siRNA and microinjection into embryos
The target sites of the ZSCAN4 transcript were selected from bovine sequences (GenBank accession number: XM_005195522.1). Then, specific siRNA for ZSCAN4 were developed using siRNA design software, BLOCK-iT RNAi Designer (Thermo Fisher Scientific, Tokyo, Japan). Both sense and antisense RNA sequences for siRNA were commercially synthesized (Table 1).
Table 1. Primers and siRNA sequences.
| Name | Nucleotide sequences (5’-3’) | Annealing temperature (°C) | Fragment size | Genbank accession no. |
|---|---|---|---|---|
| ZSCAN4 | F- GTCCCTGGAAACAGGACAAA | 60 | 179 | XM_005195522.1 |
| R- TTCTGGAGTTCCGTGGATTC | ||||
| EIF1AX | F- TTCAAAGAGGATGGGCAGGAGTATG | 53 | 287 | NM_001412.1 |
| R- GGACCAAATGTATCAGTTTCATTGA | ||||
| DPPA2 | F- CAGACCACTCTTGGCAGACA | 60 | 163 | NM_001206470.1 |
| R- GCATTCAGGGCATAACAGGT | ||||
| PIWIL2 | F- AGGCCCAGTGAGAGACAGAA | 60 | 235 | XM_010798716.1 |
| R- CCCAACGTGTCAGTTCATTG | ||||
| HcRed1 | F- GCCCGGCTTCCACTTCA | 60 | 79 | N/A |
| R- GGCCTCGTACAGCTCGAAGTA | N/A | |||
| Histone H2A | F- AGGACGACTAGCCATGGACGTGTG | 60 | 208 | NM_174809 |
| R- CCACCACCAGCAATTGTAGCCTTG | ||||
| ZSCAN4-siRNA | S- GCAUGUAUCCUGGCUCCAUTT | N/A | N/A | N/A |
| AS- AUGGAGCCAGGAUACAUGCTT | N/A | N/A | N/A | |
F, forward; R, reverse; S, sense; AS, antisense strand.
After insemination, cumulus cells and excess sperm were removed from presumptive zygotes by pipetting. These embryos were subsequently transferred to a 20 µl drop of mTALP containing 1 mg/ml BSA for microinjection. Using a Femtojet microinjector (Eppendorf, Hamburg, Germany), approximately 10 pl of 50 µM specific siRNA duplexes with 5 µM tetramethylrhodamine isothiocyanate (TRITC)-dextran (Dx; Invitrogen) were injected into the cytoplasm of a group of embryos (ZSCAN4-siRNA) during their 1-cell stage. Approximately 10 pl of 20 µM nonsilencing siRNA (AllStars Negative Control siRNA, Qiagen, Tokyo, Japan) with Dx was injected into another group of embryos (Control-siRNA) by the same method. Finally, some embryos were not injected with siRNA (Uninjected). Embryos were washed three times immediately after microinjection, and cultured as described above. Injection of siRNA into the cytoplasm of embryos was verified by the detection of red fluorescence from Dx on Day 2, and embryos devoid of red fluorescence were removed from further analysis.
Determination of the relative abundance of gene transcripts in bovine embryos
In experiment 1, total RNA was extracted from oocytes before or after IVM and from embryos developed to 1-cell (18 h after IVF), 2-cell (10–14 h after IVC), 4-cell (28 h after IVC), 8-cell (40 h after IVC), 16-cell (56 h after IVC), morula (96 h after IVC), and blastocyst (144 h after IVC) stage. The RNA was extracted from pools of 10 oocytes or embryos using the PicoPure RNA Isolation Kit (Arcturus, Mountain View, CA, USA) and treated with RNase-free DNase I (Qiagen) to remove the genomic DNA according to the manufacturer’s instructions. Before RNA isolation, each sample was spiked with 8 μl of 250 fg/μl HcRed1 cRNA, used as an exogenous control [21]. cDNA synthesis was performed using Superscript II Reverse Transcriptase with random hexamer priming, following the manufacturer’s instructions. In experiments 2 and 3, 8- to 16-cell (56 h after IVC) stage embryos were treated with 0.1% protease in 1% PVP-PBS for 5 min, and washed seven times in 1% PVP-PBS. Pools of five embryos were added to 5 µl lysis buffer [0.8% Igepal (ICN Biomedicals Inc., Aurora, OH, USA), 5 mM DTT (Invitrogen) and 1 U/µl of RNasin (Promega, Madison, WI, USA)], snap-frozen in liquid nitrogen and stored at −80°C. Extracted RNA samples were heated to 80°C for 5 min and subjected to reverse transcription (RT) using the QuantiTect Reverse Transcription Kit (Qiagen) according to the manufacturer’s instructions.
