Abstract
Several studies have suggested that long intergenic noncoding RNAs are involved in the progression of diabetic nephropathy (DN). However, the exact role and regulatory mechanism of long noncoding RNA (lncRNA) NR_038323 in diabetic nephropathy (DN) remain largely unclear. In the present study, we found that lncRNA NR_038323 overexpression ameliorated the high glucose (HG)-induced expression levels of collagen I, collagen IV, and fibronectin, whereas lncRNA NR_038323 knockdown exerted the opposite effects. Moreover, the results of bioinformatic prediction, luciferase assay, and fluorescence in situ hybridization (FISH) demonstrated that lncRNA NR_038323 directly interacted with miR-324-3p. Additionally, miR-324-3p mimic aggravated the HG-induced expression levels of collagen I, collagen IV, and fibronectin by dual-specificity protein phosphatase-1 (DUSP1) expression to activate p38 mitogen-activated protein kinase (MAPK) and ERK1/2 pathways. In contrast, overexpression of DUSP1 attenuated the HG-induced expression levels of collagen I, collagen IV, and fibronectin via inactivation of p38 MAPK and ERK1/2 pathways. In addition, lncRNA NR_038323 knockdown increased the expression levels of collagen I, collagen IV, and fibronectin by upregulating DUSP1 expression during HG treatment, which were markedly reversed by miR-324-3p inhibitor. Furthermore, these molecular changes were verified in the human kidney samples of DN patients. Finally, overexpression of lncRNA NR_038323 ameliorated the interstitial fibrosis in STZ-induced diabetic nephrology (DN) rat via miR-324-3p/DUSP1/p38MAPK and ERK1/2 axis. In conclusion, our data indicate that overexpression of lncRNA NR_038323 may suppress HG-induced renal fibrosis via the miR-324-3p/DUSP1/p38MAPK and ERK1/2 axis, which provides new insights into the pathogenesis of DN.
Keywords: diabetic nephropathy, lncRNA NR_038323, miR-324-3p, DUSP1
Introduction
Diabetic nephropathy (DN), a major cause of chronic kidney disease (CKD), is characterized by progressive renal tubulointerstitial fibrosis.1 Renal proximal tubular epithelial cells (PTECs) play a pivotal role in the pathogenesis of DN.2 A large number of studies have demonstrated that high glucose (HG) induces renal tubular cell hypertrophy and increases the levels of cytokines and growth factors by activating multiple signaling pathways and subsequently lead to fibrosis.2, 3, 4, 5 However, the pathogenetic mechanism underlying HG-induced fibrosis remains largely unclear.
Long noncoding RNAs (lncRNAs) are defined as transcripts longer than 200 nucleotides with little (or no) protein-coding ability6 and involved in a wide variety of physiological and pathological processes, including DN.4, 7, 8, 9, 10, 11, 12 We used the lncRNA chip to select the lncRNA NR_038323. It is located on chromosome 8 (Chr8:23336208–23366125), but its biological functions, regulatory mechanisms, and disease relevance remain largely unclarified. To the best of our knowledge, most lncRNAs act as competing endogenous RNAs (ceRNAs) to regulate the expression levels of targeted genes by interacting with microRNAs (miRNAs).13, 14, 15 Therefore, we hypothesized that lncRNA NR_038323 is involved in the progression of renal fibrosis in DN via miRNA regulation.
In this study, the expression levels of lncRNA NR_038323 were induced by HG treatment at different time points, as detected using real-time qPCR. In addition, HG-induced renal fibrosis was aggravated by the knockdown of lncRNA NR_038323 while ameliorated by its overexpression in HK-2 cells. Moreover, the interaction between lncRNA NR_038323 and miR-324-3p was revealed by bioinformatic analysis, dual-luciferase reporter assay and fluorescence in situ hybridization (FISH) method. The data further indicated that lncRNA NR_038323 upregulated dual-specificity protein phosphatase-1 (DUSP1) expression to inhibition the activation of p38 mitogen-activated protein kinase (p38MAPK) and ERK1/2 by sponging miR-324-3p. Additionally, the molecular mechanisms underlying lncRNA NR_038323-suppressed renal fibrosis were uncovered. Finally, overexpression of lncRNA NR_038323 ameliorated the STZ-induced progression of DN. Taken together, our results demonstrated that overexpression of lncRNA NR_038323 may inhibit renal fibrosis in DN via miR-324-3p/DUAP1/p38MAPK and ERK1/2 axis.
Results
HG Induced the Expression of lncRNA NR_038323
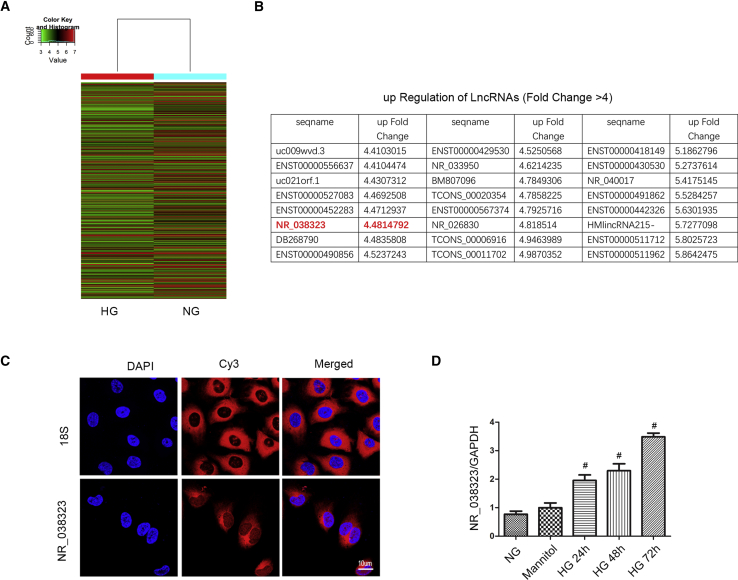
Previous findings demonstrated that lncRNA ZEB1-AS1 was suppressed by HG treatment.4 In the present study, HK-2 cells was treated with HG or normal glucose (NG) for 72 h, and then mRNA was extracted for the lncRNA chip assay. A representative heatmap of lncRNA was shown in Figure 1A. Upregulation (fold change was more than 4) of comparisons of lncRNAs between HG and NG groups was shown in Figure 1B. Among them, fold change of lncRNA NR_038323 in HG versus NG was 4.48. To further confirm the expression of it, FISH method was used, nuclei was stained by DAPI, and 18S rRNA (cytoplasmic positive) and lncRNA NR_038323 were labeled with CY3. The results showed that lncRNA NR_038323 was localized in the cytoplasm of HK-2 cells (Figure 1C). In addition, real-time qPCR analysis revealed that lncRNA NR_038323 was induced by HG at 24–72 h (Figure 1D). These data suggest that lncRNA NR_038323 may be a DN-related factor.
Figure 1.
HG Induced the Expression of lncRNA NR_038323
HK-2 cells were treated with NG, mannitol, or HG at indicated time points. (A) The heatmap of lncRNAs. (B) The sequence name and fold change of upregulation of lncRNAs. (C) Intracellular localization of lncRNA NR_038323 in HK-2 cells was carried out using RNA-FISH assay. (D) The expression levels of lncRNA NR_038323 were detected by real-time qPCR. Data are expressed as mean ± SD (n = 6). #p < 0.05, HG at 24–72 h groups versus NG or mannitol group.
