The plasmid-located mcr-9 gene, encoding a putative phosphoethanolamine transferase, was identified in a colistin-resistant human fecal Escherichia coli strain belonging to a very rare phylogroup, the D-ST69-O15:H6 clone. This MCR-9 protein shares 33% to 65% identity with the other plasmid-encoded MCR-type enzymes identified (MCR-1 to -8) that have been found as sources of acquired resistance to polymyxins in Enterobacteriaceae.
KEYWORDS: Escherichia coli, MCR, MCR-9, colistin, plasmid, polymyxin
ABSTRACT
The plasmid-located mcr-9 gene, encoding a putative phosphoethanolamine transferase, was identified in a colistin-resistant human fecal Escherichia coli strain belonging to a very rare phylogroup, the D-ST69-O15:H6 clone. This MCR-9 protein shares 33% to 65% identity with the other plasmid-encoded MCR-type enzymes identified (MCR-1 to -8) that have been found as sources of acquired resistance to polymyxins in Enterobacteriaceae. Analysis of the lipopolysaccharide of the MCR-9-producing isolate revealed a function similar to that of MCR-1 by adding a phosphoethanolamine group to lipid A and subsequently modifying the structure of the lipopolysaccharide. However, a minor impact on susceptibility to polymyxins was noticed once the mcr-9 gene was cloned and produced in an E. coli K-12-derived strain. Nevertheless, we showed here that subinhibitory concentrations of colistin induced the expression of the mcr-9 gene, leading to increased MIC levels. This inducible expression was mediated by a two-component regulatory system encoded by the qseC and qseB genes located downstream of mcr-9. Genetic analysis showed that the mcr-9 gene was carried by an IncHI2 plasmid. In silico analysis revealed that the plasmid-encoded MCR-9 shared significant amino acid identity (ca. 80%) with the chromosomally encoded MCR-like proteins from Buttiauxella spp. In particular, Buttiauxella gaviniae was found to harbor a gene encoding MCR-BG, sharing 84% identity with MCR-9. That gene was neither expressed nor inducible in its original host, which was fully susceptible to polymyxins. This work showed that mcr genes may circulate silently and remain undetected unless induced by colistin.
INTRODUCTION
Acquired resistance to polymyxins in Enterobacteriaceae may arise from two main pathways, namely, chromosomally encoded resistance mechanisms corresponding to mutations or deletions in genes involved in the biosynthesis of the lipopolysaccharide (LPS) or the acquisition of plasmid-mediated mcr genes. The latter genes encode phosphoethanolamine transferases that modify the lipid A moiety of the LPS, subsequently leading to resistance to polymyxins (1, 2). There have been eight different groups of mcr genes identified, with some variants described within some given groups (3–11). mcr-1 likely originates from a Moraxella species (12), while Moraxella pluranimalium is the exact progenitor of mcr-2 (13), Aeromonas spp. that of mcr-3-like genes (14, 15), and Shewanella spp. that of mcr-4-like genes (7). The origins of the more recently discovered mcr genes (mcr-5 to -8) remain unknown. In addition, a very recent study identified the mcr-9 gene from a colistin-susceptible Salmonella enterica serotype Typhimurium isolate recovered from a U.S. patient in 2010 (16).
The high prevalence of MCR (particularly MCR-1)-producing Escherichia coli isolates in food-producing animals, and therefore the high rate of colistin-resistant isolates, may be explained by the significant use of colistin in veterinary medicine, in particular in livestock for the treatment of poultry, swine, and cattle (17–20). In humans, the prevalence of MCR producers seems to be more limited, with many studies reporting a low prevalence of the mcr-1 gene in Enterobacteriaceae (1, 21). On the other hand, the prevalence of the mcr-2 to -9 genes remains unknown overall, even though the few reports are mainly on animal isolates.
In a recent prospective study performed in six different French university hospitals, despite the fact that the prevalence of fecal colistin-resistant E. coli carried by inpatients was found to be unexpectedly high (12.7%), the rate of MCR-1 producers was low (4.6% of the colistin-resistant isolates), and no other known MCR-like enzyme was identified (22). Two-thirds of the resistant isolates had mutations in the PmrA/PmrB chromosomally encoded two-component systems that are the likely sources of the resistance phenotype (22). Among those colistin-resistant E. coli isolates that remained negative for all known mcr-like genes (mcr-1 to mcr-8) and that did not harbor obvious mutations in chromosomal genes that might be responsible for acquired resistance, E. coli 68A, showing a colistin MIC of 8 μg/ml, was retained for further characterization.
RESULTS
Identification of the mcr-9 gene.
