Abstract
MicroRNA-191 (miR-191) has been identified as being upregulated in several types of cancers, and plays the role of oncogene. The expression of miR-191 has been found to be upregulated in prostate cancer tissues as well as cell lines. In this study, we analyzed the correlation of miR-191 expression with clinicopathologic factors and prognosis in prostate cancer.
Prostate cancer tissue samples and adjacent normal prostate tissue samples were collected from 146 patients who underwent laparoscopic radical prostatectomy between April 2013 and March 2018. Student two-tailed t-test was used for comparisons of 2 independent groups. The relationships between miR-191 expression and different clinicopathological characteristics were evaluated using the Chi-squared test. Kaplan–Meier survival plots and log-rank tests were used to assess the differences in overall survival of the different subgroups of prostate cancer patients.
miR-191 expression was significantly higher in prostate cancer tissues compared with normal adjacent prostate tissues (P < .001). miR-191 expression was observed to be significantly correlated with Gleason score (P < .001), pelvic lymph node metastasis (P = .006), bone metastases (P < .001), and T stage (P = .005). Kaplan–Meier analysis showed that patients with higher levels of miR-191 had significantly poorer survival than those with lower expression of this miRNA in prostate cancer patients (log rank test, P = .011). Multivariate analysis revealed that miR-191 expression (hazard ratio [HR] = 2.311, 95% confidence interval, [CI]: 1.666–9.006; P = .027) was independently associated with the overall survival of prostate cancer patients.
Our results demonstrated that miR-191 might serve as an independent prognostic indicator for prostate cancer patients.
Keywords: expression, microRNA-191, prognostic marker, prostate cancer, survival
1. Introduction
Prostate cancer is the most common cancer among western males and is the second-leading cause of cancer deaths among American males.[1] With the popularity of serum prostate specific antigen (PSA) screening, the incidence of prostate cancer is increasing year by year, especially in developing countries. Radical prostatectomy, including open and laparoscopic radical prostatectomy, is the standard treatment regimen for localized prostate cancer or oligometastatic prostate cancer.[2] Most patients with prostate cancer require endocrine adjuvant therapy after radical prostatectomy, and almost all of them will experience a recurrence of this disease and eventually develop into hormone-resistant prostate cancer.[3–5] Serum PSA monitoring is the most important means of postoperative management, but it cannot predict high-risk patients.[6] Therefore, we need new prognostic markers to guide postoperative treatment.
MicroRNAs (miRNAs) are a class of small, noncoding, endogenous single-stranded RNAs that modulate gene expression by binding to the 3’-untranslated region of target messenger RNAs (mRNAs).[7] miRNAs participate in many physiological processes, including cell proliferation, differentiation, apoptosis, and metabolism. Dysregulation of miRNA expression has been identified in various cancers, and miRNAs act as oncogenes or tumor suppressors in different types of cancers.[8–10]
MicroRNA-191 (miR-191) has been identified as being upregulated in several types of cancers, and plays the role of oncogene, including hepatocellular carcinoma (HCC),[11] breast cancer,[12] cholangiocarcinoma,[13] gastric adenocarcinoma,[14] and colorectal carcinoma (CRC).[15] The expression of miR-191 has been found to be upregulated in prostate cancer tissues as well as cell lines. Furthermore, miR-191 was able to regulate prostate cancer cell growth and invasion via targeting tissue inhibitor of metalloprotease 3 (TIMP3).[16] In this study, we analyzed the correlation of miR-191 expression with clinicopathologic factors and prognosis in prostate cancer.
2. Materials and methods
2.1. Patients and tissue samples
Prostate cancer tissue samples and adjacent normal prostate tissue samples were collected from 146 patients who underwent laparoscopic radical prostatectomy between April 2013 and March 2018 in the Department of Urology, Dongzhimen Hospital, Beijing University of Chinese Medicine. No patient received any preoperative chemotherapy, endocrine therapy, or radiotherapy. Samples were snap frozen in liquid nitrogen and stored at −80°C for future isolation of RNA. The prostate cancer tissue specimens were subjected to histological examination by 2 pathologists for Gleason grading and TNM staging. The pathological type of all the 146 patient samples is prostate adenocarcinoma. The use of tissues for this study has been approved by the ethics committee of Dongzhimen Hospital, Beijing University of Chinese Medicine, and each patient have given prior written informed consent and approval.
