Abstract
We aimed to investigate the correlation of long noncoding RNA nuclear enriched abundant transcript 1 (lnc-NEAT1), microRNA-124 (miR-124) and lnc-NEAT1/miR-124 axis with disease risk, severity, inflammatory cytokines, and survival of sepsis.
Eighty-two patients with sepsis and 82 healthy controls (HCs) were consecutively enrolled. Blood samples were collected for detection of lnc-NEAT1 and miR-124 expressions (using RT-qPCR) and measurement of inflammatory cytokines expressions (by ELISA). Severity and organ failure were assessed by acute physiology and chronic health evaluation II (APACHE II) score and sequential organ failure assessment (SOFA) score, and survival was assessed.
Lnc-NEAT1 expression was increased while miR-124 expression was decreased in patients with sepsis compared to HCs, and both of them were able to distinguish patients with sepsis from HCs. For disease condition, lnc-NEAT1 positively associated with APACHE II score, SOFA score, and expressions of C-reactive protein (CRP), procalcitonin, tumor necrosis factor α (TNF-α), and interleukin-1β (IL-1β), whereas miR-124 negatively correlated with APACHE II score, SOFA score and levels of serum creatinine (Scr), CRP, TNF-α, IL-1β, interleukin-6 (IL-6) and interleukin-17 (IL-17). Regarding prognosis, lnc-NEAT1 was upregulated but miR-124 was downregulated in nonsurvivors compared to survivors. Additionally, lnc-NEAT1 negatively correlated with miR-124. Besides, lnc-NEAT1/miR-124 axis was increased in patients with sepsis compared to HCs, and positively associated with APACHE II score, SOFA score, and levels of Scr, CRP, TNF-α, IL-1β, IL-6, and IL-17, while negatively correlated with survival. Most importantly, lnc-NEAT1/miR-124 axis presented numerically increased predictive value for sepsis risk and survival compared to each index alone.
Lnc-NEAT1/miR-124 axis correlates with increased sepsis risk, and associates with higher inflammation, deteriorative disease condition, and decreased survival in patients with sepsis.
Keywords: enriched abundant transcript 1, inflammatory cytokines, long noncoding RNA, sepsis, survival
1. Introduction
Sepsis is a life-threatening organ dysfunction caused by a dysregulated host response to infection, with the standardized occurrence rate of 461 cases per 100,000 populations according to a Chinese statistical report.[1–4] Currently, there are multiple recommended disease management applied for sepsis (such as fluid resuscitation, antimicrobial therapy, vasoactive mediation, and supportive therapy for sepsis-induced organ dysfunction). However, the prognosis of patients with sepsis is still dismal with an in-hospital mortality of 40%.[3,5] Thus, exploration of sensitive and specific biomarkers for early diagnosis and waring of progression is urgently needed.
Long noncoding RNA (lncRNA), characterized by a class of transcripts with more than 200 nucleotides and limited protein-coding ability, has been reported to participate in genomic regulation by various mechanisms (such as epigenetic regulation, transcription modulation, and post-transcription modulation).[6,7] LncRNA nuclear enriched abundant transcript 1 (lnc-NEAT1), located on chromosome 11q13.1, is a structure component of nuclear paraspeckles.[8–10] Based on some previous studies, lnc-NEAT1 is overexpressed in several inflammation-related diseases (such as systemic lupus erythematosus and sepsis-induced acute kidney injury), and it may participate in the disease progression and inflammatory responses in sepsis.[9–11] MicroRNA (miRNA) belongs to small noncoding RNA with 18 to 24 nucleotides, which is involved in multiple cell functions (such as cell proliferation, apoptosis, and differentiation) via targeting the 3′-untranslated region of mRNAs.[12,13] Among various miRNAs, miRNA-124 (miR-124), a tumor suppressor, is recently reported to participate in the suppression of inflammatory responses in sepsis model by several mechanisms, such as promoting peroxisome proliferator-activated receptor γ (PPARγ)-mediated anti-inflammation and targeting with signal transducer and activator of transcription 3 (STAT3) related genes.[14,15] Furthermore, miR-124 has been identified as a direct target miRNA of lnc-NEAT1.[14,16,17]
Based on these mentioned indications that
-
(1)
lnc-NEAT1 is one of the research hotspots, especially its role in inflammatory responses, and it may be involved in the pathology of progression as well as inflammation of sepsis according to previous data, while the detail evidence is seldomly reported;
-
(2)
miR-124 is a frequently-investigated miRNA and it has been reported to play a role in anti-inflammatory mechanism of sepsis according to an experiment, whereas, whether it suppresses inflammation in sepsis has not been explored in clinical practices;
-
(3)
miR-124 serves as a direct target miRNA of lnc-NEAT1, while the correlation of lnc-NEAT1 and miR-124 in sepsis is unclear, which needs further investigation.
For all these reasons, we hypothesized that lnc-NEAT1 might correlate with disease severity and inflammation in sepsis via the interaction with miR-124, while the related evidence is limited. Hence, we conducted this study to investigate the correlation of lnc-NEAT1, miR-124, and lnc-NEAT1/miR-124 axis with sepsis risk, and to further explore their association with severity, inflammatory cytokines, and survival in patients with sepsis.
