Abstract
The purposes of the study was to validate the relationship between General transcription factor II-I (GTF2I) genetic variants and kidney involvements of systemic lupus erythematosus (SLE) patients in a Chinese Han population.
Samples from 400 SLE patients and 400 age- and sex-matched healthy controls were collected and genotyped by improved multiplex ligation detection reaction technique. The relationship between gene polymorphism of rs117026326, rs73366469, and susceptibility, progression of SLE were analyzed.
The present study provided evidence that rs117026326 and rs73366469 were both associated with SLE susceptibility (both C vs T: P < .001). The analysis of dominant, recessive disease model provided us with further validation (P < .001). Both gene polymorphisms are associated with a triad of disease manifestations among SLE patients. Patients carrying genotype TT of rs117026326 had lower 24-hour urinary total protein (24 hours UTP, g/24 hours), 24-hour urinary protein level (g/L·24 hours), lower frequency of the proteinuria and lupus nephritis (LN). Patients carrying genotype TT at rs73366469 had higher 24-hour urinary protein level, higher frequency of the proteinuria, LN and positive anti-dsDNA than those with other genotypes.
This study identified the involvement of GTF2I gene polymorphisms in development of SLE, particularly in renal involvement.
Keywords: autoantibody, renal involvement, single nucleotide polymorphisms, systemic lupus erythematosus
1. Introduction
Systemic lupus erythematosus (SLE) is a prototypic, heterogeneous autoimmune disorder characterized by the presence of pathogenic autoantibodies which can affect multiple organs and systems.[1] Although the underlying mechanism of SLE remains uncertain,[2] genetic factors seem rather effective at its occurrence. Recent Genome-Wide Association studies (GWAS) revealed that some of single nucleotide polymorphisms (SNPs) in GTF2I gene were related to multiple autoimmune diseases,[3–5] including primary Sjögren syndrome (pSS), rheumatoid arthritis, and SLE. However, as 1 of main clinical challenges of SLE, the genetic background of lupus nephritis (LN) has not been completely clarified. LN occurs in 15% - 55% of SLE patients, and patients from Asia have a higher prevalence than others and always present with a more severe disease.[6] Whether GTF2I polymorphisms are associated with clinical indexes and disease manifestations among SLE patients is still uncertain, so we performed this case-control study on polymorphism rs117026326 and rs73366469, and explored the correlation of these locus to susceptibility to SLE and renal involvement in the present study.
2. Materials and methods
2.1. Patients
This study was approved by the Ethics Committee of West China Hospital, having gained the ethic approval. Total of 800 individual participates including 400 SLE cases and 400 age- and gender-matched healthy controls (HC) were recruited from the West China Hospital. Informed consents for using their results were obtained from all the study participants and any form of registration that may identify a patient were excluded from the content of the paper.
All the SLE patients in the study were hospitalized patients, fulfilling the American College of Rheumatology 1997 classification criteria. Patients with drug-induced SLE were excluded.
Total of 400 healthy individuals randomly selected from the Health Examination Center were enrolled as HC. Their physical examinations and blood tests were all within normal range. None of these healthy individuals had infectious diseases, autoimmune disorders or family history of autoimmune diseases.
2.2. Clinical and laboratory evaluation
The clinical and laboratory data of the patients including proteinuria, rash, pericarditis, pleuritis, arthritis, the serum level of anti-dsDNA, anti-Sm, anti-RNP, IL-1β, IL-4, IL-6, urea, creatinine, CYS-C, uric acid, 24 hours UTP (g/24 hours) and 24-hour urinary protein level (g/L·24 hour) were recorded. Anti-dsDNA, anti-Sm, and anti-RNP were detected in all the 400 SLE patients. Anti-dsDNA was determined through indirect immunofluorescence, while anti-Sm and anti-RNP were estimated through line immunoassays (Euroimmun, Germany). 143 of SLE patients were selected randomly for cytokine testing. IL-1β, IL-4 and IL-6 were quantitatively detected using R&D Human Inflammation Assays.
All these tests were performed strictly in accordance with the relevant guidelines and regulations. Blood samples for assessing these indexes were detected at the same time.
