Abstract
Age-associated methylation changes in genomic DNA have been recently reported in spermatozoa, and these changes can contribute to decline in fertility. In a previous study, we analyzed the genome-wide DNA methylation profiles of bull spermatozoa using a human DNA methylation microarray and identified one CpG site (CpG-1) that potentially reflects age-related methylation changes. In the present study, cryopreserved semen samples from a Japanese Black bull were collected at five different ages, which were referred to as JD1-5: 14, 19, 28, 54, and 162 months, respectively, and were used for genome-wide DNA methylation analysis and in vitro fertilization (IVF). Distinct age-related changes in methylation profiles were observed, and 77 CpG sites were found to be differently methylated between young and adult samples (JD1-2 vs. JD4-5). Using combined bisulfite restriction analysis (COBRA), nine CpG sites (including CpG-1) were confirmed to exhibit significant differences in their age-dependent methylation levels. Eight CpG sites showed an age-dependent increase in their methylation levels, whereas only one site showed age-dependent hypomethylation; in particular, these changes in methylation levels occurred rapidly at a young age. COBRA revealed low methylation levels in some CpG regions in the majority of the IVF blastocyst-stage embryos derived from spermatozoa at JD2-5. Interestingly, bulls with different ages did not show differences in their methylation levels. In conclusion, our findings indicated that methylation levels at nine CpG sites in spermatozoa changed with increasing age and that some CpG regions were demethylated after fertilization. Further studies are required to determine whether age-dependent different methylation levels in bull spermatozoa can affect fertility.
Keywords: Age-related changes, Bull spermatozoa, Combined bisulfite restriction analysis (COBRA), DNA methylation, In vitro fertilization (IVF) embryos
Impaired sperm nuclear DNA integrity and abnormal DNA methylation levels, which are considered to be caused by environmental stresses, may be important factors that lead to low rates of conception on the paternal side [1,2,3]. The DNA methylation patterns in male germ cells do not necessarily reflect the gene expression patterns in a specific cell type. However, these methylation patterns are potentially involved in the germ cell-specific chromatin organization. DNA methylation in germ cells is a unique process that is necessary for proper spermatogenesis and sperm production [4, 5]. Extensive modifications to sperm chromatin are the result of removal of histones during spermatogenesis and their replacement with protamines, chemical modifications observed in the retained histones, and methylation of the sperm DNA bound to histones [4, 5]. DNA methyltransferase (DNMT) proteins have been demonstrated to play essential roles in sperm DNA methylation and in male fertility in general [6].
Many studies have suggested that various environmental and physiological factors, including old age, can alter DNA methylation patterns in spermatozoa [7, 8]. Jenkins et al. reported an increase in both cytosine methylation and hydroxymethylation levels, as well as changes in the global methylation status, with increasing paternal age [9, 10]. Age-related global DNA hypermethylation has been observed in various species, including mice and humans; however, some genes become hypomethylated with age [7, 11, 12]. Following fertilization, active DNA demethylation occurs throughout the paternal genome prior to the first cell division, effectively erasing many methylation marks generated during spermatogenesis [4, 13,14,15]. Given that sperm epigenetic architecture is likely to contribute to the early embryonic development [3, 15, 16], DNA methylation changes in spermatozoa have been proposed to be inherited by subsequent generations [9].
Previously, we conducted a pilot study using Human Methylation array to evaluate genome-wide methylation profiles in 37,000 CpG sites using DNA samples extracted from bull spermatozoa or tissues [17, 18]. In the case of non-model organisms, such as pigs and cattle, equivalent tools for studying DNA methylation at a genome-scale are currently unavailable for commercial use. Methylation profiles of individual and age-related alterations in bull spermatozoa can be analyzed using human microarray, and methylation changes in some CpG sites can be easily visualized using combined bisulfite restriction analysis (COBRA) [17, 18]. Specifically, one CpG site was hypothesized to reflect an age-related increase in methylation levels, which was then confirmed by COBRA and bisulfite sequencing analyses. COBRA is a useful detection tool for analyzing methylation levels at targeted differentially methylated regions (DMRs) when there is a large number of samples or when the samples have small volumes, such as those for early embryos. In the present study, we aimed to verify the age-related DNA methylation changes at CpG sites in bull spermatozoa and the methylation status in in vitro fertilization (IVF) embryos using COBRA.
Materials and Methods
All chemicals used in the study were purchased from Sigma-Aldrich (St. Louis, MO, USA) unless otherwise indicated.
Semen sampling
Cryopreserved semen samples were collected from one Japanese Black bull at different ages (JD1: 14 months, JD2: 19 months, JD3: 28 months, JD4: 54 months, and JD5: 162 months) [18] and then used for genome-wide DNA methylation analysis and IVF assay. Cryopreserved semen samples were obtained from bulls (Japanese Black, n = 34; Holstein, n = 1) from six farms located in different areas of Japan. The samples were collected at different ages ranging from 10 months to 162 months and were used for subsequent COBRA.
DNA methylation analysis using Infinium EPIC BeadChip array
Thawed semen samples were washed twice in Dulbecco’s phosphate-buffered saline without calcium chloride or magnesium chloride (DPBS). Genomic DNA was extracted from the semen samples using DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions with some modifications as previously described [18].
The Infinium MethylationEPIC BeadChip (Illumina, San Diego, CA, USA) array was used for genome-wide bovine DNA methylation analysis. The EPIC array can profile more than 850,000 CpG sites of the human genome, including more than 90% of the CpGs that are also covered by the HM450 array (previous version) and more than 300,000 CpGs located in enhancer regions, representing a significant improvement over the HM450 array [19]. The processing of DNA for the methylation arrays (which involves bisulfite conversion, whole-genome amplification, labeling, hybridization to an Infinium MethylationEPIC BeadChip, scanning on a BeadArray scanner, and raw data processing) was performed according to the Illumina protocols at Takara Bio (Shiga, Japan).