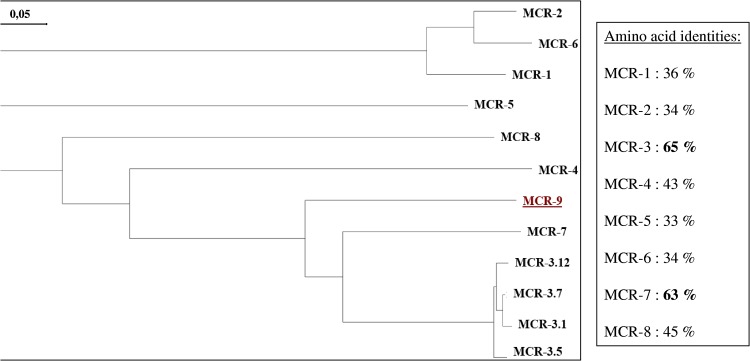
Whole-genome sequencing (WGS) of the E. coli isolate 68A (GenBank assembly accession no. GCA_900500325.1) was used in an attempt to identify the mechanism leading to this reduced susceptibility to colistin, and a gene showing significant identity to mcr-like genes was identified. It corresponded to the newly reported mcr-9 gene (16) encoding the MCR-9 enzyme, sharing 65% and 63% amino acid identity with the most closely related MCR-3 and MCR-7 enzymes, respectively, and between 33% and 45% with the other MCR-like enzymes (Fig. 1) (GenBank accession number MK070339). Phylogenetic analysis performed with MCR-9 and all other MCR-type enzymes revealed that MCR-9 was distantly related to all those enzymes and constituted a distinct cluster.
FIG 1.
Phylogenetic tree obtained for the MCR-type phosphoethanolamine transferases identified by the distance method using the Neighbor-Joining algorithm (SeaView software, version 4 [44]). Branch lengths are drawn to scale and are proportional to the number of amino acid substitutions with 500 bootstrap replications. The distance along the vertical axis has no significance. The percentage of amino acid identity compared to the MCR-9 enzyme is indicated. The tree was rooted with the MCR-3.1 enzyme. The scale along the horizontal axis is indicated.
Isolate 68A showed resistance only to penicillins (with susceptibility to penicillin-β-lactamase inhibitors) and tetracycline due to the presence of blaTEM-1 and tet(A) genes. Multilocus sequence typing analysis showed that isolate 68A belonged to the phylogroup D-ST69 and was of the O15:H6 serotype. It possessed numerous extraintestinal virulence genes either on the chromosome (fyuA, irp2, and ompT) or on a pS88-like IncFIB virulence plasmid (iro operon, iss, ompT, hlyF, and iuc operon) (23). Thus, this strain corresponds to an extraintestinal pathogenic E. coli (ExPEC) isolate.
Localization of the mcr-9 gene.
Analysis of the whole-genome sequence of E. coli 68A using PlaScope (24) showed that the mcr-9 gene was located on an IncHI2 (pMLST2) incompatibility group plasmid (p68) of 225 kb. This plasmid did not carry any other putative resistance gene. PCR-based replicon typing (PBRT) analysis (25) and Kieser extraction (26) performed from isolate 68A, followed by gel electrophoresis analysis, confirmed the whole-genome sequencing (WGS) data.
MCR-9 is a phosphoethanolamine transferase conferring reduced susceptibility to colistin.
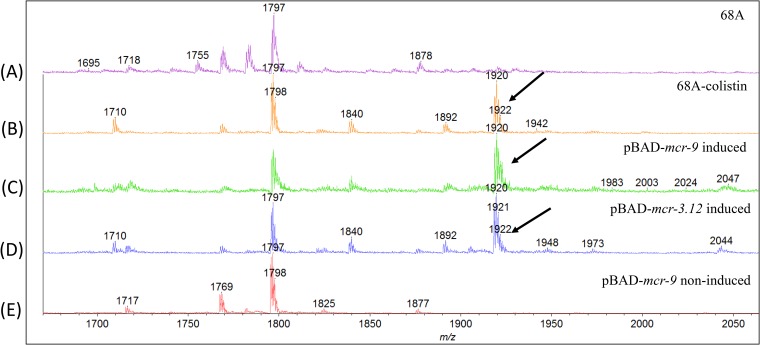
Mass spectrometry analysis of the lipid A moiety of the LPS of the MCR-9-producing E. coli isolate 68A recovered from a colistin-supplemented agar plate showed an additional peak at m/z 1,920 (Fig. 2). This modification corresponded to the addition of a phosphoethanolamine (PEtN) group to lipid A, as previously reported for MCR-1 to MCR-3 producers (27–29) (Fig. 2). Surprisingly, no peak at m/z 1,920 was observed when we performed the same experiment with isolate 68A recovered from a colistin-free medium (Fig. 2). These results strongly suggested that the isolate 68A did not produce MCR-9 at a level sufficient to significantly modify its LPS under the colistin-free condition and therefore that the expression of mcr-9 was induced in the colistin-containing plate.