In experiment 1, quantitative real-time PCR was performed in a QuantStudio 3 Real-Time PCR System (Thermo Fisher Scientific) with a final reaction volume of 20 µl containing 10 µl Fast SYBR Green Master Mix (Qiagen), 0.5 µl forward primer, 0.5 µl reverse primer (Table 1), 7 µl water, and 2 µl cDNA sample. HcRed1 (external control) abundance, which maintained a same expression level in across the different developmental stages, was determined in each sample and used to normalize for differences in RNA extraction and RT efficiency. Four samples were used for quantitative analysis, and each sample was run in duplicate for real-time PCR. In experiments 2 and 3, real-time PCR was performed using a StepOneTM system (Thermo Fisher Scientific) with a final reaction volume of 20 µl containing 10 µl QuantiTect SYBR Green Master Mix (Qiagen), 1.0 µl forward primer, 1.0 µl reverse primer (Table 1), 6 µl water, and 2 µl cDNA sample. A standard curve was generated for each amplicon based on the serial dilution amplification of a known quantity. PCR products for each gene were purified using a QIAquick PCR Purification Kit (Qiagen), quantified by measuring absorbance at 260 nm using NanoDrop (ND-1000; Thermo Fisher Scientific), and diluted as appropriate. Serial 10-fold dilutions for creating the standard curve were amplified in every real-time PCR run. The standards and cDNA samples were then co-amplified in the same reaction prepared from a master mix. Fluorescence was acquired at each cycle to determine the threshold cycle or the cycle during the log-linear phase of the reaction at which fluorescence rose above the background for each sample. Final quantification was performed using the StepOneTM quantification software (Thermo Fisher Scientific). Expression levels of the target gene in each run were normalized to the internal control Histone H2A, which is stably expressed in bovine embryos at each developmental stage [22]. Six samples were used for quantitative analysis, and each sample was run in duplicate for real-time PCR.
Statistical analysis
The expression levels of ZSCAN4, EIF1AX, DPPA2 and PIWIL2 mRNA were analyzed using the Kruskal-Wallis test, followed by multiple pairwise comparisons using Scheffé’s method (Figs. 1, 3, 5 and 6). The levels of expression of ZSCAN4 in α-amanitin-treated or untreated embryos were analyzed using the F-test, followed by Mann-Whitney’s U test (Fig. 2). Percentage data for embryo development were subjected to an arcsine transformation. The transformed values were analyzed by Bartlett’s test for the equality of multiple variances to analyze between-group differences in variance. The measures with equal variances were analyzed using one-way analysis of variance, and the measures with unequal variances were analyzed by the Kruskal-Wallis test, followed by multiple pairwise comparisons using the Scheffé method (Table 2). P value < 0.01 or 0.05 was considered statistically significant.
Results
Expression of ZSCAN4 mRNA in bovine oocytes and embryos at various stages
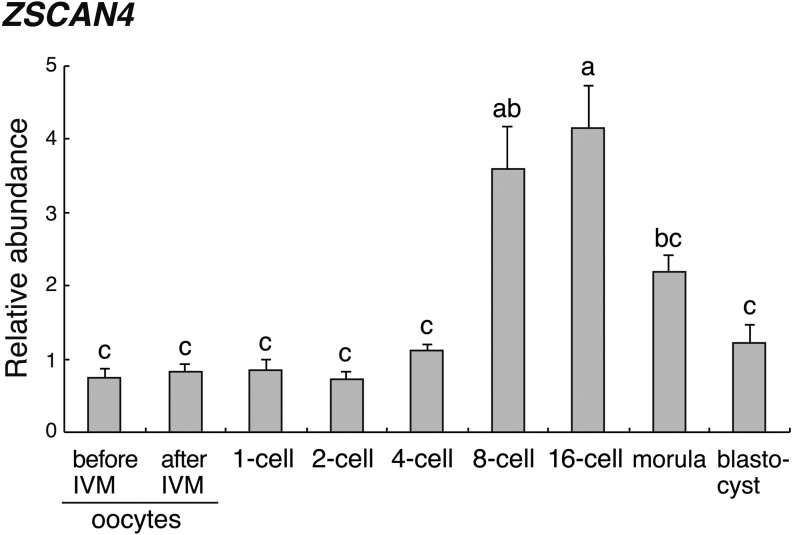
Figure 1 shows the ZSCAN4 mRNA levels in oocytes before or after IVM and in embryos at various stages. ZSCAN4 mRNA levels are low until the 4-cell stage. ZSCAN4 expression increased from the 4- to 8-cell stage, and transcript levels in the 16-cell stage embryos were significantly (P < 0.05) higher than levels in oocytes and other embryos stages, with the exception of the 8-cell stage embryos. ZSCAN4 expression was significantly (P < 0.05) decreased at the morula stage (Fig. 1).
Fig. 1.
Relative abundance (mean ± SEM) of ZSCAN4 transcripts in bovine oocytes and preimplantation embryos (n = 4). The relative abundance represents the normalized quantity compared with exogenous control (HcRed1) RNA. a, b, c Different superscript letters indicate a significant difference (P < 0.05).
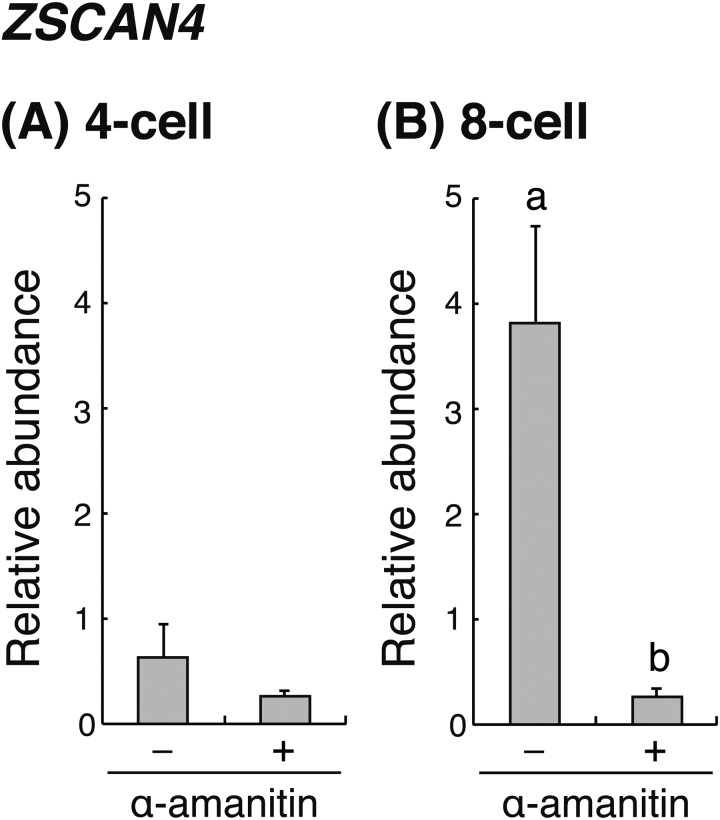
At the 4-cell stage, there was no significant difference in the relative abundance of ZSCAN4 mRNA between α-amanitin-treated and untreated embryos (Fig. 2A). However, at the 8-cell stage, the relative abundance of ZSCAN4 in the α-amanitin-treated embryos was significantly (P < 0.05) lower than that in the α-amanitin-untreated embryos (Fig. 2B). It is well known that α-amanitin is an inhibitor of RNA synthesis. This result indicated that the significant increase of ZSCAN4 transcripts at the 8-cell stage results from the de novo synthesis of zygotic ZSCAN4.