After methylation array analysis, informative CpG sites were selected as previously described [17]. Briefly, the fluorescence signal intensities of the methylated and unmethylated alleles were obtained [20]. Next, the total signal intensity (sum of these fluorescence signal intensities) and the methylation level at each CpG locus were calculated as the methylation beta-value, expressed as [signal intensity of the methylated allele / (total signal intensity + 100)]. Similarly, the detection P-values were calculated as a proportion of 636 negative control probes with signal intensities higher than those measured for each CpG site. Then, the most reliable CpG sites were checked for quality and those with detection P-values = 0 and total signal intensities > 1,000 were selected. The different methylation levels at the CpG sites were compared among different ages (< 20 months old vs. > 54 months old; JD1 and JD2 vs. JD4 and JD5).
Detection of differentially methylated regions in spermatozoa by COBRA
The BLAST program (http://blast.ncbi.nlm.nih.gov/Blast.cgi) was used to identify the sequences with high similarities for each quality-controlled CpG site on bovine genome sequences (UMD3.1.1), using human probes, as previously reported [17, 18]. Then, the CpG sites located at the restriction enzyme sites for AciI, BstUI, HpyCH4IV, and TaqI were selected within the bovine genome sequences identified by the BLAST search. Bisulfite primers were designed using MethPrimer (http://www.urogene.org/methprimer/index.html) to amplify the regions flanking the sequences of interest.
Genomic DNA was bisulfite-converted using the MethylEasy Xceed Rapid DNA Bisulphite Modification Kit (Human Genetic Signatures, New South Wales, Australia) according to the manufacturer’s instructions. The unmethylated cytosines were converted to uracil residues, whereas methylated cytosines remained unchanged upon the bisulfite treatment. The bisulfite-converted DNA was PCR-amplified using a TaKaRa EpiTaqTM HS kit (TaKaRa Bio) using the following conditions: 94°C for 2 min, followed 35 cycles of 94°C for 30 sec, 50–58°C (depending on the primer sets, see Table 1) for 30 sec, and 72°C for 30 sec. PCR products were digested with the restriction enzymes shown in Table 1. The digested fragments were analyzed by 3% agarose gel electrophoresis. The ratio of band intensities of the digested and undigested fractions indicates the DNA methylation levels at the restriction sites. The band intensities of all samples were measured by densitometry using ImageJ 1.50i (http://imagej.nih.gov/ij; National Institutes of Health, Bethesda, MD) to examine differences in methylation. The DNA band intensities were normalized by the length of the DNA fragments. Methylation levels were calculated based on the ratio between the intensities of the digested DNA band and the combined intensities of the digested and undigested DNA bands. When the restriction enzyme digested two or more sites, the band intensities were weighted with 1 for samples methylated at all sites or 0.5 for samples with both methylated and non-methylated sites. Differentially methylated regions (DMRs), including CpGs, were selected for subsequent analysis.
Table 1. Primer sequences for COBRA.
| Forward and reverse primer sequences | Product length (bp) | AT (ºC) | Restriction enzyme | Restriction fragments length* (bp) | Bovine features of the sequences including the target CpG | Bovine chromosome No. | Accession No., location | Target ID of array | |
|---|---|---|---|---|---|---|---|---|---|
| CpG-A1 | 5'TGGTTTAGTTTGTGTAATTTTTTTT3' | 248 | 54 | BstUI | 178, 37, 33 | - | 2 | AC_000159.1, 70365795 | cg03584544 |
| 5'AAACCTCCTACCATTTACATTAAAT3' | |||||||||
| CpG-A2 | 5'TAGGGTTTGTGTTTTTGTGTTTATT3' | 221 | 56 | HpyCH4IV | 126, 38, 57 | semaphorin-3B isoform | 22 | AC_000179.1, 50658598 | cg22037648 |
| 5'CCCTTTACCTATCAAAACCTCAC3' | |||||||||
| CpG-A3 | 5'GGGTAAATTGGTTATTTGAGGAAAT3' | 233 | 57 | HpyCH4IV | 66, 167 | guanine nucleotide-binding protein subunit beta-2-like 1 | 7 | AC_000164.1, 41765135 | cg23160731 |
| 5'AATAATTTCCCTACCCCTAAATACC3' | |||||||||
| CpG-A4 | 5'TGGGTATTTAATTTATTATGTTTAGT3' | 219 | 50 | HpyCH4IV | 36, 183 | - | 4 | AC_000161.1, 53059961 | cg24956561 |
| 5'AAAAAATATAATTTTCTTAAAAAAAAC3' | |||||||||
| CpG-A5 | 5'GGAGTGGGGAGATGTGTATAGTTTA3' | 245 | 58 | AciI | 169, 76 | paired mesoderm homeobox protein 2B, transmembrane protein 33 isoform 1 | 6 | AC_000163.1, 62298715 | cg01416712 |
| 5'AACTCAATTCCCCAATACCAAC3' | |||||||||
| CpG-A6 | 5'TAGGTTGGTTGGTAGGGGTTATAG3' | 248 | 58 | AciI | 33, 39, 176 | decapping and exoribonuclease protein | 23 | AC_000180.1, 27207858 | cg05626376 |
| 5'ATCAAAACAACTCTCACCTACACAC3' | |||||||||
| CpG-A7 | 5'GGGATTTGTTGTTTTATTGATGAGT3' | 221 | 57 | HpyCH4IV | 51, 159 | C10orf11 homolog | 28 | AC_000185.1, 31517505 | cg15586478 |
| 5'AAAAAAAACTCAAACCAAACCTATCT3' | |||||||||
| CpG-A8 | 5'TGTTTAAAATTTAATTTTAAAAGAAAAGAG3' | 228 | 54 | HpyCH4IV | 61,167 | COUP transcription factor 2 isoform | 21 | AC_000178.1, 10805413 | cg13848359 |
| 5'ACATAAACCCAATTCAAAAAAAA3' | |||||||||
AT, Annealing temperature. * Restriction fragments length when all CpGs located in restriction endonuclease cleavage sites are methylated.
