The gene mcr-1 conferring resistance to last-line antibiotic colistin has been reported globally. Here, we describe the first detection of plasmid-mediated colistin resistance in Russian wildlife, an isolate of Escherichia coli sequence type 2280 from a black kite (Milvus migrans) scavenging raptor. Whole-genome sequencing and plasmid transferability experiments revealed that mcr-1.1 was located on conjugative IncI2 plasmid pDR164 (59891 bp).
KEYWORDS: DNA sequencing, Escherichia coli, colistin, kite, landfill, plasmid, wildlife
ABSTRACT
The gene mcr-1 conferring resistance to last-line antibiotic colistin has been reported globally. Here, we describe the first detection of plasmid-mediated colistin resistance in Russian wildlife, an isolate of Escherichia coli sequence type 2280 from a black kite (Milvus migrans) scavenging raptor. Whole-genome sequencing and plasmid transferability experiments revealed that mcr-1.1 was located on conjugative IncI2 plasmid pDR164 (59891 bp). Migratory black kites may contribute to the global spread of mobile colistin resistance.
INTRODUCTION
Colistin is listed by World Health Organization as a critically important antibiotic of last resort for the treatment of severe human infections caused by multidrug-resistant Gram-negative bacteria (1). In veterinary medicine, apart from therapeutic purposes, colistin is used also as an in-feed growth promoter in livestock in some Asian countries, including India, Japan, and Vietnam (2). As a result of growing concern about emerging mobile colistin resistance, the use of colistin in food-producing animals is globally limited or banned (3). Plasmid-mediated colistin resistance gene mcr-1 has been described in more than 40 countries, with the majority of reports coming from inpatients and domestic animals. Until now, only a few studies observed this resistance mechanism in bacteria from wildlife (4–12). In this study, we describe an Escherichia coli isolate with mcr-1 from a migratory wild bird, the black kite (Milvus migrans), in Russia, which is to our best knowledge the first report of plasmid-mediated colistin resistance in wildlife in that country.
Black and red kites (Milvus milvus) are avian scavengers with different migration habits (13). Black kites of subspecies M. migrans migrans and M. migrans lineatus are summer residents in Europe and northern part of Asia that migrate to winter mainly in sub-Saharan Africa and southern parts of Asia, respectively. The red kite is essentially a European raptor (13). As a result of their frequent foraging in landfills, they may acquire antibiotic-resistant bacteria of human origin and spread them further over long distances (14). In 2018, a total of 168 cloacal swabs from nestlings of black and red kites were collected in Austria (6 samples), Belgium (12 samples), Czech Republic (93 samples), Germany (41 samples), and Russia (16 samples). Swabs were enriched in buffered peptone water and plated on SuperPolymyxin medium (15) to screen for Enterobacteriales isolates resistant to colistin. One single colony per plate/sample was selected, and species was identified and tested for the presence of mcr-1 to mcr-8. One isolate positive for mcr-1 was subjected to antibiotic resistance profiling and whole-genome sequencing on a MiSeq (Illumina) platform. Sequencing data were examined to identify the genetic context of mcr-1. Horizontal transfer of colistin resistance into a recipient E. coli strain was tested using the filter mating method. See the supplemental material for detailed description of sampling and methods.
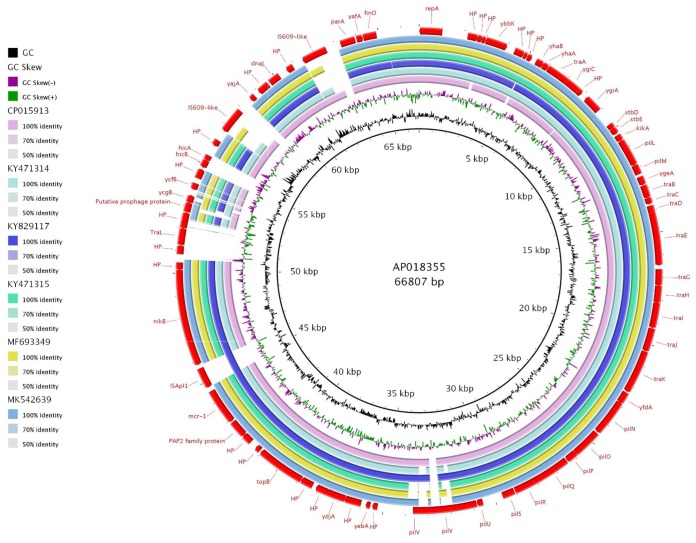
One isolate with mcr-1.1 identified as E. coli sequence type (ST) 2280 was recovered from a black kite sampled in Semenovod, Altaysky Kray, Russia (52°33′N, 85°24′E) on 7 July 2018. The isolate was susceptible to all tested antibiotics apart from colistin (MIC > 4 mg liter−1). Whole-genome sequencing analysis and transferability experiments showed that the mcr-1 gene was located on a conjugative IncI2 plasmid pDR164 (Fig. 1). The complete circular plasmid sequence was annotated according to pMCR-M19855 (GenBank accession no. KY471315 [16]) and submitted to GenBank under accession no. MK542639 (17). pDR164 sequence consists of 59,891 bp with GC content of 42.4% and 80 predicted open reading frames with a typical IncI2 backbone, including regions for stability, replication, maintenance, and horizontal gene transfer.
