Abstract
Background
Osteosarcoma (OSA), the most common primary bone malignancy, is characterized by a wide spectrum of complicated pathologies and frequent distal metastasis and causes death in adolescents and young adults worldwide. Antitumor drug treatment strategies include various cytotoxic chemotherapy drugs, while molecular targeted therapy for OSA is currently less used. The present work revealed the role played by the miR-596/Survivin axis in affecting the sensitivity of OSA cells to anlotinib, a novel molecular targeting agent.
Methods
By virtual screening, we found that miR-596 might target Survivin by using an online tool (miRDB). RNA levels of miR-596 and Survivin in clinical specimens were examined with qPCR. The effect of miR-596 on anlotinib’s antitumor effect was examined with MTT experiments, the subcutaneous tumor model, or the intramuscular tumor model.
Results
Overexpression of miR-596 via lentiviral particles repressed the protein level of Survivin in U2OS cells. Transfection of miR-596 enhanced the antitumor effect of anlotinib on U2OS cells or five cell lines derived from OSA patients.
Conclusion
miR-596 targets Survivin and enhances the antitumor effect of anlotinib on OSA cells.
Keywords: osteosarcoma cell, microRNAs, Survivin, molecular targeting agents, anlotinib
Introduction
Osteosarcoma (OSA), which is considered as the most common bone malignancy, is characterized by complicated pathologies and sometimes distal metastasis.1,2 Patients suffering from localized/primary OSA often have a good (about 60–80%) 5-year survival rate, whereas the 5-year survival rate is decreased to about 15–30% in patients with metastatic or recurrent OSA.3,4 Consequently, chemotherapeutic treatments combined with surgical resection have been widely used to treat OSA, while molecular-targeted therapy for OSA is currently less used. Although the biology and genetics of OSA have gained attention, the clinical outcomes of OSA patients have not yet significantly improved.3 It has been reported that the occurrence and progress of malignant/metastatic OSA are often driven by genetic or pathological alterations.5 Increasing evidences have confirmed that the inhibition of angiogenesis process could decelerate the progress or metastasis of OSA.5 Therefore, angiogenesis inhibitors could be used to treat advanced OSA.5
Anlotinib is an orally available, highly potent multitargeting protein-kinase inhibitor that could block the activation of some receptor tyrosine protein kinase (RTKs), eg, VEGFR2 (vascular endothelial growth factor receptor 2), platelet-derived growth factor receptors α/β (PDGFR α/β), Ret, c-Kit, c-FMS, or discoidin domain receptor 1 (DDR1).6,7 It has been reported that anlotinib revealed an antitumor effect when used in clinical trials in a variety of human solid tumors, for example non–small-cell lung cancer (NSCLC), hepatocarcinoma (HCC), gastric cancer, renal carcinoma (RC), or soft tissue sarcoma.8–10 In 2018, anlotinib was approved by the China Food and Drug Administration (CFDA) for the clinical application of NSCLC treatment.8–10 Therefore, demonstrating the therapeutic effects of anlotinib on OSA cells not only helps to deepen our understanding of anlotinib but also provides more options for the diagnosis and treatment of OSA.
Survivin is a key regulator of cellular survival and injury response. In malignant human cells, Survivin enhances cell survival and decreases apoptosis in response to cellular injury, eg, ion radiation or antitumor agents.11–13 It has been reported that Survivin could be involved in antitumor agents’ resistance.14 Thus, targeting Survivin is a potential approach to more effective antitumor treatment. MicroRNAs (miRNAs), which have emerged as post-transcriptional modulators of target genes, are endogenous small noncoding RNAs that have been found to have critical roles in functioning as tumor suppressors.15–18 In the current study, we demonstrated that Survivin was correlated with the prognosis of overall survival (OS) or progression-free survival (PFS) of OSA patients. We also demonstrated that miR-596 enhanced the antitumor effect of anlotinib by targeting Survivin. Therefore, targeting Survivin by miR-596 is a promising approach to achieve effective molecular targeting therapies in OSA treatment.
Materials and methods
Patients and specimens
The collection of clinical specimens and protocols of this work were approved by the Ethics Committee of Yantaishan Hospital. The collection and usage of clinical specimens were with written informed consent from patients, and all protocols or experiments were conducted in accordance with the Declaration of Helsinki. Total RNA samples, extracted from a cohort including 74 tumor and adjacent nontumor tissues and evaluated by accepted pathological and radiological criteria, were conserved in our laboratory and used in this study. The baseline information of the cohort was shown as Table S1. The prognoses of patients were determined as OS or PFS. The data of TCGA database were also searched following the methods provided by Cao et al (2019) and Fan et al (2019).19,20
Table S1.
The baseline characters of patients included
| Characteristics | n (%) |
|---|---|
| Total number | 74 |
| Gender | |
| Male | 41 (55.4) |
| Female | 33 (44.6) |
| Tumor size (cm) | |
| > 7 | 49 (60.8) |
| ≤ 7 | 25 (39.2) |
| TNM stage | |
| I | 32 (43.2) |
| II | 25 (33.7) |
| III | 17 (51.6) |
| Location | |
| Distal femur | 31 (41.9) |
| Proximal femur | 7 (9.4) |
| Proximal humerus | 13 (17.5) |
| Proximal tibia | 20 (27.0) |
| Others | 3 (4.0) |
| Relapse | |
| Yes | 11 (14.9) |
| No | 63 (85.1) |
| Metastasis location | |
| Lung | 21 (28.3) |
| Others | 4 (5.4) |
| None | 49 (66.2) |
Cell lines and reagents
U2OS, a typical OSA cell line, was purchased from the cell resources center of the Chinese Academy of Medical Sciences (Beijing, China). Five patient-derived cell lines (PDCs) were separated from the clinical specimens obtained from five patients with OSA during surgery as part of normal medical care. The lentivirus particles of pri-miR-596 or Survivin with a mutation of miR-596 targeted sequences located in 3′-UTR were constructed and purchased (Vigene Corporation, Jinan City, Shandong Province, China). The wild-type sequence or sequence with mutated miR-596-binding site of Survivin’s 3ʹUTR (1921–2100) was obtained by chemical synthesis and cloned into pGL4.26 plasmids to construct luciferase reporters by Vigene Corporation. The luciferase containing wild-type sequence of Survivin’s 3ʹUTR with miR-596-binding site was named as Luc, whereas the luciferase containing sequence of Survivin’s 3ʹUTR with mutated miR-596-binding site was named as LucMut. The inhibitor of the miR-596 was purchased from Thermo Fisher Scientific (Waltham, MA, USA). The antitumor agents doxorubicin (Selleck, TX, USA), cisplatin (Selleck), methotrexate (Selleck), or anlotinib (Selleck) were dissolved in DMSO and conserved in an −80°C condition.
Quantitative polymerase chain reaction
Total RNA samples were extracted from clinical specimens, and qPCR experiments were performed, following protocols provided by the manufacturer (Applied Biosystems, Thermo Fisher Scientific, ) and methods described by Ji et al (2017) and Liang et al (2017).3,15 The expression of Survivin was examined by qPCR with the following primers: Survivin (birc5), forward sequence, 5ʹ-CCACTGAGAACGAGCCA GACTT-3ʹ; reverse sequence 5ʹ-GTATTACAGGCGTAAGCCACCG-3ʹ; loading control (gapdh), forward sequence, 5ʹ-GTCTCCTCTGACTTCAACAGCG-3ʹ; reverse sequence, 5ʹ-ACCACCCTGTTGCTGTAGCCAA-3ʹ.
Luciferase experiments
Cells were transfected with control miRNA + Luc, miR-596 + Luc, miR-596 + inhibitor + Luc or miR-596 + LucMut. Then, cells were harvested for luciferase experiments. Luciferase experiments were performed by using a luciferase examination kit (Promega Corporation, Madison, WI, USA) following the instructions provided by the manufacturer and methods described by Ma et al (2016) and Wang et al (2019).21,22 The relative luciferase activation was calculated as: [(luciferase activation of control group/β-galactosidase activation of control group)/(luciferase activation of administration group/β-galactosidase administration of control group)] × 100%. Results were shown as mean ± SD of relative luciferase activation.
