Abstract
Lung cancer is a malignant tumor with high morbidity and mortality. Early diagnosis remains a great challenge for the cancer. In this study, we aimed to explore diagnostic performance of serum microRNA-520f (miR-520f) in lung cancer.
Serum specimens were collected from 139 lung cancer patients and 76 healthy volunteers. Relative expression level of serum miR-520f was detected adopting quantitative real-time polymerase chain reaction (qRT-PCR). Chi-square test was applied to evaluate the association of miR-520f with clinical parameters of the patients. Additionally, receiver operating characteristic (ROC) analysis was performed to evaluate diagnostic value of miR-520f in lung cancer.
Serum miR-520f was down-regulated in lung cancer patients compared with healthy group (P <.001). Moreover, the expression of miR-520f was significantly associated with advanced TNM stage (P = .031) and metastasis (P = .002). The area under the curve (AUC) value of ROC curve was 0.888, suggesting that miR-520f could be a diagnostic biomarker for lung cancer. The cut-off value of serum miR-520f for lung cancer diagnosis was 1.815, with a sensitivity of 79.9% and a specificity of 84.2%.
Serum miR-520f was down-regulated in lung cancer patients, and may be a candidate biomarker for non-invasive screening of the disease.
Keywords: diagnosis, lung cancer, miR-520f
1. Introduction
Lung cancer is one of the most common cancers, representing a leading cause for cancer-related deaths worldwide.[1] In China, the morbidity of lung cancer is on the rise, due to aging, pollution, and exposure to carcinogens.[2,3] Lung cancer includes 2 pathological types: non-small cell lung cancer (NSCLC) and small cell lung cancer. NSCLC is the major type, accounting for more than 80% of whole lung cancer cases.[4] Great progress has been made in the treatments of lung cancer, but long-term survival of the patients does not show significant improvements.[5,6] Delayed diagnosis may be responsible for dismal prognosis.[7] The cancer is characterized by silent growth at early stages, and most of the lung cancer patients present distant invasion or metastasis when initially diagnosed, leading to limited therapeutic effects and poor clinical outcomes.[8,9] Therefore, finding effective biomarkers for early diagnosis and treatment would be a promising approach to improve the prognosis of lung cancer patients.
MicroRNAs (miRNAs) are a group of small non-coding RNAs that play important roles in gene expression at post-transcriptional level.[10] MiRNAs are involved in a variety of biological processes, such as cell proliferation, differentiation, development, and apoptosis.[11] Abnormal expression of miRNAs may lead to various pathological states, like cancer. Expression profiles of miRNAs show close association with the development and progression of as well as treatment response to cancers, suggesting their abilities to serve as predictive biomarkers and therapeutic targets for diseases.[12] In addition, expression patterns of miRNAs are stable in body fluids and archived specimens, thus being detected through non or minimally invasive methods.[13] Circulating miRNAs are considered as promising biomarkers for early screening and supervision of cancers.
MicroRNA-520f (miR-520f), a common member of miRNA family, was reported to play important roles in the progression of human cancers. For instance, miR-520f knockdown in gastric cancer cells in vitro could lead to aggressive cellular proliferation, revealing its suppressive effects against the development and progression of the cancer.[14] Animal studies have proved that miR-520f over-expression could inhibit lung metastasis in mouse model.[15] However, diagnostic performance of serum miR-520f in lung cancer remained unclear.
In our study, we aimed to detect expression pattern of serum miR-520f in lung cancer patients, as well as its association with clinical parameters of the cases. In addition, receiver operating characteristic (ROC) curve was constituted to estimate diagnostic value of serum miR-520f in lung cancer.