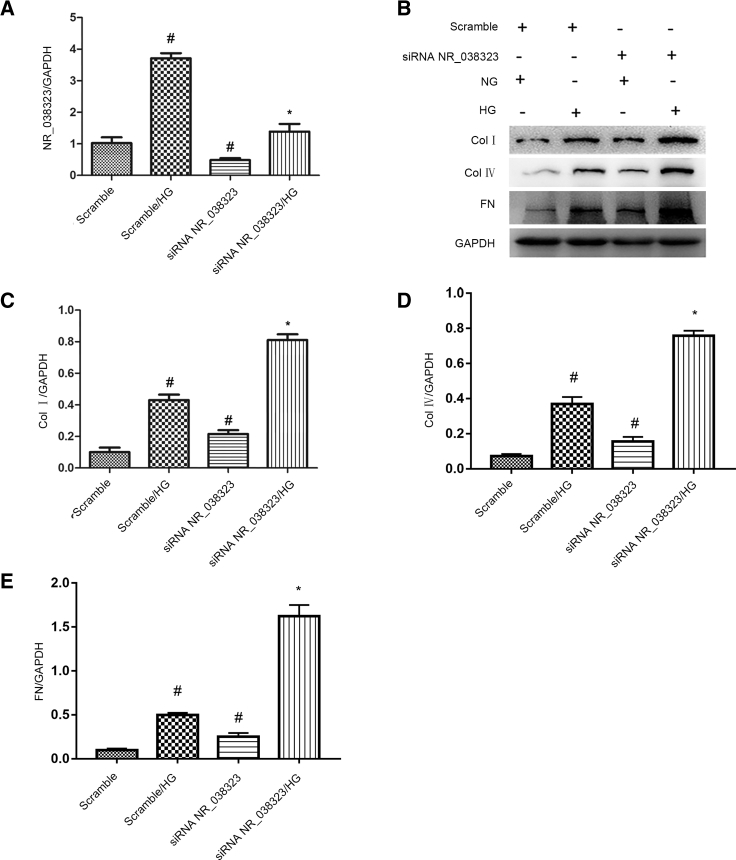
lncRNA NR_038323 Inhibition Enhanced the HG-Induced Expression Levels of Collagen I, Collagen IV, and Fibronectin
The effects of lncRNA NR_038323 on HG-induced renal fibrosis were further clarified. The HG-induced expression level of lncRNA NR_038323 was notably silenced by small interfering RNA (siRNA) lncRNA NR_038323 in HK-2 cells (Figure 2A). In addition, siRNA lncRNA NR_038323 not only increased the basal levels of collagen I, collagen IV, and fibronectin, but also markedly enhanced the HG-induced expression levels of them (Figures 2B–2E). These data indicate that knockdown of endogenous lncRNA NR_038323 enlarged the HG-induced the renal fibrosis in the HK-2 cells, which further supported that lncRNA NR_038323 has an anti-fibrosis role in HK-2 cells.
Figure 2.
lncRNA NR_038323 Knockdown Aggravated the HG-Reduced Expression Levels of Collagen I, Collagen IV, and Fibronectin
HK-2 cells were transfected with 50 nM lncRNA NR_038323 siRNA or scramble and then treated with or without HG for 72 h. (A) Real-time qPCR analysis of lncRNA NR_038323 expression. (B) Western blot analysis of collagen I, collagen IV, and fibronectin. (C–E) Densitometric measurement of western blot bands for collagen I (C), collagen IV (D), and fibronectin (E). Data are expressed as mean ± SD (n = 6). #p < 0.05, scramble with HG or lncRNA NR_038323 siRNA group versus scramble group; *p < 0.05, lncRNA NR_038323 siRNA with HG group versus scramble with HG group.
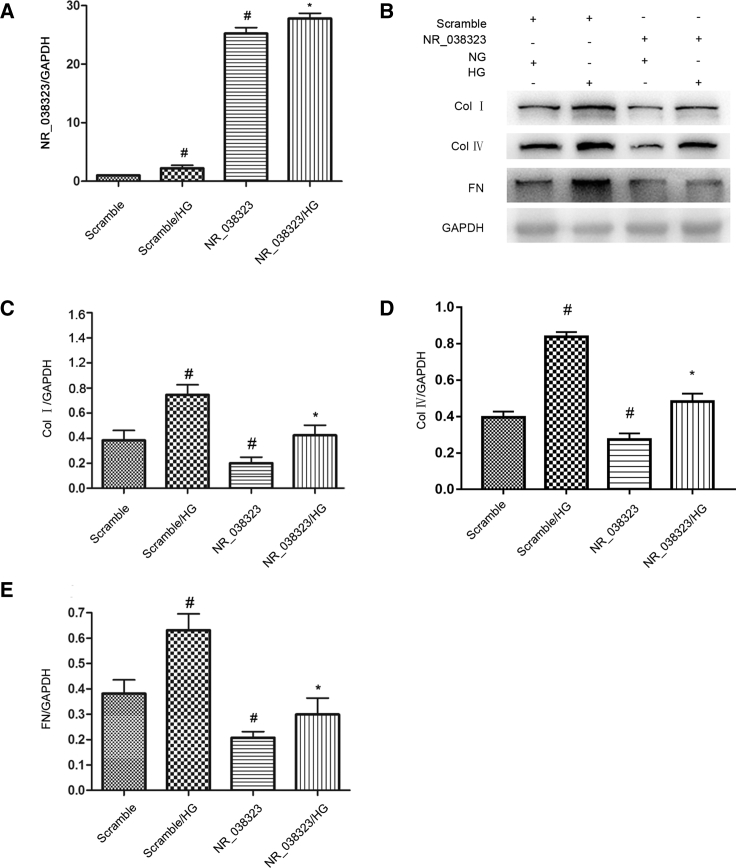
lncRNA NR_038323 Overexpression Attenuated the HG-Induced Expression Levels of Collagen I, Collagen IV, and Fibronectin
Although lncRNA NR_038323 was induced by HG treatment, the role of it in HG-induced renal fibrosis remains unclear. The HG-induced expression level of lncRNA NR_038323 in HK-2 cells was significantly enhanced by lncRNA NR_038323 overexpression (Figure 3A). Besides, lncRNA NR_038323 overexpression not only suppressed the basal levels of collagen I, collagen IV, and fibronectin, but also markedly attenuated the HG-induced expression levels of them (Figures 3B–3E). These data further suggested that endogenous lncRNA NR_038323 induced by HG was not enough to limit the HG-induced increasing of ECM; overexpression of lncRNA NR_038323 almost completely suppressed the HG-induced the ECM accumulation and supported the anti-fibrosis role of it in HK-2 cells.
Figure 3.
Overexpression of lncRNA NR_038323 Attenuated the HG-Deduced Expression Levels of Collagen I, Collagen IV, and Fibronectin
HK-2 cells were transfected with lncRNA NR_038323 plasmid or control and then treated with or without HG for 72 h. (A) Real-time qPCR analysis of lncRNA NR_038323 expression. (B) Western blot analysis of collagen I, collagen IV, and fibronectin. (C–E) Densitometric measurement of western blot bands for collagen I (C), collagen IV (D), and fibronectin (E). Data are expressed as mean ± SD (n = 6). #p < 0.05, control with HG or lncRNA NR_038323 group versus control group; *p < 0.05, lncRNA NR_038323 with HG group versus control with HG group.