2.2. RNA isolation and quantitative real-time PCR
Total RNA from prostate cancer tissue samples and adjacent normal prostate tissue samples were extracted using Trizol reagent (Invitrogen). The miR-191 expression level was measured by using TaqMan miRNA detection kit (Applied Biosystems, Foster, CA), according to the instructions provided by the manufacturer. RNA samples were reversely transcribed with miRNA-specific primers from TaqMan miRNA Reverse Transcription Kit (Applied Biosystems, New York). Real-time polymerase chain reaction (RT-PCR) was performed with a TaqMan probe, which ensures discrimination of even 1 nucleotide difference, by using 7300 sequence detection system (Applied Biosystems). The U6 was chosen as the endogenous normalizer. Each sample was analyzed in triplicate. The primers were synthesized as follows: miR-191-Fwd, 5’-TGCGGCAACGGAATCCCAA AAGC-3’; U6-Fwd, 5’-TGCGGGTGCTCGCTTCGGCAGC-3’; and a universal antisense primer reverse, 5’-CCAGTGCAGGGTCCGAGGT-3’.
2.3. Statistical analysis
Student two-tailed t-test was used for comparisons of 2 independent groups. The relationships between miR-191 expression and different clinicopathological characteristics were evaluated using the Chi-squared test. Kaplan–Meier curves were constructed to determine the patient overall survival rates. Kaplan–Meier survival plots and log-rank tests were used to assess the differences in overall survival of the different subgroups of prostate cancer patients. The associations between prostate cancer prognosis and clinical parameters were first analyzed by univariate analysis, and those significantly differing were enrolled in multiple logistic regression with a forward stepwise procedure to identify independent risk factors for prognosis in prostate cancer. P < .05 was considered statistically significant. All statistical analyses were performed using the Statistical Package of the Social Sciences software (SPSS Inc, Chicago, IL) version 22.0.
3. Results
3.1. The expression level of miR-191 in clinical prostate cancer tissues and adjacent normal tissues
We analyzed the expression levels of miR-191 in 146 pairs of prostate cancer samples and normal adjacent samples from 146 patients with prostate cancer. As revealed by quantitative RT-PCR analysis, miR-191 expression was significantly higher in prostate cancer tissues compared with normal adjacent prostate tissues (P < .001, shown in Fig. 1). The 146 prostate cancer patients were classified into 2 groups according to the median of miR-191 expression level as determined by quantitative RT-PCR. Seventy-six cases were placed in the high expression group and 70 in the low expression group.
Figure 1.
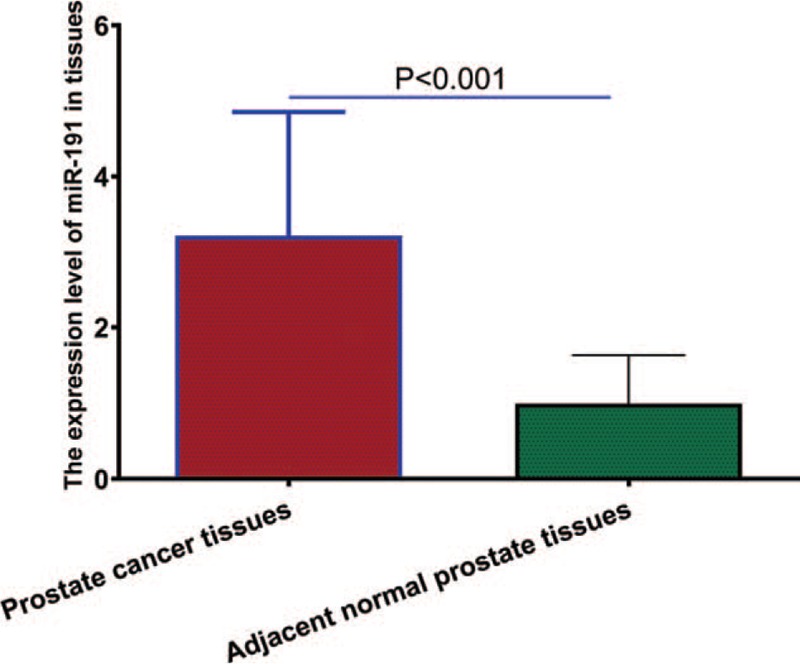
Relative miR-191 expression levels between prostate cancer tissues and adjacent normal prostate tissues. miR-191 = microRNA-191.