Analysis of methylated levels in cryopreserved semen by COBRA
DNA from cryopreserved semen samples (n = 66, collected from 35 bulls with ages ranging from 10 to 162 months old) were extracted and bisulfite-converted as previously described. For the majority of the samples, methylation levels of the selected DMRs were estimated by COBRA. Correlations between age and methylation levels of each CpG were analyzed by Spearman’s rank correlation using GraphPad Prism for Windows (version 7).
IVF
IVF tests were performed as previously described [18]. Briefly, cumulus-oocyte complexes (COCs) were collected from slaughterhouse-derived ovaries after storing overnight in physiological saline at 15°C. After 24 h of incubation to allow maturation, the COCs were transferred to BO medium containing 20 mg/ml crystallized BSA and 10 IU/ml Novo Heparin (Mochida Pharmaceutical, Tokyo, Japan). Each cryopreserved semen sample (JD1-5) was thawed and washed twice in 10 ml BO medium supplemented with 10 mM caffeine sodium benzonate. Sperm suspensions were transferred to a droplet of fertilization medium (final concentration: 5 × 106 spermatozoa/ml) and co-incubated for 5 h at 38.5°C under 5% CO2 in air with maximum humidity. After IVF, the zygotes were cultured in IVD 101 medium (Research Institute for Functional Peptides, Yamagata, Japan) for 8 days; IVF data were obtained by counting the number of embryos at the blastocyst stage per number of embryos cultured after IVF. IVF experiments were repeated twice (JD1) or thrice (JD2-5) for each ejaculate. IVF results among JD1-5 samples were analyzed by Chi-squared test. P-value < 0.05 was considered statistically significant.
Analysis of methylation levels in IVF embryos by COBRA
Bisulfite-converted DNA was directly extracted from each embryo using Blood and Tissue DNA Methylation Kit (Enzo Life Science, Lausen, Switzerland) according to the manufacturer’s instructions. Blastocyst-stage embryos were washed six times in DPBS containing 1% PVP (polyvinylpyrrolidone, average mol. wt. 40 kDa). Each embryo was placed in 1.5-ml siliconized microtubes containing 20 µl of digestion buffer. Bisulfite-converted DNA was eluted in 20 µl of elution buffer. The COBRA method was conducted as described above, and PCR amplification was performed for 45 cycles. Oocytes before IVF (n = 70) were pooled and used for COBRA as described above.
Results
DNA methylation status revealed by Infinium MethylationEPIC BeadChip array
The DNA methylation status of the bovine spermatozoa was analyzed using Infinium MethylationEPIC BeadChips. The mean number of CpG sites in the examined samples with a detection P-value of 0 was 177,186 (20.4%; 167,284 to 188,415 sites). A total of 133,388 CpG sites were associated with a detection P-value = 0 in all examined samples (15.4%), while 105,940 CpG sites fulfilled the second criterion and showed a mean total signal intensity >1,000 (12.2%).
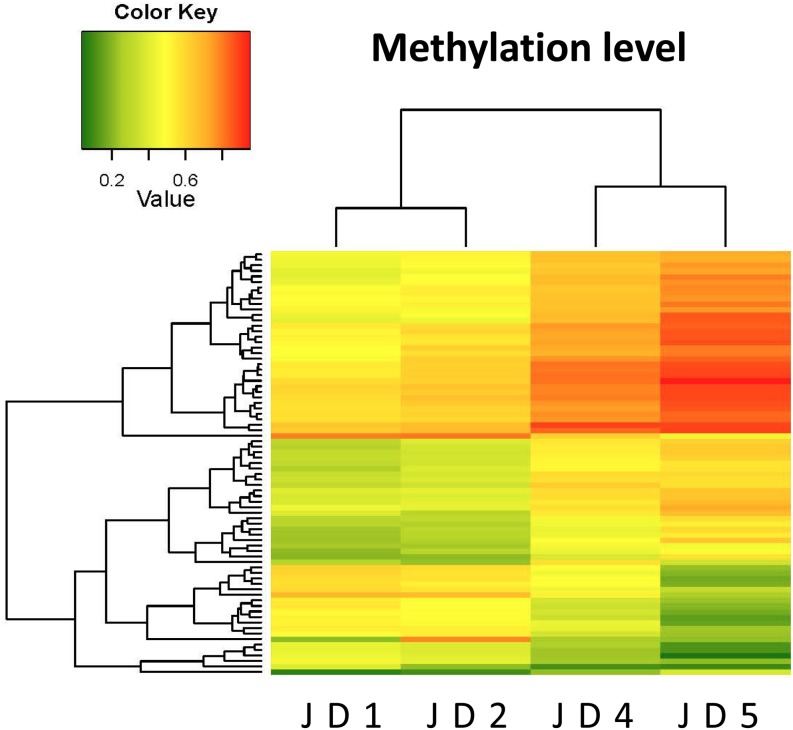
For the 105,940 probes used to analyze DNA methylation in bovine samples using the MethylationEPIC BeadChip array, the standard deviation of the beta-values of each probe among the semen samples was calculated. Using the cutoff value of 0.2 for differences in beta-values between groups (JD1 and JD2 vs. JD4 and JD5), 77 CpG sites showed significant differences in beta-values across the groups analyzed (Fig. 1).