Cell culture and survival examination
Cells were cultured in DMEM medium (Invitrogen, Carlsbad, CA, USA), with the addition of 10% FBS at 37°C with a 5% CO2 condition. Cells were seeded into a 96-well plate (Corning, Inc.) and incubated with the indicated concentrations of antitumor agents diluted by sorafenib DMSO solution (Table S1). After 48 hrs of treatment, cells were harvested for MTT experiments, following the methods described by Feng et al (2018).23 The inhibition rates of antitumor agents on cells were calculated, following the methods provided by Feng et al (2018).19 The half-maximal inhibitory concentration values (IC50 values) were calculated based on inhibition rates, following the methods described by Guan et al (2017) and Li et al (2018).24,25
Antibodies and Western blot
Antibodies to Survivin and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were obtained from Abcam (Cambridge, UK). OSA cells transfected with control, miR-596, miR-596 + SurvivinMut, or miR-596 + inhibitor were harvested 48 hrs following transfection, and total protein samples were extracted for Western blot experiments. Western blots were performed following a standard method. In brief, total protein samples extracted from OSA cells were separated using SDS-PAGE. Following electrophoresis, protein samples in SDS-PAGE gels were transferred onto polyvinylidene fluoride (PVDF) membranes and blocked, using 5% BSA diluted in tris-buffered saline with Tween 20 (TBST buffer). Membranes were subsequently incubated with the relevant primary and secondary antibodies before detection by chemiluminescence.
Animal experiments
The Ethics Committee of Yantai Hospital approved the methods of animal experiments in this study. All animal experiments were carried out in accordance with the UK Animals (Scientific Procedures) Act of 1986 and its associated guidelines.26,27 Cells (U2OS or PDC) were used to form PDX (patient-derived tumor xenograft) and were cultured (2×107 cells per inoculation point) and seeded into the subcutaneous position of nude mice (T-cell deficiency mice) aged 4–6 weeks to form subcutaneous tumor tissues. After 5–6 days, the mice received antitumor agents (anlotinib) once per 2 days. After 4–5 weeks of treatment, the mice were harvested, and subcutaneous tumors were collected.
Tumor weights were measured by the precision balance. Tumor volumes were measured by tumor width × tumor width × tumor length/2.28,29 The inhibition rates of anlotinib on OSA cells were calculated based on tumor volumes or tumor weights. The formulas used to calculate inhibition rates are as follows: 1) (control group’s tumor volumes – anlotinib group’s tumor volume)/(control group’s tumor volumes) and 2) (control group’s tumor weight – anlotinib group’s tumor volume)/(control group’s tumor volumes). The IC50 values of anlotinib on OSA cells’ subcutaneous growth were calculated based on the inhibition rates.
Next, cells were mixed with biological-medical gel (Cai-Hong-Yi-Xue-She-Bei, Kunming, China) and injected into muscle tissue located in the legs of nude mice (5×105 cells per site). After 4–6 weeks’ growth, mice were injected with F-FDG,18 received MicroPET, and were examined following the methods described by Xu et al (2013).30 Next, the mice were harvested, and the muscle tissue was collected for H&E staining.31 Images of the results from MicroPET or H&E staining were quantitatively analyzed via Image J software, according to previously detailed methods.32,33 The inhibition rate of each group was calculated following the methods provided by Wei et al (2019).34
Statistical analysis
Statistical analysis was performed by Bonferroni’s correction with or without two-way ANOVA methods using SPSS (Version No. 8.0; IBM Corporation, Armonk, NY, USA). The IC50 values of anlotinib on OSA cells were calculated by Origin Software, version 8.5 (OriginLab, Northampton, MA, USA). A P-value of <0.05 was considered statistically significant between the results from two groups.
Results
The expression of miR-596/Survivin and correlations with clinical parameters in OSA patients
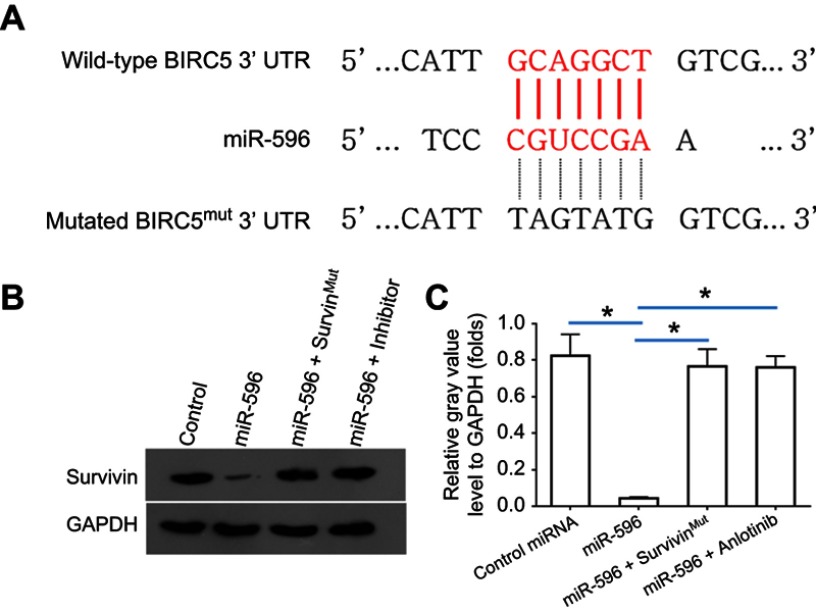
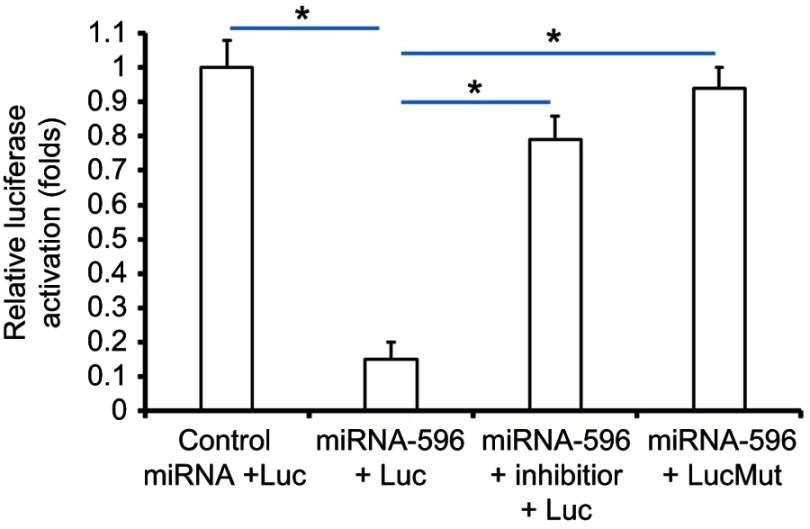
Survivin was a potential target of miR-596; the italicized font in Figure 1A indicates the potential targeted sequence within PXR mRNA’s 3′-UTR (Figure 1). Next, U2OS cells infected with control, miR-596, miR-596 + SurvivinMut, or miR-596 + inhibitor were harvested for Western blot 48 hrs after transfection. Results showed that compared to the control group, miR-596 significantly decreased the expression of Survivin (Figure 1B and C) but not SurvivinMut, which contains a mutated sequence of the miR-596-binding site (Figure 1A–C). Moreover, transfection of miR-596’s inhibitor almost blocked the effect of miR-596 on Survivin’s expression. To further examine the specificity of miR-596 on Survivin’s expression, luciferase reporters were used. As shown in Supplemental Figure 1, miR-596 inhibited the activation of luciferase reporters containing 3`UTR sequences, including the miR-596-targeting site. Transcription of the miR-596 inhibitor or mutated vectors blocked the effect of miR-596 on luciferase reporters’ activation (Figure S1).