2. Materials and methods
2.1. Patients
Eighty-two sepsis patients admitted to intensive care unit (ICU) of Zhongshan Hospital Affiliated to Xiamen University from July 2016 to June 2018 were consecutively enrolled in the present study. All enrolled patients aged above 18 years and were diagnosed as sepsis according to the “Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis” developed by the American College of Chest Physicians/Society of Critical Care Medicine.[18] Patients were excluded if they were
-
(1)
suffering from malignancies,
-
(2)
taking immunosuppressive drugs,
-
(3)
infected with human immunodeficiency virus (HIV),
-
(4)
transferred from other hospitals,
-
(5)
pregnant or lactating women.
Besides, 82 healthy subjects (whose age and gender were matched to the enrolled patients) underwent physical examination in the Hospital were recruited as healthy controls (HCs), and all HCs did not have any symptoms of active infection or any history of immune compromise such as HIV, cancer, pregnancy, or on current chronic steroid therapy. The approval for this study was acquired from the Ethics Committee of the Zhongshan Hospital Affiliated to Xiamen University (ethics approval number: 2016.008), and the information consents were obtained from all patients or their guardians.
2.2. Collection of blood samples and data
The blood samples of sepsis patients were collected within 24 hours after admitted to ICU, and the demographic features including age and gender were recorded. Meanwhile, biochemical indexes such as serum creatinine (Scr), albumin, white blood cell (WBC) count, C-reactive protein (CRP) as well as procalcitonin (PCT) were measured routinely. Additionally, the acute physiology and chronic health evaluation (APACHE) II score and sequential organ failure assessment (SOFA) score were recorded within 24 hours after patients were admitted to ICU. All patients’ vital signs were monitored daily, and conventional treatment was administered as Guidelines of sepsis. All patients were followed up for 28 days. The 28-day survival rate was recorded, and accumulating survival was calculated from the day of ICU admission to the day of death in the hospital or last follow up. In addition, the blood samples of HCs were collected during the physical examination.
2.3. Detection of baseline lnc-NEAT1 and miR-124
The plasma (which was taken within 24 hours from admission) was isolated from blood samples by centrifugation at 4°C 2000 g for 10 minutes. Using RNeasy Protect Mini Kit (Qiagen, Duesseldorf, Nordrhein-Westfalen, German), total RNA was extracted from plasma samples. Then, reverse transcription to complementary DNA was conducted using PrimeScript RT reagent Kit (Takara, Dalian, Liaoning, China), and subsequently qPCR was performed by TB Green Fast qPCR Mix (Takara). The results were calculated by the 2−ΔΔCt formula. For lnc-NEAT1, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as the internal reference. As for miR-124, U6 was applied as the internal reference. Primers used in the RT-qPCR were as follows: lnc-NEAT1, forward: 5′-TGTCCCTCGGCTATGTCAGA-3′, reverse: 5′-GAGGGGACGTGTTTCCTGAG-3′; miR-124, forward: 5′-ACACTCCAGCTGGGCGTGTTCACAGCGGACCT-3′, reverse: 5′-TGTCGTGGAGTCGGCAATTC-3′; GAPDH, forward: 5′-TGACCACAGTCCATGCCATCAC-3′, reverse: 5′-GCCTGCTTCACCACCTTCTTGA-3′; U6, forward: 5′-CTCGCTTCGGCAGCACATATACTA-3′; reverse: 5′-ACGAATTTGCGTGTCATCCTTGC-3′.
2.4. Detection of inflammatory cytokines
For the patients with sepsis, plasma concentrations of tumor necrosis factor (TNF-α), interleukin-1β (lL-1β), interleukin-6 (IL-6), and interleukin-17 (IL-17) were determined using the TNF-α, IL-1β IL-6, and IL-17 human ELISA kits (Abcam, Cambridge, MA). Standard curves were made by using standards provided in the kits, and the cytokine concentrations were appointed from the standard curves by use of linear regression analysis. The detection limits of these parameters were consistent with the manufacturer's instructions. All processes were performed according to the protocols provided by the manufacturer.
2.5. Statistical analysis
All continuous variables were checked for normality by using the Kolmogorov-Smirnov test. Normal distributed or approximatively normal distributed continuous variable was expressed as mean value (standard deviation), skewed or unknown distributed continuous variable were displayed as median (interquartile range), and categorized variable was presented as count (percentage). Differences between 2 groups were assessed by the Wilcoxon rank-sum test; the correlation between variables was determined by Spearman rank correlation test. Receiver operating characteristic (ROC) curve analysis and the derived area under the curve (AUC) statistic were used to assess the accuracy of a variable in distinguishing different subjects. Kaplan–Meier (K–M) curves were plotted to display the survival profiles, and the log-rank test was used to determine the survival difference between 2 groups. The values of P < .05 were considered statistically significant. SPSS 19.0 statistical software (SPSS Inc, Chicago, IL) was used for statistical data processing, and the GraphPad Prism 6.00 software (GraphPad Software, La Jolla, CA) was used for plotting.