FIG 2.
Mass spectrometry analysis of lipid A from isolate E. coli 68A after growth on a colistin-free LB plate (A), E. coli 68A after growth on a colistin-supplemented (2 μg/ml) LB plate (B), E. coli TOP10(pBAD-mcr-9) recombinant strain induced by l-arabinose (C), E. coli TOP10(pBAD-mcr-3.12) recombinant strain induced by l-arabinose (D), and E. coli TOP10(pBAD-mcr-9) (not induced by l-arabinose) (E). The addition of phosphoethanolamine is indicated by an arrow.
The mcr-9 gene alone then was cloned into the arabinose-inducible pBAD vector, leading to E. coli TOP10(pBAD-mcr-9), in which the expression of mcr-9 is high in the presence of l-arabinose. Mass spectrometry analysis of the lipid A moiety of the LPS of both MCR-9- and MCR-3.12-producing recombinant E. coli TOP10 pBAD-mcr-9 and pBAD-mcr-3.12 recombinant strains, both recovered from an l-arabinose-supplemented agar plate, showed additional peaks at m/z 1,920 not seen for the E. coli TOP10(pBAD-mcr-9) strain recovered from an l-arabinose-free plate (Fig. 2).
Apart from colistin induction, and since the mcr-9 gene seemed to be poorly transcribed in its natural host, we evaluated the level of resistance (or reduced susceptibility) it might confer by comparing it to that of the mcr-1 gene. After induction with arabinose, E. coli TOP10(pBAD-mcr-9) showed a colistin MIC of 0.15 μg/ml (susceptibility range), whereas the same noninduced strain presented a MIC of 0.03 μg/ml, corresponding to a 5-fold increase of the colistin MIC. Conversely, the MIC of colistin for the E. coli(pBAD-mcr-1) recombinant strain was 4 μg/ml. These data confirmed the impact of the MCR-9 enzyme on colistin susceptibility but to a lower extent than that of MCR-1.
Genetic context of the mcr-9 gene.
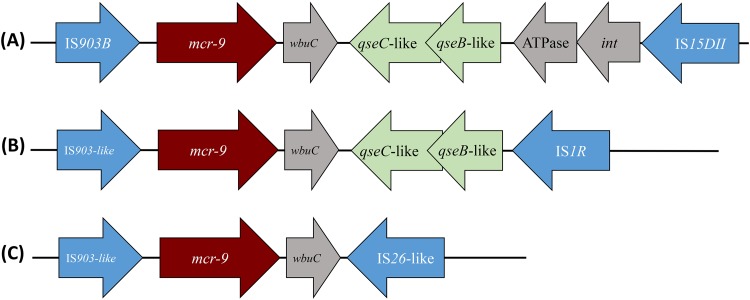
Analysis of the genomic data of E. coli 68A showed that the mcr-9 gene was bracketed by two insertion sequences (IS). Upstream of mcr-9, IS15DII (IS5 family) was identified, in which no obvious promoter sequences that might be responsible for mcr-9 expression could be identified (Fig. 3). Downstream of mcr-9, another IS, namely, IS903B (IS6 family), was identified (Fig. 3). No target site duplication was identified at the extremities of each IS or when looking at the left-hand extremity of IS903B and the right-hand extremity of IS903B. The latter observation and the fact that those two IS are distantly related (with totally different inverted repeat sequences) likely rule out the hypothesis that mcr-9 was acquired through a composite transposon bracketed by those two IS.
FIG 3.
Different genetic structure surrounding the mcr-9 gene. The wbuC, qseC, qseB, and ATPase-encoding genes located downstream of mcr-9 are represented, as well as the different insertion sequences (IS) bracketing that DNA fragment, namely, IS903B, IS1R, IS26-like, and IS15DII. Arrows indicate the sense of transcription of those different genes. (A) Structure surrounding the mcr-9 gene in E. coli isolate 68A (present study) and those identified in silico in Leclercia species (GenBank accession no. CP031102.1), Enterobacter hormaechei (CP031575.1), Salmonella enterica (CP029037.1 and CP006057.1), and Enterobacter kobei (CP032893.1). (B) Structure surrounding the mcr-9 gene identified in silico in Leclercia adecarboxylata (MH909331.1) and Salmonella enterica (CP026661.1). (C) Structure surrounding the mcr-9 gene identified in silico in Enterobacter cloacae (CP020529.1), Salmonella enterica (MK191844.1), Cronobacter sakazakii (CP028975.1), and Enterobacter hormaechei (CP031575.1).