2. Materials and methods
2.1. Patients and specimen collection
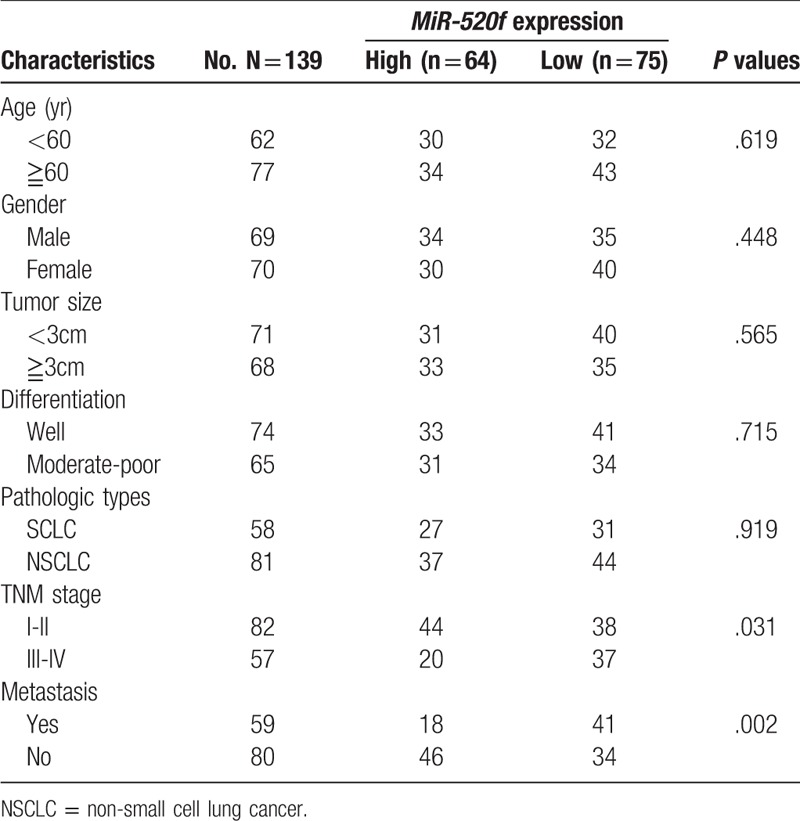
A total of 139 lung cancer patients who were diagnosed by pathologists between October 2015 and November 2017 were finally recruited from Ningbo Medical Center Lihuili Hospital. None of the patients had received any treatments in advance. Additionally, 76 healthy volunteers who were matched with the cases in gender and age were collected as the control group. In the healthy group, no one had been diagnosed with any malignancies. This study was approved by the ethics committee of the hospital. Informed consents were also obtained from all patients and healthy individuals. Clinicopathological features of the lung cancer patients were listed in Table 1, including age, gender, tumor size, differentiation, pathologic types, TNM stage, and metastasis.
Table 1.
Association of miR-520f expression with clinicopathological parameters of lung cancer patients.
A total of 5 mL fasting blood samples was taken from each of 139 lung cancer patients and 76 healthy volunteers. Then serum specimens were isolated from the blood through centrifugation and stored at −80°C until RNA extraction.
2.2. RNA extraction and quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was isolated from serum specimens using Trizol (Invitrogen). Then the first-strand cDNA was synthesized employing TaqMan MicroRNA Reverse Transcription Kit (Applied biosystems, Carlsbad, CA). In this study relative level of serum, miR-520f was determined using the SYBR Green Realtime PCR Master Mix (QPK-201, Toyobo Co, Ltd, Osaka, Japan). U6 acted as an internal control. Relative expression level of miR-520f was normalized to U6 and calculated using the 2−ΔΔCt method.
2.3. Statistical analysis
All statistical analyses were performed with SPSS 19.0 (SPSS Inc., Chicago, IL) and GraphPad Prism 5 (GraphPad Software, Inc., La Jolla, CA). Expression level of miR-520f was expressed as mean ± SD. Student t tests was used to analyze differences in miR-520f expression levels between the case and control groups. The relationship between miR-520f expression and various clinicopathological characteristics was assessed using Chi-square tests. To determine diagnostic performance of miR-520f expression in lung cancer, we performed ROC (receiver operating characteristic) analysis. The sensitivity and misjudgment rate (1-Specificity) can be obtained through the movement of cut off point/cut off value using SPSS 19.0 to draw ROC curve. With sensitivity as the vertical axis and misjudgment rate as the horizontal axis, the curve can be drawn via connecting the points. Then the area under the curve (AUC) an be calculated. The larger the area appears, the higher the judgment value is. The best cut off value is commonly used to determine the “Youden Index”. Maximum value of the index is the best boundary value. P <.05 in this study was considered statistically significant.