3. Results
3.1. Baseline characteristics
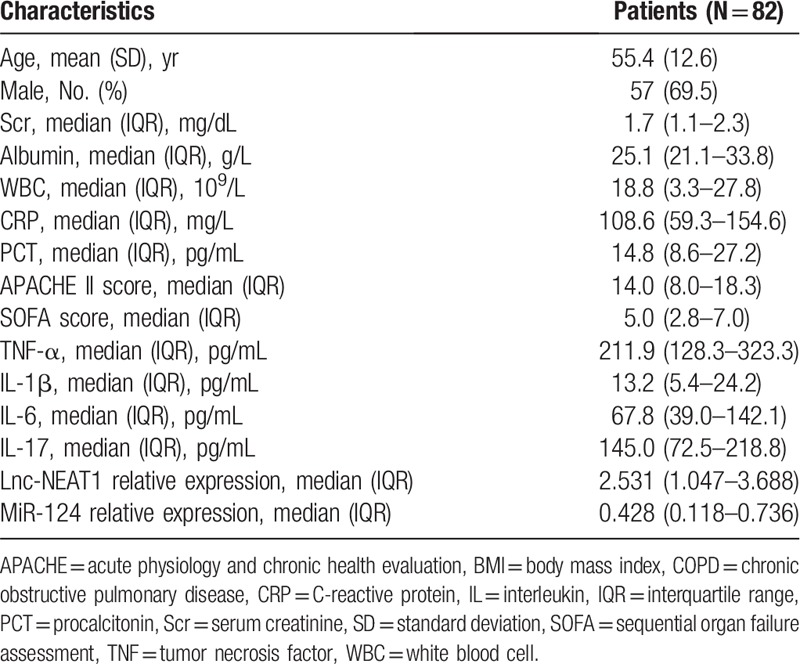
A total of 82 patients with sepsis were enrolled in this study, with mean age of 55.4 ± 12.6 years and 57 (69.5%) males (Table 1). For routine biochemical indexes, median values of Scr, albumin, WBC, CRP, and PCT were 1.7 (1.1–2.3) mg/dL, 25.1 (21.1–33.8) g/L, 18.8 (3.3–27.8) × 109/L, 108.6 (59.3–154.6) mg/L, and 14.8 (8.6–27.2) pg/mL, respectively. Regarding disease severity, median values of APACHE II score and SOFA score were 14.0 (8.0–18.3) and 5.0 (2.8–7.0) respectively. Besides, median values of inflammatory cytokines including TNF-α, IL-1β, IL-6, and IL-17 were 211.9 (128.3–323.3) pg/mL, 13.2 (5.4–24.2) pg/mL, 67.8 (39.0–142.1) pg/mL, and 145.0 (72.5–218.8) pg/mL, respectively. In addition, median values of lnc-NEAT1 relative expression and miR-124 relative expression were 2.531 (1.047–3.688) and 0.428 (0.118–0.736), respectively (Table 1).
Table 1.
Sepsis patients’ characteristics.
3.2. Correlation of lnc-NEAT1, miR-124, and lnc-NEAT1/miR-124 axis with sepsis risk
Lnc-NEAT1 expression was increased in patients with sepsis compared to HCs (P < .001) (Fig. 1A), and it had a good predictive value for increased sepsis risk with an AUC of 0.785 (95% confidence interval [CI]: 0.716–0.853) (Fig. 1D). Whereas miR-124 expression was decreased in patients with sepsis compared to HCs (P < .001) (Fig. 1B), and it presented well predictive value for decreased sepsis risk (AUC: 0.804, 95% CI: 0.737–0.871) (Fig. 1E). For lnc-NEAT1/miR-124 axis, it was raised in patients with sepsis compared to HCs (P < .001) (Fig. 1C), which could also distinguish patients with sepsis from HCs (AUC: 0.829, 95% CI: 0.767–0.891) (Fig. 1F), and the AUC was numerically elevated compared to lnc-NEAT1 (AUC 0.785) or miR-124 (AUC 0.804) alone. To further assess the correlation of lnc-NEAT1 expression with miR-124 expression, Spearman rank correlation test was performed, and the results showed a negative correlation of lnc-NEAT1 expression with miR-124 expression in patients with sepsis (P = .002, r = −0.341) (Fig. 1G), as well as in HCs (P < .001, r = −0.540) (Fig. 1H).
Figure 1.