Fig. 1.
Heatmap showing the different methylation levels at the detected CpG sites between the samples JD1, 2 vs. JD4, 5 that are mapped on the human chromosomes (n = 77 CpGs). The dendrograms above and to the left of the heatmap show hierarchical clustering results among animal individuals (JD1, JD2, JD4, JD5) and among the CpG sites based on the methylation data, respectively. Scale on the upper side of the heatmap indicates the methylation status, ranging from the lowest (green, 0.0) to the highest (red, 1.0) methylation levels.
Age-related DMRs detected by COBRA
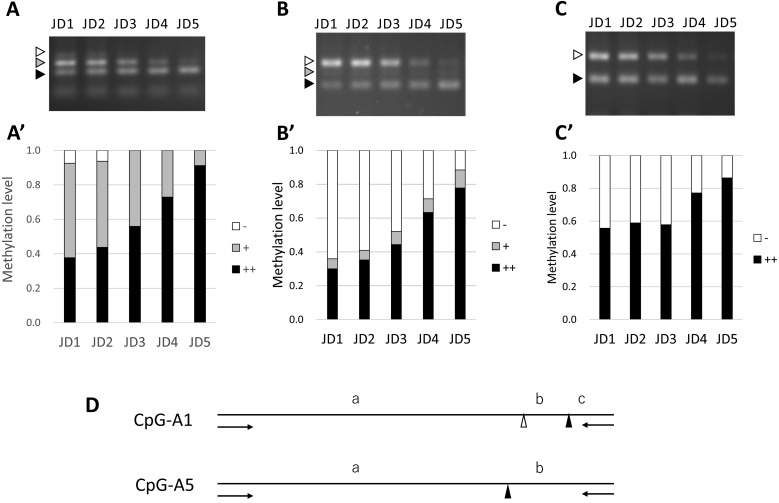
The comparison between the semen samples obtained from bulls at different ages revealed 77 differentially methylated CpG sites; furthermore, 55 matching bovine DNA sequences (71.4%) were selected after BLAST analysis. Thirteen CpG sites, including restriction enzyme sites, were analyzed and primer sets were designed for 12 of these regions. Changes in DNA methylation levels were confirmed by COBRA, and nine CpGs (CpG-1, CpG-A1-A8) were found to reflect age-related changes in methylation levels (Table 2). One of these CpGs (CpG-1) was identified in our previous study [18]. Figure 2 shows the agarose gel electrophoresis results for three of the DMRs (CpG-1, CpG-A1, and CpG-A5) analyzed by COBRA. The methylation level of the CpG-1 region (Fig. 2A) was calculated using the three DNA bands visualized at 181 bp (0% methylation, indicated by a white arrow head in Fig. 2A), 140–151 bp (indicated by a gray arrow head in Fig. 2A), and 101–112 bp (indicated by a black arrow head in Fig. 2A) reported in our previous study [18]. In the CpG-A1 region, two methylated CpG sites, including CpG-A1, were digested using BstUI (Table 1, Fig. 2D), which produced three DNA bands. The methylation patterns could be visualized based on the presence of bands at 248 bp (non-methylated, indicated by a white arrow head in Fig. 2B, a + b + c in Fig. 2D), 215 bp (including methylated and non-methylated, indicated by a gray arrow head in Fig. 2B, a + b in Fig. 2D), and 178 bp (methylated, indicated by a black arrow head in Fig. 2B, a in Fig. 2D). The CpG-A5 region (Fig. 2C) contains only one targeted methylated CpG site (Table 1, Fig. 2D). For regions with two or more digestion sites (CpG-1, A1, A2, and A6), DNA methylation status in DMRs containing multiple CpGs are also considered informative and biologically important. Thus, for CpG-1 and CpG-A1, the formula described below was used to estimate the DNA methylation status of multiple CpGs as DMRs in the present study. To determine the methylation levels of each DMR with multiple CpGs in Figs. 3, 4, and 5, the band intensities were weighted as 1, 0.5, and 0, for the black, gray, and white arrow heads, respectively, in Fig. 2A and 2B. For instance, CpG-1 methylation levels were calculated using the formula (black/106.5)/(black/106.5 + gray/145.5 + white/181) + (gray/145.5)/(black/106.5 + gray/145.5 + white/181) × 0.5, and those of CpG-A1 were calculated using the expression (black/178)/(black/178 + gray/215 + white/248) + (gray/215)/(black/178 + gray/215 + white/248) × 0.5. For regions with only one digestion site (CpG-A3, A4, A5, A7 and A8), the DNA methylation level of the CpG in the digestion site was calculated using the formula (black/169)/(black/169 + white/245) as exemplified for CpG-A5 using the two bands with white and black arrow heads in Fig. 2C
Table 2. Methylation levels at the target CpG sites determined by microarray and COBRA.