Fig. 2.
Relative abundance (mean ± SEM) of ZSCAN4 transcripts in bovine (A) 4-cell and (B) 8-cell embryos treated with or without α-amanitin (n = 4). The relative abundance represents the normalized quantity compared with exogenous control (HcRed1) RNA. a, b Different superscript letters indicate a significant difference (P < 0.05).
Effect of siRNA injection on ZSCAN4 expression in bovine embryos
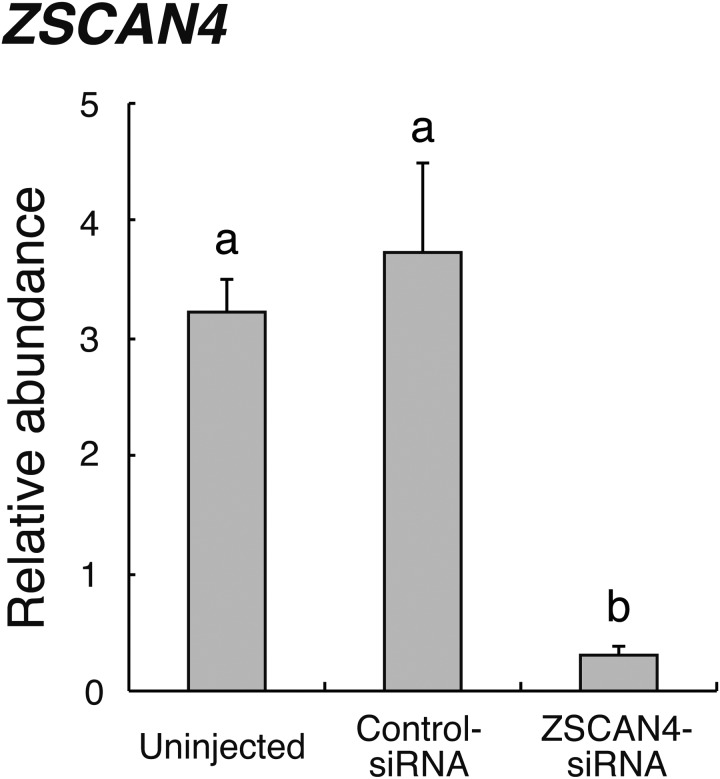
The expression levels of ZSCAN4 mRNA in 8- to 16-cell stage embryos obtained from Uninjected, Control-siRNA-injected, or ZSCAN4-siRNA-injected were evaluated (Fig. 3). ZSCAN4 gene expression was significantly (P < 0.01) lower in embryos injected with ZSCAN4-siRNA than in Control-siRNA injected or Uninjected embryos.
Fig. 3.
Relative abundance (mean ± SEM) of ZSCAN4 transcripts at the 8- to 16-cell stage in Uninjected, Control-siRNA-injected, or ZSCAN4-siRNA-injected embryos (n = 6). The relative abundance represents the normalized quantity compared with Histone H2A. a, b Different superscript letters indicate a significant difference (P < 0.01).
Effect of ZSCAN4 downregulation on the development of bovine embryos
In vitro developmental competence of ZSCAN4-siRNA-injected embryos was evaluated (Table 2). No difference in developmental rates for the 2-cell ≤ (Day 2) and 8-cell ≤ (Day 3) stages were observed between ZSCAN4-siRNA-injected and control (Uninjected or Control-siRNA-injected) embryos. However, the rate of ZSCAN4-siRNA-injected embryos that developed to 16-cell ≤ stage (Day 4; 40.5%) was significantly (P < 0.05) reduced than that of control embryos (59.7–64.4%). Remarkable developmental arrest occurred in the ZSCAN4-siRNA-injected embryos at the 32-cell ≤ (Day 4), the morula ≤ (Day 5), and the blastocyst stage (Day 7). The developmental ratios of ZSCAN4-siRNA-injected embryos (2.0%, 3.4%, and 2.7%, respectively) were significantly (P < 0.01) lower than those of Uninjected (52.9%, 40.3%, and 45.0%, respectively) and Control-siRNA-injected embryos (45.5%, 36.9%, and 46.0%, respectively).