2.3. GTF2I polymorphisms genotyping and linkage disequilibrium evaluation
All the samples were genotyped for rs117026326 and rs73366469 using improved multiplex ligation detection reaction (iMLDR) (Genesky Biotechnologies Inc, China). Polymerase chain reaction was performed at 95°C for 2 minutes followed by 11 cycles of denaturation (94°C, 20 seconds), annealing (65°C–0.5°C/cycle 40 seconds) and 24 cycles of 94°C 20 seconds, 59°C 30 seconds, 72°C 1 minutes 30 seconds, plus 1 cycle of 72°C for 2 minutes. During detection, some of samples were randomly selected for direct sequencing to confirm the results genotyped using iMLDR. The Haploview software version 4.2 was applied to perform linkage disequilibrium (LD) evaluation for combination of SNPs by calculating the pairwise r2 coeffcient.
2.4. Statistical analysis
Hardy–Weinberg equilibrium (HWE) was performed for the polymorphisms by the 2-sided Chi-Squared (χ2) test. Median and interquartile, mean ± SD or number and percentage were used to describe variables. Student t test or Mann–Whitney U test for quantitative variables were used to compare demographic and clinical data between patients and controls as appropriate. Chi-Square (χ2) test or Fisher exact test were applied to analyze differences in allelic and genotypic frequencies. If allele x is the major allele of the SNP, y is the minor allele, recessive model is known as xx + xy vs yy, dominant model as xx vs xy + yy, co-dominant model as xx vs xy vs yy. The association between genetic polymorphisms and LN, proteinuria was evaluated using the logistic regression model, calculated odds ratios (ORs) and 95% confidence intervals (CIs).
The statistical analyses were applied by using the Statistical Package for the Social Sciences (SPSS) software (version 19.0). Statistical power was calculated using “Power and Sample Size Calculation” software. A two-sided P value less than .05 was considered as statistically significant.
3. Results
3.1. The main characteristics of the study population
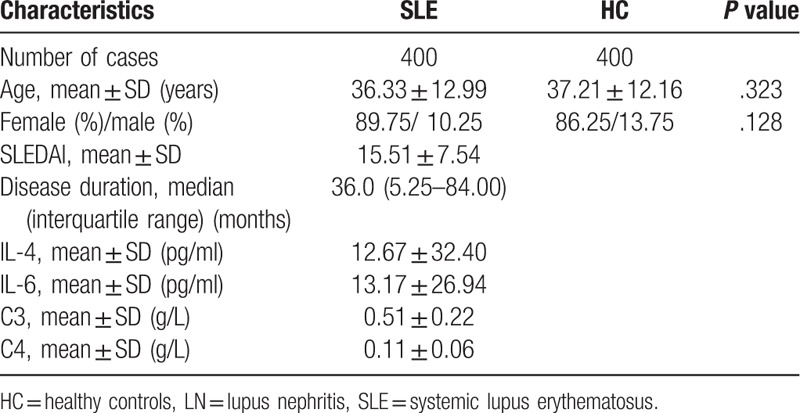
The demographic and clinical characteristics of all the included subjects were illustrated in Table 1. The age (P = .323) and sex (P = .128) of SLE patients and controls were adequately matched. The average age of SLE patients and healthy controls were 36.33 ± 12.99 and 37.21 ± 12.16 years old, and the gender ratio (Female/male) were 359/41 and 345/55, respectively. Among all the SLE patients, the median disease duration is 36 months, the average SLEDAI score is 15.51 ± 7.54 and the average level of IL-4, IL-6, C3, C4 in SLE patients were 12.67 ± 32.40 pg/ml, 13.17 ± 26.94 pg/ml, 0.51 ± 0.22 g/L, 0.11 ± 0.06 g/L, respectively (Table 1).
Table 1.
Demographic and clinical features of the study participants.