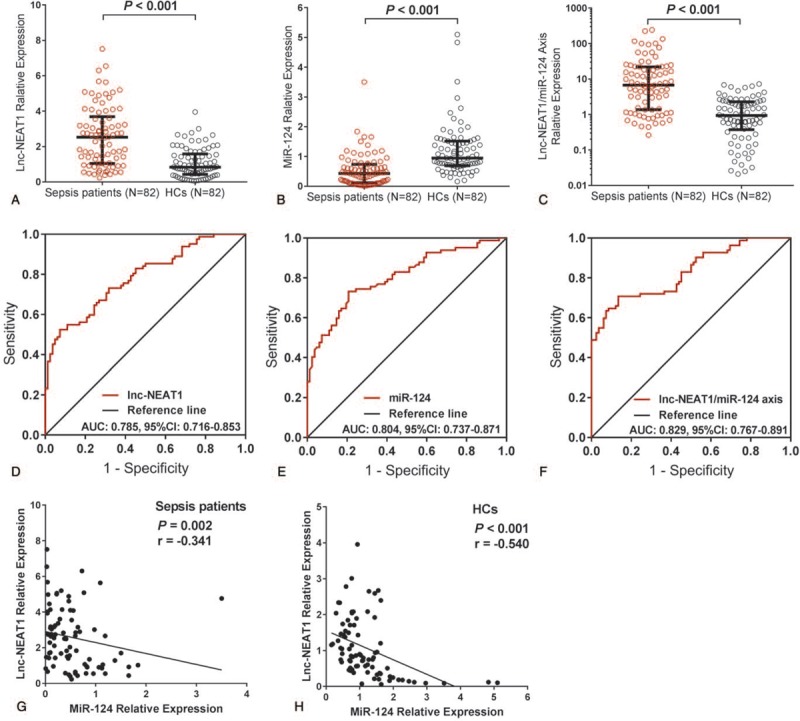
Predictive value of lnc-NEAT1, miR-124, and lnc-NEAT1/miR-124 axis for sepsis risk. Lnc-NEAT1 expression was higher in patients with sepsis compared to HCs (A). miR-124 expression was decreased in patients with sepsis compared to HCs (B). Lnc-NEAT1/miR-124 axis was increased in patients with sepsis compared to HCs (C). Lnc-NEAT1 presented a good predictive value for sepsis risk (D). miR-124 presented well predictive value for sepsis risk (E). Lnc-NEAT1/miR-124 axis presented a good predictive value for sepsis risk (F). Lnc-NEAT1 expression was negatively correlated with miR-124 expression in patients with sepsis (G). Lnc-NEAT1 expression was negatively correlated with miR-124 in HCs (H). ROC curves were used to assess the predictive value of lnc-NEAT1 for sepsis. Comparison between 2 groups was determined by the Wilcoxon rank-sum test or Spearman test. P < .05 was considered significant. Lnc-NEAT1 = long noncoding RNA nuclear enriched abundant transcript 1, HCs = healthy controls, miR-124 = microRNA-124, ROC curves = receiver operating characteristic curves.
3.3. Correlation of baseline lnc-NEAT1, miR-124, and lnc-NEAT1/miR-124 axis with disease condition in patients with sepsis
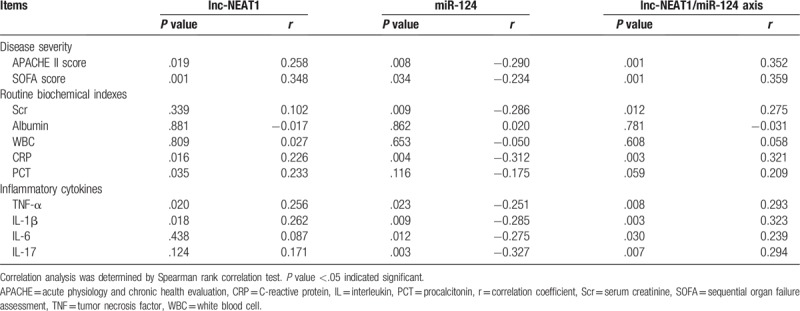
Baseline lnc-NEAT1 expression was positively correlated with disease severity (including APACHE II score [P = .019, r = 0.258] and SOFA score [P = .001] [r = 0.348] in patients with sepsis) (Table 2), and it was also positively associated with levels of CRP (P = .016, r = 0.226), PCT (P = .035, r = 0.233), TNF-α (P = .020, r = 0.256), and IL-1β (P = .018, r = 0.262). As for miR-124 expression, it was negatively correlated with APACHE II score (P = .008, r = −0.290) as well as SOFA score (P = .034, r = −0.234), and it was negatively associated with Scr (P = .009, r = −0.286), CRP (P = .004, r = −0.312), TNF-α (P = .023, r = −0.251), IL-1β (P = .009, r = −0.285), IL-6 (P = .012, r = −0.275), and IL-17 (P = .003, r = −0.327). Regarding lnc-NEAT1/miR-124 axis, it was positively correlated with APACHE II score (P = .001, r = 0.352) as well as SOFA score (P = .001, r = 0.359), and it also was positively associated with Scr (P = .012, r = 0.275), CRP (P = .003, r = 0.321), TNF-α (P = .008, r = 0.293), IL-1β (P = .003, r = 0.323), IL-6 (P = .030, r = 0.239), and IL-17 (P = .007, r = 0.294), while numerically positively correlated with PCT (P = .059, r = 209).
Table 2.
Correlation of lnc-NEAT1, miR-124, and lnc-NEAT1/miR-124 axis with disease severity, routine biochemical indexes, and inflammatory cytokines.
3.4. Correlation of baseline lnc-NEAT1, miR-124, and lnc-NEAT1/miR-124 axis with 28-day survival in patients with sepsis
All patients were followed up for 28 days, and the 28-day survival rate was recorded. Besides, patients were categorized as survivors group (n = 54) and nonsurvivors group (n = 28). Baseline lnc-NEAT1 expression was lower in survivors group compared to non-survivors group (P = .001) (Fig. 2A), and ROC curves showed that lnc-NEAT1 was able to distinguish survivors from nonsurvivors (AUC: 0.726, 95% CI: 0.615–0.837) (Fig. 2D). Whereas miR-124 expression was increased in survivors group compared to nonsurvivors group (P < .001) (Fig. 2B), and it also had the ability to distinguish survivors from nonsurvivors in patients with sepsis (AUC: 0.784, 95% CI: 0.688–0.900) (Fig. 2E). For lnc-NEAT1/miR-124 axis, it was decreased in survivors group compared to nonsurvivors group (P < .001) (Fig. 2C), and ROC curves disclosed that it was able to distinguish survivors from non-survivors with an AUC of 0.830 (95% CI: 0.738–0.922) (Fig. 2F), which was numerically larger compared to lnc-NEAT1 and miR-124 alone, indicating that lnc-NEAT1/miR-124 axis might exhibit increased predictive value for survival in patients with sepsis.