Table 2. Effect of ZSCAN4-siRNA injection on in vitro development of bovine embryos *.
| Treatment | Numbers of embryos cultured †† |
No. (%) † of embryos develop to |
|||||
|---|---|---|---|---|---|---|---|
| Day 2 |
Day 3 |
Day 4 |
Day 5 |
Day 7 |
|||
| 2-cell ≤ | 8-cell ≤ | 16-cell ≤ | 32-cell ≤ | morula ≤ | blastocyst ≤ | ||
| Uninjected | 191 | 153 (80.1) | 148 (77.5) | 123 (64.4) a | 101 (52.9) a | 77 (40.3) a | 86 (45.0) a |
| Control-siRNA | 176 | 142 (80.7) | 129 (73.3) | 105 (59.7) a | 80 (45.5) a | 65 (36.9) a | 81 (46.0) a |
| ZSCAN4-siRNA | 148 | 122 (82.4) | 107 (72.3) | 60 (40.5) b | 3 (2.0) b | 5 (3.4) b | 4 (2.7) b |
* Experiments were replicated five times. † Percentages of the number of embryos cultured. †† Control-siRNA, ZSCAN4-siRNA; number of embryos with Dx signals. a, b Values with different superscripts within each column differ significantly (P < 0.01 or P < 0.05).
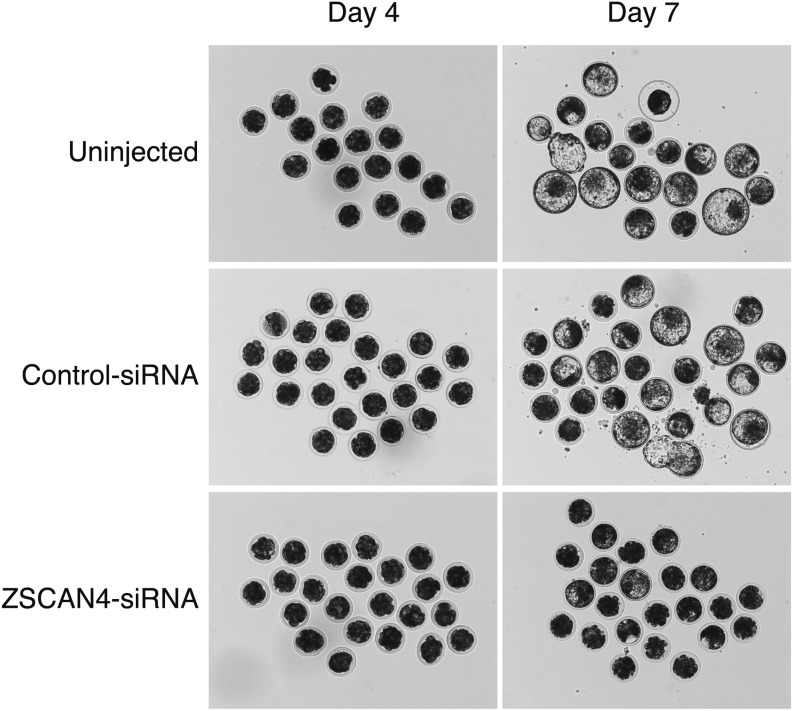
Representative photographs of embryos morphology are shown in Fig. 4. In the Uninjected and Control-siRNA groups, embryos had developed to the blastocyst stage at Day 7 and the expanded blastocysts are shown. However, the ZSCAN4-siRNA-injected embryos showed for the majority a developmental arrest at the 16-cell stage, with only a few embryos reaching the blastocyst stage (Fig. 4).
Fig. 4.
Representative photographs showing the developmental morphology in late culture periods of bovine embryos obtained from Uninjected, Control-siRNA-injected, or ZSCAN4-siRNA-injected embryos. Embryos obtained from each treatment were cultured continuously until Day 7.
Gene expressions in bovine embryos derived from ZSCAN4-siRNA injection
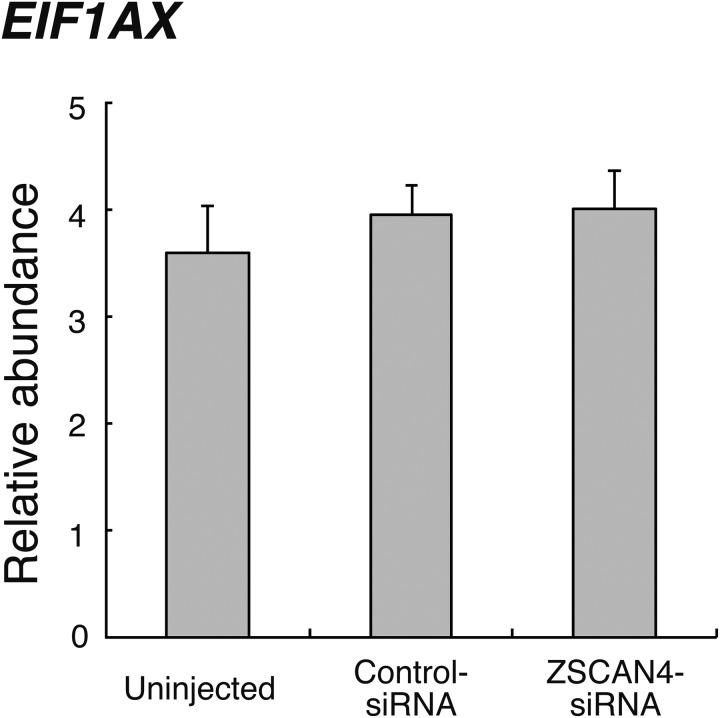
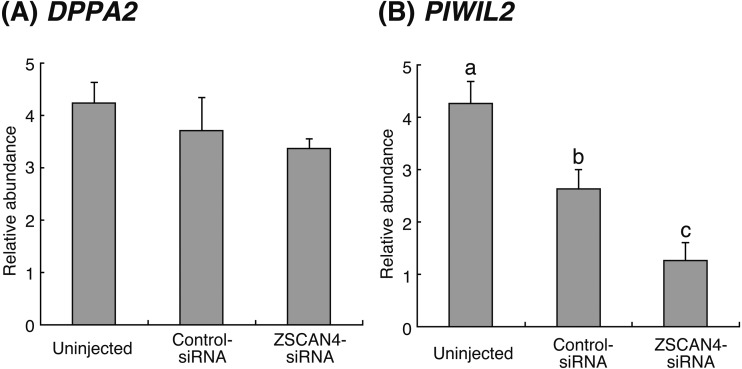
The expression levels of the ZGA marker, eukaryotic translation initiation factor 1a X-linked (EIF1AX), were not significantly different between groups, as shown in Fig. 5. To elucidate the effect of ZSCAN4 downregulation on gene transcripts related to reprogramming, we examined the mRNA levels of developmental pluripotency associated 2 (DPPA2) and piwi-like RNA-mediated gene silencing 2 (PIWIL2) in 16-cell stage embryos. As shown in Fig. 6A, the relative abundance of DPPA2 did not differ between treatment groups. On the other hand, PIWIL2 transcript level in ZSCAN4-siRNA-injected embryos was significantly (P < 0.05) lower than that in Uninjected and Control-siRNA-injected embryos (Fig. 6B).