Figure 1.
miR-596 targets Survivin’s 3′-UTR. (A) The bold, italicized type image indicates the potential miR-596-binding sites or mutated sites located in Survivin’s 3′-UTR. (B) U2OS cells, which were transfected with control (control miRNA), miR-596, miR-596 + SurvivinMut, or miR-596c + its inhibitor, were harvested for Western blot experiments. The protein level of Survivin or GAPDH in U2OS cells was examined by their antibodies, and GAPDH was chosen as the loading control. Results were shown as images from Western blot (B) or quantitative analysis results (C). *P<0.05.
Abbreviations: miR, microRNA; Mut, mutation.
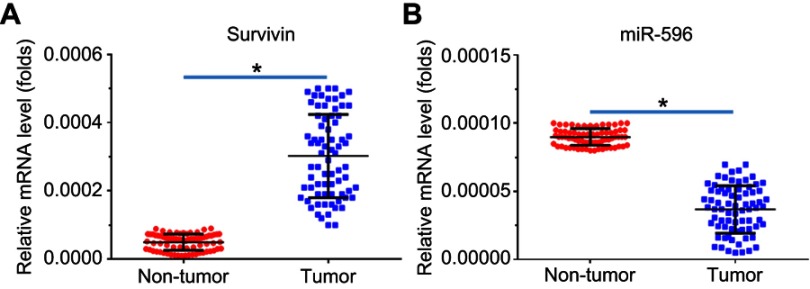
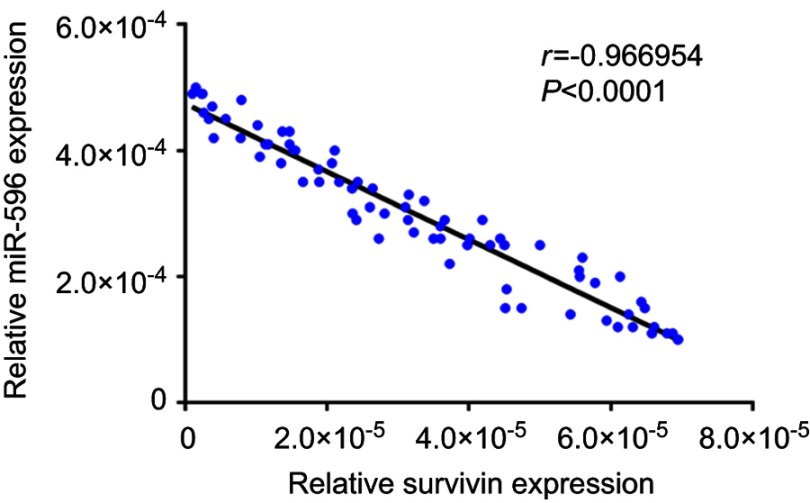
Next, the expression of Survivin or miR-596 in clinical specimens was examined. As shown in Figure 2A and B, the expression of Survivin is much higher in OSA specimens than in paired nontumor specimens, whereas the expression of miR-596 is much lower in OSA specimens than in paired nontumor specimens. To further investigate the clinical significance of miR-596/Survivin, the relationship between Survivin or miR-596 levels and prognoses of patients was examined. As shown in Figure 3 and Tables 1 and 2, Kaplan–Meier survival analysis indicated that patients with high levels of Survivin had a poorer OS and PFS than those with low Survivin levels, whereas patients with high levels of miR-596 showed better OS and PFS than those with low miR-596 levels. Moreover, results shown in Figure 4 further confirm that the expression of Survivin is negatively correlated with miR-596 in OSA specimens. Therefore, miR-596 could repress the expression of Survivin by targeting Survivin mRNA’s 3′-UTR directly.
Figure 2.
Expression of Survivin and miR-596 in clinical specimens. RNA levels of Survivin (A) or miR-596 were examined in 74 paired nontumor/tumor clinical tissues. *P<0.05 versus nontumor group with tumor group.
Abbreviation: miR, microRNA.
Figure 3.
Expression of Survivin/miR-596 and correlation with patients’ prognosis (A–B). Kaplan–Meier survival curves and log-rank tests were used to compare the OS (A) or PFS (B) of low-Survivin group and high-Survivin group patients (C-D). Kaplan–Meier survival curves and log-rank tests were used to compare the OS (C) or PFS (D) of low-miR-596 group and high-miR-596 group patients. (E) The representative image of clinical specimens by qPCR and DNA electrophoresis. *P<0.05.
Abbreviations: miR, microRNA; OS, overall survival; PFS, progression-free survival.
Table 1.
miR-596 expression and clinical outcome of osteosarcoma patients
| miR-596 mRNA expression | P | ||
|---|---|---|---|
| High (n=37) | Low (n=37) | ||
| PFS | 66.8 | 50.3 | 0.041 |
| 56.8–76.8 (M) | 41.2–59.4 (M) | ||
| OS | 70.2 | 52.7 | 0.036 |
| 60.2–80.2 (M) | 42.8–62.0 (M) | ||
Abbreviations: PFS, progress free survival; OS, overall survival; M, months.
Table 2.
Survivin expression and clinical outcome of osteosarcoma patients
| Survivin mRNA expression | P | ||
|---|---|---|---|
| High (n=37) | Low (n=37) | ||
| PFS | 50.5 | 65.3 | 0.035 |
| 41.2–59.7 (M) | 56.0–74.6 (M) | ||
| OS | 51.1 | 70.3 | 0.022 |
| 41.2–61.0 (M) | 60.7–79.9 (M) | ||
Abbreviations: PFS, progress free survival; OS, overall survival; M, months.
Figure 4.
Correlation of RNA levels of miR-596 and Survivin in clinical tumor specimens from osteosarcoma patients. The relationship between the RNA levels of miR-596 and Survivin was assessed by Spearman’s rank correlation analysis. The symbols represent individual samples (a total of 74 specimens).
Abbreviation: miR, microRNA.
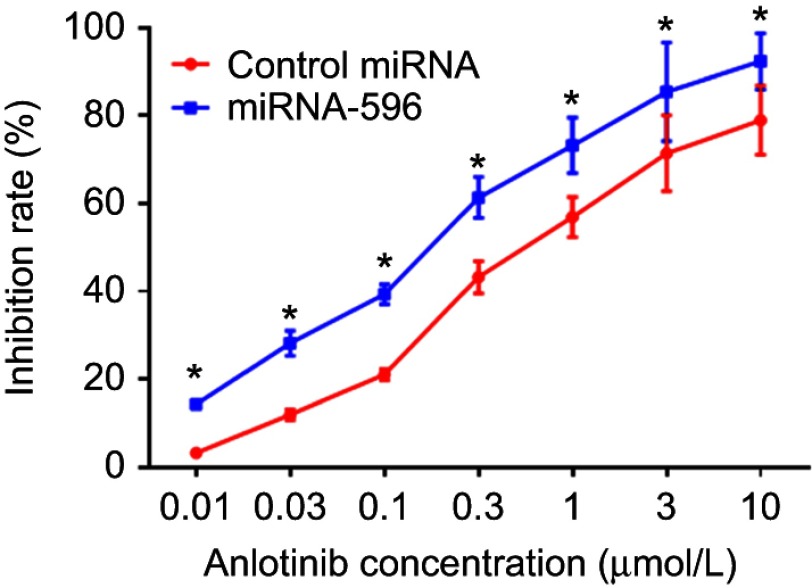
miR-596 enhances the sensitivity of OSA cells to anlotinib
MTT assays were performed to determine whether miR-596 enhances the in vitro antitumor effect of anlotinib on OSA cells. As shown in Figure 5 and Table 3, anlotinib inhibited the survival of U2OS cells. Transfection of miR-596 enhanced the sensitivity of OSA cells to anlotinib, and the IC50 values of anlotinib were, respectively, decreased (Table 3). The effects of anlotinib treatment on OSA cell lines from the five patients (PDCs) are also shown in Table 3. Anlotinib inhibited the survival of the OSA cells (PDCs) in a dose-dependent manner, and the IC50 values of anlotinib on PDCs’ survival were, respectively, decreased.
