Abstract
It has been proposed that cytotoxic T-lymphocyte antigen 4 (CTLA-4) may attenuate the T-cell activation threshold, thereby decreasing the antitumor response and conferring susceptibility to hepatocellular carcinoma (HCC).
In the present study, we selected CTLA-4 tagging single nucleotide polymorphisms (SNPs) and explored the relationship between these polymorphisms and susceptibility to HCC. A hospital-based case-control study, comprising 584 cases with HCC and 923 controls, was performed in an eastern Chinese Han population. CTLA-4 SNPs were genotyped using a custom-by-design 48-Plex SNPscan Kit.
We found that the CTLA-4 rs3087243 G>A polymorphism might be associated with increased risk of HCC (GA vs GG: adjusted odds ratio [OR], 1.38; 95% confidence interval [CI], 1.04–1.85; P = .028 and AA/GA vs GG: adjusted OR, 1.43; 95% CI, 1.08–1.89; P = .012). After using Bonferroni correction, this association remained (P = .012 for the AA/GA vs GG genetic model). In addition, the power value was 0.904 in the AA/GA versus GG genetic model. Haplotype analysis showed that CTLA4 Crs16840252Ars231775Ars3087243Trs733618, Crs16840252Grs231775Ars3087243Trs733618, and other haplotypes might increase the risk of HCC risk (P = .018, <.001, and .017, respectively). However, we found that CTLA4 Trs16840252A rs231775Grs3087243Trs733618 decreased the risk of HCC (P = .020).
Our results suggest that the CTLA-4 rs3087243 G>A polymorphism increases susceptibility to HCC in an eastern Chinese Han population. CTLA-4 haplotypes may influence the development of HCC. In the future, a population-based fine-mapping study with functional assessment should be performed to further determine these potential correlations.
Keywords: CTLA-4, haplotype, hepatocellular carcinoma, polymorphism, risk
1. Introduction
In 2012, an estimated 782,500 new liver cancer cases were diagnosed worldwide, accounting for approximately 5.55% of all cancer patients.[1] In addition, there were 745,500 liver cancer-related deaths.[1] China has a high rate of hepatocellular carcinoma (HCC) accounting for more than 50% of liver cancer cases and deaths.[2] Most primary liver cancers are HCC. The high incidence of HCC in Asia and sub-Saharan Africa mainly reflects the elevated levels of hepatitis B virus (HBV) infection. The high HBV infection rate is thought to be associated with the development of HCC. However, chronic HBV infection does not account for all of the etiology of HCC. Recently, investigations have focused on the association between heredity factors and the risk of HCC. Thus, the exploration of potential genetic factors might be beneficial to the diagnosis and prevention of HCC.
Chronic virus infection and inflammation might contribute to the development of HCC. Cytotoxic T-lymphocyte antigen 4 (CTLA4) is considered a vital negative regulator of immune responses.[3] CTLA4, a member of the immunoglobulin superfamily (IgSF), is expressed on activated T cells and inhibits their function. CTLA4 is homologous to CD28, both of which bind to CD80 and CD86. CTLA-4 has greater affinity and avidity than CD28 when binding with CD80 and CD86. CTLA4 inhibits the function of T cells[4]; however, CD28 transmits a stimulatory signal to T cells. In addition, CTLA4 is also found on the surface of regulatory T cells and leads to its inhibitory function. CTLA-4 binds with B7 to represses T cells at the G1 phase and decreases the expression of interleukin-2 (IL-2) and the IL-2 receptor.[5] CTLA-4 can also induce Fas cell surface death receptor-independent apoptosis of activated T lymphocytes, and then further restrain T cells.[6] It has been proposed that, during the development of malignancy, CTLA-4 may attenuate the T-cell activation threshold, thereby decreasing the antitumor response and conferring susceptibility to cancer.
Recently, case-control studies have focused on the association of the CTLA-4 single-nucleotide polymorphism (SNP) rs231775 G>A with the risk of HCC.[7–9] However, Hu et al found that CTLA-4 rs231775 G>A did not confer susceptibility to HCC.[8] Thus, because of the limited numbers of case-control study, the relationship between CTLA-4 rs231775 G>A with HCC susceptibility is not well established. In addition, other important SNPs in the CTLA-4 gene may be associated with the risk of HCC, whose potential correlations are unknown. Therefore, in the present study, we selected CTLA-4 tagging SNPs (rs733618 T>C, rs231775 G>A, rs3087243 G>A, and rs16840252 C>T) and explored the relationship between these polymorphisms and susceptibility to HCC.