Fig. 5.
Relative abundance (mean ± SEM) of EIF1AX transcripts at the 8- to 16-cell stage in Uninjected, Control-siRNA-injected, or ZSCAN4-siRNA-injected embryos (n = 6). The relative abundance represents the normalized quantity compared with Histone H2A.
Fig. 6.
Relative abundance (mean ± SEM) of (A) DPPA2 and (B) PIWIL2 transcripts at the 16-cell stage in Uninjected, Control-siRNA-injected, or ZSCAN4-siRNA-injected embryos (n = 6). The relative abundance represents the normalized quantity compared with Histone H2A. a, b, c Different superscript letters indicate a significant difference (P < 0.05).
Discussion
Zscan4 has been reported as a reprogramming factor in the generation of murine iPSCs, and it may play a role in the development of mouse embryos after implantation [12, 14]. In the present study, we report the ZSCAN4 expression status at different developmental stages and the necessity of ZSCAN4 transcription for the preimplantation development of bovine embryos.
Zscan4 is a DNA-binding protein that is specifically expressed in 2-cell stage embryos during mouse development [12]. The expression of Zscan4 in mouse ESCs is transient and reversible, resulting in a unique expression pattern with expression limited 1–5% of ESCs [12, 13, 23]. In human preimplantation embryos, ZSCAN4 transcripts are significantly upregulated between the 4- and 8-cell stages of embryo development, with a mean expression level peaking at the 8-cell stage [24, 25]. In our study, the level of ZSCAN4 mRNA in bovine preimplantation embryos was low in the oocyte through the 4-cell stage embryos and increased at the 8-cell stage. ZSCAN4 transcript level was kept high in 16-cell stage embryos and decreased to low levels from the morula stage. Furthermore, in the present study, we indicated that the higher levels of ZSCAN4 transcripts at 8- to 16-cell stages result from the de novo synthesis of ZSCAN4. In the present study, the ZSCAN4 expression period in bovine embryos was in concordance with the timing of ZGA, like it is the case in murine and human embryos.
In the present study, due to the lack of bovine-specific ZSCAN4 antibodies and the low conservation of ZSCAN4 proteins across species, with only 35.3–42.1% between mouse and bovine and 56.5% between human and bovine according to HomoloGene [26], we could not assess the effect of ZSCAN4-siRNA injection on ZSCAN4 protein levels. However, we clearly demonstrated the downregulation efficiency of ZSCAN4 mRNA by siRNA injection at the 8- and 16-cell stage. In mouse embryos, the reduction of Zscan4 transcripts delayed the progression from the 2- to 4-cell stage and produced blastocysts that failed implantation or did not proliferate in blastocyst outgrowth cultures [12]. In the present study, downregulation of ZSCAN4 expression in bovine embryos had no effects on development to the 8-cell stage. However, in the ZSCAN4 downregulated embryos, developmental competence at the 16-cell stage was reduced, and embryo development to the 32-cell stage was clearly inhibited. The effects of ZSCAN4 downregulation also appeared in the development to the morula and blastocyst formation, and developmental competences to both stages were very low.
As described above, ZSCAN4 shows high expression during ZGA period in mouse [12], human [24, 25] and bovine embryos (present study). In our study, embryo development from the 16- to 32-cell stage was inhibited by ZSCAN4 downregulation; i.e., developmental arrest was observed just after ZGA. Therefore, to test the hypothesis that bovine embryos obtained from ZSCAN4-siRNA injection are defective in terms of ZGA, we evaluated the expression level of ZGA related gene. EIF1AX is a transiently expressed endogenous marker of ZGA found in mouse and bovine embryos [27, 28]. Furthermore, Eif1a is found to be co-upregulated with Zscan4 in mouse 2-cell embryos [29,30,31] and is one of the most upregulated genes in mouse ESCs, which are enriched for Zscan4 [17]. Moreover, many preimplantation-specific genes, including Eif1a, were activated during the early phase of mouse iPSCs formation generated with Zscan4 [14]. Therefore, we used the EIF1AX transcript level as a ZGA marker in ZSCAN4 downregulated embryos. However, there was no significant difference in EIF1AX transcripts between ZSCAN4 downregulated embryos and control embryos. These findings suggest that ZGA in ZSCAN4 downregulated embryos occurs normally. Moreover, ZGA is not related to the developmental arrest of bovine embryos obtained from ZSCAN4-siRNA injection. Alternatively, EIF1AX expression might be not regulated by ZSCAN4 in bovine preimplantation embryos.