3.2. Correlations of miR-191 expression with clinicopathologic features of patients with prostate cancer
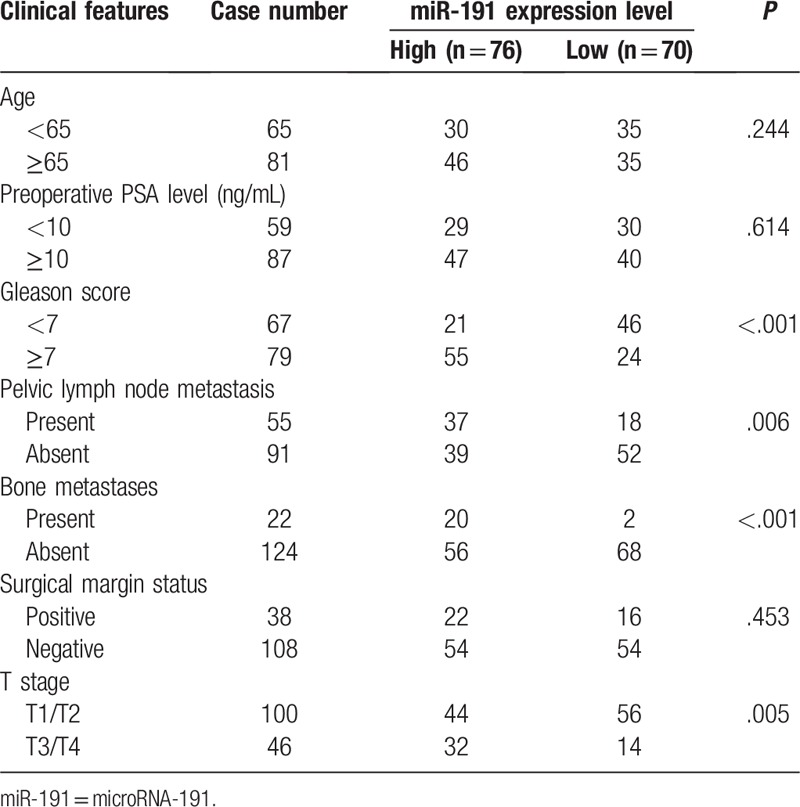
The correlations of miR-191 expression with clinicopathologic features of patients with prostate cancer were statistically analyzed. As shown in Table 1, miR-191 expression level was observed to be significantly correlated with Gleason score (P < .001), pelvic lymph node metastasis (P = .006), bone metastases (P < .001), and T stage (P = .005). However, there were no significant correlations between miR-191 expression level and other clinicopathologic factors, including age, preoperative PSA level, and surgical margin status (all P > .05).
Table 1.
Association between microRNA-191 expression and clinicopathological parameters of prostate cancer.