3. Results
3.1. Down-regulated expression of miR-520f in lung cancer
To examine miR-520f expression in 139 lung cancer patients and 76 healthy controls, qRT-PCR was performed. The results shown in Figure 1 revealed that the expression of miR-520f was significantly lower in lung cancer patients than in healthy volunteers (P <.001).
Figure 1.
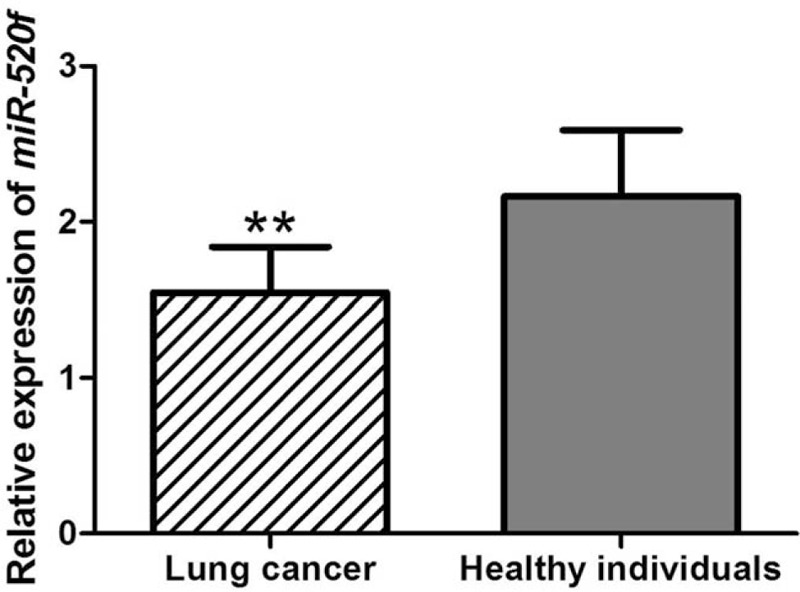
Serum miR-520f expression levels evaluated by qRT-PCR in lung cancer patients and healthy controls. Analysis results demonstrated that lung cancer patients exhibited significantly up-regulated miR-520f level, compared with healthy individuals. ∗∗: P <.01. qRT-PCR = quantitative real-time polymerase chain reaction.
3.2. Association between miR-520f and clinicopathological parameters of lung cancer patients
In this study, the lung cancer cases were categorized into high expression group (n = 64) and low expression group (n = 75), according to their average expression value of miR-520f. Chi-square test was used to detect the correlation between miR-520f expression level and clinicopathological data of lung cancer patients. MiR-520f expression was negatively associated with TNM stage (P = .031) and metastasis (P = .002). However, expression pattern of miR-520f did not show significant association with age, gender, differentiation or pathologic types among the patients with lung cancer (all P >.05) (Table 1).
3.3. Diagnostic value of miR-520f expression in lung cancer
ROC curves were plotted based on serum levels of miR-520f in lung cancer cases and healthy individuals in our study. The curve showed that lung cancer patients could be distinguished from healthy volunteers according to their serum levels of miR-520f with an AUC value of 0.888. The cut-off value for miR-520f diagnosing lung cancer was 1.815, with a sensitivity of 79.9% and a specificity of 84.2% (Fig. 2).
Figure 2.