2. Materials and methods
2.1. Study population and patient selection
A hospital-based case-control study comprising 584 patients with HCC (age range, 20–83 years) and 923 controls (age range, 21–80 years) was performed between January 2002 and December 2016 in an eastern Chinese Han population. All the HCC cases and controls were recruited from Fuzong Clinical Medical College and Union Clinical Medical College of Fujian Medical University. The major enrollment criteria for the patients with HCC were:
-
(1)
Pathologically diagnosed HCC;
-
(2)
living in eastern China for more than 10 years, and
-
(3)
not treated with chemoradiotherapy.
The major exclusion criteria were:
-
(1)
Patients with HCC who received prior chemoradiotherapy;
-
(2)
cases of HCC with autoimmune disease;
-
(3)
cases only diagnosed using ultrasound; and
-
(4)
patients with a history of another malignancy.
In total, 923 healthy subjects who attended the hospital for a routine physical examination were enrolled as noncancer controls. The eligibility criteria for the controls were:
-
(1)
No chronic liver disease;
-
(2)
no autoimmune disease; and
-
(3)
no history of malignancy.
All subjects were from the Chinese Han population and were unrelated. All participants were informed about the aim of study in an interview and provided written consent. Information on the demographic factors, risk factors, and clinical characteristics of the HCC cases and controls was collected using a questionnaire. The study protocol was approved by the Ethical Committee of Fujian Medical University, and conformed to the principles of the Helsinki Declaration. HCC cases and controls were full-matched by age and sex (Table 1). The criteria for “ever smokers” and “ever drinkers” were presented in our previous study.[10]
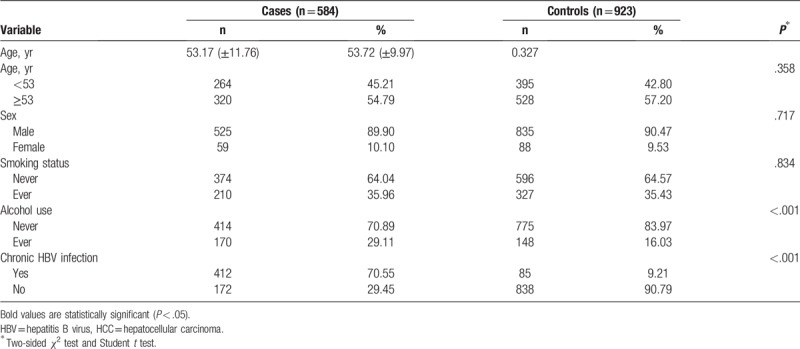
Table 1.
Distribution of selected demographic variables and risk factors in HCC cases and controls.
2.2. Selection of CTLA-4 tagging SNPs
Using the Genome Variation Server data (http://gvs.gs.washington.edu/GVS147/), CTLA-4 tagging SNPs were selected. The major criterion were:
-
(1)
the data came from a Chinese Han in Beijing cohort;
-
(2)
a P value for the Hardy–Weinberg equilibrium (HWE) of no less than .05;
-
(3)
the minor allele frequency was more than 0.05; and
-
(4)
a pairwise linkage disequilibrium r2 threshold of 0.8 between SNPs (r2 > 0.8); and
- (5)
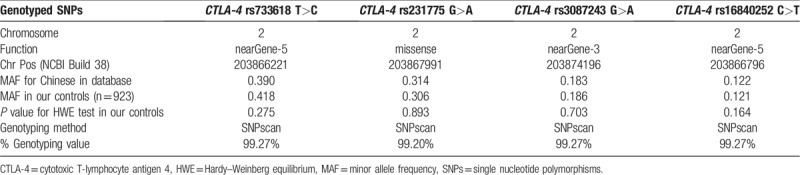
Finally, 4 CTLA-4 tagging SNPs (rs733618 T>C, rs231775 G>A, rs3087243 G>A, and rs16840252 C>T) were selected as eligible for the study. Table 2 presents important information for these polymorphisms.
Table 2.
Primary information for CTLA-4 polymorphisms (rs3087243 G>A, rs16840252 C>T, rs733618 T>C, and rs231775 G>A).