3.3. Relationship between miR-191 expression and prostate cancer patients’ survival
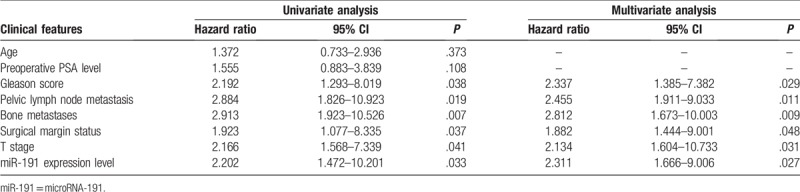
To evaluate whether miR-191 expression can predict prostate cancer prognosis, we next performed survival analysis. Kaplan–Meier analysis showed that patients with higher levels of miR-191 had significantly poorer survival than those with lower expression of this miRNA in prostate cancer patients (log rank test, P = .011; shown in Fig. 2). However, Kaplan–Meier analysis showed that the preoperative PSA levels could not predict survival in prostate cancer patients (log rank test, P = .275; shown in Fig. 3). Univariate and multivariate analyses were utilized to evaluate whether the miR-191 expression level and various clinicopathological features were independent prognostic parameters of prostate cancer patient outcomes. Multivariate analysis revealed that miR-191 expression (HR = 2.311, 95% CI: 1.666–9.006; P = .027) was independently associated with the overall survival of prostate cancer patients (shown in Table 2).
Figure 2.
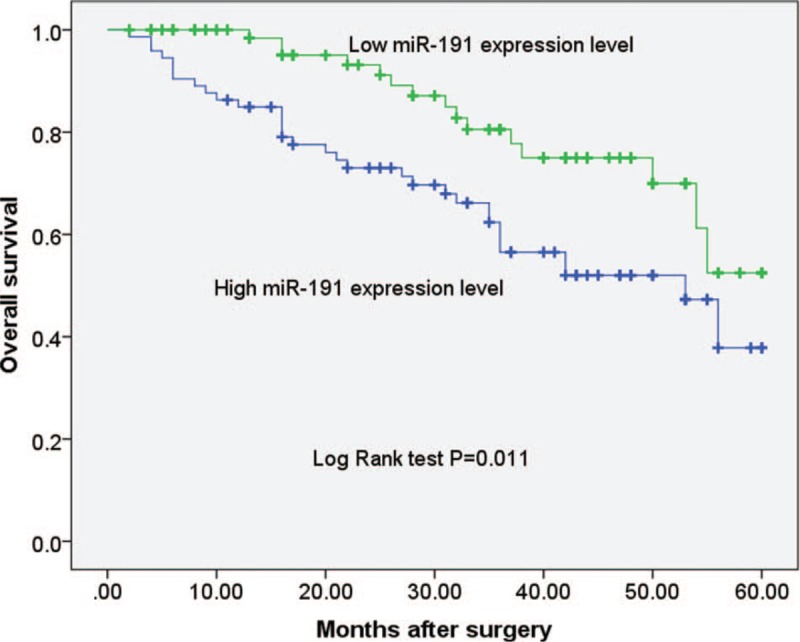
Kaplan–Meier survival curves for patients with prostate cancer expressing high and low levels of miR-191. miR-191 = microRNA-191.
Figure 3.
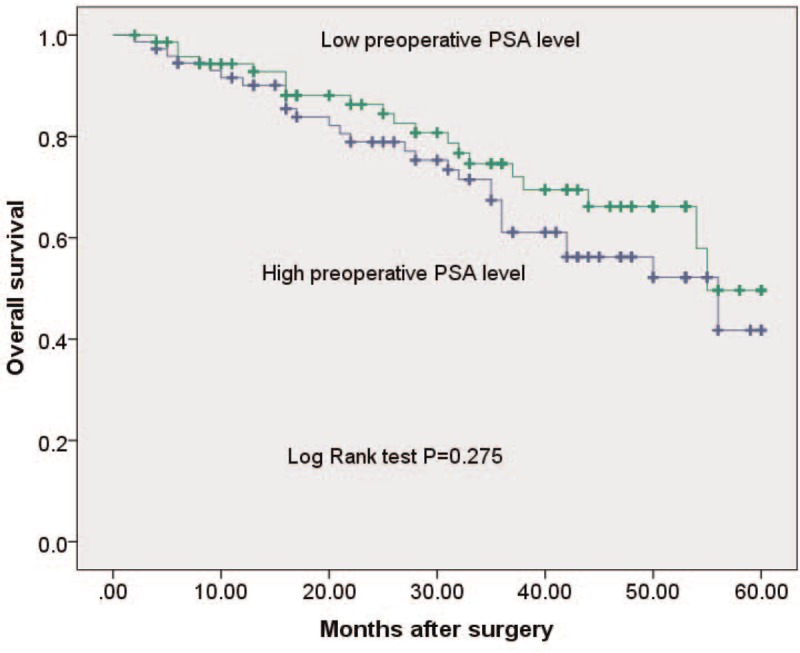
Kaplan–Meier survival curves for prostate cancer patients with high and low levels of preoperative PSA level. miR-191 = microRNA-191.