FIG 1.
BRIG comparison of mcr-1-positive IncI2 plasmid pDR164 (GenBank accession no. MK542639 [17]) with similar plasmid sequences retrieved from GenBank. Plasmids used for comparison include pMCR-M19736 (25), pMCR-M19855 (KY471315 [16]), and pG3216 (MF693349 [26]) originating from human clinical samples in Argentina, pSLy1 (CP015913 [27]) from a swine sample in the United States, pVE362 (AP018355 [19]) from poultry in Vietnam, and pMCR_WCHEC1604-IncI2 (KY829117 [18]) from sewage in China.
For the comparison of pDR164 with similar plasmid sequences, a BLAST search was performed (on 15 March 2019), and complete plasmid sequences with identity thresholds of ≥99% and query coverage thresholds of ≥96% were selected. The five IncI2 plasmid sequences that met the criteria are listed in Table S1, all of which carried mcr-1 with the absence of any other antibiotic resistance gene. A high level of similarity within the conserved IncI2 plasmid backbone and the variable region carrying mcr was observed (Fig. 1). pDR164 did not contain insertion sequence ISApl1 flanking the mcr-1 region, unlike pMCR_WCHEC1604-IncI2 (KY829117 [18]) and pVE362 (AP018355 [19]). The mcr-1 gene can be mobilized in a conjugative plasmid with two ISApaI1 sequences flanking it as a composite transposon (Tn6330) or with one ISApl1. However, the absence of ISApl1 from the vicinity of the mcr-1 gene is common and may occur after the insertion of mcr-1 in a conjugative plasmid and following subsequent recombination events (20).
Seven reports of mcr-1-positive E. coli isolates in wildlife were published. Most of them originated from migrating birds such as European herring gull (Larus argentatus) in Lithuania (4), kelp gull (Larus dominicanus) in Argentina (5), Eurasian coot (Fulica atra) from Pakistan (6), and swallows from China (10). The mcr-1 gene was commonly detected on conjugative IncI2 and IncX4 plasmids in human, domestic animal (21), and wildlife E. coli isolates of diverse sequence types and antibiotic resistance profiles. As in our study, IncI2 plasmids carrying mcr-1 were reported in wild birds from Argentina (5) and Pakistan (6). IncX4 plasmids, which appear to be a globally major source of mobility and dissemination of mcr-1 gene (2), were described in Enterobacterales in penguins in Brazil (7), common blowflies from urban and rural communities in Thailand (11), and stable flies in Germany (12). At the time of writing this article, eleven mcr-1-positive E. coli isolates from clinical sources with phenotypic colistin resistance (MIC > 4 mg liter−1) have been reported in Russia (22).
Black kites have a wide range of human-made and natural diets (14). Based on the available food opportunities and the reported food diet of black kites in the vicinity of the sampled nestling (23), two possible sources of mcr-1 isolates can be hypothesized. The first source could be Biysk municipal landfill located 16 km northwest of the sampled nest, which is commonly used by local black kites for foraging, while the second source could be natural food (carrions, rodents, birds, or small mammals) of black kites from flooded area along the Biya River where the nest was located.
In conclusion, we describe the first report of mobile colistin resistance from wildlife in Russia and present the first complete sequence of an mcr-1-harboring IncI2 plasmid from wildlife. Detection of a plasmid-mediated gene for resistance to a last-line drug in black kites is worrisome. Black kites can act as reservoirs and vectors of antibiotic-resistant bacteria that can be disseminated through their long migratory pathways. Since its discovery in 2015 (24), the mcr-1 gene appears to be spread globally in veterinary, clinical, and environmental sectors (2). Based on our finding and on previous reports of colistin-resistant E. coli in wildlife, it appears that mcr-1 found its way to the environment. Thus, continued surveillance of bacteria with transferrable colistin resistance mediated by mcr genes in wildlife is desirable.
(Part of this work was presented at the Central European Institute of Technology[CEITEC] PhD retreat, Kouty, Czech Republic, 2019 [talk number T18], and the 8th Symposium on Antimicrobial Resistance in Animals and the Environment [ARAE], Institut national de la recherche agronomique, Tours, France, 2019 [talk number O7].)