| Microarray |
COBRA |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CpG ID | JD1 | JD2 | JD4 | JD5 | Dif.* | JD1 | JD2 | JD3 | JD4 | JD5 | Dif.* |
| CpG-1 | 0.51 | 0.61 | 0.74 | 0.86 | 0.24 | 0.65 | 0.69 | 0.78 | 0.86 | 0.96 | 0.24 |
| CpG-A1 | 0.51 | 0.55 | 0.74 | 0.87 | 0.28 | 0.36 | 0.41 | 0.52 | 0.71 | 0.88 | 0.41 |
| CpG-A2 | 0.54 | 0.62 | 0.8 | 0.89 | 0.26 | 0.66 | 0.66 | 0.69 | 0.74 | 0.85 | 0.14 |
| CpG-A3 | 0.41 | 0.46 | 0.65 | 0.73 | 0.25 | 0.29 | 0.34 | 0.36 | 0.43 | 0.67 | 0.24 |
| CpG-A4 | 0.44 | 0.49 | 0.64 | 0.76 | 0.23 | 0.31 | 0.3 | 0.38 | 0.46 | 0.67 | 0.26 |
| CpG-A5 | 0.6 | 0.64 | 0.79 | 0.89 | 0.22 | 0.56 | 0.59 | 0.58 | 0.77 | 0.86 | 0.24 |
| CpG-A6 | 0.47 | 0.54 | 0.65 | 0.77 | 0.21 | 0.69 | 0.7 | 0.71 | 0.83 | 0.9 | 0.17 |
| CpG-A7 | 0.48 | 0.54 | 0.65 | 0.78 | 0.2 | 0.40 | 0.42 | 0.48 | 0.54 | 0.75 | 0.23 |
| CpG-A8 | 0.56 | 0.5 | 0.34 | 0.2 | –0.25 | 0.35 | 0.31 | 0.29 | 0.17 | 0.09 | –0.20 |
* Differences between group (JD1-2 vs. JD4-5).
Fig. 2.
Age-related increase in the methylations at three CpG regions in spermatozoa confirmed by COBRA. Samples with fragments digested at all sites (black arrow heads), partially digested fragments (gray arrow heads), and undigested (white arrow heads) fragments were subjected to 3% agarose gel electrophoresis (A: CpG-1, B: CpG-A1, C: CpG-A5). Methylation levels at the differentially methylated regions (DMRs), including the target CpGs, are expressed as ratios of DNA band intensity between digested (black: ++, gray: +) and undigested (white: –) fragments (A': CpG-1, B': CpG-A1, C': CpG-A5). Methylation levels were measured by densitometry using ImageJ 1.50i. D: Positions of the restriction enzyme recognition sites (BstUI or AciI) of the PCR products amplified using CpG-A1 or CpG A5 primers (arrows) for COBRA. The methylated CpG sites, including CpG-A1 and CpG-A5 (indicated by black triangles), were digested by BstUI or AciI, respectively, and are indicated by triangles. The DNA bands at 248 bp (a + b + c, a white arrow head in B, – in B'), 215 bp (a + b, a gray arrow head in B, + in B'), and 178 bp (a, a black arrow head in B, ++ in B') can be obtained by BstUI digestion. The DNA bands at 245 bp (a + b, a white arrow head in C, – in C'), and 169 bp (a, a black arrow head in C, ++ in C') can be obtained by AciI digestion.
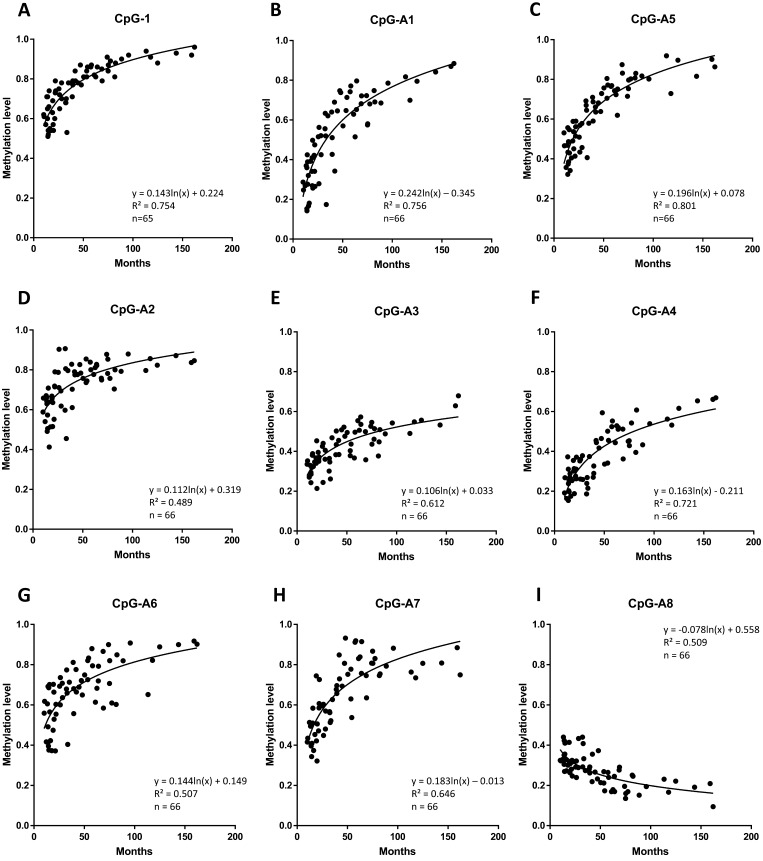
Fig. 3.
Age-related methylation changes at nine DMRs (A: CpG-1, B: CpG-A1, C: CpG-A5, D: CpG-A2, E: CpG-A3, F: CpG-A4, G: CpG-A6, H: CpG-A7, I: CpG-A8) analyzed by COBRA. For regions containing two or more digestion sites (CpG-1, A1, A2, and A6), the DNA band intensities were weighted as, 1, 0.5, and 0, for the black, gray, and white arrow head in Fig. 2. For regions with only one digestion site (CpG-A3, A4, A5, A7 and A8), the DNA methylation level at the CpG was calculated using the two bands with white and black arrow heads in Fig. 2. Semen samples were obtained from 35 bulls at different ages (10–162 months, n = 66, x axis). The equation and R value are reported in the graph. The correlation between the methylation level of each DMR and the age of the sample was statistically significant (P < 0.01).