Table 2.
Univariate and multivariate survival analysis of overall survival in 146 prostate cancer patients.
4. Discussion
PSA testing is widely used in the clinic for the diagnosis and postoperative follow-up of prostate cancer, but it is not sufficient for an accurate early diagnosis and prognosis. Moreover, some factors including benign prostatic hyperplasia, glandular inflammation, drug therapy, and the finger test can directly affect serum PSA levels.[17,18] Current techniques are limited to distinguish the prostate cancer patients that are most at risk of metastasis and death. Therefore, the novel, specific, and efficient diagnostic and prognostic biomarkers for prostate cancer are needed for prediction of aggressive prostate cancer and improvement of clinical outcome of prostate cancer patients. miRNAs can contribute to cancer development and progression and are differentially expressed in normal tissues and cancer tissues.[19] Deregulated miRNA expression has been reported in many cancer types, including prostate cancer. Many studies have shown that the specific expression of miRNAs in cancer tissues can be used as a prognostic factor for prostate cancer.[20,21]
miR-191 has been viewed as an oncomiR in several types of cancers. For example, Elyakim et al[11] found that miR-191 was upregulated in HCC tissues and inhibition of miR-191 decreased cell proliferation and induced apoptosis in vitro and significantly reduced tumor masses in vivo in an orthotopic xenograft mouse model of HCC. Additionally, miR-191 was found to be upregulated by a dioxin, a known liver carcinogen, and was found to be a regulator of a variety of cancer-related pathways.[11] Nagpal et al[12] showed that miR-191 functioned as an oncogenic regulator in human breast cancer. miR-191-mediated enhanced cell proliferation and migration were partly dependent on targeted downregulation of special AT rich sequence binding protein 1.[12] Kang et al[13] found that miR-191 was upregulated in cholangiocarcinoma cell lines and patients. Knockdown of miR-191 by transfection of its inhibitor sequence blocked cholangiocarcinoma cells viability and induced apoptosis of cholangiocarcinoma cells. Furthermore, miR-191 regulated Wnt/β-catenin signaling pathway via secreted frizzled-related protein-1.[13] Shi et al[14] found that miR-191 was at a high-expression level in human gastric adenocarcinoma cell line and human gastric cancer tissues. miR-191 could promote cell growth and suppress apoptosis of gastric adenocarcinoma cells. The N-deacetylase/N-sulfotransferase 1 was confirmed to be a direct target gene of miR-191 by enhanced green fluorescent protein reporter experiment.[14] Qin et al[15] demonstrated that high miR-191 expression was associated with clinical stage, lymph node metastasis, liver metastasis, and depth of tumor invasion of CRC. Furthermore, they found that TIMP3 was a direct target of miR-191 in colorectal cancer SW620 cells.[15] The prognostic value of miR-191 in various cancer types have also been investigated. For example, in the study by Gao et al,[22] Kaplan–Meier and Cox regression analyses demonstrated that overexpression of miR-191 was an independent and significant predictor of esophageal squamous cell carcinoma prognosis. Song et al[23] found that the expression of miR-191 closely associated with the tumor size, pTNM stage, lymph node metastasis and perineural invasion, and poor prognosis of pancreatic cancer. In the study by Qin et al,[15] Kaplan–Meier analysis indicated that patients with high miR-191 expression had a poor overall survival in colorectal cancer. Moreover, multivariate analysis showed that miR-191 was an independent prognostic factor in patients with colorectal cancer.[15]