3.2. Polymorphisms association with SLE susceptibility
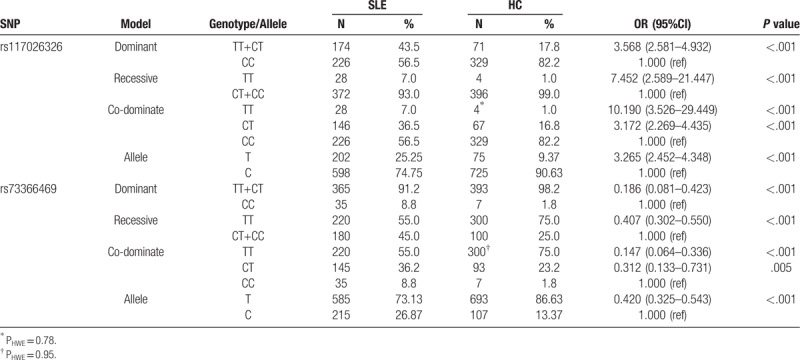
Cohorts showed no significant deviation from HWE for genotyped SNPs (all P > .05). When comparing allele frequency of rs117026326 and rs73366469 between SLE patients and HC, significant differences were indicated (both P < .001, statistical power = 1.000) with rs117026326 T allele correlated to an increased risk of SLE and rs73366469 T allele correlated to a decreased risk of SLE (Table 2). The odds ratio and 95% confidence interval ((OR) 95% CI) were 3.265 (2.452–4.348) and 0.420 (0.325–0.543), respectively. When comparing genotype frequency, we observed that TT and CT genotype at rs117026326 and rs73366469 were associated with SLE susceptibility (TT at rs117026326: OR (95% CI) = 10.190 (3.526–29.449); CT at rs117026326: OR (95%CI) = 3.172 (2.269–4.435); TT at rs73366469: OR (95%CI) = 0.147 (0.064–0.336); CT at rs73366469: OR (95%CI) = 0.312 (0.133–0.731), all P < .05). These results were also supported by further disease model analysis (rs117026326: Dominant model: OR (95% CI) = 3.568 (2.581–4.932); Recessive model: OR (95% CI) = 7.452 (2.589–21.447); rs73366469: Dominant model: OR (95% CI) = 0.186 (0.081–0.423); Recessive model: OR (95% CI) = 0.407 (0.302–0.550), All P < .001).
Table 2.
Genotype and allele frequencies of SNPs within GTF2IRD1-GTF2I region in SLE patients and controls.
3.3. Polymorphisms association with clinical characteristics of SLE patients
SLE is a heterogeneous autoimmune disease. Patients with SLE always have variations in disease severity and clinical involvement. Thus, we tried to examine the effect of gene polymorphisms on clinical parameters and phenotypes among SLE patients, summarized in Table 3. The results suggested that there was association between these polymorphisms and renal involvement in SLE patients.
Table 3.
Association between polymorphisms with clinical features in SLE patients.
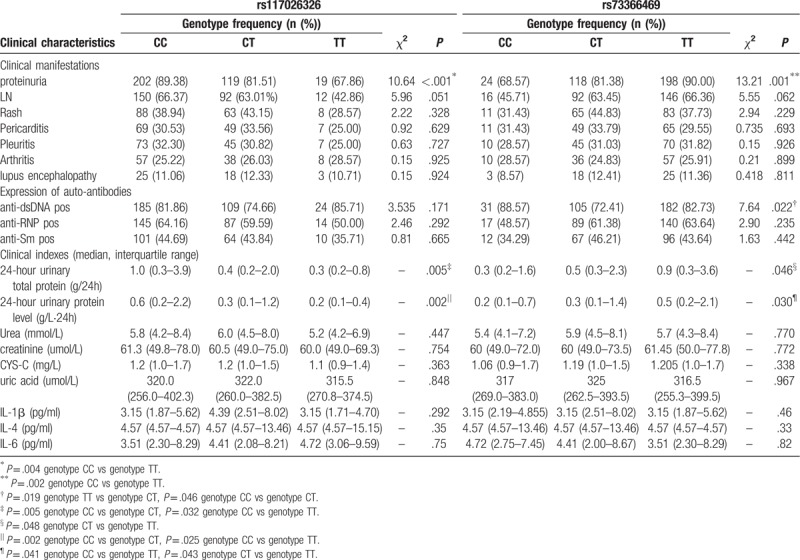
Although there was no significant association between the polymorphisms and levels of urea, creatinine, cystatin-C (CYS-C), uric acid, IL-4, IL-6, IL-1β in SLE patients, we found that the 24 hours UTP and 24-hour urinary protein level had significant difference in SLE patients with different genotypes of rs117026326 (P = .005, P = .030, respectively) or rs73366469 (P = .046, P = .002, respectively). SLE cases with CC genotype at rs117026326 had higher 24 hours UTP and 24-hour urinary protein level than patients with other genotypes, while cases with TT genotype at rs73366469 had significant increased 24 hours UTP and 24-hour urinary protein level than patients with other genotypes.