Zscan4 has also been shown to enhance the efficiency of generating mouse iPSCs and their quality [14, 15]. Thus, there was the possibility that ZSCAN4 has a role in regulating gene expression involved in reprogramming the somatic cell nuclei [14]. During early development, epigenetic reprogramming occurs in order to remove gamete-specific epigenetic patterns [32, 33]. Therefore, to examine the ability of ZSCAN4 to act as a regulator of gene transcription in bovine embryos, we focused on the genes which were highly expressed in iPSCs generated using Zscan4. Compared with iPSCs generated using conventional methods, Dppa2 and Piwil2 mRNA transcripts showed higher levels of expression in the mouse iPSCs generated using Zscan4 [14]. Dppa2 is involved in the proliferation of mouse ESCs [34, 35] and has a role in maintaining their pluripotency and self-renewal in vitro [36]. Piwil2, a member of the Piwi gene family, promotes proliferation and inhibits apoptosis in tumor cells [37]. Notably, when Piwil2 was knocked down, the proliferation and invasion of mouse cervical cancer cell lines were significantly inhibited [38]. In the present study, although the DPPA2 transcripts were not different among the experimental groups, PIWIL2 transcript levels in ZSCAN4 downregulated embryos were significantly lower than that in control embryos. Piwi proteins and their associated small RNAs, Piwi-interacting RNA (piRNAs), repress transcription of transposable elements in metazoan germ cells [39]. It is also reported that knock out of Piwil2 led to significant overexpression of long terminal repeat retrotransposons in mouse male germline [40]. A large number of retrotransposons are expressed when the zygotic genome is first transcribed, including the endogenous retroviruses [41]. It is reported that both piRNA-like RNAs [42, 43] and retrotransposons [44] were expressed in bovine preimplantation embryos. Furthermore, recent in silico analysis showed that piRNA-like RNAs were markedly reduced in bovine 8-cell stage embryos as compared with oocytes, but appeared to partially rebound at the blastocyst stage [43]. From these observations, PIWI-piRNA associated regulation of retrotransposons expression may prove critical for successful ZGA and embryo development in mammals [43]. Additionally, piRNAs have been shown to be essential for the targeted elimination of mRNA transcripts during pachytene spermatogenesis [45]. It is worth noting that Zscan4 protein was highly detected in late pachytene or diplotene spermatogenesis [46]. We examined the expression levels of retrotransposons, BERV-K1 and BERV-K2 [44] in ZSCAN4 downregulated bovine embryos (data not shown). In ZSCAN4 downregulated embryos, aberrant transcript levels of both retrotransposons were not observed. However, it is well known that various retrotransposons express in mammalian preimplantation embryos [44]. It is possible that the developmental arrest after ZSCAN4-siRNA injection is due to failed gene transcript regulation including other retrotransposons in bovine embryos caused by PIWIL2 downregulation. However, the interaction between ZSCAN4 and PIWI-piRNA pathway in bovine embryos remain to be clarified. Further studies are necessary to clarify the relationship between ZSCAN4 and the epigenetic status including retrotransposons expression in bovine embryos.
In conclusion, we found that ZSCAN4 is essential for early development of bovine embryos. The present study is the first to demonstrate the critical importance of ZSCAN4 in bovine embryos and provide new insights for understanding the mechanisms of gene expression after fertilization.
Acknowledgments
This work was supported by JSPS KAKENHI Grant Number 16J07000 and 26292162.
References
- 1.Sawai K, Kageyama S, Moriyasu S, Hirayama H, Minamihashi A, Onoe S. Analysis of mRNA transcripts for insulin-like growth factor receptors and binding proteins in bovine embryos derived from somatic cell nuclear transfer. Cloning Stem Cells 2005; 7: 189–198. [DOI] [PubMed] [Google Scholar]
- 2.Wrenzycki C, Herrmann D, Lucas-Hahn A, Korsawe K, Lemme E, Niemann H. Messenger RNA expression patterns in bovine embryos derived from in vitro procedures and their implications for development. Reprod Fertil Dev 2005; 17: 23–35. [DOI] [PubMed] [Google Scholar]
- 3.Sawai K, Takahashi M, Fujii T, Moriyasu S, Hirayama H, Minamihashi A, Hashizume T, Onoe S. DNA methylation status of bovine blastocyst embryos obtained from various procedures. J Reprod Dev 2011; 57: 236–241. [DOI] [PubMed] [Google Scholar]
- 4.Schultz RM, Davis W, Jr, Stein P, Svoboda P. Reprogramming of gene expression during preimplantation development. J Exp Zool 1999; 285: 276–282. [DOI] [PubMed] [Google Scholar]
- 5.Kidder GM, McLachlin JR. Timing of transcription and protein synthesis underlying morphogenesis in preimplantation mouse embryos. Dev Biol 1985; 112: 265–275. [DOI] [PubMed] [Google Scholar]