The expression of miR-191 has been found to be upregulated in prostate cancer tissues as well as cell lines. Furthermore, miR-191 has been found to be able to regulate prostate cancer cell growth and invasion via targeting TIMP3.[16] However, the clinicopathologic value and prognosis of miR-191 expression in prostate cancer have not been investigated. In the present study, we analyzed the expression levels of miR-191 in 146 pairs of prostate cancer samples and normal adjacent samples from 146 patients with prostate cancer. As revealed by quantitative RT-PCR analysis, miR-191 expression was significantly higher in prostate cancer tissues compared with normal adjacent prostate tissues. The correlations of miR-191 expression with clinicopathologic features of patients with prostate cancer were statistically analyzed. miR-191 expression level was observed to be significantly correlated with Gleason score, pelvic lymph node metastasis, bone metastases, and T stage. However, there were no significant correlations between miR-191 expression level and other clinicopathologic factors, including age, preoperative PSA level, and surgical margin status. To evaluate whether miR-191 expression can predict prostate cancer prognosis, we next performed survival analysis. Kaplan–Meier analysis showed that patients with higher levels of miR-191 had significantly poorer survival than those with lower expression of this miRNA in prostate cancer patients. Univariate and multivariate analyses were utilized to evaluate whether the miR-191 expression level and various clinicopathological features were independent prognostic parameters of prostate cancer patient outcomes. Multivariate analysis revealed that miR-191 expression was independently associated with the overall survival of prostate cancer patients. The limitation of the present study was that we have not shown the miRNA in situ hybridization to clarify miR-191 expression in prostate cancer tissue compared with normal adjacent tissue, and only statistics data was not enough.
In conclusions, our results demonstrated that miR-191 may serve as an important prognostic indicator for prostate cancer patients and more studies are needed to confirm our findings in the future.
Author contributions
Conceptualization: Jing-bo Liu, Lin-feng Dai.
Data curation: Jing-bo Liu, Jing Shi, Ya-bing Wu, Lin-feng Dai.
Formal analysis: Jing-bo Liu, Jing Shi.
Funding acquisition: Xue-tao Ma.
Investigation: Jing-bo Liu, Yong-ji Yan, Jing Shi, Ya-bing Wu.
Methodology: Jing-bo Liu, Yong-ji Yan, Jing Shi, Yan-feng Li.
Project administration: Jing-bo Liu.
Resources: Jing-bo Liu, Ya-bing Wu.
Software: Jing-bo Liu, Yong-ji Yan, Ya-bing Wu.
Supervision: Jing-bo Liu.
Validation: Jing-bo Liu, Yan-feng Li.
Visualization: Jing-bo Liu.
Writing – original draft: Jing-bo Liu, Jing Shi, Lin-feng Dai.
Writing – review & editing: Xue-tao Ma.
Footnotes
Abbreviations: HCC = hepatocellular carcinoma, miRNAs= microRNAs.
The authors have no conflicts of interest to disclose.