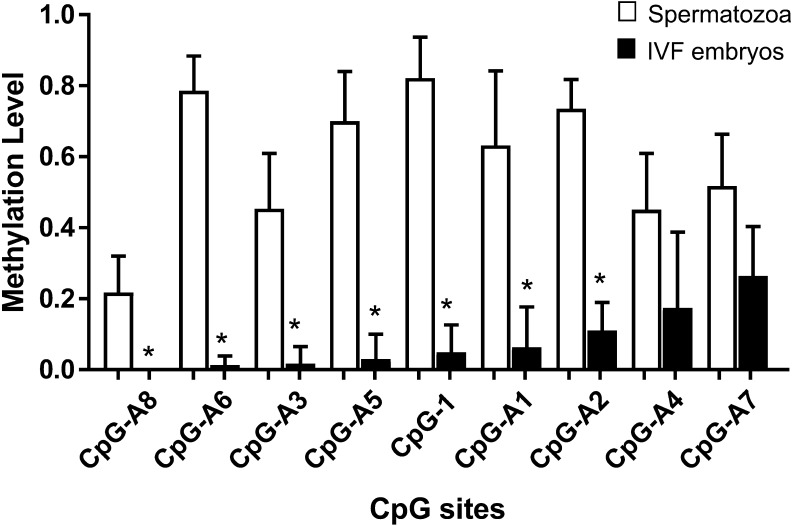
Fig. 4.
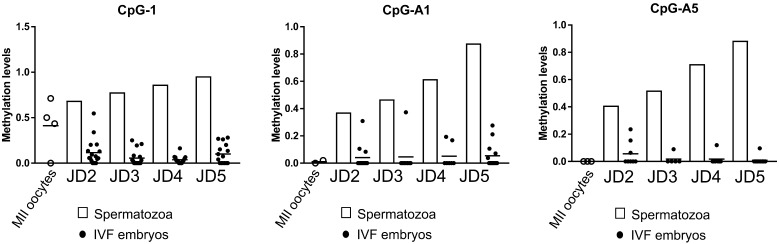
Methylation levels at the DMRs, with age-dependent differential methylation levels in the IVF blastocyst-stage embryos analyzed by COBRA. Blastocyst-stage embryos (n = 16) were obtained from IVF using JD2–5 spermatozoa (Tables 2 and 3) and nine DMRs (CpG-1, CpG-A1-A8) were analyzed individually. The DMRs are arranged in descending order of average methylation degree. The methylation levels of IVF embryos (black bars, right side) at the seven DMRs were significantly lower (*, t-test, P < 0.05) than the half value of methylation levels of spermatozoa (white bars, left side). Bars indicate the standard errors.
Fig. 5.
Methylation changes as revealed by COBRA at three DMRs (A: CpG-1, B: CpG-A1, C: CpG-A5) in MII oocytes before IVF (white circles, pooled n = 70), spermatozoa (white bars, JD2–5), and IVF-derived blastocyst embryos (black circles, n = 5–17, analyzed one by one) using JD2–5 spermatozoa. Bars indicate average methylation levels.
Age-related methylation changes in the identified nine DMRs were confirmed by COBRA using independent samples (n = 66, Fig. 3). A logarithmic approximation curve was generated, considering that the methylation rate at the DMR rapidly increased or decreased in bulls with young age and then gradually became stable at old age. We observed a significant correlation between the methylation level at each DMR and the age of the sample (P-value < 0.01).
Methylation levels in IVF embryos
The developmental rates of IVF embryos are shown in Table 3. Blastocyst-stage embryos failed to develop when JD1 spermatozoa were used, which was attributed to impaired/very weak motility after thawing (5++). IVF embryos derived from JD4 and JD5 showed lower cleaved rates than those derived from JD3 (P < 0.01). There were no differences in the rates of developmental to the blastocyst stage among IVF embryos derived from JD2-5. Considering that the zygote inherits DNA from both spermatozoa and oocyte, we calculated the contribution of IVF embryos with methylation levels of spermatozoa/oocytes as 0.5. The differences in methylation levels were considered when the methylation levels in the embryos were less than 50% of the methylation levels of spermatozoa/oocytes. Results of the COBRA revealed that the methylation levels of blastocyst-stage embryos (n = 16) obtained from IVF using JD2-5 spermatozoa were less than 50% of those obtained for JD2-5 spermatozoa at the seven DMRs (CpG-1, A1, A2, A3, A5, A6, A8, t-test, P-value < 0.05) (Table 2, Fig. 4). The methylation levels of oocytes before IVF (pooled 70 oocytes) showed high variability among the samples at CpG-1 DMR (Fig. 5). On the other hand, the oocytes before IVF showed hypomethylation values of 0.01 and 0.00 at the CpG-A1 and A5 DMRs, respectively (Fig. 5). The majority of the IVF-derived blastocyst-stage embryos were hypomethylated at the CpG-1, A1, and A5 DMRs, whereas some of the embryos were hypermethylated at these DMRs (Fig. 5). The semen samples collected from bulls at different ages showed no significant differences in methylation levels at these DMRs in the embryos (P-value > 0.05).