- 6.Braude P, Bolton V, Moore S. Human gene expression first occurs between the four- and eight-cell stages of preimplantation development. Nature 1988; 332: 459–461. [DOI] [PubMed] [Google Scholar]
- 7.Hyttel P, Laurincik J, Viuff D, Fair T, Zakhartchenko V, Rosenkranz C, Avery B, Rath D, Niemann H, Thomsen PD, Schellander K, Callesen H, Wolf E, Ochs RL, Greve T. Activation of ribosomal RNA genes in preimplantation cattle and swine embryos. Anim Reprod Sci 2000; 60–61: 49–60. [DOI] [PubMed] [Google Scholar]
- 8.Camous S, Kopecný V, Fléchon JE. Autoradiographic detection of the earliest stage of [3H]-uridine incorporation into the cow embryo. Biol Cell 1986; 58: 195–200. [DOI] [PubMed] [Google Scholar]
- 9.Frei RE, Schultz GA, Church RB. Qualitative and quantitative changes in protein synthesis occur at the 8-16-cell stage of embryogenesis in the cow. J Reprod Fertil 1989; 86: 637–641. [DOI] [PubMed] [Google Scholar]
- 10.Svoboda P. Mammalian zygotic genome activation. Semin Cell Dev Biol 2018; 84: 118–126. [DOI] [PubMed] [Google Scholar]
- 11.Mann MR, Chung YG, Nolen LD, Verona RI, Latham KE, Bartolomei MS. Disruption of imprinted gene methylation and expression in cloned preimplantation stage mouse embryos. Biol Reprod 2003; 69: 902–914. [DOI] [PubMed] [Google Scholar]
- 12.Falco G, Lee SL, Stanghellini I, Bassey UC, Hamatani T, Ko MS. Zscan4: a novel gene expressed exclusively in late 2-cell embryos and embryonic stem cells. Dev Biol 2007; 307: 539–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zalzman M, Falco G, Sharova LV, Nishiyama A, Thomas M, Lee SL, Stagg CA, Hoang HG, Yang HT, Indig FE, Wersto RP, Ko MS. Zscan4 regulates telomere elongation and genomic stability in ES cells. Nature 2010; 464: 858–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirata T, Amano T, Nakatake Y, Amano M, Piao Y, Hoang HG, Ko MS. Zscan4 transiently reactivates early embryonic genes during the generation of induced pluripotent stem cells. Sci Rep 2012; 2: 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang J, Lv W, Ye X, Wang L, Zhang M, Yang H, Okuka M, Zhou C, Zhang X, Liu L, Li J. Zscan4 promotes genomic stability during reprogramming and dramatically improves the quality of iPS cells as demonstrated by tetraploid complementation. Cell Res 2013; 23: 92–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amano T, Hirata T, Falco G, Monti M, Sharova LV, Amano M, Sheer S, Hoang HG, Piao Y, Stagg CA, Yamamizu K, Akiyama T, Ko MS. Zscan4 restores the developmental potency of embryonic stem cells. Nat Commun 2013; 4: 1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Akiyama T, Xin L, Oda M, Sharov AA, Amano M, Piao Y, Cadet JS, Dudekula DB, Qian Y, Wang W, Ko SB, Ko MS. Transient bursts of Zscan4 expression are accompanied by the rapid derepression of heterochromatin in mouse embryonic stem cells. DNA Res 2015; 22: 307–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hung SS, Wong RC, Sharov AA, Nakatake Y, Yu H, Ko MS. Repression of global protein synthesis by Eif1a-like genes that are expressed specifically in the two-cell embryos and the transient Zscan4-positive state of embryonic stem cells. DNA Res 2013; 20: 391–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abe H, Yamashita S, Itoh T, Satoh T, Hoshi H. Ultrastructure of bovine embryos developed from in vitro-matured and -fertilized oocytes: comparative morphological evaluation of embryos cultured either in serum-free medium or in serum-supplemented medium. Mol Reprod Dev 1999; 53: 325–335. [DOI] [PubMed] [Google Scholar]
- 20.Bavister BD, Leibfried ML, Lieberman G. Development of preimplantation embryos of the golden hamster in a defined culture medium. Biol Reprod 1983; 28: 235–247. [DOI] [PubMed] [Google Scholar]
- 21.Bettegowda A, Patel OV, Ireland JJ, Smith GW. Quantitative analysis of messenger RNA abundance for ribosomal protein L-15, cyclophilin-A, phosphoglycerokinase, beta-glucuronidase, glyceraldehyde 3-phosphate dehydrogenase, beta-actin, and histone H2A during bovine oocyte maturation and early embryogenesis in vitro. Mol Reprod Dev 2006; 73: 267–278. [DOI] [PubMed] [Google Scholar]
- 22.Ross PJ, Wang K, Kocabas A, Cibelli JB. Housekeeping gene transcript abundance in bovine fertilized and cloned embryos. Cell Reprogram 2010; 12: 709–717. [DOI] [PubMed] [Google Scholar]
- 23.Carter MG, Stagg CA, Falco G, Yoshikawa T, Bassey UC, Aiba K, Sharova LV, Shaik N, Ko MS. An in situ hybridization-based screen for heterogeneously expressed genes in mouse ES cells. Gene Expr Patterns 2008; 8: 181–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vassena R, Boué S, González-Roca E, Aran B, Auer H, Veiga A, Izpisua Belmonte JC. Waves of early transcriptional activation and pluripotency program initiation during human preimplantation development. Development 2011; 138: 3699–3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shaw L, Sneddon SF, Brison DR, Kimber SJ. Comparison of gene expression in fresh and frozen-thawed human preimplantation embryos. Reproduction 2012; 144: 569–582. [DOI] [PubMed] [Google Scholar]