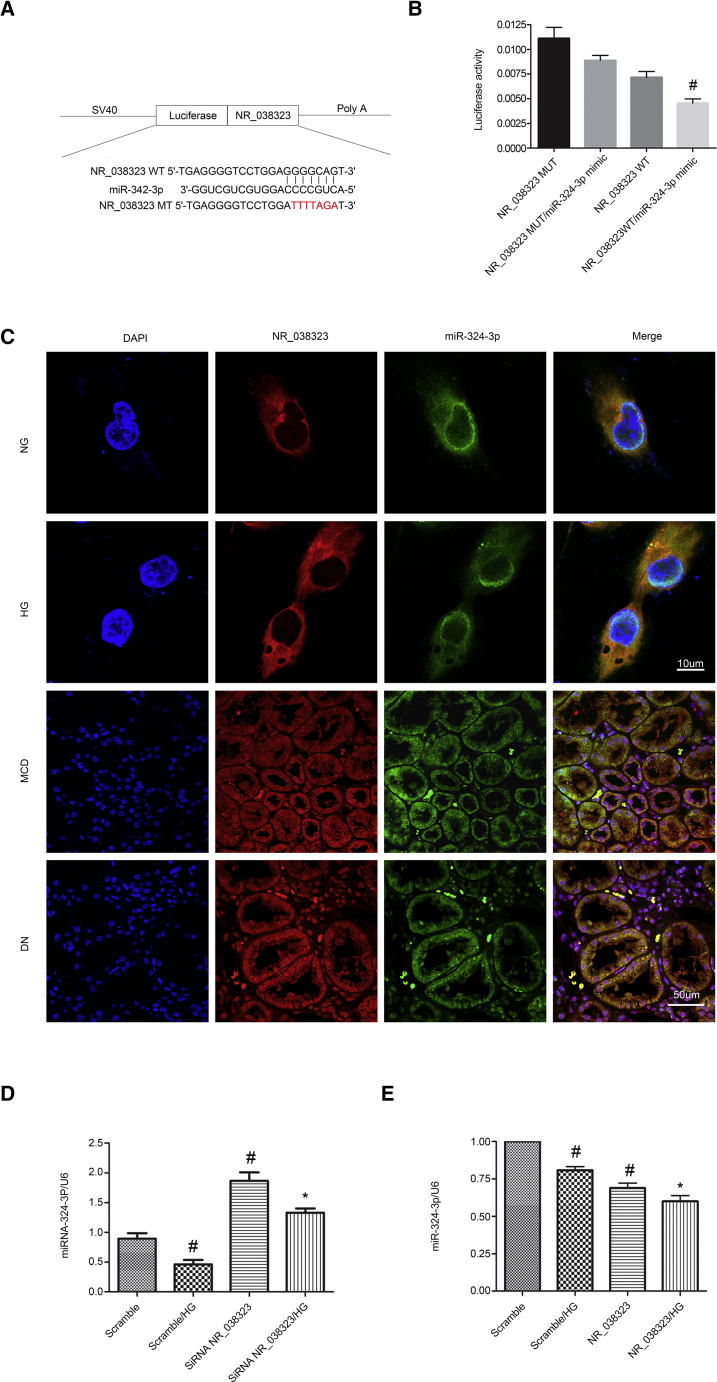
lncRNA NR_038323 Suppressed the Expression and Activity of miR-324-3p
To our knowledge, miRNAs could be sponged by lncRNAs. We predicated that miR-324-3p was a potential downstream target of lncRNA NR_038323 by using RegRNA 2.0 software. As shown in Figure 4A, lncRNA NR_038323 contained the binding site of miR-324-3p. The results of luciferase reporter assay showed that miR-324-3p mimic inhibited the luciferase activity of lncRNA NR_038323-WT, but not lncRNA NR_038323-MUT (Figure 4B). Besides, the results of intracellular co-localization demonstrated that lncRNA NR_038323 interacted with miR-324-3p in the cytoplasm of HK-2 cells treated with or without HG as well as the renal tubular cells of minimal change disease (MCD) and DN patients (Figure 4C). In addition, the HG-suppressed expression of miR-324-3 was enhanced by lncRNA NR_038323 overexpression, but this effect was reversed by lncRNA NR_038323 knockdown (Figures 4D and 4E). Collectively, these results suggest that miR-324-3p is a direct target of lncRNA NR_038323.
Figure 4.
lncRNA NR_038323 Directly Bound to miR-324-3p
(A) Sequence alignment analysis revealed that lncRNA NR_038323 contained the complementary strand to miR-324-3p. (B) Detection of luciferase activities after co-transfection with lncRNA NR_038323-WT or lncRNA NR_038323-MUT and miR-324-3p or scramble. (C) Intracellular co-localization of lncRNA NR_038323 and miR-324-3p in HK-2 cells and human DN kidney samples. (D and E) Real-time qPCR analysis of lncRNA NR_038323 expression. Data are expressed as mean ± SD (n = 6). #p < 0.05, Scramble with HG group versus Scramble group; * p < 0.05, siRNA NR_038323 or NR_038323 with HG group versus Scramble with HG group or NR_038323 WT/miR-324-3p mimic versus other groups.
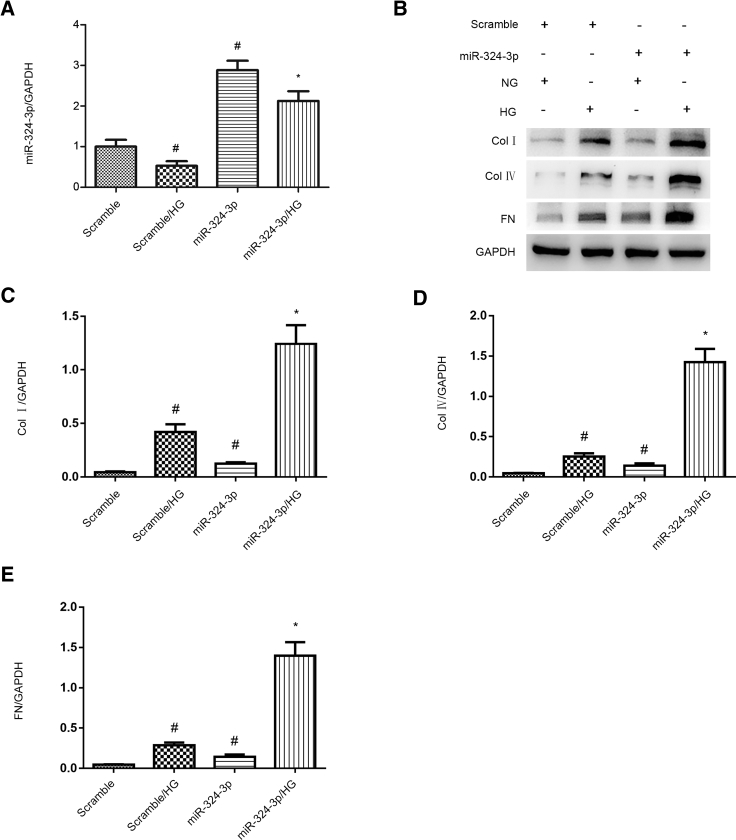
miR-324-3p Mimic Enhanced the HG-Induced Expression Levels of Collagen I, Collagen IV, and Fibronectin
Previous findings have suggested that miR-324-3p promotes the development of renal fibrosis in progressive proteinuric nephropathy.16 In the present study, we hypothesized that miR-324-3p could mediate HG-induced renal fibrosis. The results showed that the expression levels of miR-324-3p was elevated by miR-324-3p mimic (Figure 5A). Furthermore, miR-324-3p mimic enhanced the HG-induced expression levels of collagen I, collagen IV, and fibronectin (Figures 5B–5E). These data suggest that miR-324-3p can mediate HG-induced renal fibrosis.
Figure 5.
Overexpression of miR-324-3p Aggravated the HG-Induced Expression Levels of Collagen I, Collagen IV, and Fibronectin
HK-2 cells were transfected with 100 nM miR-324-3p mimics or scramble and then treated with or without HG for 72 h. (A) The mRNA expression levels of miR-324-3p were detected by real-time qPCR. (B) The protein expression levels of collagen I, collagen IV, and fibronectin were analyzed by western blotting. (C–E) Densitometric measurement of western blot bands for collagen I (C), collagen IV (D), and fibronectin (E). Data are expressed as mean ± SD (n = 6). #p < 0.05, scramble with HG group versus scramble group; *p < 0.05, miR-324-3p mimics with HG group versus scramble with HG group.
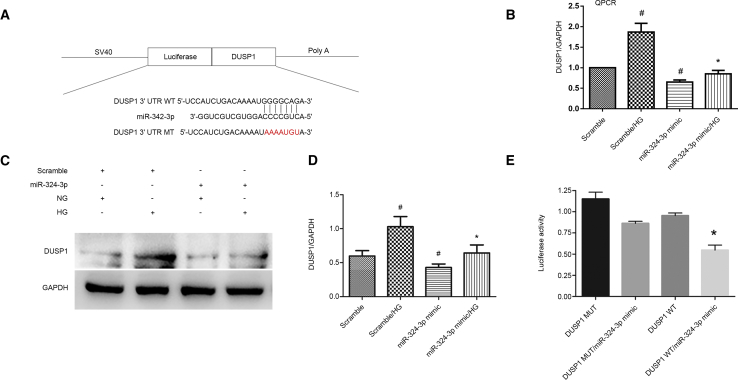
DUSP1 Was a Direct Target Gene of miR-324-3p
DUSP1 ameliorates microvascular fibrosis and inflammation via dephosphorylation of MAPK.17, 18, 19 In this study, DUSP1 was identified as the target gene of miR-324-3p, as predicted by miRbase (Figure 6A). Moreover, miR-324-3p mimic significantly suppressed the HG-induced expression of DUSP1 (Figures 6B–6D). The results of luciferase assay showed that miR-324-3p mimic inhibited the luciferase activity of DUSP1-WT, but not DUSP1-MUT (Figure 6E). Taken together, these data demonstrate that DUSP1 is a target gene of miR-324-3p.
Figure 6.
DUSP1 Was Identified as a Target Gene of miR-324-3p
HK-2 cells were transfected with miR-324-3p analog (100 nM) and then treated with HG for 72 h. (A) Putative miR-301a-5p complementary binding sites in the 3′ UTR of human DUSP1 mRNA. (B) The mRNA expression levels of DUSP1 were detected by RT-qPCR. (C) Western blot analysis of DUSP1 and GAPDH. (D) Densitometric analysis of proteins signals. (E) Measurement of luciferase activities after co-transfection with the 3′ UTR luciferase reporter vector of human WT or MUT-DUSP1 and miR-324-3p or miR-NC. Data are expressed as mean ± SD (n = 6). #p < 0.05, scramble with HG group versus scramble group; *p < 0.05, miR-324-3p mimics with HG group versus scramble with HG group or DUSP1 WT/miR-324-3p mimic versus other groups.