2.3. DNA extraction and genotyping
Genomic DNA was carefully extracted from 2 ml of EDTA-anticoagulated peripheral blood using a Promega DNA kit (Promega, Madison, WI) according to the manufacturer's instructions (www.promega.com/protocols/). The CTLA-4 tagging SNPs were genotyped using a custom-by-design 48-Plex SNPscan Kit (Genesky Biotechnologies Inc, Shanghai, China) as described in previous studies.[12–14] Sixty (4%) DNA samples were randomly selected and genotyped again by another laboratory technician, and the reproducibility was 100%.
2.4. Statistical analysis
The differences in demographic or risk factors were calculated using Student t test or Pearson 2-sided χ2 test when appropriate. Consistency of the genotype frequencies with the HWE for each CTLA-4 tagging SNP among the controls was measured using an online Pearson 2-sided χ2 test (http://ihg.gsf.de/cgi-bin/hw/hwa1.pl).[15] Pearson χ2 or Fisher's exact test were also used to assess the differences in the genotype frequencies between 2 groups. The distribution of CTLA-4 tagging SNPs genotype frequencies in the different groups was compared. To adjust for confounding factors (eg, the status of chronic HBV infection, age, sex, smoking, and alcohol use), logistic regression analysis was also performed. The odds ratios (ORs) were calculated to assess the associations and for all adjusted ORs, the 95% confidence intervals (95% CI) were calculated. SAS software (version 9.4, SAS Inc. Cary, NC) was used for the statistical analysis. Significance was assumed for P < .05 (2-tailed). In this study, a Bonferroni correction test was applied to conduct multiple testing.[16,17] We assessed the power of the present study (α = 0.05) using the PS Calculator (http://biostat.mc.vanderbilt.edu/twiki/bin/view/Main/PowerSampleSize). We used an expectation-maximization algorithm (SHESIS program [Bio-X Inc, Shanghai, China, http://analysis.bio-x.cn/myAnalysis.php])[18] to conduct the haplotype analysis.
3. Results
3.1. Demographic characteristics
The demographic characteristics of the participants are summarized in Table 1. A total of 584 patients with HCC (male:female = 525:59; mean age: 53.17 ± 11.76 years) and 923 control subjects (male:female = 835:88; mean age: 53.72 ± 9.97 years) participated in this case-control study. The status of chronic HBV infection, smoking, and alcohol use of both groups are presented in Table 1. There were no significant differences in the distribution of sex, age, and smoking between the patients with HCC and the controls (P = .717, .327, and .834, respectively). The rate of chronic HBV infection was higher for patients with HCC than for the controls (70.55% vs 9.21%; P < .001). In addition, alcohol use was significantly higher for the patients with HCC than for the controls (29.11% vs 16.03%; P < .001, respectively). As shown in Table 2, the genotyping success rates ranged from 99.20% to 99.27%. The distribution of the CTLA-4 rs733618 T>C, rs231775 G>A, rs3087243 G>A, and rs16840252 C>T genotypes among the controls conformed to the HWE.
3.2. Association of CTLA-4 rs733618 T>C, rs231775 G>A, rs3087243 G>A, and rs16840252 C>T polymorphisms with susceptibility to HCC
The CTLA-4 rs733618 T>C, rs231775 G>A, rs3087243 G>A, and rs16840252 C>T genotypes are listed in Table 3.
Table 3.
Genotypes of CTLA-4 rs3087243 G>A, rs16840252 C>T, rs733618 T>C, and rs231775 G>A polymorphisms.