Figure 5.
miR-596 enhances the antitumor effect of anlotinib on cultured U2OS cells. U2OS cells transfected with control miRNA or miR-596 were treated with the indicated concentration of anlotinib. Cells were harvested for MTT experiments, and the inhibition rates of anlotinib were calculated by OD490 nm. *P<0.05.
Abbreviation: miR, microRNA.
Table 3.
The IC50 values of anlotinib on osteosarcoma cell lines
| Cell lines | Control | miR-596 | miR-596 + SurvivinMut | miR-596 + inhibitor |
|---|---|---|---|---|
| IC50 values (μmol/L) of anlotinib | ||||
| U2OS | 0.66±0.11 | 0.21±0.02 | 0.58±0.15 | 0.49±0.08 |
| PDC-1 | 0.75±0.22 | 0.15±0.01 | 0.70±0.20 | 0.55±0.10 |
| PDC-2 | 0.80±0.25 | 0.28±0.05 | 0.78±0.23 | 0.65±0.13 |
| PDC-3 | 0.65±0.30 | 0.10±0.03 | 0.67±0.33 | 0.56±0.15 |
| PDC-4 | 0.71±0.33 | 0.20±0.04 | 0.53±0.25 | 0.60±0.07 |
| PDC-5 | 0.75±0.10 | 0.35±0.02 | 0.72±0.24 | 0.63±0.14 |
Abbreviation: PDC, patient-derived cells.
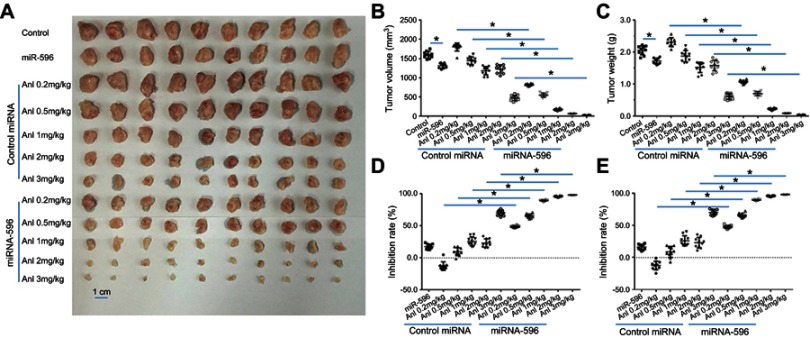
Next, the effect of miR-596 and anlotinib coadministration on OSA cells was examined in a subcutaneous model. As shown in Figure 6 and Table 4, anlotinib inhibited the subcutaneous growth of U2OS cells in nude mice. Transfection of miR-596 enhanced the antitumor effect of anlotinib on OSA cells’ subcutaneous growth, and the IC50 values of anlotinib on OSA cells’ subcutaneous growth were, respectively, decreased. The effect of anlotinib treatment on the PDX model, formed by PDC cell lines from the five patients, is shown in Table 4. Anlotinib inhibited the survival of PDC cells in a dose-dependent manner. To further examine the antitumor effect of anlotinib on OSA cells, U2OS cells were injected into the muscle tissues of nude mice. As shown in Figures 7 and 8, U2OS cells can form tumor lesions in the muscle tissues of nude mice. Anlotinib inhibited the growth of U2OS cells in nude mice muscle tissues, and miR-596 enhanced the antitumor effects of anlotinib on U2OS cells.
Figure 6.
miR-596 enhances the antitumor effect of anlotinib on U2OS cell subcutaneous growth. U2OS cells transfected with control or miR-596 were injected into nude mice to form subcutaneous tumors. Four days to 5 days after injection, mice received orally administrated concentrations of anlotinib once per 2 days. After 10 treatments (about 3 weeks), mice were harvested, and tumor tissues were harvested. Results are shown as images of subcutaneous tumors (A), tumor volumes (B), tumor weights (C), inhibition rates of anlotinib calculated by tumor volumes (D), or inhibition rates of anlotinib calculated by tumor weights (E). *P<0.05.
Abbreviations: miR, microRNA; Mut, mutation; Anl, anlotinib.
Table 4.
The IC50 values of anlotinib on osteosarcoma cell lines in nude mice
| Cell lines | Control | miR-596 |
|---|---|---|
| IC50 values (mg/kg) of anlotinib | ||
| U2OS | 2.50±0.75 | 0.33±0.11 |
| PDX-1 | 2.88±0.55 | 0.50±0.05 |
| PDX-2 | 1.97±0.30 | 0.44±0.10 |
| PDX-3 | 2.32±0.51 | 0.76±0.22 |
| PDX-4 | 1.62±0.29 | 0.35±0.10 |
| PDX-5 | 1.98±0.46 | 0.55±0.25 |
Abbreviation: PDX, patient-derived tumor xenograft.
Figure 7.
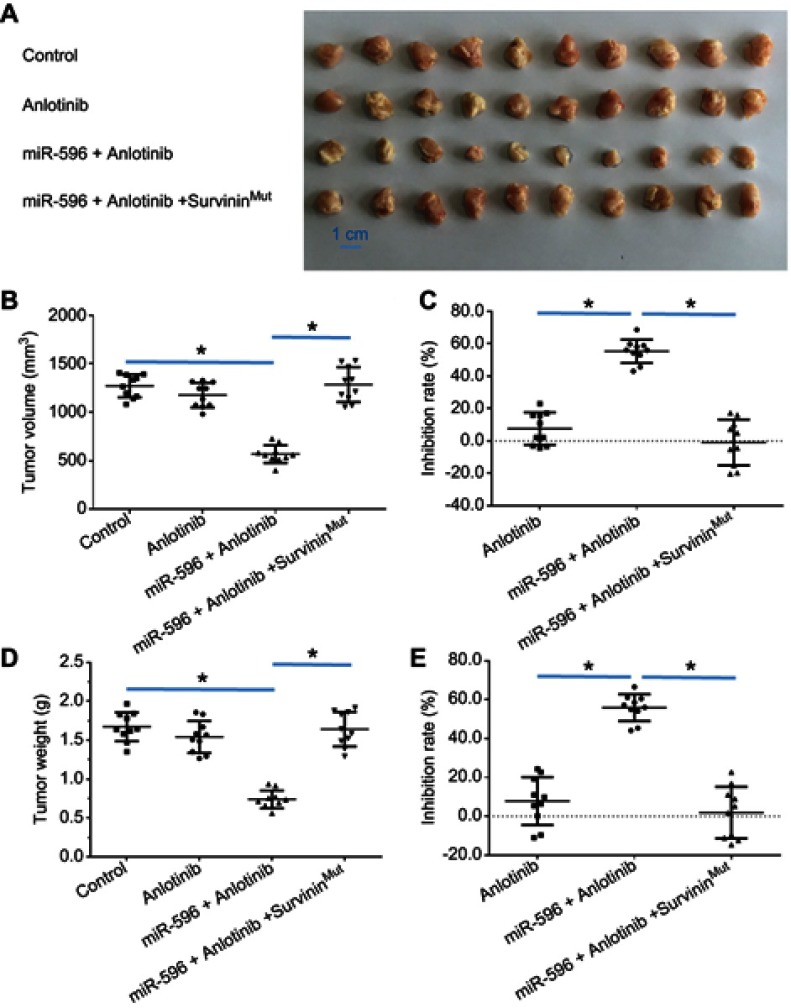
miR-596 enhanced the antitumor activation of anlotinib on U2OS cell subcutaneous growth by targeting Survivin. U2OS cells transfected with control miRNA, miR-596, or miR-596 + SurvivinMut were seeded into nude mice to form subcutaneous tumors. The mice received a 0.5 mg/kg dose of anlotinib (a dose without significant antitumor effect) by oral administration. The results are shown as (A) images of subcutaneous tumors, (B) tumor volumes, and (C) or tumor weight (D) tumor weights. The inhibition rate calculated by tumor volume (E) is shown. *P<0.05.