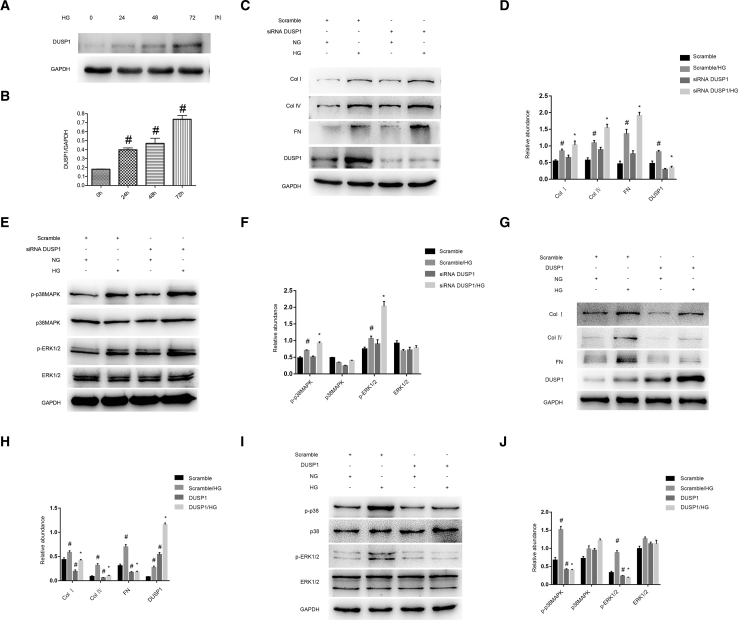
DUSP1 Mediated the HG-Induced Expression Levels of Collagen I, Collagen IV, and Fibronectin via Regulation of p38MAPK and ERK1/2
Although DUSP1 is involved in microvascular fibrosis, its role in HG-induced fibrosis remains largely unclear. Initially, DUSP1 was induced by HG treatment at indicated time points (Figures 7A and 7B). DUSP1 knockdown enhanced the HG-induced expression levels of collagen I, collagen IV, and fibronectin (Figures 7C and 7D). Moreover, DUSP1 knockdown triggered the activation of p38MAPK and ERK1/2 pathways (Figures 7E and 7F). In contrast, overexpression of DUSP1 ameliorated the HG-induced expression levels of collagen I, collagen IV, and fibronectin via inactivation of p38MAPK and ERK1/2 pathways (Figures 7G–7J). Collectively, these data indicate that DUSP1 plays an anti-fibrotic role in HK-2 cell by inactivating both p38MAPK and ERK1/2 pathways.
Figure 7.
DUSP1-Mediated HG-Induced Expression Levels of Collagen I, Collagen IV, and Fibronectin via MAPK Signaling
HK-2 cells were transfected with siRNA DUSP1 (100 cnM) or DUSP1 plasmid and then treated with HG for 72 h. (A) Western blot analysis of DUSP1 and GAPDH at indicated time points. (B) Densitometric analysis of proteins signals. (C and G) Western blot analysis of collagen I, collagen IV, fibronectin, and DUSP1. (E and I) Western blot analysis of p-p38MAPK/p38MAPK and p-ERK1/2/ERK1/2. (D, F, H, and J) Densitometric measurement of western blot bands. Data are expressed as mean ± SD (n = 6). #p < 0.05, scramble with HG group or HG at 24–72 h versus scramble group or HG at 0 h; *p < 0.05, siRNA DUSP1 or DUSP1 plasmid with HG group versus scramble with HG group.
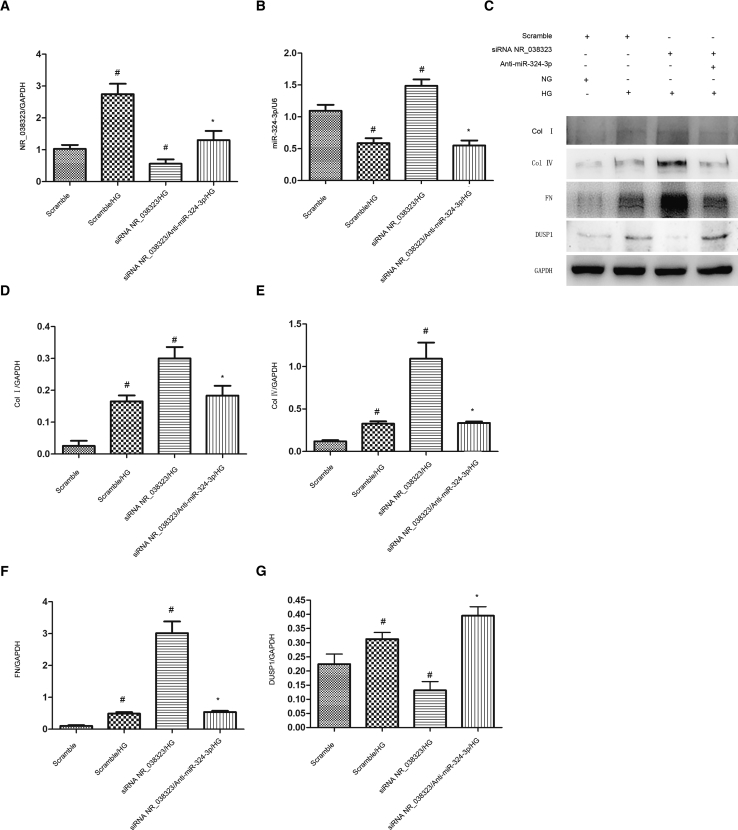
miR-324-3p Mediated the Anti-fibrotic Effects of lncRNA NR_038323
Further, we investigated whether miR-324-3p could mediate the anti-fibrotic properties of lncRNA NR_038323 during HG treatment. Real-time qPCR results demonstrated that the transfection of siRNA lncRNA NR_038323 and miR-324-3p was proved to be effective in HK-2 cells (Figures 8A and 8B). Immunoblot analysis revealed that siRNA lncRNA NR_038323 enhanced the HG-induced expression levels of collagen I, collagen IV, and fibronectin, which were reversed by miR-324-3p inhibitor (Figures 8C–8G). These findings provide a strong evidence that lncRNA NR_038323 suppresses the renal fibrosis by downregulating miR-324-3p expression.
Figure 8.
lncRNA NR_038323 Knockdown Aggravated the HG-Induced Expression Levels of ECM-Related Genes, Which Were Reversed by miR-324-3p Inhibitor
HK-2 cells were co-transfected with siRNA lncRNA NR_038323 (100 cnM) and anti-miR-324-3p or scramble and then treated with HG for 72 h. (A) Real-time qPCR analysis of lncRNA NR_038323 expression. (B) Real-time qPCR analysis of miR-324-3p expression. (C) Western blot analysis of collagen I, collagen IV, fibronectin, and DUSP1. (D–G) Densitometric measurement of protein signals in collagen I (D), collagen IV (E), fibronectin (F), and DUSP1 (G). Data are expressed as mean ± SD (n = 6). #p < 0.05, scramble with HG group versus scramble group, lncRNA NR_038323 siRNA with HG group versus scramble with HG group; *p < 0.05, lncRNA NR_038323 plus anti-miR-324-3p with HG group versus lncRNA NR_038323 siRNA with HG group.