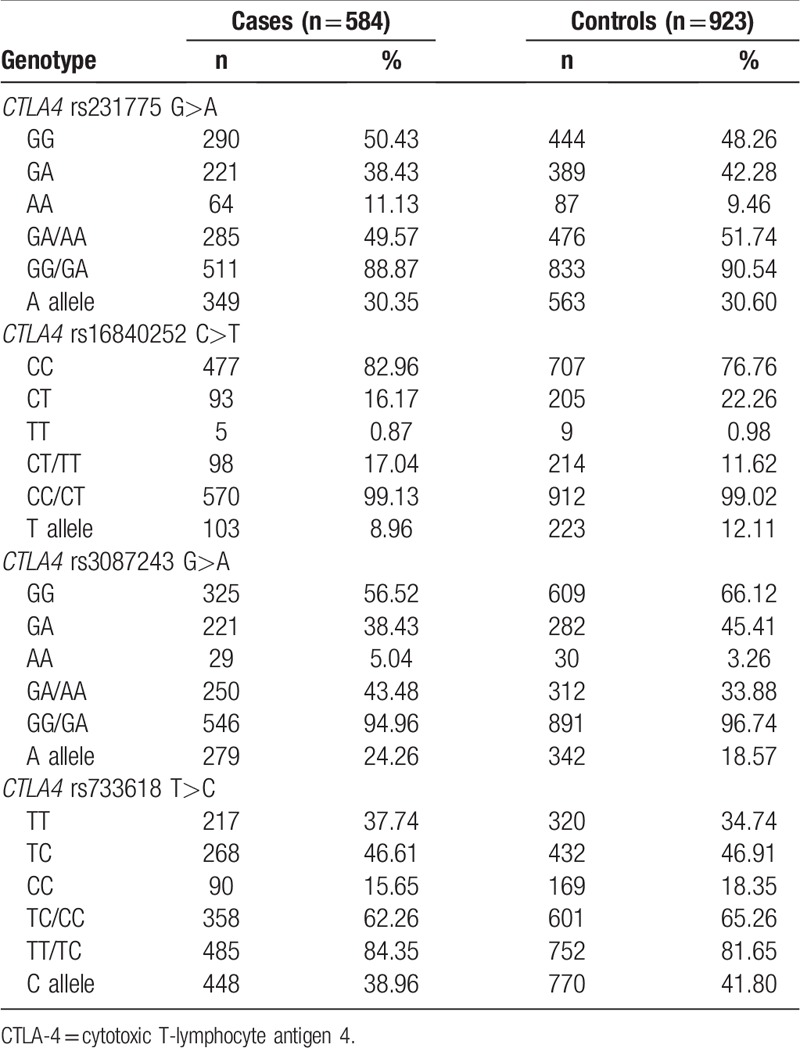
The frequencies of the CTLA-4 rs231775 GG, GA, and AA genotypes were 50.43%, 38.43% and 11.13% in the HCC cases and 48.26%, 42.28%, and 9.46% in the controls, respectively. After adjusting for chronic HBV infection status, age, sex, smoking, and drinking, we found that the CTLA-4 rs231775 G>A polymorphism might decrease the risk of HCC (GA vs GG: adjusted OR = 0.74, 95% CI: 0.55–0.99, P = .042; AA vs GG: adjusted OR = 1.16, 95% CI: 0.73–1.84, P = .519; GA/AA vs GG: adjusted OR = 0.83, 95% CI: 0.63–1.09, P = .168 and AA vs GG/GA: adjusted OR = 1.34, 95% CI: 0.86–2.08, P = .193; Table 4).
Table 4.
Logistic regression analyses of associations between CTLA-4 rs3087243 G>A, rs16840252 C>T, rs733618 T>C, and rs231775 G>A polymorphisms and the risk of HCC.
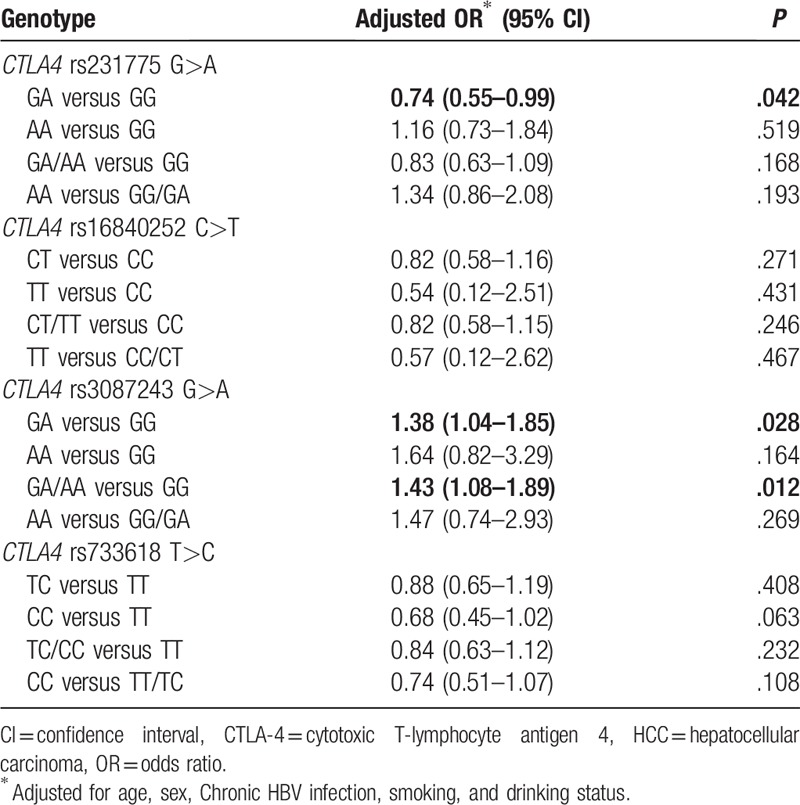
The frequencies of the CTLA-4 rs3087243 GG, GA, and AA genotypes were 56.52%, 38.43%, and 5.04% in the HCC cases and 66.12%, 45.41%, and 3.26% in the controls, respectively. After adjusting for chronic HBV infection status, age, sex, smoking, and drinking, an association between the CTLA-4 rs3087243 G>A polymorphism and increased risk of HCC was found (GA vs GG: adjusted OR, 1.38; 95% CI, 1.04–1.85; P = .028 and AA/GA vs GG: adjusted OR, 1.43; 95% CI, 1.08–1.89; P = .012; Table 4). For the CTLA-4 rs3087243 G>A polymorphism, the power value was 0.904 in the AA/GA vs GG genetic model. After using Bonferroni correction for multiple tests, we found that the CTLA-4 rs3087243 G>A polymorphism was still associated with an increased risk of HCC (P = .012 for the AA/GA vs GG genetic model). Other results of the Bonferroni correction test are not shown.
By contrast, we found that the CTLA-4 rs16840252 C>T and rs733618 T>C polymorphisms were not associated with susceptibility to HCC (Table 4).
3.3. SNP haplotypes
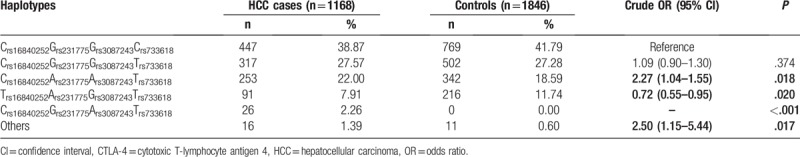
Results of haplotype analysis suggested that CTLA4 Crs16840252Ars231775Ars3087243Trs733618, Crs16840252Grs231775Ars3087243Trs733618, and other haplotypes significantly increased the risk of HCC (P = .018, <.001, and .017, respectively, Table 5). However, we also found that CTLA4 Trs16840252Ars231775Grs3087243Trs733618 decreased the risk of HCC (P = .020, Table 5).
Table 5.
CTLA4 haplotype frequencies (%) in cases and controls and risk of HCC.