Abbreviation: miR, microRNA.
Figure 8.
miR-596 enhances the antitumor effect of anlotinib on U2OS cell growth in muscle tissues of mice via microPET screening. U2OS cells transfected with control, miR-596, or miR-596 + SurvivinMut were injected into muscle tissues to form tumor lesions. Four days to 5 days after intramuscular injection of U2OS cells, mice received oral administration of a 1mg/kg dose of anlotinib once per 2 days. After 10 treatments with anlotinib, mice received microPET screening. Results were shown as microPET images: (A) intensity of 18F-FDG of microPET images and (B) radioactivation of tumor tissues to blood. (C) The radio-activation of tumor tissues to blood was shown as quantitative results. *P<0.05.
Abbreviations: miR, microRNA; PET, positron emission computed tomography; 18F-FDG, 18F-Fludeoxyglucose.
Moreover, the specificity of miR-596 on anlotinib-inhibiting OSA cell survival or growth was examined. As shown in Figures 7–9, and Table 1, SurvivinMut -expressed cells and tumors could not be affected by miR-596. Therefore, miR-596 enhances the sensitivity of OSA cells to anlotinib.
Figure 9.
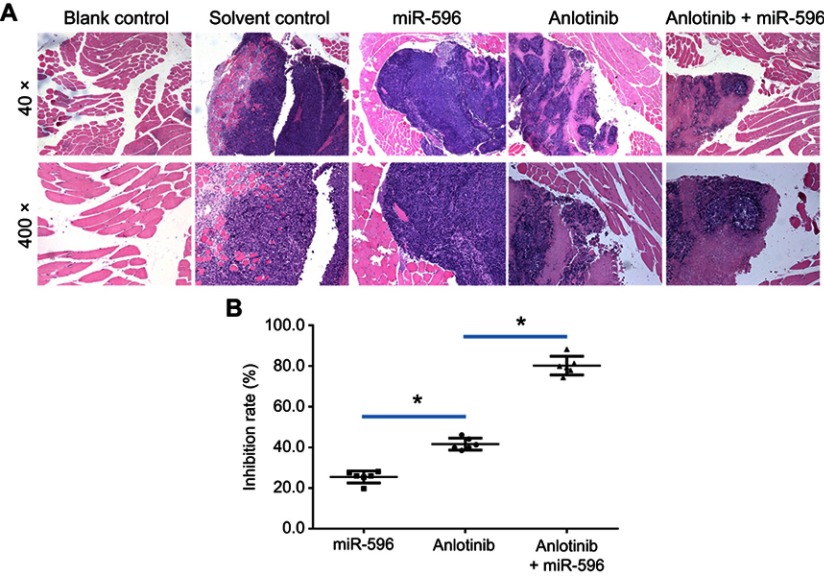
miR-596 enhances the antitumor effect of anlotinib on U2OS cell growth in muscle tissues. U2OS cells transfected with control, miR-596, or miR-596 + SurvivinMut were injected into muscle tissues to form tumor lesions. Four days to 5 days after intramuscular injection of U2OS cells, mice received oral administration of 1mg/kg dose of anlotinib once per 2 days. After 10 treatments, mice were harvested, and muscle tissues were harvested for H&E staining. Results were shown as (A) images from H&E staining and (B) inhibition rates of anlotinib on intramuscular growth of U2OS cells. *P<0.05.
Abbreviation: miR, microRNA.
miR-596 enhances the sensitivity of OSA cells to cytotoxic chemotherapies
To further determine whether miR-596 enhances the in vitro antitumor effect of other antitumor agents, MTT assays were performed. As shown below, the cytotoxic chemotherapies doxorubicin (Table 5), cisplatin (Table 6), or methotrexate (Table 7) inhibited the survival of U2OS cells in a dose-dependent manner. Transfection of miR-596 enhanced the sensitivity of OSA to other antitumor agents, and the IC50 values of anlotinib were, respectively, decreased. Therefore, miR-596 enhances the sensitivity of OSA cells to cytotoxic chemotherapies. The effects of anlotinib treatment on the OSA cell lines from the five patients are shown in Table 2. Anlotinib inhibited the survival of OSA cells in a dose-dependent manner, and miR-596 enhanced the antitumor effect of anlotinib on OSA cells.
Table 5.
The IC50 values of Doxorubicin on osteosarcoma cell lines
| Cell lines | Control | miR-596 | miR-596 + SurvivinMut | miR-596 + inhibitor |
|---|---|---|---|---|
| IC50 values (μmol/L) of doxorubicin | ||||
| U2OS | 0.25±0.04 | 0.05±0.01 | 0.19±0.01 | 0.22±0.04 |
| PDC-1 | 0.40±0.10 | 0.06±0.02 | 0.30±0.05 | 0.25±0.03 |
| PDC-2 | 0.35±0.06 | 0.10±0.02 | 0.33±0.10 | 0.26±0.05 |
| PDC-3 | 0.41±0.12 | 0.21±0.05 | 0.36±0.08 | 0.35±0.02 |
| PDC-4 | 0.50±0.03 | 0.08±0.01 | 0.48±0.05 | 0.40±0.03 |
| PDC-5 | 0.15±0.00 | 0.01±0.00 | 0.10±0.01 | 0.12±0.01 |
Abbreviation: PDCs, patient-devived cells.
Table 6.
The IC50 values of cisplatin on osteosarcoma cell lines
| Cell lines | Control | miR-596 | miR-596 + SurvivinMut | miR-596 + inhibitor |
|---|---|---|---|---|
| IC50 values (μmol/L) of cisplatin | ||||
| U2OS | 0.44±0.14 | 0.11±0.03 | 0.35±0.04 | 0.29±0.01 |
| PDC-1 | 0.60±0.09 | 0.15±0.01 | 0.50±0.10 | 0.46±0.16 |
| PDC-2 | 0.46±0.04 | 0.10±0.02 | 0.43±0.05 | 0.36±0.04 |
| PDC-3 | 0.38±0.10 | 0.10±0.03 | 0.35±0.07 | 0.56±0.15 |
| PDC-4 | 0.59±0.09 | 0.20±0.04 | 0.53±0.25 | 0.60±0.07 |
| PDC-5 | 0.36±0.04 | 0.35±0.02 | 0.33±0.24 | 0.28±0.07 |
Abbreviation: PDCs, patient-derived cells.
Table 7.
The IC50 values of methotrexate on osteosarcoma cell lines
| Cell lines | Control | miR-596 | miR-596 + SurvivinMut | miR-596 + inhibitor |
|---|---|---|---|---|
| IC50 values (μmol/L) methotrexate | ||||
| U2OS | 0.58±0.11 | 0.12±0.03 | 0.54±0.06 | 0.51±0.15 |
| PDC-1 | 0.45±0.06 | 0.09±0.02 | 0.35±0.13 | 0.40±0.04 |
| PDC-2 | 0.32±0.22 | 0.07±0.01 | 0.25±0.10 | 0.30±0.06 |
| PDC-3 | 0.31±0.02 | 0.16±0.05 | 0.40±0.20 | 0.56±0.15 |
| PDC-4 | 0.45±0.09 | 0.25±0.04 | 0.53±0.25 | 0.35±0.16 |
| PDC-5 | 0.32±0.08 | 0.08±0.01 | 0.25±0.06 | 0.22±0.05 |
Abbreviation: PDCs, patient-derived cells.