lncRNA NR_038323/miR-324-3p/DUSP1 Axis and the Expression Levels of Collagen I, Collagen IV, and Fibronectin in Patients with DN
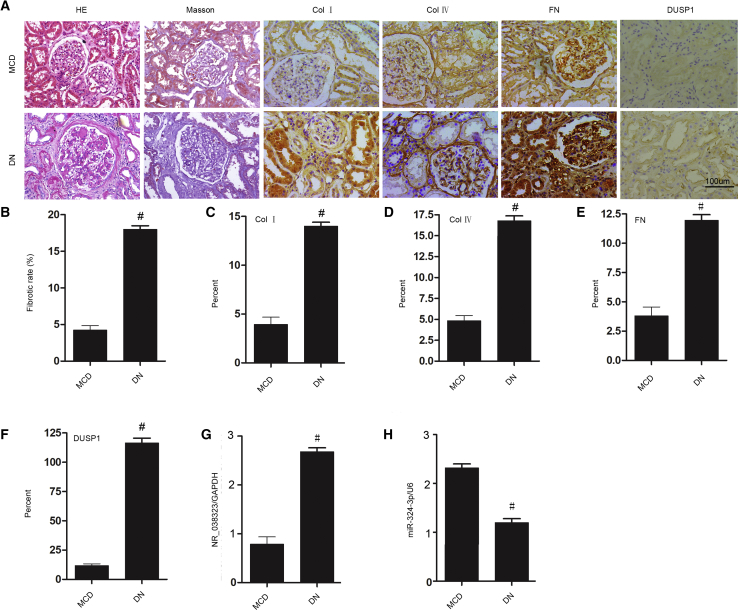
To further verify our in vitro findings, the renal expression levels of lncRNA NR_038323, miR-324-3p, and DUSP1, as well as the expression levels of collagen I, collagen IV, and fibronectin were detected in patients with DN. The patients’ basic information of MCD (n = 10) and DN (n = 9) was described in Table S1. Notably, the renal samples of DN patients demonstrated loss of glomerular function and tubulointerstitial fibrosis compared to those of MCD patients (Figures 9A and 9B). Moreover, the expression levels of collagen I, collagen IV, fibronectin, and DUSP1 were higher in DN patients than in MCD patients (Figure 9A). These findings were confirmed by semiquantitative immunohistochemistry scoring analysis (Figures 9C–9F). Furthermore, the expression levels of lncRNA NR_038323 and miR-324-3p were increased and decreased, respectively, in patients with DN (Figures 9G and 9H). Taken all together, these data indicate that lncRNA NR_038323/miR-324-3p/DUSP1 axis may be involved in human DN.
Figure 9.
lncRNA NR_038323/miR-324-3p/DUSP1 Axis in Patients with DN
(A) Renal tissues were stained with Masson’s trichrome for fibrosis analysis and immunohistochemical staining for the levels of DUSP1- and ECM-related proteins. (B) Quantitative analysis of tubulointerstitial fibrosis in the kidney cortex. (C–F) Quantification of immunohistochemical staining. (G and H) Real-time qPCR analysis of the expression levels of miR-324-3p and lncRNA NR_038323. Original magnification, ×200. Data are expressed as mean ± SD (n = 6). #p < 0.05, DN group versus MCD group.
Overexpression of lncRNA NR_038323 Ameliorated the Renal Fibrosis in STZ-Induced DN Rats
To further in vitro findings, we further investigated whether overexpression of lncRNA NR_038323 can ameliorate rat kidney injury caused by STZ. The male Wistar rats were subjected to STZ treatment to produced diabetes. After 8 weeks, STZ-treated rats indicated higher levels of blood glucose and albumin and creatinine ratio (ACR) but lower body weight; however, only ACR was improved by overexpression of lncRNA NR_038323 (Figures S1A–S1C). Furthermore, overexpression of lncRNA NR_038323 markedly ameliorated the STZ-induced tubular epithelial disruption, renal fibrosis, and glomerular hypertrophy (Figures S1D–S1G).
Overexpression of lncRNA NR_038323 Attenuated ECM Accumulation via Targeting miR-324-3p/DUSP1 Axis in STZ-Induced DN Rats
To further investigate antifibrosis molecular mechanism of lncRNA NR_038323, we first detected the expression of lncRNA NR_038323. The real-time qPCR results indicated that lncRNA NR_038323 only expressed in vein injection of lncRNA NR_038323 plasmids group but not control group (Figure S2A). The real-time qPCR also showed that STZ-induced the downregulation of miR-324-3p, which was further suppressed by lncRNA NR_038323. The immunoblotting indicated that overexpression of lncRNA NR_038323 markedly reduced the STZ-increased the expressions of collagen I, collagen IV, and fibronectin; however, it further increased the DUSP1 induced by STZ treatment (Figures S2C and S2D). The immunoblotting further detected that overexpression of lncRNA NR_038323 significantly suppressed the STZ-induced the activation of p38MAPK and ERK1/2 signaling (Figures S2E and S2F). The immunohistochemistry staining of collagen I, collagen IV, and fibronectin results further confirmed the results of Masson staining and immunoblotting (Figure S3). The data supplied a strong evidence that lncRNA NR_038323 prevented the progression of STZ-induced DN rats via targeting miR-324-3p/DUSP1/p38MAPK and ERK1/2 axis.
Discussion
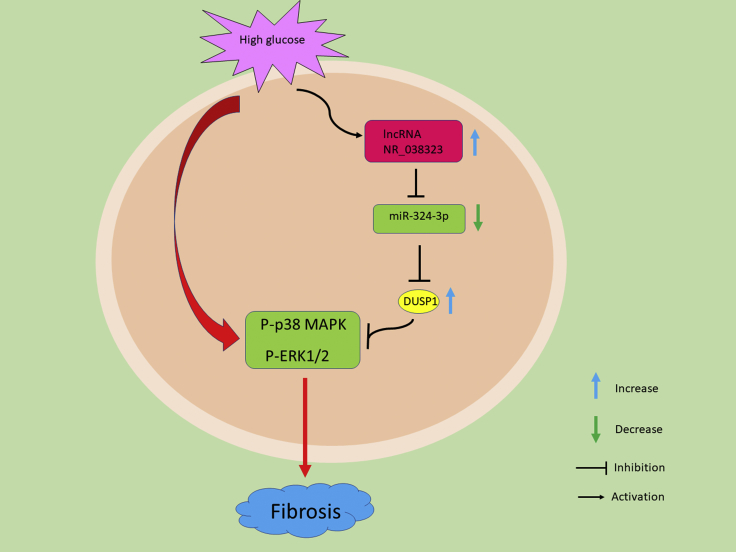
The results of the present study demonstrated, for the first time, that lncRNA NR_038323 was induced by HG treatment. In addition, HG-induced renal fibrosis was attenuated by the overexpression of lncRNA NR_038323, while aggravated by its knockdown. Moreover, we found that overexpression of lncRNA NR_038323 exerted anti-renal fibrotic effects via lncRNA NR_038323/miR-324-3p/DUSP1/p38MAPK/ERK1/2 axis. Furthermore, similar findings were observed in the renal samples of DN patients. Finally, overexpression of lncRNA NR_038323 attenuated the STZ-induced the progression of rat DN. Collectively, these findings provide new insights into the pathogenesis and treatment of DN (Figure 10).
Figure 10.
The Role and Molecular Mechanism of lncRNA NR_038323 in HG-Induced Renal Fibrosis
The expression of lncRNA NR_038323 was induced by HG treatment. Mechanistically, lncRNA NR_038323 sponged miR-324-3p to upregulate DUSP1 expression and consequently suppressed the HG-induced activation of p38MAPK and ERK1/2 pathways in renal fibrosis.
Several lines of evidence indicate that lncRNAs are responsible for renal cell apoptosis in DN.11, 12, 14, 20 However, recent evidence demonstrates that lncRNAs also mediate renal fibrosis in DN. For instance, lncRNA NEAT1 promotes renal fibrosis in DN by activating Akt/mTOR signaling pathway.21 lncRNA 1700020I14Rik attenuates renal fibrosis in DN via miR-34a-5p/Sirt1/HIF-1α signaling. lncRNA ASncmtRNA-2 enhances HG-induced renal fibrosis in mesangial cells through upregulation of pro-fibrotic factors.22 lncRNA-Gm4419 mediates HG-induced renal fibrosis and inflammation in mesangial cells via activation of nuclear factor κB (NF-κB)/NLRP3 pathway.23 Our previous findings showed that lncRNA ZEB1-AS exhibited an anti-fibrotic role in DN by regulating ZEB1 expression.4 In the present study, we found that lncRNA NR_038323 was significantly induced by HG treatment via lncRNA chip analysis and located in the cytoplasm of HK-2 cells, and its expression was induced by HG treatment (Figure 1). In addition, overexpression of lncRNA NR_038323 alleviated the HG-induced renal fibrosis (Figure 2), while the silencing of lncRNA NR_038323 aggravated the fibrosis (Figure 3). Overall, these data suggest that endogenous lncRNA NR_038323 exerts an anti-fibrotic potential; however, its anti-fibrosis ability is lower than that of HG-induced fibrosis; hence, overexpression of lncRNA NR_038323 has an anti-fibrosis role in HK-2 cells.