4. Discussion
CTLA-4, an IgSF receptor, plays an important role in immune regulation by feeding a negative signal to T cells once an immune response has been initiated and completed.[19] In this case-control study, the role of CTLA-4 tagging polymorphisms (rs733618 T>C, rs231775 G>A, rs3087243 G>A, and rs16840252 C>T) in HCC susceptibility was assessed. We found that the CTLA-4 rs3087243 G>A polymorphism was associated with an increased risk of HCC, and this association remained after Bonferroni correction.
Polymorphism rs3087243 G>A is located in the 3′-untranslated region of the CTLA-4 gene. It has also been named as CTLA4 60G/A. A meta-analysis reported that the CTLA-4 rs3087243 G>A polymorphism increased susceptibility to skin cancer.[20] However, a pooled-analysis also suggested that CTLA-4 rs3087243 G>A might decrease the risk of breast cancer.[21] Thus, these previous results were conflicting and ambiguous. Considering the common variants conferring a low penetrance susceptibility to the development of cancer, we enrolled 1507 participants and conducted a case-control study to obtain a more precise evaluation. In this study, we found that the CTLA-4 rs3087243 A allele was associated with an increased risk of HCC. To further confirm this potential association, Bonferroni correction was performed, after which this association remained valid. In addition, the power of this case-control study (α = 0.05) was evaluated using an internet-based Power and Sample Size Calculator. The power value was 0.904 in the AA/GA versus GG genetic model, which suggested a potential association. Recently, a functional study found lower expression of the soluble (s)-CTLA4 isoform compared with the membrane-bound CTLA4 in lymphocytes from patients with inflammatory bowel disease with the CTLA-4 rs3087243 GG genotype compared with that of the AA genotype.[22] These results indicated that the CTLA-4 rs3087243 G→A variant might increase the expression of CTLA4, thereby elevating the T-cell activation threshold, leading to a weakened antitumor response and conferring a risk of HCC. However, in the present study, because of the moderate sample size, these primary findings should be explained with very caution. In the future, further investigations should be performed to verify these correlations.