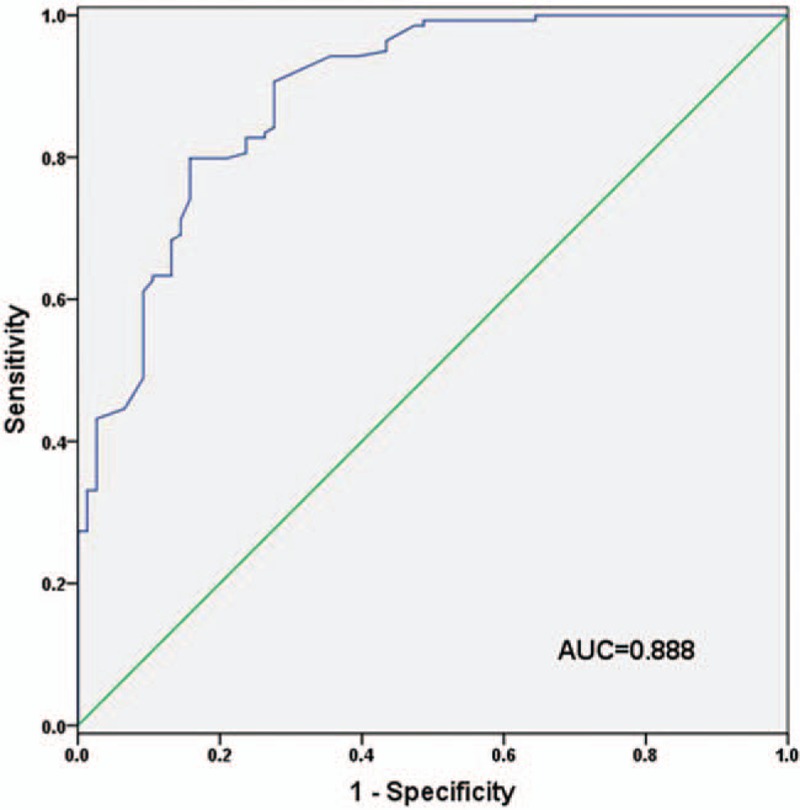
ROC analysis evaluating diagnostic accuracy of serum miR-520f in lung cancer. The curve suggested that serum miR-520f could discriminate between lung cancer cases and healthy volunteers with an AUC value of 0.888, combing with a sensitivity of 79.9% and a specificity of 84.2%. AUC = area under the curve, ROC = receiver operating characteristic.
4. Discussion
Lung cancer is a fatal cancer with high morbidity and mortality.[16] Low diagnosis rate may be responsible for dismal prognosis of the cancer.[17] Until now, early screening of lung cancer mainly depends on computer tomography (CT)-based strategies. However, those approaches are high cost and invasive, and their accuracy is limited by tumor type and size.[18,19] Therefore, reliable biomarkers which can be employed as minimally invasive tools for early detection of lung cancer are in urgent need in clinic. In this study, we investigated diagnostic performance of serum miR-520f in lung cancer. ROC analysis suggested that serum miR-520f might be a diagnostic biomarker for lung cancer.
MiRNAs are a group of small endogenous RNA molecules that can mediate gene expression by binding to the 3’ untranslated region (3’ UTR) of their target genes.[20] MiRNAs as oncogenes or tumor suppressors play an important role in tumorigenesis, and their expression profiles show specific to the development and progression of malignancies.[21] Yu et al reported that plasma levels of miR-92a-2 were significantly different between small cell lung cancer patients and healthy individuals, suggesting its function as a biomarker for early detection of the disease.[22] The study carried out by Shi et al reported that serum levels of miR-22 and miR-15b could be employed as reference factors for early detection of NSCLC, thus enhancing diagnostic accuracy of serum CEA.[23] MiRNAs might be used for prognosis evaluation as well. The meta-analysis constructed by Zhan et al demonstrated that expression profiles of miR-21, miR-200c, miR-125b, miR-148b, miR-365, miR-124, miR-32, miR-146a, and miR-375 showed close association with clinical outcomes of NSCLC patients, revealing their abilities as prognostic biomarkers for the cancer.[24] MiRNAs may be promising biomarkers for early screening and prognosis evaluation in human malignancies.