What's more, the frequency of proteinuria had significant difference in SLE patients with different genotypes of rs117026326 or rs73366469 (χ2 = 10.64, P < .001; χ2 = 13.21, P = .001, respectively). When detailed analysis was performed on co-dominant model, results revealed that cases with TT genotype at rs117026326 (67.86%) had decreased frequency of the proteinuria than patients with CC genotype (89.38%) (P = .004), while cases with TT genotype at rs73366469 (90.00%) had significant increased frequency of the proteinuria than patients with CC genotype (68.57%) (P = .002). Furthermore, there was a strong trend towards significance when comparing the frequency of LN in patients with different genotypes of rs117026326 or rs73366469 (P = .051, .062, respectively). We also observed that the positive frequency of anti-dsDNA in patients with TT genotype at rs73366469 (82.73%) was significantly higher than those with CT genotype (72.41%) (χ2 = 5.532, P = .019).
Further analysis performed on recessive model indicted that patients with genotype TT at rs117026326 (42.85%) had significantly lower prevalence of LN than those with other genotypes (65.05%) (P = .019), while patients with genotype TT at rs73366469 (65.77%) had significantly higher prevalence of LN than patients with other genotypes (50.23%) (P = .001) (Table 4).
Table 4.
Association between polymorphisms with LN in SLE patients performed on recessive model.

3.4. Linkage analysis
We constructed the LD analysis of rs117026326 and rs73366469. The result showed that they were in strong linkage disequilibrium with r2 = 0.71 (Fig. 1).
Figure 1.

Linkage disequilibrium for rs117026326 and rs73366469 in GTF2I gene. The LD status is expounded by the r2 value. There was strong LD between two SNPs (r2 = 0.71).
4. Discussion
In this case-control study, we focused on the influence of GTF2I gene polymorphisms on susceptibility and clinical characteristics of SLE in a Chinese Han population. We described, for the first time, an interesting relationship between rs117026326, rs73366469 and kidney involvement of SLE patients. Results suggested that there was a highly significant association between rs117026326 and proteinuria, while rs73366469 was associated with proteinuria as well as expression of anti-dsDNA. Moreover, the association between two SNPs and the frequency of LN was also observed on recessive model.
GTF2I gene is clustered on chromosome 7 within a 1.8 Mb region. It can encode the transcription factor IIi (TFII-I) family, involving in regulation of growth factor signaling, immune cell signaling, and cell proliferation.[7] As a signal-induced transcription, TFII-I can not only regulate transcription,[8] signal transduction and development of bone, neural tissue,[9] but also participate in the process of immune regulation. It can act as a cellular regulator of transcription from the promoter of cytomegalovirus, and its transcriptional regulation can be induced by immune signaling of lymphocytes. The activity of TFII-I is significantly related to expression of β-globin gene in erythroid cells.[10] Previous study indicated that GTF2I gene can contribute to regulation of immunoglobulin heavy chain transcription in B lymphocytes.[11] All these recent studies emphasized the validity and immune relevance of this gene.