Table 3. IVF results of frozen semen samples of the same Japanese Black bull at different ages (JD1~5).
| Semen | Age | Motility | IVF |
||||
|---|---|---|---|---|---|---|---|
| (months) | Total (r) | Cleaved (%) | Blastocyst (%) | ||||
| JD1 | 14.0 | 5 | 86 (2) | 7 | (8.1) a | 0 | (0.0) a |
| JD2 | 19.3 | 50 | 141 (3) | 87 | (61.7) bc | 24 | (33.3) b |
| JD3 | 28.1 | 43 | 122 (3) | 91 | (72.8) b | 23 | (33.6) b |
| JD4 | 54.2 | 37 | 158 (3) | 88 | (55.7) c | 25 | (29.1) b |
| JD5 | 162.0 | 57 | 133 (3) | 70 | (52.6) c | 15 | (24.1) b |
Motility: determined under microscope just after thawing. Total: number of oocytes cultured after in vitro fertilization. r: repeat number for IVF experiment. a ,b, c The values with different superscripts within the same column are significantly different (P < 0.05).
Discussion
The present study confirmed that the methylation levels at nine CpG sites in spermatozoa showed age-related changes throughout the bull’s life, and seven of these CpG regions were found to be hypomethylated after fertilization. In the DMRs analyzed, we observed rapid changes in methylation levels until the animals reached four years of age; however, the methylation changes were less pronounced with advancing age (Fig. 3). The methylation state of sperm DNA in bulls appeared to be influenced by the age of breeding maturation. Recent studies revealed that the overall DNA methylation levels of the genome increase during the first year of life and then stabilize through adulthood to advanced age in human blood leucocytes [21]. Potential correlations between the methylation status and the breeding maturation could be used as markers to evaluate the fertile period in bulls.
The methylation levels of the analyzed DMRs can also be used as markers to estimate the age of the bull at which fresh semen was collected. Recently, many studies have developed age prediction models based on DNA methylation in various tissues and body fluids [7, 22]. Methylation levels at multiple CpG sites are highly associated with age and could therefore be used to predict chronological age in humans [7]. In the present study, the CpGs with age-dependent different methylation levels (ADDM) are common across the so-called epigenetic clock and can be used to predict an individual’s age [23, 24]. We deduced an age-estimation formula for the bull based on the methylation levels at three DMRs (CpG-1, CpG-A1, and CpG-A5). Indeed, when the methylation levels of the sperm were used in this formula to estimate the age of the bulls, the predictions showed 80% accuracy (data not shown).
We identified target CpG sites that showed ADDM and used COBRA to analyze the methylation levels at the DMRs for a large number of samples or a small volume of sample, such as the blastocyst-stage embryos. Methylation levels showed an age-dependent increase or decrease in the semen samples (Fig. 3). However, it is not clear whether the bovine features of the sequences, which include the target CpGs (indicated in Table 1), could be affected by aging. Interestingly, COBRA results showed that the methylation levels were different among individuals of the same age (Fig. 3). In particular, these data showed that the methylation levels were more variable among younger bulls. The samples used in this study were obtained from six farms located in different areas of Japan; however, it is not clear whether pedigree or environmental factors can contribute to the observed differences in methylation levels among the individual bulls.
Male age at conception has long been recognized to be associated with an increased risk for de novo mutations, which can cause some disorders [25]. Epigenetic changes, mainly DNA methylation, have been associated with age-related diseases in humans [26, 27]. Certain ADDM regions can be potentially associated with bull fertility. The paternal epigenome is required not only for spermatogenesis and mature sperm function, but also for embryonic development. COBRA revealed low methylation levels at seven ADDM regions in the IVF-derived blastocyst-stage embryos (Fig. 4). The above results suggested that these ADDM regions in spermatozoa could be demethylated during early embryonic development. We observed no differences in the rates of developmental to the blastocyst stage among IVF embryos derived from the spermatozoa of bulls at different ages (19–162 months, Table 3). The different methylation levels at these ADDM regions in sperm DNA did not affect the methylation status of the IVF-derived embryos; however, some of the IVF-derived blastocyst stage embryos showed hypermethylation at these ADDM regions (Fig. 5). The methylation levels of the spermatozoa can be preserved in the embryos. If the abnormal methylation levels are confirmed to reflect the methylation error in the early-stage embryos, it will be interesting to analyze the correlations between different methylation levels and the developmental abilities of IVF-derived embryos.
As shown in Table 3, after warming the frozen semen sample, low sperm motility was seen to be associated with poor IVF results. It is possible that the results of IVF reflect poor motility after freezing and thawing. However, whether the results shown in Table 3 are related to ADDM methylation levels remain unknown. To effectively predict fertility from methylation levels, evaluation of the fertility of young bulls during periods of severe changes in ADDM methylation levels should be considered in subsequent studies to identify useful markers for fertility from the ADDM regions. If the methylation levels accurately reflect the maturity of sperm, abnormal methylation levels of these CpG sites can be used to identify immature sperm and associated fertilization characteristics.
In conclusion, our findings revealed that the methylation levels at some DMRs in spermatozoa are altered throughout the bull’s life and that certain DMRs are hypomethylated after fertilization. These DMRs can serve as epigenetic biomarkers for predicting bull age. Further studies are required to determine whether these age-dependent changes in methylation levels can affect bull fertility.
Acknowledgments
This work was supported by Ito Foundation Grants (H28-Ken42) and JSPS KAKENHI Grant Number JP16K07999. The authors are grateful to the members of the Livestock Research Support Center in NARO for their help with the semen collection.