Discussion
Currently, one of the main strategies for molecular targeted therapy of malignant tumors is the oral administration of small-molecule multitarget protein kinase inhibitors.35,36 These drugs can directly inhibit the RTK and mitogen-activated protein kinase as well as other signaling pathways to inhibit malignant tumors delaying cell proliferation or decelerating tumor tissue growth. Molecular targeting agents also can inhibit tumor cell metastasis, invasion, and tumor angiogenesis by inhibiting RTK, such as in VEGFR.37–41 Anlotinib is a new molecular targeted drug with intellectual property rights in China and has strong inhibitory effects on VEGFR and c-Kit.8–10 However, these molecularly targeted drugs also face many challenges in clinical use: molecular targeted drugs are expensive and drug resistant.42 At the same time, the toxic side effects of these drugs cannot be ignored.43 Therefore, how to achieve more safe and effective molecular targeted therapy and to achieve lower doses of drugs that achieve antitumor effect is an urgent problem to be solved. The results of this study indicate that the use of miR-596 can significantly increase the killing effect of anlotinib on OSA cells, using a lower dose to achieve an equal, or even greater, antitumor effect. This study uses not only existing cell lines but also patient-derived tumor cells, which helps to increase our understanding of anlotinib and also provides an experimental basis for establishing safer, more effective molecular-targeted therapy.
Survivin, also called baculoviral inhibitor of apoptosis repeat-containing (BIRC5), API4, EPR-1, or baculoviral IAP repeat-containing 5, is a protein that, in humans, is encoded by the BIRC5 gene.44–46 Survivin is currently found to be the most potent cell survival antiapoptotic regulator.47,48 Antitumor drugs used in molecular targeted therapy can damage and kill tumor cells, or damage and cause death due to injury factors.49,50 Survivin can act as a major regulator of the tumor cell damage response and can play a protective role for cells.51,52 This study found that the expression of Survivin in adjacent tissues was significantly lower than that in cancer tissues, while the trend of miR-596 was exactly the opposite. At the same time, the use of miR-596 to downregulate the expression of Survivin can upregulate the sensitivity of OSA cells to anlotinib. The use of miR-596 to downregulate the expression of Survivin also upregulates the sensitivity of OSA cells to some cytotoxic chemotherapeutic drugs. Moreover, the BCL-xL, BAX, Casp 9, Casp 3, and p21 are the negative regulators of cancer cell proliferation or cell cycle. It is valuable to examine the expression of these factors and develop the therapeutic strategies based on these factors.53 Some other pathways, such as Notch pathway, can function as regulating hub that modulates the expression of Survivin, CL-xL, BAX, Casp 9, Casp 3, and p21.54–57 Therefore, it is valuable to examine the effect of Notch pathway on anlotinib.
Overexpression of miRNA via virus vectors has been considered as a promising approach to inhibit the proliferation or metastasis of cancer cells.58–64 In the present work, miR-596 was identified in clinical specimens and was prepared as lentivirus particles. Besides miR-596, microRNA-330-5p, microRNA-218, miR-542-3p, or miR-494 could also target Survivin and inhibit the proliferation or metastasis in some other cancers, such as gastric cancer.65–68 It is valuable to examine the expression of the miRs in clinical tissues. TCGA is a common database; however, there are no data about Survivin-OSA in TCGA. Therefore, we examined the expression level of miR-596 and Survivin in clinical specimens.
Furthermore, subcutaneous tumors are a common research model for the antitumor effects of drugs. This study established a subcutaneous tumor research model to examine the role of anlotinib. However, it is difficult to inspect the invasion and destruction of OSA cells on muscle and other tissues through simple subcutaneous tumors. For this reason, OSA cells were directly injected into the muscle tissue of nude mice for PET imaging in small animals. The growth of OSA cells in the muscle tissues was examined by the pathological analysis of tissues. The quantitative analysis of the images of pathological analysis finally determined the damage of OSA cells in muscle tissues.
Conclusion
miR-596 targets Survivin and enhances the antitumor effect of anlotinib on OSA cells.
Supplementary materials
Acknowledgment
We thank Prof. Xiaojie Xu in Department of Medical Molecular Biology, Beijing Institute of Biotechnology, Palo Alto, Beijing, China, and Dr. Fan Feng in Research Center for Clinical and Translational Medicine, the Fifth Medical Center of the General Hospital of the Chinese People’s Liberation Army, Beijing, China, for their help and advice.
Abbreviations
OSA, osteosarcoma; miR, microRNAs; VEGFR2, vascular endothelial growth factor receptor 2; platelet-derived growth factor receptors α/β, PDGFR α/β; discoidin domain receptor 1, DDR1; non–small-cell lung cancer (NSCLC); HCC, hepatocarcinoma; OS, survival; PFS, progression-free survival; RTK, receptor tyrosine protein kinase; MAPK, mitogen-activated protein kinase; BIRC5, baculoviral inhibitor of apoptosis repeat-containing.
Author contributions
All authors made substantial contributions to the design and conception, acquisition, analysis, or interpretation of data. All authors took part in either drafting or revising the manuscript. All authors also gave final approval of the version to be published and agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Edelmann MN, Daryani VM, Bishop MW, et al. Neurocognitive and patient-reported outcomes in adult survivors of childhood osteosarcoma. JAMA Oncol. 2016;2:201–208. doi: 10.1001/jamaoncol.2015.4398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Whelan JS, Davis LE. Osteosarcoma, chondrosarcoma, and chordoma. J Clin Oncol. 2018;36:188–193. doi: 10.1200/JCO.2017.75.1743 Epub 2017 Dec 8. [DOI] [PubMed] [Google Scholar]
- 3.Ji Q, Xu X, Li L, et al. miR-216a inhibits osteosarcoma cell proliferation, invasion and metastasis by targeting CDK14. Cell Death Dis. 2017;8:e3103. doi: 10.1038/cddis.2017.499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lagmay JP, Krailo MD, Dang H, et al. Outcome of patients with recurrent osteosarcoma enrolled in seven Phase II trials through children’s cancer group, pediatric oncology group, and children’s oncology group: learning from the past to move forward. J Clin Oncol. 2016;34:3031–3038. doi: 10.1200/JCO.2015.65.5381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang G, Sun M, Jiang Y, et al. Anlotinib, a novel small molecular tyrosine kinase inhibitor, suppresses growth and metastasis via dual blockade of VEGFR2 and MET in osteosarcoma. Int J Cancer. 2019;145:979–993. doi: 10.1002/ijc.32180 [DOI] [PubMed] [Google Scholar]
- 6.Si X, Zhang L, Wang H, et al. Management of anlotinib-related adverse events in patients with advanced non-small cell lung cancer: experiences in ALTER-0303. Thorac Cancer. 2019;10:551–556. doi: 10.1111/1759-7714.12977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen HM, Feng G. Use of anlotinib in intra-abdominal desmoplastic small round cell tumors: a case report and literature review. Onco Targets Ther. 2018;12:57–61. doi: 10.2147/OTT.S190333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen XZ. Anlotinib for refractory advanced non-small cell lung cancer in China. JAMA Oncol. 2019;5:116–117. doi: 10.1001/jamaoncol.2018.5526 [DOI] [PubMed] [Google Scholar]