Recent GWAS study identified susceptibility locus in GTF2I for pSS.[5] Given the close clinical relationship and similar pathophysiological features between pSS and SLE, and given the fact that GTF2I gene is one of genetic susceptibility factors of SLE, GTF2I gene may play an essential role in SLE. rs117026326 is located in intron of GTF2I gene on chromosome 7q11.23 and was recently identified as a risk gene loci for pSS,[12] RA[13] and SLE.[14,15] rs73366469 is located within conserved enhancers, overlaps transcription start sites for GTF2I gene. It was also one of autoimmune susceptibility loci for RA[13] and SLE.[15] Our study indicated that rs117026326 and rs73366469 were candidate risk factors for SLE in a Chinese Han population, which further confirmed previously reported associations. What is more, differences exist in linkage disequilibrium in different populations. When only enrolling in West Chinese population, associations between rs73366469 and rs117026326 were consistently replicated, but the r2 in our study was higher than those in Kim and Sun, et al's study with Asian ancestry.[13,15]
Although there was no correlation between rs117026326 genotypes and GTF2I expression,[13] our study showed that the GTF2I polymorphisms are correlative with clinical parameters and phenotypes of SLE. As one of the most known devastating kidney-threatening manifestation of SLE patients,[16,17] LN affects about 40% to 70% of SLE patients and Asian, African populations have relatively higher incidence.[18] Some genes have been shown to possess the potential of predisposing susceptible population to LN.[19,20] Proteinuria can indicate potential kidney disorders, suggestive for the presence of LN.[21] With regard to autoimmune antibodies, anti-dsDNA as one of nephrotoxic autoantibodies can directly involve in renal pathology of SLE[22,23] and increased incidence of LN,[24] of which one of the reasons is immune complex with anti-dsDNA can lead to renal inflammation if deposited in kidney,[25] and may result in activation of complement, reactive oxygen species and some cytokines which are also causative factors of LN.[26] Moreover, positive anti-Sm, anti-RNP[27–29] and biomarkers for inflammation such as interleukin[30–32] are also associated with renal involvement. Both IL-4 and IL-6 play important roles in kidney injury, IL-4 has function in polarization of macrophages to M2 phenotype, essential for recovery from acute kidney injury,[33] while IL-6 is one of markers of chronic kidney damage. In addition, upregulation of IL-1β caused by TLR7 stimulation can lead to development of LN.[34] In order to investigate relationship between GTF2I polymorphisms and kidney injury, we measured the frequency of proteinuria, LN, serum levels of clinical indexes which are related to kidney including urea, creatinine, CYS-C, uric acid, IL-4, IL-6, IL-1β and positivity of anti-dsDNA, anti-Sm, and anti-RNP. The result of our study showed that GTF2I polymorphisms were related to severity of kidney impairment of SLE.
Last year, an international group developed a new classification criteria for SLE patients. The criteria was based on a scoring system for clinical and laboratory domains. A positive antinuclear antibody (ANA) at a titer 1:80 or greater by immunofluorescence (IFA) is used as the entry criterion. The data in our study took years to collect, so we did not change the inclusion criteria in order to keep condition of enrolled patients in accordance with each other. All the SLE patients we enrolled have positive ANA, and the HC are from Health Examination Center of our hospital, so the diagnosis will not change by newer criteria and the result will hold true.
Ultimately, this is the first study carried out in Chinese SLE patients which is tempting to suppose that GTF2I polymorphisms were possibly useful predictors for kidney impairment in SLE patients. The present study suggested that 24 hours UTP, 24-hour urinary protein level, frequency of the proteinuria and prevalence of LN were significantly lower in cases with rs117026326 TT genotype, while patients with rs73366469 TT genotype had higher 24-hour urinary protein level, frequency of the proteinuria, prevalence of LN, and positive frequency of anti-dsDNA.
Author contributions
Conceptualization: Min Yang, Yongkang Wu.
Data curation: Yanming Meng.
Formal analysis: Yanming Meng, Yao He.
Funding acquisition: Yongkang Wu.
Investigation: Yanming Meng, Qibing Xie.
Methodology: Yao He, Yongkang Wu.
Project administration: Junlong Zhang, Min Yang.
Resources: Junlong Zhang, Qibing Xie.
Software: Yanming Meng, Yao He.
Supervision: Junlong Zhang, Qibing Xie.
Writing - original draft: Yanming Meng.
Writing - review & editing: Yanming Meng, Yuning Chen, Yongkang Wu.
Yanming Meng orcid: 0000-0003-1490-2622.