As summarized in Table 5, the haplotype analysis indicated that the frequency of CTLA4 Crs16840252Ars231775Ars3087243Trs733618, Crs16840252Grs231775Ars3087243Trs733618, and other haplotypes significantly increased the risk of HCC. However, we also found that CTLA4 Trs16840252Ars231775Grs3087243Trs733618 decreased the risk of HCC. We first explored the potential relationship of these CTLA-4 haplotypes with the development of HCC. A previous study highlighted that the CTLA4 Crs16840252Grs231775Ars3087243Trs733618 haplotype might increase susceptibility to gastric cardia adenocarcinoma,[15] which was similar to the results of the present study.
The present study has some merits. First, we selected tagging SNPs to explore the potential relationship of CTLA-4 variants with HCC susceptibility. Second, to the best of our knowledge, this was the first case-control study to explore the association of CTLA-4 rs733618 T>C, rs3087243 G>A, and rs16840252 C>T polymorphisms with HCC. Finally, the distribution of CTLA-4 rs733618 T>C, rs231775 G>A, rs3087243 G>A, and rs16840252 C>T genotypes among controls conformed to HWE, which suggested our findings were less prone to bias.
Some limitations of the study should be acknowledged. First, our case-control study only included eastern Chinese Han population; thus, the findings are only applicable to this ethnicity. Second, the sample size of the present study was moderate. Further large-scale and well-designed studies taking into account detailed environmental factors are needed to confirm this relationship in various populations. Third, we lacked sufficient information on the prognosis of HCC among the patients; therefore, we could not further analyze the potential role of CTLA-4 variants in HCC survival. Finally, although we took several risk factors into consideration, such as chronic HBV infection status, sex, age, drinking, and smoking status, many other environmental factors and lifestyle parameters that might be related to the risk of HCC, were not assessed.
In conclusion, our results highlighted that the CTLA-4 rs3087243 G>A polymorphism was associated with susceptibility to HCC in an eastern Chinese Han population. We also found that CTLA-4 haplotypes might influence the development of HCC. In the future, a population-based fine-mapping study with functional assessment should be performed to further verify these correlations, especially for gene-environment and gene-gene interactions.
Acknowledgments
We appreciate all subjects who participated in this study. We wish to thank Dr Yan Liu (Genesky Biotechnologies Inc, Shanghai, China) for technical support.
Author contributions
Conceptualization: Yuling Sun.
Data curation: Chao Liu.
Methodology: Yu Chen.
Project administration: Jianping Chen.
Resources: Jianping Chen.
Software: Jiaochun Liu.
Writing – original draft: Jing Yang.
Writing – review and editing: Weifeng Tang, Jing Yang.
Weifeng Tang orcid: 0000-0002-4157-4057.
Footnotes
Abbreviations: CI = confidence interval, CTLA-4 = cytotoxic T-lymphocyte antigen 4, HBV = hepatitis B virus, HCC = hepatocellular carcinoma, HWE = Hardy–Weinberg equilibrium, IgSF = immunoglobulin superfamily, IL-2 = interleukin-2, OR = odds ratio, SNP = single nucleotide polymorphisms.
JY and JL contributed equally to this study.
This study was supported in part by Young Talent Training Project of Health Development Planning Commission of Changzhou (QN201706).
The authors have no conflicts of interest to disclose.