References
- 1.Rahman MB, Schellander K, Luceño NL, Van Soom A. Heat stress responses in spermatozoa: Mechanisms and consequences for cattle fertility. Theriogenology 2018; 113: 102–112. [DOI] [PubMed] [Google Scholar]
- 2.Tunc O, Tremellen K. Oxidative DNA damage impairs global sperm DNA methylation in infertile men. J Assist Reprod Genet 2009; 26: 537–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aston KI, Uren PJ, Jenkins TG, Horsager A, Cairns BR, Smith AD, Carrell DT. Aberrant sperm DNA methylation predicts male fertility status and embryo quality. Fertil Steril 2015; 104: 1388–1397.e1–5. [DOI] [PubMed] [Google Scholar]
- 4.Jenkins TG, Carrell DT. Dynamic alterations in the paternal epigenetic landscape following fertilization. Front Genet 2012; 3: 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carrell DT. Epigenetics of the male gamete. Fertil Steril 2012; 97: 267–274. [DOI] [PubMed] [Google Scholar]
- 6.Kaneda M, Okano M, Hata K, Sado T, Tsujimoto N, Li E, Sasaki H. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature 2004; 429: 900–903. [DOI] [PubMed] [Google Scholar]
- 7.Jung SE, Shin KJ, Lee HY. DNA methylation-based age prediction from various tissues and body fluids. BMB Rep 2017; 50: 546–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jenkins TG, Aston KI, Carrell DT. Sperm epigenetics and aging. Transl Androl Urol 2018; 7 (Suppl 3): S328–S335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jenkins TG, Aston KI, Pflueger C, Cairns BR, Carrell DT. Age-associated sperm DNA methylation alterations: possible implications in offspring disease susceptibility. PLoS Genet 2014; 10: e1004458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jenkins TG, Aston KI, Cairns BR, Carrell DT. Paternal aging and associated intraindividual alterations of global sperm 5-methylcytosine and 5-hydroxymethylcytosine levels. Fertil Steril 2013; 100: 945–951. [DOI] [PubMed] [Google Scholar]
- 11.Kobayashi N, Okae H, Hiura H, Chiba H, Shirakata Y, Hara K, Tanemura K, Arima T. Genome-Scale Assessment of Age-Related DNA Methylation Changes in Mouse Spermatozoa. PLoS One 2016; 11: e0167127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones MJ, Goodman SJ, Kobor MS. DNA methylation and healthy human aging. Aging Cell 2015; 14: 924–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park JS, Jeong YS, Shin ST, Lee KK, Kang YK. Dynamic DNA methylation reprogramming: active demethylation and immediate remethylation in the male pronucleus of bovine zygotes. Dev Dyn 2007; 236: 2523–2533. [DOI] [PubMed] [Google Scholar]
- 14.Seisenberger S, Peat JR, Hore TA, Santos F, Dean W, Reik W. Reprogramming DNA methylation in the mammalian life cycle: building and breaking epigenetic barriers. Philos Trans R Soc Lond B Biol Sci 2013; 368: 20110330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang L, Zhang J, Duan J, Gao X, Zhu W, Lu X, Yang L, Zhang J, Li G, Ci W, Li W, Zhou Q, Aluru N, Tang F, He C, Huang X, Liu J. Programming and inheritance of parental DNA methylomes in mammals. Cell 2014; 157: 979–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kropp J, Carrillo JA, Namous H, Daniels A, Salih SM, Song J, Khatib H. Male fertility status is associated with DNA methylation signatures in sperm and transcriptomic profiles of bovine preimplantation embryos. BMC Genomics 2017; 18: 280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kobayashi E, Takeda K. A data driven approach to utilizing Human Methylation arrays in genome-wide study for bovine DNA methylation. Journal of Animal Genetics 2016; 44: 45–52. [Google Scholar]
- 18.Takeda K, Kobayashi E, Akagi S, Nishino K, Kaneda M, Watanabe S. Differentially methylated CpG sites in bull spermatozoa revealed by human DNA methylation arrays and bisulfite analysis. J Reprod Dev 2017; 63: 279–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pidsley R, Zotenko E, Peters TJ, Lawrence MG, Risbridger GP, Molloy P, Van Djik S, Muhlhausler B, Stirzaker C, Clark SJ. Critical evaluation of the Illumina MethylationEPIC BeadChip microarray for whole-genome DNA methylation profiling. Genome Biol 2016; 17: 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bibikova M, Barnes B, Tsan C, Ho V, Klotzle B, Le JM, Delano D, Zhang L, Schroth GP, Gunderson KL, Fan JB, Shen R. High density DNA methylation array with single CpG site resolution. Genomics 2011; 98: 288–295. [DOI] [PubMed] [Google Scholar]
- 21.Alisch RS, Barwick BG, Chopra P, Myrick LK, Satten GA, Conneely KN, Warren ST. Age-associated DNA methylation in pediatric populations. Genome Res 2012; 22: 623–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Paoli-Iseppi R, Deagle BE, McMahon CR, Hindell MA, Dickinson JL, Jarman SN. Measuring animal age with DNA methylation: from humans to wild animals. Front Genet 2017; 8: 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horvath S. DNA methylation age of human tissues and cell types. Genome Biol 2013; 14: R115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Freire-Aradas A, Phillips C, Lareu MV. Forensic individual age estimation with DNA: From initial approaches to methylation tests. Forensic Sci Rev 2017; 29: 121–144. [PubMed] [Google Scholar]
- 25.Sharma R, Agarwal A, Rohra VK, Assidi M, Abu-Elmagd M, Turki RF. Effects of increased paternal age on sperm quality, reproductive outcome and associated epigenetic risks to offspring. Reprod Biol Endocrinol 2015; 13: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Atsem S, Reichenbach J, Potabattula R, Dittrich M, Nava C, Depienne C, Böhm L, Rost S, Hahn T, Schorsch M, Haaf T, El Hajj N. Paternal age effects on sperm FOXK1 and KCNA7 methylation and transmission into the next generation. Hum Mol Genet 2016; 25: 4996–5005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Curley JP, Mashoodh R, Champagne FA. Epigenetics and the origins of paternal effects. Horm Behav 2011; 59: 306–314. [DOI] [PMC free article] [PubMed] [Google Scholar]