Figure 2.
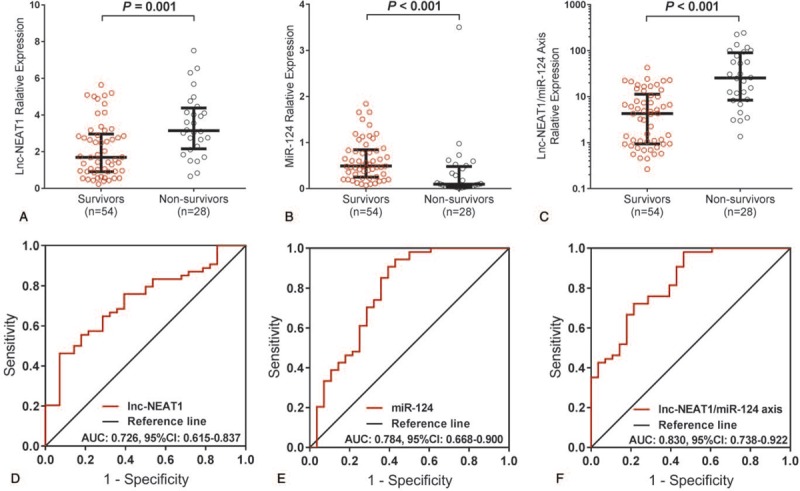
Predictive value of lnc-NEAT1, miR-124, and lnc-NEAT1/miR-124 axis for 28-d survival in patients with sepsis. Lnc-NEAT1 expression was reduced in survivors group compared to nonsurvivors group (A). miR-124 expression was elevated in survivors group compare to nonsurvivors group (B). Lnc-NEAT1/miR-124 axis expression was raised in survivors group compared to nonsurvivors group (C). Lnc-NEAT1 could distinguish survivors from nonsurvivors (D). miR-124 was able to distinguish survivors from nonsurvivors (E). Lnc-NEAT1/miR-124 axis could distinguish survivors from nonsurvivors (F). ROC curves were used to evaluate the value of Lnc-NEAT1 to distinguish survivors from non-survivors. Comparison between 2 groups was determined by the Wilcoxon rank-sum test. P < .05 was considered significant. Lnc-NEAT1 = long non-coding RNA nuclear enriched abundant transcript 1, miR-124 = microRNA-124, ROC curves = receiver operating characteristic curves.
3.5. Correlation of baseline lnc-NEAT1, miR-124, and lnc-NEAT1/miR-124 axis with accumulating survival by K–M curve analysis in patients with sepsis
In the 82 patients with sepsis, the accumulating 28-day survival rate was 65.9% (Fig. 3A). Additionally, K–M curve analysis revealed that lnc-NEAT1 expression was negatively associated with accumulating survival in patients with sepsis (P = .006) (Fig. 3B), while miR-124 expression was positively associated with accumulating survival (P = .004) (Fig. 3C). For lnc-NEAT1/miR-124 axis, it was negatively correlated with accumulating survival as well (P < .001), and the difference of accumulating survival between lnc-NEAT1/miR-124 axis high and low group seemed to increased compared to division by lnc-NEAT1 or miR-124 alone (Fig. 3D).
Figure 3.
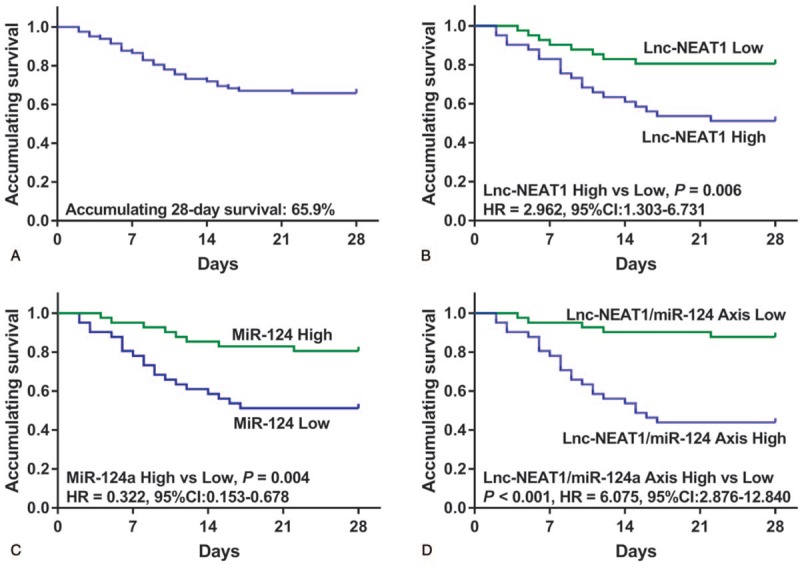
Predictive value of lnc-NEAT1, miR-124, and lnc-NEAT1/miR-124 axis for accumulating survival in patients with sepsis. Accumulating 28-d survival rate in totally sepsis patients was 65.9% (A). Lnc-NEAT1 high expression was associated with shorter accumulating survival (B). miR-124 expression was positively correlated with accumulating survival (C). Lnc-NEAT1/miR-124 axis high expression was associated with reduced accumulating survival (D). K–M curves were applied to display survival. Comparison between 2 groups was determined by log-rank test. P < .05 was considered significant. Lnc-NEAT1 = long non-coding RNA nuclear enriched abundant transcript 1, K–M curves = Kaplan-Meier curves, miR-124 = microRNA-124.