In our study, we investigated the function of miR-520f in lung cancer. Serum level of miR-520f mRNA was significantly down-regulated in lung cancer cases, compared with healthy volunteers. Moreover, its expression level was negatively correlated with TNM stage and metastasis. Patients with low serum level of miR-520f were more likely to present advanced TNM stage and positive metastasis. The data might reveal that miR-520f acted as a tumor suppressor gene, and that its inhibition could promote the development and progression of lung cancer. In vitro studies have demonstrated that miR-520 could regulate invasive and metastatic abilities of cancer cells. Moreover, the down-regulating of miR-520f might contribute to drug resistance via weakening apoptosis inhibition.[15,25] Taken together, miR-520f might be a tumor suppressor gene in malignancies. Studies indicated that miR-520f inhibited the proliferation and invasion of hepatocellular carcinoma cells through targeting TM4SF1.[26]MiR-520f also suppressed the invasion of cancer cells via targeting ADAM9 and TGFBR2, had the anti-invasion and anti-metastasis effects in vitro and in vivo.[27] All these findings may provide therapeutic approaches for cancer treatment.
Consequently, we investigated diagnostic value of serum miR-520f in lung cancer. The results showed that the expression of miR-520f could distinguish lung cancer patients from healthy individuals with high sensitivity and specificity. Serum miR-520f might be a non-invasive biomarker in early detection of lung cancer. However, there were still several limitations in the present study. First, we proved that miR-520f was down-regulated in lung cancer, but its specific molecular mechanism is still unknown. Second, our results might be limited by the relatively small sample size. Well-designed researches with larger sample sizes are required to verify diagnostic significance of serum miR-520f in lung cancer.
In conclusion, serum miR-520f is downregulated in lung cancer, and its expression level shows negative association with advanced tumor stage and metastasis. Serum miR-520f may be a biomarker in early detection of lung cancer.
Author contributions
Conceptualization: Yingyan Zhou.
Data curation: Yingyan Zhou.
Formal analysis: Yingyan Zhou.
Resources: Shimo Shen.
Software: Shimo Shen.
Supervision: Shimo Shen.
Validation: Shimo Shen.
Visualization: Shimo Shen.
Writing – original draft: Shimo Shen.
Writing – review & editing: Shimo Shen.
Footnotes
Abbreviations: AUC = area under the curve, CT = computer tomography, miR-520f = microRNA-520f, NSCLC = non-small cell lung cancer, qRT-PCR = quantitative real-time polymerase chain reaction, ROC = receiver operating characteristic.
The authors have no conflicts of interest to disclose.
References
- [1].Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA: Cancer J Clin 2013;63:11–30. [DOI] [PubMed] [Google Scholar]
- [2].Wen C, Dehnel T. China wrestles with lung cancer. Lancet Oncol 2011;12:15. [DOI] [PubMed] [Google Scholar]
- [3].Gyoba J, Shan S, Roa W, et al. Diagnosing lung cancers through examination of micro-RNA biomarkers in blood, plasma, serum and sputum: a review and summary of current literature. Int J Mol Sci 2016;17:494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Rezaei MK, Nolan NJ, Schwartz AM. Surgical pathology of lung cancer. Semin Respir Crit Care Med 2013;34:770–86. [DOI] [PubMed] [Google Scholar]
- [5].Reck M, Heigener DF, Mok T, et al. Management of non-small-cell lung cancer: recent developments. Lancet 2013;382:709–19. [DOI] [PubMed] [Google Scholar]