Increasing evidence has demonstrated that lncRNAs can act as ceRNAs to regulate targeted gene expression.10, 23, 24 Hence, we focused on miRNAs as targets of lncRNA NR_038323. Using RegRNA 2.0 software, we predicated that lncRNA NR_038323 transcript contained the binding site of miR-324-3p, suggesting that miR-324-3p is a putative target of lncRNA NR_038323. The following findings provide strong support for this prediction. First, we demonstrated that lncRNA NR_038323 directly bound to miR-324-3p through dual-luciferase reporter assay (Figure 4B). Second, FISH co-localization assay revealed that lncRNA NR_038323 interacted with miR-324-3p both in vitro and in vivo (Figure 4C). Finally, real-time qPCR data indicated that HG suppressed the expression of miR-324-3p, which was enhanced by lncRNA NR_038323 overexpression and reversed by lncRNA NR_038323 knockdown (Figures 4D and 4E). Taken together, these data suggest that miR-324-3p is a direct target of lncRNA NR_038323.
A growing body of research has demonstrated that miR-324-3p promotes tumor growth in various types of cancer.25, 26, 27, 28, 29, 30 However, little is known about the role of miR-324-3p in renal fibrosis. Only one study has reported that miR-324-3p triggers renal fibrosis in proteinuric nephropathy via suppression of angiotensin I-converting enzyme (ACE).16 Hence, the role of miR-324-3p in DN and its underlying mechanisms need to be further explored. In this study, miR-324-3p overexpression aggravated HG-induced renal fibrosis, in which its function was opposed to lncRNA NR_038323 (Figure 5). Besides, DUSP1 was identified as a target of miR-324-3p, as indicated by the following evidence. First, the dual-luciferase reporter assay revealed that miR-324-3p interacted with DUSP1 (Figures 6A and 6E). Second, miR-324-3p overexpression markedly suppressed the HG-induced expression of DUSP1 (Figures 6B–6D). A previous study has reported that DUSP1 is expressed in multiple cell lines and serves as a negative regulator for the MAPK signaling pathway.31 Another more recent study demonstrates that DUSP1 inhibits glomerular cell apoptosis in DN through the inactivation of the JNK-Mff-mitochondrial fission pathway.17 However, the effects of DUSP1 on renal fibrosis in DN and its regulatory mechanisms have yet to be investigated. Interestingly, we found that DUSP1 was induced by HG treatment (Figures 7A and 7B), and the knockdown of DUSP1 enhanced HG-induced renal fibrosis via p38MAPK and ERK1/2 pathway activation (Figures 7C–7F). Furthermore, we verified that lncRNA NR_038323 knockdown aggravated HG-induced renal fibrosis via downregulation of DUSP1, which could be reversed by miR-324-3p inhibitor (Figure 8). All these molecular changes were confirmed using human kidney samples of DN patients (Figure 9). Finally, overexpression of lncRNA NR_038323 attenuated STZ-induced the rat renal fibrosis via targeting miR-324-3p/DUSP1/p38MAPK and ERK1/2 axis (Figures S1–S3). These data support that high expression of lncRNA NR_038323 suppresses HG-induced renal fibrosis by reducing miR-324-3p expression and results in increased DUSP1 expression to inactivate p38MAPK and ERK1/2 pathways (Figure 10).
In sum, our findings reveal that HG induces the expression of lncRNA NR_038323. Interestingly, lncRNA NR_038323 inhibition aggravates HG-induced renal fibrosis, whereas its overexpression attenuates the fibrosis. Mechanistically, overexpression of lncRNA NR_038323 directly interacts with miR-324-3p to upregulate DUSP1 expression to inactive p38MAPK and ERK1/2 pathways, leading to the suppression of renal fibrosis. Taken all together, our data suggest that overexpression of lncRNA NR_038323 plays an anti-fibrotic role, which may serve a novel therapeutic target for DN.
Materials and Methods
Antibodies and Reagents
Collagen І (cat. no. 14695-1-AP) and IV (cat. no. 55131-1-AP), fibronectin (cat. no. 15613-1-AP), ERK1/2 (cat. no. 6619-1-Ig), and GAPDH (cat. no. 60004-1-Ig) were purchased from Proteintech North America (Rosemont, IL, USA), whereas DUSP1 (cat. no. Ab61201) and p38MAPK (cat. no. ab31828) were obtained from Abcam (Cambridge, MA, USA); phospho-p38MAPK (cat. no. 4511) and phospho-ERK1/2 (cat. no. 3470) were from Cell Signaling Technology (Danvers, MA, USA). The luciferase assay kit was purchased from BioVision (Milpitas, CA, USA).
Cell Culture and Treatments
HK-2 cells were cultured in DMEM (Sigma-Aldrich) supplemented with 10% fetal bovine serum at 37°C in a humidified atmosphere of 5% CO2, followed by NG (5 mmol/L D-glucose), HG (25 mmol/L D-glucose), or mannitol (25 mmol/L) treatment for 24–72 h. For gene disruption experiments, the cells were incubated with miR-324-3p mimic (100 nM), miR-324-3p antagomir (100 nM), siRNA lncRNA NR_038323 (100 nM), DUSP1 siRNA (100 nM), or negative control (Ruibo, Guangzhou, China) using Lipofectamine 2000 (Life Technologies, Carlsbad, CA, USA).
Luciferase Reporter Assays
Luciferase reporter assays were performed as previously described.24, 32, 33 The pmirGLO dual-luciferase miRNA target expression vector was used to assess miRNA activity via the insertion of miRNA target sites 3′ of the firefly luciferase gene (luc2). The luciferase vectors of DUSP1-3′ UTR (WT-Luc-DUSP1) or lncRNA NR_038323 (WT-Luc- lncRNA NR_038323) containing response elements of DUSP1 or lncRNA NR_038323 interaction with miR-324-3p, as well as the mutant plasmids of DUSP1 (MU-Luc-DUSP1) or lncRNA NR_038323 (MU-Luc-lncRNA NR_038323) lacking the response elements, were respectively used. Meanwhile, PGMLR-TK luciferase reporter was used as a control vector. All plasmids were constructed by RuQi Biotechnology (Guanzhou, Guandong, China). In brief, the plasmids of WT-Luc, Mut-Luc, or pGMLR-TK and miR-324-3p mimics were transfected into HK-2 cells for 48 h. Luciferase activities were then measured using a SpectraMax M5 (Molecular Devices, Sunnyvale, CA, USA) and normalized according to pGMLR-TK activity.
Animal Models
The Wistar rats were purchased from Shanghai Animal Center (Shanghai, People’s Republic of China). Animal experiments were performed according to the guidelines established by the Animal Care Ethics Committee of Second Xiangya Hospital, People’s Republic of China. After ethics approval was acquired, the rats were housed in a 12-h light/dark cycle and had free to access to food and water. For the STZ-induced the model of diabetes, rats (250–300 g) were injected with 50 mg/kg body weight STZ for 5 consecutive days, with sodium citrate (SC) as a control. Fasting blood glucose levels of more than 200 mg/dL for two consecutive readings were considered as diabetic. Rats were injected with lncRNA NR_038323 or control plasmid via tail vein at 25 μg each 3 weeks for 8 weeks.
Analysis of Physiological Parameters
The body weights and blood glucose levels of rats were measured. The urine albumin and creatinine levels were analyzed as previously described.4 The ACR was calculated using the method described previously.34
Histology and Immunohistochemistry
Histological analysis was analyzed by H&E staining. The glomerular and tubular damage score of kidney according to the method previously described.35, 36 Masson’s trichrome staining was used to assess the fibrosis degree. Immunohistochemical analysis was performed using anti-α-collagen I (1:100 dilution), collagen IV (1:100 dilution), FN (1:50 dilution), anti-α-SMA (1:100 dilution), and anti-DUSP1 (1:50 dilution) according to the previous protocol.35 Stained samples were analyzed using an Olympus microscope equipped with UV epi-illumination.