4. Discussion
In this study, we observed that:
-
(1)
lnc-NEAT1 correlated with increased sepsis risk and severe disease condition, and disclosed a good predictive value for decreased survival in patients with sepsis;
-
(2)
miR-124 correlated with decreased sepsis risk and attenuated disease condition, and predicted increased survival in patients with sepsis;
-
(3)
lnc-NEAT1 expression negatively correlated with miR-124 expression in both patients with sepsis and HCs, and lnc-NEAT1/miR-124 axis also associated with increased sepsis risk and deteriorative disease conditions, and predicted reduce survival in patients with sepsis. Most importantly, lnc-NEAT1/miR-124 axis presented with a numerically increased predictive value for sepsis risk and survival compared to each index alone.
Lnc-NEAT1 is widely expressed in mammalian cells and participates in many biological activities.[19] Recently, it has been reported to be involved in the pathological mechanism of several inflammatory-related diseases.[13,19,20] For example, an in vivo experiment shows that lnc-NEAT1 is overexpressed in spinal cord tissues of chronic constriction injury rat model, and its downregulation represses neuroinflammation through inhibiting cytokine expressions such as IL-6, IL-1β, and TNF-α.[13] Also, a study displays that lnc-NEAT1 facilitates oxidized low-density lipoprotein-induced inflammation and oxidative stress in atherosclerosis development through sponging miR-128 in RAW264.7 cells.[19] Besides, another study discloses that lnc-NEAT1 downregulation inhibits inflammation response via targeting miR-342–3p in human macrophages THP-1 cells.[20] These data reveal that lnc-NEAT1 may participate in the pathology of these inflammation-related diseases, which may support further investigations of lnc-NEAT1 in sepsis.
In clinical practices, lnc-NEAT1 dysregulation has been investigated in some previous studies, which display that lnc-NEAT1 is overexpressed and correlates with deteriorative disease condition in some inflammatory diseases.[9–11] For example, a study shows that lnc-NEAT1 is overexpressed and correlates with raised IL-8 level in fibroblasts derived from smokers with chronic obstructive pulmonary disease (COPD) compared to smokers without COPD.[11] Moreover, a previous study displays that lnc-NEAT1 high expression is associated with severer disease degrees in patients with sepsis-induced acute kidney injury.[10] Additionally, a study reveals that lnc-NEAT1 is overexpressed in peripheral blood mononuclear cells of patients with systemic lupus erythematosus (SLE), and positively correlates with disease activity of SLE.[9] These studies indicate the overexpression of lnc-NEAT1 in several inflammation-related diseases, as well as a positive correlation of lnc-NEAT1 expression with disease severity and inflammation.[9–11] To further investigate the role of lnc-NEAT1 in sepsis, we enrolled 82 patients with sepsis and 82 HCs to explore the association of lnc-NEAT1 expression with sepsis risk and the correlation of lnc-NEAT1 expression with disease condition in patients with sepsis. We found that lnc-NEAT1 was able to predict increased sepsis risk, and its high expression correlated with increased disease severity (APACHE II score and SOFA score), elevated routine biochemical indexes (CRP and PCT), and raised inflammatory cytokines (TNF-α and IL-1β) in patients with sepsis. The results might be explained by that: lnc-NEAT1 might contribute to a high reactive oxygen species level, which initiated inflammatory cascades and enhanced production of inflammatory cytokines as well as promoted dysfunction of endothelial cells by several mechanisms (such as protein kinase pathways, transcription factors, and genomic expression of proinflammatory regulators), thereby increased proinflammatory cytokine levels and aggravated organ dysfunction.[21] As to the prognostic value of lnc-NEAT1 in these diseases, limited investigations have been discovered.[11] In our study, we found that lnc-NEAT1 expression negatively correlated with survival in patients with sepsis. The possible reason might be that lnc-NEAT1 raised the levels of proinflammatory cytokines and further accelerated disease progression, thereby reduced the survival in patients with sepsis.
MiRNAs are evolutionarily conserved noncoding RNA oligonucleotides that regulate the expression of numerous gene targets, and a variety of studies have indicated that miRNAs play crucial roles in mechanisms of human inflammatory diseases.[22–25] For miR-124, some in vivo or in vitro experiments disclose that it may facilitate the suppression of inflammatory responses in sepsis.[14,15,26] For instance, a study discovers that miR-124 downregulation attenuates the PPARγ-mediated inhibition of pro-inflammatory cytokines in RAW264.7 cells.[15] Besides, a study discloses the decreased serum levels of TNF-α, IL-1β, and IL-6 in the sepsis mice treated with miR-124 mimics.[26] Also, another study shows that miR-124 attenuates production of inflammatory cytokines through targeting related genes of STAT3, TNF-α converting enzyme and acetylcholinesterase in LPS-induced sepsis mice and LPS-exposed cells.[14] These studies reveal that miR-124 may play a role in the anti-inflammatory mechanism of sepsis, and indicate that miR-124 may serve as a diagnostic or prognostic marker in sepsis, while the related evidence in clinical practice is rarely seen. In our study, we found that miR-124 correlated with decreased sepsis risk as well as attenuated disease condition. Notably, survivors have increased miR-124 expression compared to the nonsurvivors, and the reason was that: the patients with sepsis with baseline miR-124 low expression might have deteriorated disease progression, which further led to poor survival, while sepsis patients with miR-124 high level might have milder disease progression, which led to better survival. Therefore, the survivors have increased miR-124 expression compared to the non-survivors. These data might expand the evidence about the correlation of miR-124 with disease condition and prognosis in patients with sepsis.