References
- [1].Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87–108. [DOI] [PubMed] [Google Scholar]
- [2].Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115–32. [DOI] [PubMed] [Google Scholar]
- [3].Teft WA, Kirchhof MG, Madrenas J. A molecular perspective of CTLA-4 function. Annu Rev Immunol 2006;24:65–97. [DOI] [PubMed] [Google Scholar]
- [4].Waterhouse P, Penninger JM, Timms E, et al. Lymphoproliferative disorders with early lethality in mice deficient in Ctla-4. Science 1995;270:985–8. [DOI] [PubMed] [Google Scholar]
- [5].Appleman LJ, Berezovskaya A, Grass I, et al. CD28 costimulation mediates T cell expansion via IL-2-independent and IL-2-dependent regulation of cell cycle progression. J Immunol 2000;164:144–51. [DOI] [PubMed] [Google Scholar]
- [6].Scheipers P, Reiser H. Fas-independent death of activated CD4(+) T lymphocytes induced by CTLA-4 crosslinking. Proc Natl Acad Sci U S A 1998;95:10083–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Liu G, Lu GX. Association of CTLA4 gene +49G/A polymorphism with HBV infection and HBV-related hepatocellular carcinoma in Hunan Han population. Nan Fang Yi Ke Da Xue Xue Bao 2010;30:1838–40. [PubMed] [Google Scholar]
- [8].Hu L, Liu J, Chen X, et al. CTLA-4 gene polymorphism +49 A/G contributes to genetic susceptibility to two infection-related cancers-hepatocellular carcinoma and cervical cancer. Hum Immunol 2010;71:888–91. [DOI] [PubMed] [Google Scholar]
- [9].Gu X, Qi P, Zhou F, et al. +49G > A polymorphism in the cytotoxic T-lymphocyte antigen-4 gene increases susceptibility to hepatitis B-related hepatocellular carcinoma in a male Chinese population. Hum Immunol 2010;71:83–7. [DOI] [PubMed] [Google Scholar]
- [10].Tang W, Zhang S, Qiu H, et al. Genetic variations in MTHFR and esophageal squamous cell carcinoma susceptibility in Chinese Han population. Med Oncol 2014;31:915. [DOI] [PubMed] [Google Scholar]
- [11].Carlson CS, Eberle MA, Kruglyak L, et al. Mapping complex disease loci in whole-genome association studies. Nature 2004;429:446–52. [DOI] [PubMed] [Google Scholar]
- [12].Zheng L, Yin J, Wang L, et al. Interleukin 1B rs16944 G>A polymorphism was associated with a decreased risk of esophageal cancer in a Chinese population. Clin Biochem 2013;46:1469–73. [DOI] [PubMed] [Google Scholar]
- [13].Yin J, Wang L, Shi Y, et al. Interleukin 17A rs4711998 A>G polymorphism was associated with a decreased risk of esophageal cancer in a Chinese population. Dis Esophagus 2014;27:87–92. [DOI] [PubMed] [Google Scholar]
- [14].Qiu H, Lin X, Tang W, et al. Investigation of TCF7L2, LEP and LEPR polymorphisms with esophageal squamous cell carcinomas. Oncotarget 2017;8:109107–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Tang W, Wang Y, Chen S, et al. Investigation of cytotoxic T-lymphocyte antigen 4 polymorphisms in gastric cardia adenocarcinoma. Scand J Immunol 2016;83:212–8. [DOI] [PubMed] [Google Scholar]
- [16].Bland JM, Altman DG. Multiple significance tests: the Bonferroni method. BMJ 1995;310:170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Lesack K, Naugler C. An open-source software program for performing Bonferroni and related corrections for multiple comparisons. J Pathol Inform 2011;2:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Shi YY, He L. SHEsis a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res 2005;15:97–8. [DOI] [PubMed] [Google Scholar]
- [19].Walunas TL, Bakker CY, Bluestone JA. CTLA-4 ligation blocks CD28-dependent T cell activation. J Exp Med 1996;183:2541–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Yan Q, Chen P, Lu A, et al. Association between CTLA-4 60G/A and -1661A/G polymorphisms and the risk of cancers: a meta-analysis. PloS One 2013;8:e83710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Dai Z, Tian T, Wang M, et al. CTLA-4 polymorphisms associate with breast cancer susceptibility in Asians: a meta-analysis. PeerJ 2017;5:e2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Repnik K, Potocnik U. CTLA4 CT60 single-nucleotide polymorphism is associated with Slovenian inflammatory bowel disease patients and regulates expression of CTLA4 isoforms. DNA Cell Biol 2010;29:603–10. [DOI] [PubMed] [Google Scholar]