- 26.Coordinators NR, NCBI Resource Coordinators. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 2016; 44 (D1): D7–D19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Sousa PA, Watson AJ, Schultz RM. Transient expression of a translation initiation factor is conservatively associated with embryonic gene activation in murine and bovine embryos. Biol Reprod 1998; 59: 969–977. [DOI] [PubMed] [Google Scholar]
- 28.Davis W, Jr, De Sousa PA, Schultz RM. Transient expression of translation initiation factor eIF-4C during the 2-cell stage of the preimplantation mouse embryo: identification by mRNA differential display and the role of DNA replication in zygotic gene activation. Dev Biol 1996; 174: 190–201. [DOI] [PubMed] [Google Scholar]
- 29.Hamatani T, Carter MG, Sharov AA, Ko MS. Dynamics of global gene expression changes during mouse preimplantation development. Dev Cell 2004; 6: 117–131. [DOI] [PubMed] [Google Scholar]
- 30.Wang QT, Piotrowska K, Ciemerych MA, Milenkovic L, Scott MP, Davis RW, Zernicka-Goetz M. A genome-wide study of gene activity reveals developmental signaling pathways in the preimplantation mouse embryo. Dev Cell 2004; 6: 133–144. [DOI] [PubMed] [Google Scholar]
- 31.Zeng F, Baldwin DA, Schultz RM. Transcript profiling during preimplantation mouse development. Dev Biol 2004; 272: 483–496. [DOI] [PubMed] [Google Scholar]
- 32.Monk M, Boubelik M, Lehnert S. Temporal and regional changes in DNA methylation in the embryonic, extraembryonic and germ cell lineages during mouse embryo development. Development 1987; 99: 371–382. [DOI] [PubMed] [Google Scholar]
- 33.Oswald J, Engemann S, Lane N, Mayer W, Olek A, Fundele R, Dean W, Reik W, Walter J. Active demethylation of the paternal genome in the mouse zygote. Curr Biol 2000; 10: 475–478. [DOI] [PubMed] [Google Scholar]
- 34.Nakamura T, Nakagawa M, Ichisaka T, Shiota A, Yamanaka S. Essential roles of ECAT15-2/Dppa2 in functional lung development. Mol Cell Biol 2011; 31: 4366–4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Du J, Chen T, Zou X, Xiong B, Lu G. Dppa2 knockdown-induced differentiation and repressed proliferation of mouse embryonic stem cells. J Biochem 2010; 147: 265–271. [DOI] [PubMed] [Google Scholar]
- 36.Watabe T. Roles of Dppa2 in the regulation of the present status and future of pluripotent stem cells. J Biochem 2012; 152: 1–3. [DOI] [PubMed] [Google Scholar]
- 37.Shahali M, Kabir-Salmani M, Nayernia K, Soleimanpour-Lichaei HR, Vasei M, Mowla SJ, Ranaie E, Shakibaie M, Modaresi MH. A novel in vitro model for cancer stem cell culture using ectopically expressed piwil2 stable cell line. Cell J 2013; 15: 250–257. [PMC free article] [PubMed] [Google Scholar]
- 38.Feng D, Yan K, Zhou Y, Liang H, Liang J, Zhao W, Dong Z, Ling B. Piwil2 is reactivated by HPV oncoproteins and initiates cell reprogramming via epigenetic regulation during cervical cancer tumorigenesis. Oncotarget 2016; 7: 64575–64588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siomi MC, Sato K, Pezic D, Aravin AA. PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol 2011; 12: 246–258. [DOI] [PubMed] [Google Scholar]
- 40.Manakov SA, Pezic D, Marinov GK, Pastor WA, Sachidanandam R, Aravin AA. MIWI2 and MILI have differential effects on piRNA biogenesis and DNA methylation. Cell Reports 2015; 12: 1234–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peaston AE, Evsikov AV, Graber JH, de Vries WN, Holbrook AE, Solter D, Knowles BB. Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev Cell 2004; 7: 597–606. [DOI] [PubMed] [Google Scholar]
- 42.Russell S, Patel M, Gilchrist G, Stalker L, Gillis D, Rosenkranz D, LaMarre J. Bovine piRNA-like RNAs are associated with both transposable elements and mRNAs. Reproduction 2017; 153: 305–318. [DOI] [PubMed] [Google Scholar]
- 43.Cuthbert JM, Russell SJ, White KL, Benninghoff AD. The maternal-to-zygotic transition in bovine in vitro-fertilized embryos is associated with marked changes in small non-coding RNAs. Biol Reprod 2019; 100: 331–350. [DOI] [PubMed] [Google Scholar]
- 44.Khazaee E, Farzaneh N, Mirshokraei P, Tabatabaeizadeh SE, Dehghani H. Expression of endogenous retroviruses in pre-implantation stages of bovine embryo. Reprod Domest Anim 2018; 53: 1405–1414. [DOI] [PubMed] [Google Scholar]
- 45.Gou LT, Dai P, Yang JH, Xue Y, Hu YP, Zhou Y, Kang JY, Wang X, Li H, Hua MM, Zhao S, Hu SD, Wu LG, Shi HJ, Li Y, Fu XD, Qu LH, Wang ED, Liu MF. Pachytene piRNAs instruct massive mRNA elimination during late spermiogenesis. Cell Res 2014; 24: 680–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ishiguro KI, Monti M, Akiyama T, Kimura H, Chikazawa-Nohtomi N, Sakota M, Sato S, Redi CA, Ko SB, Ko MS. Zscan4 is expressed specifically during late meiotic prophase in both spermatogenesis and oogenesis. In Vitro Cell Dev Biol Anim 2017; 53: 167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]