Considering that lnc-NEAT1 might participate in the pathology of progression as well as inflammatory responses in sepsis, and that miR-124 was also involved in the anti-inflammatory mechanisms of sepsis, we hypothesized that lnc-NEAT1 might correlate with disease condition in sepsis through interacting with miR-124. However, the relevant investigation was seldom reported.[9–11,14,15,26] Thus, we performed this study to explore the association of lnc-NEAT1 with miR-124, and further investigate the correlation of lnc-NEAT1/miR-124 axis with sepsis risk and severity, inflammatory cytokines levels as well as survival in patients with sepsis. We observed that lnc-NEAT1 expression was negatively associated with miR-124 expression in patients with sepsis as well as in HCs, and lnc-NEAT1/miR-124 axis correlated with elevated sepsis risk and severer disease condition, and predicted decreased survival in patients with sepsis. Moreover, compared to lnc-NEAT1 and miR-124 alone, lnc-NEAT1/miR-124 axis showed numerically higher predictive value for sepsis risk and survival, indicating that lnc-NEAT1/miR-124 axis might serve as a more convincing biomarker for diagnosis and prognosis of sepsis.
Some limitations existed in this study:
-
(1)
sample size of patients with sepsis (N = 82) was relatively small, thus the statistical power might be relatively poor in our study;
-
(2)
this was a single-center study, which might have limited representativeness;
-
(3)
underlying mechanisms of lnc-NEAT1 in sepsis and the detailed function of lnc-NEAT1/miR-124 axis remained unclear.
Our results cannot be generalized to a bigger population at this time due to single center study, and further study is needed to verify our results and explore the pathology of lnc-NEAT1/miR-124 axis in sepsis.
5. Conclusion
In conclusion, lnc-NEAT1/miR-124 axis correlates with increased sepsis risk, and associates with higher inflammation, deteriorative disease condition and decreased survival in patients with sepsis.
Author contributions
Conceptualization: Fuyun He, Chengju Zhang.
Data curation: Fuyun He, Chengju Zhang, Qinghe Huang.
Formal analysis: Fuyun He, Chengju Zhang, Qinghe Huang.
Investigation: Fuyun He, Chengju Zhang, Qinghe Huang.
Methodology: Fuyun He, Chengju Zhang, Qinghe Huang.
Project administration: Fuyun He, Chengju Zhang, Qinghe Huang.
Resources: Fuyun He, Chengju Zhang, Qinghe Huang.
Software: Chengju Zhang, Qinghe Huang.
Supervision: Fuyun He, Chengju Zhang, Qinghe Huang.
Validation: Fuyun He, Chengju Zhang, Qinghe Huang.
Visualization: Fuyun He, Chengju Zhang, Qinghe Huang.
Writing – original draft: Fuyun He, Chengju Zhang, Qinghe Huang.
Writing – review and editing: Fuyun He, Chengju Zhang, Qinghe Huang.
Footnotes
Abbreviations: APACHE = acute physiology and chronic health evaluation, AUC = area under the curve, COPD = chronic obstructive pulmonary disease, CRP = C-reactive protein, GAPDH = glyceraldehyde-3-phosphate dehydrogenase, HCs = healthy controls, HIV = human immunodeficiency virus, ICU = intensive care unit, IL-6 = interleukin-6, IL-7 = interleukin-7, lL-1β = interleukin-1β, lnc-NEAT1 = lncRNA nuclear enriched abundant transcript 1, lncRNA = long non-coding RNA, miR-124 = miRNA-124, miRNA = microRNA, PCT = procalcitonin, PPARγ = peroxisome proliferator-activated receptor γ, ROC = receiver operating characteristic, Scr = serum creatinine, SOFA = sequential organ failure assessment, STAT3 = signal transducer and activator of transcription 3, TNF-α = tumor necrosis factor α, WBC = white blood cell.
The authors have no conflicts of interest to disclose.