- [6].Xu CH, Yang Y, Wang YC, et al. Prognostic significance of serum chemerin levels in patients with non-small cell lung cancer. Oncotarget 2017;8:22483–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Agababaoglu I, Onen A, Demir AB, et al. Chaperonin (HSP60) and annexin-2 are candidate biomarkers for non-small cell lung carcinoma. Medicine 2017;96:e5903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lv S, Xue J, Wu C, et al. Identification of a panel of serum microRNAs as biomarkers for early detection of lung adenocarcinoma. J Cancer 2017;8:48–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Ulivi P, Zoli W. miRNAs as non-invasive biomarkers for lung cancer diagnosis. Molecules 2014;19:8220–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer 2015;15:321–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Barciszewska AM. MicroRNAs as efficient biomarkers in high-grade gliomas. Folia Neuropathol 2016;54:369–74. [DOI] [PubMed] [Google Scholar]
- [12].Iorio MV, Croce CM. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med 2012;4:143–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Wang J, Chen J, Sen S. MicroRNA as biomarkers and diagnostics. J Cell Physiol 2016;231:25–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hong S, Bi M, Chen S, et al. MicroRNA-520f suppresses growth of gastric carcinoma cells by target ATPase family AAA domain-containing protein 2 (ATAD2). Neoplasma 2016;63:873–9. [DOI] [PubMed] [Google Scholar]
- [15].van Kampen JG, van Hooij O, Jansen CF, et al. microRNA-520f reverses epithelial-tomesenchymal transition by targeting ADAM9 and TGFBR2. Cancer Res 2017;77:2008–17. [DOI] [PubMed] [Google Scholar]
- [16].Tseng TS, Park JY, Zabaleta J, et al. Role of nicotine dependence on the relationship between variants in the nicotinic receptor genes and risk of lung adenocarcinoma. PloS one 2014;9:e107268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Al Mohammad B, Brennan PC, Mello-Thoms C. A review of lung cancer screening and the role of computer-aided detection. Clin Radiol 2017;72:433–42. [DOI] [PubMed] [Google Scholar]
- [18].Kohler J, Schuler M, Gauler TC, et al. Circulating U2 small nuclear RNA fragments as a diagnostic and prognostic biomarker in lung cancer patients. J Cancer Res Clin Oncol 2016;142:795–805. [DOI] [PubMed] [Google Scholar]
- [19].Jun W, Xia L, Di D, et al. Prediction of malignant and benign of lung tumor using a quantitative radiomic method. Conference proceedings: Annual International Conference of the IEEE Engineering in Medicine and Biology Society. IEEE Eng Med Biol Soc Ann Conference 2016;2016:1272–5. [DOI] [PubMed] [Google Scholar]
- [20].Kobayashi M, Saito A, Tanaka Y, et al. MicroRNA expression profiling in canine prostate cancer. J Veterinary Med Sci/Jpn Soc Vet Sci 2017;79:719–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Bertoli G, Cava C, Castiglioni I. MicroRNAs: new biomarkers for diagnosis, prognosis, therapy prediction and therapeutic tools for breast cancer. Theranostics 2015;5:1122–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Yu Y, Zuo J, Tan Q, et al. Plasma miR-92a-2 as a biomarker for small cell lung cancer. Cancer Biomark Sect A Dis Mark 2017;18:319–27. [DOI] [PubMed] [Google Scholar]
- [23].Shi GL, Chen Y, Sun Y, et al. Significance of serum MicroRNAs in the auxiliary diagnosis of non-small cell lung cancer. Clin Lab 2017;63:133–40. [DOI] [PubMed] [Google Scholar]
- [24].Zhan B, Lu D, Luo P, et al. Prognostic value of expression of MicroRNAs in non-small cell lung cancer: a systematic review and meta-analysis. Clin Lab 2016;62:2203–11. [DOI] [PubMed] [Google Scholar]
- [25].Harvey H, Piskareva O, Creevey L, et al. Modulation of chemotherapeutic drug resistance in neuroblastoma SK-N-AS cells by the neural apoptosis inhibitory protein and miR-520f. Int J Cancer J Int Cancer 2015;136:1579–88. [DOI] [PubMed] [Google Scholar]
- [26].Du X, Fan W, Chen Y. microRNA-520f inhibits hepatocellular carcinoma cell proliferation and invasion by targeting TM4SF1. Gene 2018;657:30–8. [DOI] [PubMed] [Google Scholar]
- [27].van Kampen JGM, van Hooij O, Jansen CF, et al. miRNA-520f reverses epithelial-to-mesenchymal transition by targeting ADAM9 and TGFBR2. Cancer Res 2017;77:2008–17. [DOI] [PubMed] [Google Scholar]