Supplementary Material
ACKNOWLEDGMENTS
We thank L. Cavaco (Technical University of Denmark, Kongens Lyngby, Denmark), B. B. Xavier (University of Antwerp, Antwerp, Belgium), M. Ugarte Ruiz (Foodborne Zoonoses and Antimicrobial Resistance Unit, Complutense University, Madrid, Spain), A. Carattoli (Istituto Superiore di Sanita, Rome, Italy), and Marcela Krutova (Charles University, 2nd Faculty of Medicine, Motol University Hospital, Prague, Czech Republic) for providing positive-control strains. We also thank Katerina Rehakova and Martin Klvana from the University of Veterinary and Pharmaceutical Sciences Brno, Czech Republic, for their help in the laboratory. We thank Karel Makon, Hynek Matusik, and Roman Bachtin for the cooperation in the field.
The project was supported by Czech Health Research Council (grant number NV18-09-00605) and the National Sustainability Programs II (NPU II; grant LQ1601) provided by the Ministry of Education Youth and Sports of the Czech Republic. The work was partially financed by Internal Grant Agency of University of Veterinary and Pharmaceutical Sciences Brno (204/2018/FVHE) to M.D. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01266-19.
REFERENCES
- 1.WHO. 2016. Critically important antibiotics for human medicine. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 2.Sun J, Zhang H, Liu YH, Feng Y. 2018. Towards understanding MCR-like colistin resistance. Trends Microbiol 29:794–808. doi: 10.1016/j.tim.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 3.Webb HE, Angulo FJ, Granier SA, Scott HM, Loneragan GH. 2017. Illustrative examples of probable transfer of resistance determinants from food animals to humans: streptothricins, glycopeptides, and colistin. F1000Res 6:1805. doi: 10.12688/f1000research.12777.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ruzauskas M, Vaskeviciute L. 2016. Detection of the mcr-1 gene in Escherichia coli prevalent in the migratory bird species Larus argentatus. J Antimicrob Chemother 71:2333–2334. doi: 10.1093/jac/dkw245. [DOI] [PubMed] [Google Scholar]
- 5.Liakopoulos A, Mevius DJ, Olsen B, Bonnedahl J. 2016. The colistin resistance mcr-1 gene is going wild. J Antimicrob Chemother 71:2335–2336. doi: 10.1093/jac/dkw262. [DOI] [PubMed] [Google Scholar]
- 6.Mohsin M, Raza S, Roschanski N, Schaufler K, Guenther S. 2016. First description of plasmid-mediated colistin-resistant extended-spectrum beta-lactamase-producing Escherichia coli in a wild migratory bird from Asia. Int J Antimicrob Agents 48:463–464. doi: 10.1016/j.ijantimicag.2016.07.001. [DOI] [PubMed] [Google Scholar]
- 7.Sellera FP, Fernandes MR, Sartori L, Carvalho MP, Esposito F, Nascimento CL, Dutra GH, Mamizuka EM, Perez-Chaparro PJ, McCulloch JA, Lincopan N. 2017. Escherichia coli carrying IncX4 plasmid-mediated mcr-1 and blaCTX-M genes in infected migratory Magellanic penguins (Spheniscus magellanicus). J Antimicrob Chemother 72:1255–1256. doi: 10.1093/jac/dkw543. [DOI] [PubMed] [Google Scholar]
- 8.Bachiri T, Lalaoui R, Bakour S, Allouache M, Belkebla N, Rolain JM, Touati A. 2018. First report of the plasmid-mediated colistin resistance gene mcr-1 in Escherichia coli ST405 isolated from wildlife in Bejaia. Microb Drug Resist 24:890–895. doi: 10.1089/mdr.2017.0026. [DOI] [PubMed] [Google Scholar]
- 9.Wu J, Huang Y, Rao D, Zhang Y, Yang K. 2018. Evidence for environmental dissemination of antibiotic resistance mediated by wild birds. Front Microbiol 9:745. doi: 10.3389/fmicb.2018.00745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y, Zhang R, Li J, Wu Z, Yin W, Schwarz S, Tyrrell JM, Zheng Y, Wang S, Shen Z, Liu Z, Liu J, Lei L, Li M, Zhang Q, Wu C, Zhang Q, Wu Y, Walsh TR, Shen J. 2017. Comprehensive resistome analysis reveals the prevalence of NDM and MCR-1 in Chinese poultry production. Nat Microbiol 2:16260. doi: 10.1038/nmicrobiol.2016.260. [DOI] [PubMed] [Google Scholar]