References
- [1].Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394–424. [DOI] [PubMed] [Google Scholar]
- [2].Droz JP, Albrand G, Gillessen S, et al. Management of prostate cancer in elderly patients: recommendations of a Task Force of the International Society of Geriatric Oncology. Eur Urol 2017;72:521–31. [DOI] [PubMed] [Google Scholar]
- [3].Fletcher RH. Guideline: Experts recommend against prostate cancer screening with prostate-specific antigen test. Ann Intern Med 2019;170:JC2. [DOI] [PubMed] [Google Scholar]
- [4].Cornford P, Bellmunt J, Bolla M, et al. EAU-ESTRO-SIOG Guidelines on prostate cancer. Part II: Treatment of relapsing, metastatic, and castration-resistant prostate cancer. Eur Urol 2017;71:630–42. [DOI] [PubMed] [Google Scholar]
- [5].Mottet N, Bellmunt J, Bolla M, et al. EAU-ESTRO-SIOG Guidelines on prostate cancer. Part 1: screening, diagnosis, and local treatment with curative intent. Eur Urol 2017;71:618–29. [DOI] [PubMed] [Google Scholar]
- [6].Gillessen S, Attard G, Beer TM, et al. Management of patients with advanced prostate cancer: the report of the Advanced Prostate Cancer Consensus Conference APCCC 2017. Eur Urol 2018;73:178–211. [DOI] [PubMed] [Google Scholar]
- [7].Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009;136:215–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kohlhapp FJ, Mitra AK, Lengyel E, Peter ME. MicroRNAs as mediators and communicators between cancer cells and the tumor microenvironment. Oncogene 2015;34:5857–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Le XF, Merchant O, Bast RC, et al. The roles of MicroRNAs in the cancer invasion-metastasis cascade. Cancer Microenviron 2010;3:137–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Farazi TA, Spitzer JI, Morozov P, et al. miRNAs in human cancer. J Pathol 2011;223:102–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Elyakim E, Sitbon E, Faerman A, et al. hsa-miR-191 is a candidate oncogene target for hepatocellular carcinoma therapy. Cancer Res 2010;70:8077–87. [DOI] [PubMed] [Google Scholar]
- [12].Nagpal N, Ahmad HM, Molparia B, et al. MicroRNA-191, an estrogen-responsive microRNA, functions as an oncogenic regulator in human breast cancer. Carcinogenesis 2013;34:1889–99. [DOI] [PubMed] [Google Scholar]
- [13].Kang PC, Leng KM, Liu YP, et al. miR-191 inhibition induces apoptosis through reactivating secreted frizzled-related protein-1 in cholangiocarcinoma. Cell Physiol Biochem 2018;49:1933–42. [DOI] [PubMed] [Google Scholar]
- [14].Shi X, Su S, Long J, et al. MicroRNA-191 targets N-deacetylase/N-sulfotransferase 1 and promotes cell growth in human gastric carcinoma cell line MGC803. Acta Biochim Biophys Sin 2011;43:849–56. [DOI] [PubMed] [Google Scholar]
- [15].Qin S, Zhu Y, Ai F, et al. MicroRNA-191 correlates with poor prognosis of colorectal carcinoma and plays multiple roles by targeting tissue inhibitor of metalloprotease 3. Neoplasma 2014;61:27–34. [PubMed] [Google Scholar]
- [16].Wang X, Shi Z, Liu X, et al. Upregulation of miR-191 promotes cell growth and invasion via targeting TIMP3 in prostate cancer. J BUON 2018;23:444–52. [PubMed] [Google Scholar]
- [17].Hamdy FC, Donovan JL, Lane JA, et al. 10-year outcomes after monitoring, surgery, or radiotherapy for localized prostate cancer. N Engl J Med 2016;375:1415–24. [DOI] [PubMed] [Google Scholar]
- [18].Sox HC. Quality of life and guidelines for PSA screening. N Engl J Med 2012;367:669–71. [DOI] [PubMed] [Google Scholar]
- [19].Liu HN, Wu H, Tseng YJ, et al. Serum microRNA signatures and metabolomics have high diagnostic value in gastric cancer. BMC cancer 2018;18:415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Lu L, Mao X, Shi P, et al. MicroRNAs in the prognosis of triple-negative breast cancer: a systematic review and meta-analysis. Medicine 2017;96:e7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Tutar Y. miRNA and cancer; computational and experimental approaches. Curr Pharm Biotechnol 2014;15:429. [DOI] [PubMed] [Google Scholar]
- [22].Gao X, Xie Z, Wang Z, et al. Overexpression of miR-191 predicts poor prognosis and promotes proliferation and invasion in esophageal squamous cell carcinoma. Yonsei Med J 2017;58:1101–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Song Z, Ren H, Gao S, et al. The clinical significance and regulation mechanism of hypoxia-inducible factor-1 and miR-191 expression in pancreatic cancer. Tumour Biol 2014;35:11319–28. [DOI] [PubMed] [Google Scholar]