Footnotes
Abbreviations: 24 h UTP = 24-hour urinary total protein, CI = confidence interval, CYS-C = cystatin-C, GTF2I = general transcription factor II-I, HC = healthy controls, HWE = Hardy–Weinberg Equilibrium, LN = lupus nephritis, OR = odds ratio, pSS = primary Sjögren syndrome, SLE = systemic lupus erythematosus, SNPs = single nucleotide polymorphisms.
The Department of Science and Technology of Sichuan Province (Grant No. 2016JY0035) and National Clinical Research Center for Geriatrics, West China Hospital, Sichuan University (Grant No. Z2018C03).
The authors confirm that there is no conflict of interest.
References
- [1].Zhu TY, Tam LS, Li EK. Labour and non-labour market productivity in Chinese patients with systemic lupus erythematosus. Rheumatology (Oxford) 2012;51:284–92. [DOI] [PubMed] [Google Scholar]
- [2].Long H, Yin H, Wang L, et al. The critical role of epigenetics in systemic lupus erythematosus and autoimmunity. J Autoimmun 2016;74:118–38. [DOI] [PubMed] [Google Scholar]
- [3].Song IW, Chen HC, Lin YF, et al. Identification of susceptibility gene associated with female primary Sjögren's syndrome in Han Chinese by genome-wide association study. Hum Genet 2016. [DOI] [PubMed] [Google Scholar]
- [4].Fang K, Zhang K, Wang J. Network-assisted analysis of primary Sjogren's syndrome GWAS data in Han Chinese. Sci Rep 2015;5:18855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Li Y, Zhang K, Chen H, et al. A genome-wide association study in Han Chinese identifies a susceptibility locus for primary Sjögren's syndrome at 7q11.23. Nat Genet 2013;45:1361–5. [DOI] [PubMed] [Google Scholar]
- [6].Bolin K, Sandling JK, Zickert A, et al. Association of STAT4 polymorphism with severe renal insufficiency in lupus nephritis. PLoS One 2013;8:e84450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Carmona-Mora P, Widagdo J, Tomasetig F, et al. The nuclear localization pattern and interaction partners of GTF2IRD1 demonstrate a role in chromatin regulation. Hum Genet 2015. [DOI] [PubMed] [Google Scholar]
- [8].Cetkovska K, Sustova H, Kosztyu P, et al. A novel interaction between TFII-I and Mdm2 with a negative effect on TFII-I transcriptional activity. PLoS One 2015;10:e0144753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Poitras L, Yu M, Lesage-Pelletier C, et al. An SNP in an ultraconserved regulatory element affects Dlx5/Dlx6 regulation in the forebrain. Development 2010;137:3089–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Roy AL. Biochemistry and biology of the inducible multifunctional transcription factor TFII-I: 10 years later. Gene 2012;492:32–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Rajaiya J, Nixon JC, Ayers N, et al. Induction of immunoglobulin heavy-chain transcription through the transcription factor Bright requires TFII-I. Mol Cell Biol 2006;26:4758–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Zheng J, Huang R, Huang Q, et al. The GTF2I rs117026326 polymorphism is associated with anti-SSA-positive primary Sjogren's syndrome. Rheumatology (Oxford) 2015;54:562–4. [DOI] [PubMed] [Google Scholar]
- [13].Kim K, Bang SY, Ikari K, et al. Association-heterogeneity mapping identifies an Asian-specific association of the GTF2I locus with rheumatoid arthritis. Sci Rep 2016;6:27563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Li Y, Li P, Chen S, et al. Association of GTF2I and GTF2IRD1 polymorphisms with systemic lupus erythematosus in a Chinese Han population. Clin Exp Rheumatol 2015. [PubMed] [Google Scholar]
- [15].Sun C, Molineros JE, Looger LL, et al. High-density genotyping of immune-related loci identifies new SLE risk variants in individuals with Asian ancestry. Nat Genet 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Liu Z, Zhang H, Liu Z, et al. Multitarget therapy for induction treatment of lupus nephritis: a randomized trial. Ann Intern Med 2015;162:18–26. [DOI] [PubMed] [Google Scholar]