- 11.Yang QE, Tansawai U, Andrey DO, Wang S, Wang Y, Sands K, Kiddee A, Assawatheptawee K, Bunchu N, Hassan B, Walsh TR, Niumsup PR. 2019. Environmental dissemination of mcr-1 positive Enterobacteriaceae by Chrysomya spp. (common blowfly): an increasing public health risk. Environ Int 122:281–290. doi: 10.1016/j.envint.2018.11.021. [DOI] [PubMed] [Google Scholar]
- 12.Guenther S, Falgenhauer L, Semmler T, Imirzalioglu C, Chakraborty T, Roesler U, Roschanski N. 2017. Environmental emission of multiresistant Escherichia coli carrying the colistin resistance gene mcr-1 from German swine farms. J Antimicrob Chemother 72:1289–1292. doi: 10.1093/jac/dkw585. [DOI] [PubMed] [Google Scholar]
- 13.Del Hoyo J, Elliot A, Sargatal J. 1994. Handbook of the birds of the world, vol 2 Lynx Edicions, Barcelona, Spain. [Google Scholar]
- 14.De Giacomo U, Guerrieri G. 2008. The feeding behavior of the black kite (Milvus migrans) in the rubbish dump of Rome. J Raptor Res 42:110–118. doi: 10.3356/JRR-07-09.1. [DOI] [Google Scholar]
- 15.Nordmann P, Jayol A, Poirel L. 2016. A universal culture medium for screening polymyxin-resistant Gram-negative isolates. J Clin Microbiol 54:1395–1399. doi: 10.1128/JCM.00446-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tijet N, Melano RG. 2017. Escherichia coli strain M19855 plasmid pMCR-M19855, complete sequence. https://www.ncbi.nlm.nih.gov/nuccore/KY471315. Accessed 11 June 2019.
- 17.Tarabai H, Valcek A, Jamborova I, Literak I, Dolejska M. 2019. Escherichia coli strain DR164 plasmid pDR164, complete sequence. https://www.ncbi.nlm.nih.gov/nuccore/MK542639. Accessed 11 June 2019.
- 18.Zhao F, Feng Y, Zong Z. 2017. Escherichia coli strain WCHEC1604 plasmid pMCR_WCHEC1604-IncI2, complete sequence. https://www.ncbi.nlm.nih.gov/nuccore/KY829117. Accessed 20 June 2019.
- 19.Yamaguchi T, Kawahara R, Harada K, Teruya S, Nakayama T, Motooka D, Nakamura S, Nguyen PD, Kumeda Y, Van DC, Hirata K, Yamamoto Y. 2018. Escherichia coli plasmid pVE362 DNA, complete genome, strain: E362. https://www.ncbi.nlm.nih.gov/nuccore/AP018355. Accessed 20 June 2019.
- 20.Snesrud E, McGann P, Chandler M. 2018. The birth and demise of the ISApl1-mcr-1-ISApl1 composite transposon: the vehicle for transferable colistin resistance. mBio 9:e02381-17. doi: 10.1128/mBio.02381-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matamoros S, van Hattem JM, Arcilla MS, Willemse N, Melles DC, Penders J, Vinh TN, Thi Hoa N, COMBAT Consortium, de Jong MD, Schultsz C. 2017. Global phylogenetic analysis of Escherichia coli and plasmids carrying the mcr-1 gene indicates bacterial diversity but plasmid restriction. Sci Rep 7:15364. doi: 10.1038/s41598-017-15539-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Azizov I, Sheck E, Sukhorukova M, Edelstein M. 2019. Plasmid-mediated resistance to colistin in clinical isolates of Klebsiella spp. and Escherichia coli: results of large retrospective surveillance in Russia, p 13–16. abstr 29th European Congress of Clinical Microbiology & Infectious Diseases, Amsterdam, Netherlands. [Google Scholar]
- 23.Bachtin RFV, Makarov AV. 2010. Ecology of synanthropic populations of the black kite in the vicinities of Biysk, Altai Kray, Russia. Raptors Conserv 20:68–83. doi: 10.19074/1814-8654-2016-32-49-58. [DOI] [Google Scholar]
- 24.Liu Y-Y, Wang Y, Walsh TR, Yi L-X, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu L-F, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu J-H, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 25.Tijet N, Melano RG. 2017. Escherichia coli strain M19736 plasmid pMCR-M19736, complete sequence. https://www.ncbi.nlm.nih.gov/nuccore/KY471314. Accessed 11 June 2019.
- 26.Elena A, Cejas D, Magarinos F, Gutkind G, Di Conza J, Radice M. 2017. Escherichia coli strain 3216 plasmid pG3216, complete sequence. https://www.ncbi.nlm.nih.gov/nuccore/MF693349. Accessed 20 June 2019.
- 27.Meinersmann R, Ladely S, Hall C, Simpson S, Ballard L, Scheffler B. 2016. Escherichia coli strain 210205630 plasmid pSLy1, complete sequence. https://www.ncbi.nlm.nih.gov/nuccore/CP015913. Accessed 20 June 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.