- 9.Shen G, Zheng F, Ren D, et al. Anlotinib: a novel multi-targeting tyrosine kinase inhibitor in clinical development. J Hematol Oncol. 2018;11:120. doi: 10.1186/s13045-018-0664-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu W, Jiang Q, Zhou Y. Advances of systemic treatment for adult soft-tissue sarcoma. Chin Clin Oncol. 2018;7:42. doi: 10.21037/cco.2018.08.02 [DOI] [PubMed] [Google Scholar]
- 11.Wirries A, Jabari S, Jansen EP, et al. Panobinostat mediated cell death: a novel therapeutic approach for osteosarcoma. Oncotarget. 2018;9:32997–33010. doi: 10.18632/oncotarget.26038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soleimanpour E, Babaei E. Survivin as a potential target for cancer therapy. Asian Pac J Cancer Prev. 2015;16:6187–6191. doi: 10.7314/apjcp.2015.16.15.6187 [DOI] [PubMed] [Google Scholar]
- 13.Bashash D, Safaroghli-Azar A, Dadashi M, Safa M, Momeny M, Ghaffari SH. Anti-tumor activity of PI3K-δ inhibitor in hematologic malignant cells: shedding new light on resistance to idelalisib. Int J Biochem Cell Biol. 2017;85:149–158. doi: 10.1016/j.biocel.2017.02.007 [DOI] [PubMed] [Google Scholar]
- 14.Lyu H, Wang S, Huang J, Wang B, He Z, Liu B. Survivin-targeting miR-542-3p overcomes HER3 signaling-induced chemoresistance and enhances the antitumor activity of paclitaxel against HER2-overexpressing breast cancer. Cancer Lett. 2018;420:97–108. doi: 10.1016/j.canlet.2018.01.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liang Y, Xu X, Wang T, et al. The EGFR/miR-338-3p/EYA2 axis controls breast tumor growth and lung metastasis. Cell Death Dis. 2017;8:e2928. doi: 10.1038/cddis.2017.325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li L, Kang L, Zhao W, et al. miR-30a-5p suppresses breast tumor growth and metastasis through inhibition of LDHA-mediated Warburg effect. Cancer Lett. 2017;400:89–98. doi: 10.1016/j.canlet.2017.04.034 [DOI] [PubMed] [Google Scholar]
- 17.Li J, Zhao J, Wang H, et al. MicroRNA-140-3p enhances the sensitivity of hepatocellular carcinoma cells to sorafenib by targeting pregnenolone X receptor. Onco Targets Ther. 2018;11:5885–5894. doi: 10.2147/OTT.S179509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khan NM, Haqqi TM. Epigenetics in osteoarthritis: potential of HDAC inhibitors as therapeutics. Pharmacol Res. 2018;128:73–79. doi: 10.1016/j.phrs.2017.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fan Z, Duan J, Wang L, et al. PTK2 promotes cancer stem cell traits in hepatocellular carcinoma by activating Wnt/β-catenin signaling. Cancer Lett. 2019;450:132–143. doi: 10.1016/j.canlet.2019.02.040 [DOI] [PubMed] [Google Scholar]
- 20.Cao Y, Trillo-Tinoco J, Sierra RA, et al. ER stress-induced mediator C/EBP homologous protein thwarts effector T cell activity in tumors through T-bet repression. Nat Commun. 2019;10:1280. doi: 10.1038/s41467-019-09263-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang L, Zhao L, Jia X, et al. Aminophenols increase proliferation of thyroid tumor cells by inducing the transcription factor activity of estrogen receptor α. Biomed Pharmacother. 2019;109:621–628. doi: 10.1016/j.biopha.2018.10.168 [DOI] [PubMed] [Google Scholar]
- 22.Ma H, Yao Y, Wang C, et al. Transcription factor activity of estrogen receptor α activation upon nonylphenol or bisphenol A treatment enhances the in vitro proliferation, invasion, and migration of neuroblastoma cells. Onco Targets Ther. 2016;9:3451–3463. doi: 10.2147/OTT.S105745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feng F, Jiang Q, Cao S, et al. Pregnane X receptor mediates sorafenib resistance in advanced hepatocellular carcinoma. Biochim Biophys Acta Gen Subj. 2018;1862:1017–1030. doi: 10.1016/j.bbagen.2018.01.011 [DOI] [PubMed] [Google Scholar]
- 24.Guan F, Ding R, Zhang Q, et al. WX-132-18B, a novel microtubule inhibitor, exhibits promising anti-tumor effects. Oncotarget. 2017;8:71782–71796. doi: 10.18632/oncotarget.17710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li F, Wei A, Bu L, et al. Procaspase-3-activating compound 1 stabilizes hypoxia-inducible factor 1α and induces DNA damage by sequestering ferrous iron. Cell Death Dis. 2018;9:1025. doi: 10.1038/s41419-018-1038-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y, Tang Z. A novel long-sustaining system of apatinib for long-term inhibition of the proliferation of hepatocellular carcinoma cells. Onco Targets Ther. 2018;11:8529–8541. doi: 10.2147/OTT.S188209 eCollection 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu M, Zhao G, Zhuang X, et al. Triclosan treatment decreased the antitumor effect of sorafenib on hepatocellular carcinoma cells. Onco Targets Ther. 2018;11:2945–2954. doi: 10.2147/OTT.S165436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y, Li D, Jiang Q, et al. Novel ADAM-17 inhibitor ZLDI-8 enhances the in vitro and in vivo chemotherapeutic effects of Sorafenib on hepatocellular carcinoma cells. Cell Death Dis. 2018;9:743. doi: 10.1038/s41419-018-0804-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li L, Liang Y, Kang L, et al. Transcriptional regulation of the warburg effect in cancer by SIX1. Cancer Cell. 2018;33:368–385.e7. doi: 10.1016/j.ccell.2018.01.010 [DOI] [PubMed] [Google Scholar]
- 30.Xu X, Fan Z, Kang L, et al. Hepatitis B virus X protein represses miRNA-148a to enhance tumorigenesis. J Clin Invest. 2013;123:630–645. doi: 10.1172/JCI64265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shao Z, Li Y, Dai W, et al. ETS-1 induces Sorafenib-resistance in hepatocellular carcinoma cells via regulating transcription factor activity of PXR. Pharmacol Res. 2018;135:188–200. doi: 10.1016/j.phrs.2018.08.003 [DOI] [PubMed] [Google Scholar]
- 32.Meng D, Meng M, Luo A, et al. Effects of VEGFR1 + hematopoietic progenitor cells on pre-metastatic niche formation and in vivo metastasis of breast cancer cells. J Cancer Res Clin Oncol. 2019;145:411–427. doi: 10.1007/s00432-018-2802-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meng D, Lei M, Han Y, et al. MicroRNA-645 targets urokinase plasminogen activator and decreases the invasive growth of MDA-MB-231 triple-negative breast cancer cells. Onco Targets Ther. 2018;11:7733–7743. doi: 10.2147/OTT.S187221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wei L, Lun Y, Zhou X, et al. Novel urokinase-plasminogen activator inhibitor SPINK13 inhibits growth and metastasis of hepatocellular carcinoma in vivo. Pharmacol Res. 2019;143:73–85. doi: 10.1016/j.phrs.2019.03.009 [DOI] [PubMed] [Google Scholar]
- 35.Lacal PM, Graziani G. Therapeutic implication of vascular endothelial growth factor receptor-1 (VEGFR-1) targeting in cancer cells and tumor microenvironment by competitive and non-competitive inhibitors. Pharmacol Res. 2018;136:97–107. doi: 10.1016/j.phrs.2018.08.023 [DOI] [PubMed] [Google Scholar]
- 36.Rajagopal C, Lankadasari MB, Aranjani JM, Harikumar KB. Targeting oncogenic transcription factors by polyphenols: a novel approach for cancer therapy. Pharmacol Res. 2018;130:273–291. doi: 10.1016/j.phrs.2017.12.034 [DOI] [PubMed] [Google Scholar]
- 37.Jr RR, Sadeghi-Nejad A. Role of RET protein-tyrosine kinase inhibitors in the treatment RET-driven thyroid and lung cancers. Pharmacol Res. 2018;128:1–17. doi: 10.1016/j.phrs.2017.12.021 [DOI] [PubMed] [Google Scholar]