- [17].Zhang CX, Chen J, Cai L, et al. DNA induction of MDM2 promotes proliferation of human renal mesangial cells and alters peripheral B cells subsets in pediatric systemic lupus erythematosus. Mol Immunol 2018;94:166–75. [DOI] [PubMed] [Google Scholar]
- [18].Feldman CH, Hiraki LT, Liu J, et al. Epidemiology and sociodemographics of systemic lupus erythematosus and lupus nephritis among US adults with Medicaid coverage, 2000-2004. Arthritis Rheum 2013;65:753–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Lech M, Anders HJ. The pathogenesis of lupus nephritis. J Am Soc Nephrol 2013;24:1357–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Taylor KE, Remmers EF, Lee AT, et al. Specificity of the STAT4 genetic association for severe disease manifestations of systemic lupus erythematosus. PLoS Genet 2008;4:e1000084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Melderis S, Wiech T, Iking-Konert C, et al. Lupus nephritis. Z Rheumatol 2018;77:593–608. [DOI] [PubMed] [Google Scholar]
- [22].Sarfaraz S, Anis S, Ahmed E, et al. Clinical significance of anti-ribosomal P protein antibodies in patients with lupus nephritis. Rev Recent Clin Trials 2018;13:281–6. [DOI] [PubMed] [Google Scholar]
- [23].Tanha N, Hansen RB, Nielsen CT, et al. Clinical and serological associations with the development of incident proteinuria in Danish patients with systemic lupus erythematosus. J Rheumatol 2018;45:934–41. [DOI] [PubMed] [Google Scholar]
- [24].Ren Y, Xie J, Lin F, et al. Serum human epididymis protein 4 is a predictor for developing nephritis in patients with systemic lupus erythematosus: a prospective cohort study. Int Immunopharmacol 2018;60:189–93. [DOI] [PubMed] [Google Scholar]
- [25].He J, Sun M, Tian S. Procyanidin B2 prevents lupus nephritis development in mice by inhibiting NLRP3 inflammasome activation. Innate Immun 2018;1753425918780985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Rovin BH, Parikh SV. Lupus nephritis: the evolving role of novel therapeutics. Am J Kidney Dis 2014;63:677–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Meng Y, Deng S, Huang Z, et al. Evaluating the diagnostic and prognostic value of lone anti-Sm for autoimmune diseases using Euroimmun line immunoassays. Clin Rheumatol 2018. [DOI] [PubMed] [Google Scholar]
- [28].Alba P, Bento L, Cuadrado MJ, et al. Anti-dsDNA, anti-Sm antibodies, and the lupus anticoagulant: significant factors associated with lupus nephritis. Ann Rheum Dis 2003;62:556–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Ishizaki J, Saito K, Nawata M, et al. Low complements and high titre of anti-Sm antibody as predictors of histopathologically proven silent lupus nephritis without abnormal urinalysis in patients with systemic lupus erythematosus. Rheumatology (Oxford) 2015;54:405–12. [DOI] [PubMed] [Google Scholar]
- [30].Fathy SA, Mohamed MR, Ali MAM, et al. Influence of IL-6, IL-10, IFN-( and TNF-( genetic variants on susceptibility to diabetic kidney disease in type 2 diabetes mellitus patients. Biomarkers 2018;1–38. [DOI] [PubMed] [Google Scholar]
- [31].Kolber W, Dumnicka P, Maraj M, et al. Does the automatic measurement of interleukin 6 allow for prediction of complications during the first 48 h of acute pancreatitis? Int J Mol Sci 2018;19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].El-Shereef RR, Lotfi A, Abdel-Naeam EA, et al. Serum and urinary interleukin-6 in assessment of renal activity in egyptian patients with systemic lupus erythematosus. Clin Med Insights Arthritis Musculoskelet Disord 2016;9:29–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Raghav A, Ahmad J, Alam K. Preferential recognition of advanced glycation end products by serum antibodies and low-grade systemic inflammation in diabetes mellitus and its complications. Int J Biol Macromol 2018. [DOI] [PubMed] [Google Scholar]
- [34].Wolf SJ, Theros J, Reed TJ, et al. TLR7-mediated lupus nephritis is independent of type I IFN signaling. J Immunol 2018;201:393–405. [DOI] [PMC free article] [PubMed] [Google Scholar]