Real-Time qPCR
Total RNA was extracted from HK-2 cells using Trizol Reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. Approximately 40 ng of total RNA was reverse-transcribed by M-MLV reverse transcriptase (Invitrogen). To detect the expression levels of miRNA, mRNA, and lncRNA, real-time qPCR was performed using Bio-Rad (Hercules, CA, USA) IQ SYBR green supermix with Opticon (MJ Research, Waltham, MA, USA) in accordance with the manufacturer’s instructions. The sequences of lncRNA NR_038323 and miR-324-3p were retrieved from the GenBank database (gen ID: 100507156 and 442898, respectively). The primers used were as follows: lncRNA NR_038323, 5′-TGCATTCTACTGCTTCCACGA-3′ (forward) and 5′-CCAAGGTGCTGTCGTTTGAG-3′ (reverse); miR-324-3p, 5′-ACTGCCCCAGGTGCTG-3′ (forward) and 5′-CAGTGCGTGTCGTGGAGT-3′ (reverse); and GAPDH, 5′-GGTCTCCTCTGACTTCAACA-3′ (forward) and 5′-GTGAGGGTCTCTCTCTTCCT-3′ (reverse), U6 primers were described in a previous report.37 The relative quantification was carried out by determining ΔCt values.
Immunoblot Analysis
Equal amounts of proteins were loaded into each lane and separated by SDS-PAGE.35, 37, 38, 39, 40 After electrophoresis, the separated proteins were transferred onto a nitrocellulose membrane (Amersham, Buckinghamshire, UK). The blots were then probed with primary antibodies against collagen І and IV, fibronectin, DUSP1, p-p38MAPK, p38MAPK, p-ERK1/2, and ERK1/2, followed by incubation with secondary antibody and detection reagents. GAPDH was used as an internal control.
FISH
For FISH analysis, HK-2 cells were cultured in NG medium containing the fluorescence probes of miR-324-3p and lncRNA NR_038323 (Ruibo, Guangzhou, China) as described previously.10 18S rRNA was the probe for cytoplasmic control. In brief, the slides were fixed in 4% paraformaldehyde (Sigma), hybridized overnight with the probes of miR-324-3p and lncRNA NR_038323, and then stained with DAPI. Fluorescence imaging was performed using a laser scanning confocal microscope.
Human Samples
The research protocol was approved by the Hospital Review Board. Written informed consent was obtained from all participants. Human kidney biopsy samples were collected from patients with MCD (n = 10) and DN (n = 9) at the Second Xiangya Hospital, People’s Republic of China. Part of the tissue samples were fixed with 4% buffered paraformaldehyde and subjected to H&E staining, Masson’s trichrome staining, and immunohistochemical analysis, according to previous protocol.4, 36, 37, 38, 41 Meanwhile, the remainders of the specimens were steeped in RNAlater solution (Ambion) and stored at −80°C for real-time qPCR analysis.
Statistical Analyses
Two-tailed Student t tests were used for comparing the difference between two groups. One-way ANOVA was performed for multiple group comparison. Quantitative data were expressed as mean ± SD. p < 0.05 was considered statistically significant.
Author Contributions
D.Z. and H.L. designed this study; Y.G. performed the experiments; J.W., D.W., Y.Z., and S.Q. collected the data; statistical analysis and reagent purchasing were carried out by J.C., X.Z., and X.X.; D.Z. wrote the manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
The National Natural Science Foundation of China (81870475, 81570646, and 81770951), the Hunan Province Natural Science Foundation (2018JJ2568), and the excellent Youth Foundation of Hu’nan Scientific Committee (2017JJ1035) supported this study.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.07.007.
Contributor Information
Huiling Li, Email: lihuiling1120@163.com.
Dongshan Zhang, Email: dongshanzhang@csu.edu.cn.
Supplemental Information
References
- 1.Loeffler I., Wolf G. Epithelial-to-Mesenchymal Transition in Diabetic Nephropathy: Fact or Fiction? Cells. 2015;4:631–652. doi: 10.3390/cells4040631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tang S.C., Lai K.N. The pathogenic role of the renal proximal tubular cell in diabetic nephropathy. Nephrol. Dial. Transplant. 2012;27:3049–3056. doi: 10.1093/ndt/gfs260. [DOI] [PubMed] [Google Scholar]
- 3.Liakopoulos V., Roumeliotis S., Gorny X., Eleftheriadis T., Mertens P.R. Oxidative Stress in Patients Undergoing Peritoneal Dialysis: A Current Review of the Literature. Oxid. Med. Cell. Longev. 2017;2017:3494867. doi: 10.1155/2017/3494867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang J., Pan J., Li H., Long J., Fang F., Chen J., Zhu X., Xiang X., Zhang D. lncRNA ZEB1-AS1 Was Suppressed by p53 for Renal Fibrosis in Diabetic Nephropathy. Mol. Ther. Nucleic Acids. 2018;12:741–750. doi: 10.1016/j.omtn.2018.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu X., Zhang Y., Shi M., Wang Y., Zhang F., Yan R., Liu L., Xiao Y., Guo B. Notch1 regulates PTEN expression to exacerbate renal tubulointerstitial fibrosis in diabetic nephropathy by inhibiting autophagy via interactions with Hes1. Biochem. Biophys. Res. Commun. 2018;497:1110–1116. doi: 10.1016/j.bbrc.2018.02.187. [DOI] [PubMed] [Google Scholar]
- 6.Ma L., Bajic V.B., Zhang Z. On the classification of long non-coding RNAs. RNA Biol. 2013;10:925–933. doi: 10.4161/rna.24604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huarte M. The emerging role of lncRNAs in cancer. Nat. Med. 2015;21:1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 8.Uchida S., Dimmeler S. Long noncoding RNAs in cardiovascular diseases. Circ. Res. 2015;116:737–750. doi: 10.1161/CIRCRESAHA.116.302521. [DOI] [PubMed] [Google Scholar]
- 9.Briggs J.A., Wolvetang E.J., Mattick J.S., Rinn J.L., Barry G. Mechanisms of Long Non-coding RNAs in Mammalian Nervous System Development, Plasticity, Disease, and Evolution. Neuron. 2015;88:861–877. doi: 10.1016/j.neuron.2015.09.045. [DOI] [PubMed] [Google Scholar]
- 10.Li A., Peng R., Sun Y., Liu H., Peng H., Zhang Z. LincRNA 1700020I14Rik alleviates cell proliferation and fibrosis in diabetic nephropathy via miR-34a-5p/Sirt1/HIF-1α signaling. Cell Death Dis. 2018;9:461. doi: 10.1038/s41419-018-0527-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Long J., Badal S.S., Ye Z., Wang Y., Ayanga B.A., Galvan D.L., Green N.H., Chang B.H., Overbeek P.A., Danesh F.R. Long noncoding RNA Tug1 regulates mitochondrial bioenergetics in diabetic nephropathy. J. Clin. Invest. 2016;126:4205–4218. doi: 10.1172/JCI87927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kato M., Wang M., Chen Z., Bhatt K., Oh H.J., Lanting L., Deshpande S., Jia Y., Lai J.Y., O’Connor C.L. An endoplasmic reticulum stress-regulated lncRNA hosting a microRNA megacluster induces early features of diabetic nephropathy. Nat. Commun. 2016;7:12864. doi: 10.1038/ncomms12864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu L., Yang J., Zhu X., Li D., Lv Z., Zhang X. Long noncoding RNA H19 competitively binds miR-17-5p to regulate YES1 expression in thyroid cancer. FEBS J. 2016;283:2326–2339. doi: 10.1111/febs.13741. [DOI] [PubMed] [Google Scholar]