References
- [1].Singer M, Deutschman CS, Seymour CW, et al. The third international consensus definitions for sepsis and septic shock (sepsis-3). JAMA 2016;315:801–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Seymour CW, Liu VX, Iwashyna TJ, et al. Assessment of clinical criteria for sepsis: for the third international consensus definitions for sepsis and septic shock (sepsis-3). JAMA 2016;315:762–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Rhodes A, Evans LE, Alhazzani W, et al. Surviving sepsis campaign: international guidelines for management of sepsis and septic shock: 2016. Intensive Care Med 2017;43:304–77. [DOI] [PubMed] [Google Scholar]
- [4].Zhou J, Tian H, Du X, et al. Population-based epidemiology of sepsis in a subdistrict of Beijing. Crit Care Med 2017;45:1168–76. [DOI] [PubMed] [Google Scholar]
- [5].Zhang TN, Li D, Xia J, et al. Non-coding RNA: a potential biomarker and therapeutic target for sepsis. Oncotarget 2017;8:91765–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Zhang H, Sun D, Li D, et al. Long non-coding RNA expression patterns in lung tissues of chronic cigarette smoke induced COPD mouse model. Sci Rep 2018;8:7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Colvin LA, Dougherty PM. Peripheral neuropathic pain: signs, symptoms, mechanisms, and causes: are they linked? Br J Anaesth 2015;114:361–3. [DOI] [PubMed] [Google Scholar]
- [8].Huang Q, Huang C, Luo Y, et al. Circulating lncRNA NEAT1 correlates with increased risk, elevated severity and unfavorable prognosis in sepsis patients. Am J Emerg Med 2018;36:1659–63. [DOI] [PubMed] [Google Scholar]
- [9].Zhang F, Wu L, Qian J, et al. Identification of the long noncoding RNA NEAT1 as a novel inflammatory regulator acting through MAPK pathway in human lupus. J Autoimmun 2016;75:96–104. [DOI] [PubMed] [Google Scholar]
- [10].Chen Y, Qiu J, Chen B, et al. Long non-coding RNA NEAT1 plays an important role in sepsis-induced acute kidney injury by targeting miR-204 and modulating the NF-kappaB pathway. Int Immunopharmacol 2018;59:252–60. [DOI] [PubMed] [Google Scholar]
- [11].Imamura K, Imamachi N, Akizuki G, et al. Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol Cell 2014;53:393–406. [DOI] [PubMed] [Google Scholar]
- [12].Frixa T, Donzelli S, Blandino G. Oncogenic microRNAs: key players in malignant transformation. Cancers (Basel) 2015;7:2466–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Xia LX, Ke C, Lu JM. NEAT1 contributes to neuropathic pain development through targeting miR-381/HMGB1 axis in CCI rat models. J Cell Physiol 2018;233:7103–11. [DOI] [PubMed] [Google Scholar]
- [14].Sun Y, Li Q, Gui H, et al. MicroRNA-124 mediates the cholinergic anti-inflammatory action through inhibiting the production of pro-inflammatory cytokines. Cell Res 2013;23:1270–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Wang D, Shi L, Xin W, et al. Activation of PPARgamma inhibits pro-inflammatory cytokines production by upregulation of miR-124 in vitro and in vivo. Biochem Biophys Res Commun 2017;486:726–31. [DOI] [PubMed] [Google Scholar]
- [16].Liu X, Liang Y, Song R, et al. Long non-coding RNA NEAT1-modulated abnormal lipolysis via ATGL drives hepatocellular carcinoma proliferation. Mol Cancer 2018;17:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Cheng N, Guo Y. Long noncoding RNA NEAT1 promotes nasopharyngeal carcinoma progression through regulation of miR-124/NF-kappaB pathway. Onco Targets Ther 2017;10:5843–53. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [18].Bone RC, Balk RA, Cerra FB, et al. Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. The ACCP/SCCM Consensus Conference Committee. American College of Chest Physicians/Society of Critical Care Medicine. Chest 1992;101:1644–55. [DOI] [PubMed] [Google Scholar]
- [19].Chen DD, Hui LL, Zhang XC, et al. NEAT1 contributes to ox-LDL-induced inflammation and oxidative stress in macrophages through inhibiting miR-128. J Cell Biochem 2018. [DOI] [PubMed] [Google Scholar]
- [20].Wang L, Xia JW, Ke ZP, et al. Blockade of NEAT1 represses inflammation response and lipid uptake via modulating miR-342-3p in human macrophages THP-1 cells. J Cell Physiol 2019;234:5319–26. [DOI] [PubMed] [Google Scholar]
- [21].Dua K, Malyla V, Singhvi G, et al. Increasing complexity and interactions of oxidative stress in chronic respiratory diseases: an emerging need for novel drug delivery systems. Chem Biol Interact 2019;299:168–78. [DOI] [PubMed] [Google Scholar]
- [22].Xiao YT, Wang J, Lu W, et al. Downregulated expression of microRNA-124 in pediatric intestinal failure patients modulates macrophages activation by inhibiting STAT3 and AChE. Cell Death Dis 2016;7:e2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Essandoh K, Li Y, Huo J, et al. MiRNA-mediated macrophage polarization and its potential role in the regulation of inflammatory response. Shock 2016;46:122–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Yang Z, Zeng B, Wang C, et al. MicroRNA-124 alleviates chronic skin inflammation in atopic eczema via suppressing innate immune responses in keratinocytes. Cell Immunol 2017;319:53–60. [DOI] [PubMed] [Google Scholar]
- [25].Liu Y, Zhang J, Han R, et al. Downregulation of serum brain specific microRNA is associated with inflammation and infarct volume in acute ischemic stroke. J Clin Neurosci 2015;22:291–5. [DOI] [PubMed] [Google Scholar]
- [26].Li XY, Zhang YQ, Xu G, et al. miR-124/MCP-1 signaling pathway modulates the protective effect of itraconazole on acute kidney injury in a mouse model of disseminated candidiasis. Int J Mol Med 2018;41:3468–76. [DOI] [PubMed] [Google Scholar]