- 38.Tewari D, Nabavi SF, Nabavi SM, et al. Targeting activator protein 1 signaling pathway by bioactive natural agents: possible therapeutic strategy for cancer prevention and intervention. Pharmacol Res. 2018;128:366–375. doi: 10.1016/j.phrs.2017.09.014 [DOI] [PubMed] [Google Scholar]
- 39.Roskoski R Jr. Properties of FDA-approved small molecule protein kinase inhibitors. Pharmacol Res. 2019;144:19–50. doi: 10.1016/j.phrs.2019.03.006 [DOI] [PubMed] [Google Scholar]
- 40.Roskoski R Jr. Targeting oncogenic Raf protein-serine/threonine kinases in human cancers. Pharmacol Res. 2018;135:239–258. doi: 10.1016/j.phrs.2018.08.013 [DOI] [PubMed] [Google Scholar]
- 41.Roskoski R Jr. The role of small molecule platelet-derived growth factor receptor (PDGFR) inhibitors in the treatment of neoplastic disorders. Pharmacol Res. 2018;129:65–83. doi: 10.1016/j.phrs.2018.01.021 [DOI] [PubMed] [Google Scholar]
- 42.Zhu YJ, Zheng B, Wang HY, Chen L. New knowledge of the mechanisms of sorafenib resistance in liver cancer. Acta Pharmacol Sin. 2017;38:614–622. doi: 10.1038/aps.2017.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Feng F, Jiang Q, Jia H, et al. Which is the best combination of TACE and Sorafenib for advanced hepatocellular carcinoma treatment? A systematic review and network meta-analysis. Pharmacol Res. 2018;135:89–101. doi: 10.1016/j.phrs.2018.06.021 [DOI] [PubMed] [Google Scholar]
- 44.Frassanito MA, Saltarella I, Vinella A, et al. Survivin overexpression in head and neck squamous cell carcinomas as a new therapeutic target (Review). Oncol Rep. 2019;41:2615–2624. doi: 10.3892/or.2019.7082 [DOI] [PubMed] [Google Scholar]
- 45.Dinesh P, Rasool M. uPA/uPAR signaling in rheumatoid arthritis: Shedding light on its mechanism of action. Pharmacol Res. 2018;134:31–39. doi: 10.1016/j.phrs.2018.05.016 [DOI] [PubMed] [Google Scholar]
- 46.Shojaei F, Yazdani-Nafchi F, Banitalebi-Dehkordi M, Chehelgerdi M, Khorramian-Ghahfarokhi M. Trace of survivin in cancer. Eur J Cancer Prev. 2019;28:365–372. doi: 10.1097/CEJ.0000000000000453 [DOI] [PubMed] [Google Scholar]
- 47.Li D, Hu C, Li H. Survivin as a novel target protein for reducing the proliferation of cancer cells. Biomed Rep. 2018;8:399–406. doi: 10.3892/br.2018.1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Khan Z, Khan AA, Yadav H, Prasad GBKS, Bisen PS. Survivin, a molecular target for therapeutic interventions in squamous cell carcinoma. Cell Mol Biol Lett. 2017;22:8. doi: 10.1186/s11658-017-0038-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jia H, Yang Q, Wang T, et al. Rhamnetin induces sensitization of hepatocellular carcinoma cells to a small molecular kinase inhibitor or chemotherapeutic agents. Biochim Biophys Acta. 2016;1860:1417–1430. doi: 10.1016/j.bbagen.2016.04.007 [DOI] [PubMed] [Google Scholar]
- 50.Kang J, Kim E, Kim W, et al. Rhamnetin and cirsiliol induce radiosensitization and inhibition of epithelial-mesenchymal transition (EMT) by miR-34a-mediated suppression of Notch-1 expression in non-small cell lung cancer cell lines. J Biol Chem. 2013;288:27343–27357. doi: 10.1074/jbc.M113.490482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Braný D, Dvorská D, Slávik P, Školka R, Adamkov M. Survivin and gynaecological tumours. Pathol Res Pract. 2017;213:295–300. doi: 10.1016/j.prp.2017.02.009 [DOI] [PubMed] [Google Scholar]
- 52.Xie Y, Ma X, Gu L, et al. Prognostic and clinicopathological significance of survivin expression in renal cell carcinoma: a systematic review and meta-analysis. Sci Rep. 2016;6:29794. doi: 10.1038/srep29794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.De Vita A, Recine F, Mercatali L, et al. Primary culture of undifferentiated pleomorphic sarcoma: molecular characterization and response to anticancer agents. Int J Mol Sci. 2017;18pii:E2662. doi: 10.3390/ijms18122662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang X, Xia W, Chen L, et al. Synergistic antitumor effect of a γ-secretase inhibitor PF-03084014 and sorafenib in hepatocellular carcinoma. Oncotarget. 2018;9:34996–35007. doi: 10.18632/oncotarget.26209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.An L, Li DD, Chu HX, et al. Terfenadine combined with epirubicin impedes the chemo-resistant human non-small cell lung cancer both in vitro and in vivo through EMT and Notch reversal. Pharmacol Res. 2017;124:105–115. doi: 10.1016/j.phrs.2017.07.021 [DOI] [PubMed] [Google Scholar]
- 56.Zhang Y, Li D, Feng F, et al. Progressive and prognosis value of notch receptors and ligands in hepatocellular carcinoma: a systematic review and meta-analysis. Sci Rep. 2017;7:14809. doi: 10.1038/s41598-017-14897-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li DD, Zhao CH, Ding HW, et al. A novel inhibitor of ADAM17 sensitizes colorectal cancer cells to 5-Fluorouracil by reversing Notch and epithelial-mesenchymal transition in vitro and in vivo. Cell Prolif. 2018;51:e12480. doi: 10.1111/cpr.12480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu L, Tian YC, Mao G, Zhang YG, Han L. MiR-675 is frequently overexpressed in gastric cancer and enhances cell proliferation and invasion via targeting a potent anti-tumor gene PITX1. Cell Signal. 2019;62:109352. doi: 10.1016/j.cellsig.2019.109352 [DOI] [PubMed] [Google Scholar]
- 59.Kochan-Jamrozy K, Króliczewski J, Moszyńska A, Collawn JF, Bartoszewski R. miRNA networks modulate human endothelial cell adaptation to cyclic hypoxia. Cell Signal. 2019;54:150–160. doi: 10.1016/j.cellsig.2018.11.020 [DOI] [PubMed] [Google Scholar]
- 60.Xiong H, Yan T, Zhang W, et al. miR-613 inhibits cell migration and invasion by downregulating Daam1 in triple-negative breast cancer. Cell Signal. 2018;44:33–42. doi: 10.1016/j.cellsig.2018.01.013 [DOI] [PubMed] [Google Scholar]
- 61.Momen-Heravi F, Bala S. Emerging role of non-coding RNA in oral cancer. Cell Signal. 2018;42:134–143. doi: 10.1016/j.cellsig.2017.10.009 [DOI] [PubMed] [Google Scholar]
- 62.Luo M, Wu L, Zhang K, et al. miR-216b enhances the efficacy of vemurafenib by targeting Beclin-1, UVRAG and ATG5 in melanoma. Cell Signal. 2018;42:30–43. doi: 10.1016/j.cellsig.2017.09.024 [DOI] [PubMed] [Google Scholar]
- 63.Sanchez V, Golyardi F, Mayaki D, et al. Negative regulation of angiogenesis by novel micro RNAs. Pharmacol Res. 2019;139:173–181. doi: 10.1016/j.phrs.2018.11.010 [DOI] [PubMed] [Google Scholar]
- 64.Hasanpourghadi M, Pandurangan AK, Mustafa MR. Modulation of oncogenic transcription factors by bioactive natural products in breast cancer. Pharmacol Res. 2018;128:376–388. doi: 10.1016/j.phrs.2017.09.009 [DOI] [PubMed] [Google Scholar]
- 65.Wang H, Liu L, Fang S. MicroRNA‑330‑5p inhibits osteosarcoma cell growth and invasion by targeting the proto‑oncogene survivin. Mol Med Rep. 2019. doi: 10.3892/mmr.2019.10447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tong X, Yang P, Wang K, et al. Survivin is a prognostic indicator in glioblastoma and may be a target of microRNA-218. Oncol Lett. 2019;18:359–367. doi: 10.3892/ol.2019.10335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vahidi Manesh P, Farazmand A, Gharibdoost F, et al. Downregulation of miR-542-3p contributes to apoptosis resistance in dermal fibroblasts from systemic sclerosis patients via survivin overexpression. Iran J Allergy Asthma Immunol. 2019;18:173–181. [PubMed] [Google Scholar]
- 68.Xu S, Li D, Li T, et al. miR-494 sensitizes gastric cancer cells to TRAIL treatment through downregulation of survivin. Cell Physiol Biochem. 2018;51:2212–2223. doi: 10.1159/000495867 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.