- 14.Tsai Y.C., Kuo P.L., Hung W.W., Wu L.Y., Wu P.H., Chang W.A., Kuo M.C., Hsu Y.L. Angpt2 Induces Mesangial Cell Apoptosis through the MicroRNA-33-5p-SOCS5 Loop in Diabetic Nephropathy. Mol. Ther. Nucleic Acids. 2018;13:543–555. doi: 10.1016/j.omtn.2018.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu B., Wu S., Ma J., Yan S., Xiao Z., Wan L., Zhang F., Shang M., Mao A. lncRNA GAS5 Reverses EMT and Tumor Stem Cell-Mediated Gemcitabine Resistance and Metastasis by Targeting miR-221/SOCS3 in Pancreatic Cancer. Mol. Ther. Nucleic Acids. 2018;13:472–482. doi: 10.1016/j.omtn.2018.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Macconi D., Tomasoni S., Romagnani P., Trionfini P., Sangalli F., Mazzinghi B., Rizzo P., Lazzeri E., Abbate M., Remuzzi G., Benigni A. MicroRNA-324-3p promotes renal fibrosis and is a target of ACE inhibition. J. Am. Soc. Nephrol. 2012;23:1496–1505. doi: 10.1681/ASN.2011121144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sheng J., Li H., Dai Q., Lu C., Xu M., Zhang J., Feng J. DUSP1 recuses diabetic nephropathy via repressing JNK-Mff-mitochondrial fission pathways. J. Cell. Physiol. 2019;234:3043–3057. doi: 10.1002/jcp.27124. [DOI] [PubMed] [Google Scholar]
- 18.Dauletbaev N., Herscovitch K., Das M., Chen H., Bernier J., Matouk E., Bérubé J., Rousseau S., Lands L.C. Down-regulation of IL-8 by high-dose vitamin D is specific to hyperinflammatory macrophages and involves mechanisms beyond up-regulation of DUSP1. Br. J. Pharmacol. 2015;172:4757–4771. doi: 10.1111/bph.13249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carver K.A., Smith T.L., Gallagher P.E., Tallant E.A. Angiotensin-(1-7) prevents angiotensin II-induced fibrosis in cremaster microvessels. Microcirculation. 2015;22:19–27. doi: 10.1111/micc.12159. [DOI] [PubMed] [Google Scholar]
- 20.Chen S., Dong C., Qian X., Huang S., Feng Y., Ye X., Miao H., You Q., Lu Y., Ding D. Microarray analysis of long noncoding RNA expression patterns in diabetic nephropathy. J. Diabetes Complications. 2017;31:569–576. doi: 10.1016/j.jdiacomp.2016.11.017. [DOI] [PubMed] [Google Scholar]
- 21.Huang S., Xu Y., Ge X., Xu B., Peng W., Jiang X., Shen L., Xia L. Long noncoding RNA NEAT1 accelerates the proliferation and fibrosis in diabetic nephropathy through activating Akt/mTOR signaling pathway. J. Cell. Physiol. 2019;234:11200–11207. doi: 10.1002/jcp.27770. [DOI] [PubMed] [Google Scholar]
- 22.Gao Y., Chen Z.Y., Wang Y., Liu Y., Ma J.X., Li Y.K. Long non-coding RNA ASncmtRNA-2 is upregulated in diabetic kidneys and high glucose-treated mesangial cells. Exp. Ther. Med. 2017;13:581–587. doi: 10.3892/etm.2017.4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yi H., Peng R., Zhang L.Y., Sun Y., Peng H.M., Liu H.D., Yu L.J., Li A.L., Zhang Y.J., Jiang W.H., Zhang Z. LincRNA-Gm4419 knockdown ameliorates NF-κB/NLRP3 inflammasome-mediated inflammation in diabetic nephropathy. Cell Death Dis. 2017;8:e2583. doi: 10.1038/cddis.2016.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li P., Zhang X., Wang L., Du L., Yang Y., Liu T., Li C., Wang C. lncRNA HOTAIR Contributes to 5FU Resistance through Suppressing miR-218 and Activating NF-κB/TS Signaling in Colorectal Cancer. Mol. Ther. Nucleic Acids. 2017;8:356–369. doi: 10.1016/j.omtn.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun G.L., Li Z., Wang W.Z., Chen Z., Zhang L., Li Q., Wei S., Li B.W., Xu J.H., Chen L. miR-324-3p promotes gastric cancer development by activating Smad4-mediated Wnt/beta-catenin signaling pathway. J. Gastroenterol. 2018;53:725–739. doi: 10.1007/s00535-017-1408-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tuo H., Wang Y., Wang L., Yao B., Li Q., Wang C., Liu Z., Han S., Yin G., Tu K., Liu Q. MiR-324-3p promotes tumor growth through targeting DACT1 and activation of Wnt/β-catenin pathway in hepatocellular carcinoma. Oncotarget. 2017;8:65687–65698. doi: 10.18632/oncotarget.20058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu C., Li G., Yang N., Su Z., Zhang S., Deng T., Ren S., Lu S., Tian Y., Liu Y., Qiu Y. miR-324-3p suppresses migration and invasion by targeting WNT2B in nasopharyngeal carcinoma. Cancer Cell Int. 2017;17:2. doi: 10.1186/s12935-016-0372-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuo W.T., Yu S.Y., Li S.C., Lam H.C., Chang H.T., Chen W.S., Yeh C.Y., Hung S.F., Liu T.C., Wu T. MicroRNA-324 in Human Cancer: miR-324-5p and miR-324-3p Have Distinct Biological Functions in Human Cancer. Anticancer Res. 2016;36:5189–5196. doi: 10.21873/anticanres.11089. [DOI] [PubMed] [Google Scholar]
- 29.Gao X., Wang Y., Zhao H., Wei F., Zhang X., Su Y., Wang C., Li H., Ren X. Plasma miR-324-3p and miR-1285 as diagnostic and prognostic biomarkers for early stage lung squamous cell carcinoma. Oncotarget. 2016;7:59664–59675. doi: 10.18632/oncotarget.11198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu J., Ai Q., Cao H., Liu Q. MiR-185-3p and miR-324-3p Predict Radiosensitivity of Nasopharyngeal Carcinoma and Modulate Cancer Cell Growth and Apoptosis by Targeting SMAD7. Med. Sci. Monit. 2015;21:2828–2836. doi: 10.12659/MSM.895660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Korhonen R., Moilanen E. Mitogen-activated protein kinase phosphatase 1 as an inflammatory factor and drug target. Basic Clin. Pharmacol. Toxicol. 2014;114:24–36. doi: 10.1111/bcpt.12141. [DOI] [PubMed] [Google Scholar]
- 32.Sun C.C., Zhang L., Li G., Li S.J., Chen Z.L., Fu Y.F., Gong F.Y., Bai T., Zhang D.Y., Wu Q.M., Li D.J. The lncRNA PDIA3P Interacts with miR-185-5p to Modulate Oral Squamous Cell Carcinoma Progression by Targeting Cyclin D2. Mol. Ther. Nucleic Acids. 2017;9:100–110. doi: 10.1016/j.omtn.2017.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu X., Zheng J., Xue Y., Qu C., Chen J., Wang Z., Li Z., Zhang L., Liu Y. Inhibition of TDP43-Mediated SNHG12-miR-195-SOX5 Feedback Loop Impeded Malignant Biological Behaviors of Glioma Cells. Mol. Ther. Nucleic Acids. 2018;10:142–158. doi: 10.1016/j.omtn.2017.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhan M., Usman I.M., Sun L., Kanwar Y.S. Disruption of renal tubular mitochondrial quality control by Myo-inositol oxygenase in diabetic kidney disease. J. Am. Soc. Nephrol. 2015;26:1304–1321. doi: 10.1681/ASN.2014050457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu L., Li X., Zhang F., Wu L., Dong Z., Zhang D. EGFR drives the progression of AKI to CKD through HIPK2 overexpression. Theranostics. 2019;9:2712–2726. doi: 10.7150/thno.31424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang D., Liu Y., Wei Q., Huo Y., Liu K., Liu F., Dong Z. Tubular p53 regulates multiple genes to mediate AKI. J. Am. Soc. Nephrol. 2014;25:2278–2289. doi: 10.1681/ASN.2013080902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang J., Li H., Qiu S., Dong Z., Xiang X., Zhang D. MBD2 upregulates miR-301a-5p to induce kidney cell apoptosis during vancomycin-induced AKI. Cell Death Dis. 2017;8:e3120. doi: 10.1038/cddis.2017.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu X., Wang J., Yang R., Dong Z., Zhang D. Genetic or pharmacologic inhibition of EGFR ameliorates sepsis-induced AKI. Oncotarget. 2017;8:91577–91592. doi: 10.18632/oncotarget.21244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang R., Xu X., Li H., Chen J., Xiang X., Dong Z., Zhang D. p53 induces miR199a-3p to suppress SOCS7 for STAT3 activation and renal fibrosis in UUO. Sci. Rep. 2017;7:43409. doi: 10.1038/srep43409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peng J., Li X., Zhang D., Chen J.K., Su Y., Smith S.B., Dong Z. Hyperglycemia, p53, and mitochondrial pathway of apoptosis are involved in the susceptibility of diabetic models to ischemic acute kidney injury. Kidney Int. 2015;87:137–150. doi: 10.1038/ki.2014.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang D., Pan J., Xiang X., Liu Y., Dong G., Livingston M.J., Chen J.K., Yin X.M., Dong Z. Protein Kinase Cδ Suppresses Autophagy to Induce Kidney Cell Apoptosis in Cisplatin Nephrotoxicity. J. Am. Soc. Nephrol. 2017;28:1131–1144. doi: 10.1681/ASN.2016030337. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.