Analysis of the DNA fragment bracketed by the two above-mentioned IS showed that four open reading frames were present downstream of mcr-9, including wbuC, qseC, qseB, and an ATPase gene (Fig. 3). Interestingly, although wbuC encodes a putative enzyme of the cupin superfamily of unknown function, the qseC and qseB genes encode a putative two-component system corresponding to a histidine kinase sensor (QseC) and its cognate partner (QseB). Similar proteins have been shown to play a role in the signaling network inducing resistance to colistin (30). In order to evaluate whether those two genes located downstream of mcr-9 play a role in colistin resistance, the whole structure encompassing the mcr-9, qseC, and qseB genes along with the two IS903B and IS15DII elements was cloned, leading to the E. coli TOP10(pBAD-Tot) strain. The MIC of colistin for this recombinant strain was compared with that of the isogenic recombinant E. coli TOP10(pBAD-mcr-9), and only a slight increase was observed for E. coli TOP10(pBAD-Tot) (0.3 versus 0.15 μg/ml).
Inducibility of mcr-9 gene expression.
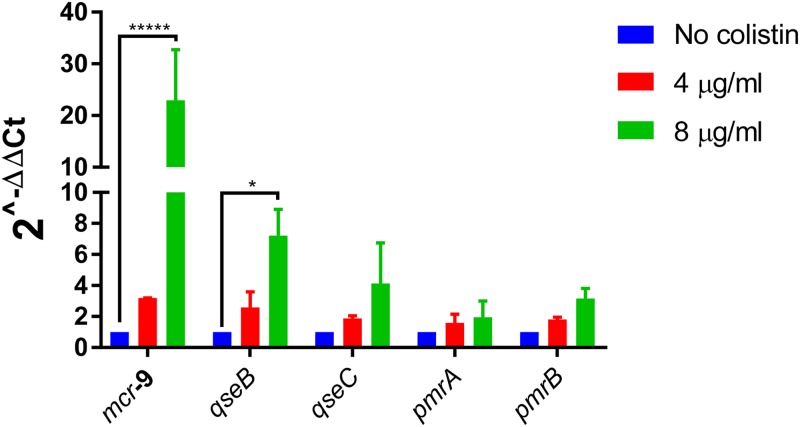
In order to evaluate whether the expression of mcr-9 was constitutive or inducible in its original host, induction assays were performed using isolate 68A as the template for mRNA extraction. It is noteworthy that a 3-fold increase of the mcr-9 mRNA amount was detected once the culture was induced with 4 μg/ml colistin, and a 24-fold increase was detected when using a concentration of 8 μg/ml (Fig. 4). In parallel, the transcription levels of the chromosomal pmrA and pmrB genes of E. coli 68A, encoding a response regulator and a sensor kinase, respectively, which may be involved in acquired resistance to colistin, remained almost unchanged upon induction (Fig. 4). Also, the amount of transcripts for the IncHI2 plasmid replicase gene remained unchanged, ruling out the possibility that induction assays have led to an increased plasmid copy number, thereby biasing the measurement of the mcr-9 transcription level (data not shown). Of note, no induction of mcr-9 expression was observed when performing the induction assay with ampicillin (50 μg/ml), suggesting that the colistin inducer effect was quite specific. In parallel, the same experiment with colistin (4 μg/ml) as the inducer was conducted with E. coli isolate Af23 harboring the mcr-1 gene (31) and exhibiting a colistin MIC of 4 μg/ml, but no induction of mcr-1 was noticed, highlighting the specific effect observed in E. coli 68A.
FIG 4.
Expression of five selected genes in clinical isolate 68A with and without colistin exposure. The genes are the plasmid-located mcr-9, qseB, and qseC as well as the chromosome-located pmrA (DNA-binding response regulator) and pmrB (sensor protein) genes. Fold changes in mRNA levels were determined by RT-qPCR. The data have been normalized to values for reference 16S rRNA genes. Three independent replicates were performed; the data shown represent the means, and the error bars represent standard deviations. The asterisks indicate statistical significance at different levels by analysis of variance: *, P ≤ 0.1; **, P ≤ 0.05; ***, P ≤ 0.01; ****, P ≤ 0.001; *****, P ≤ 0.0001. The different colistin concentrations used in our assays are shown by color, with blue indicating no colistin, red indicating 4 μg/ml colistin, and green indicating 8 μg/ml colistin.
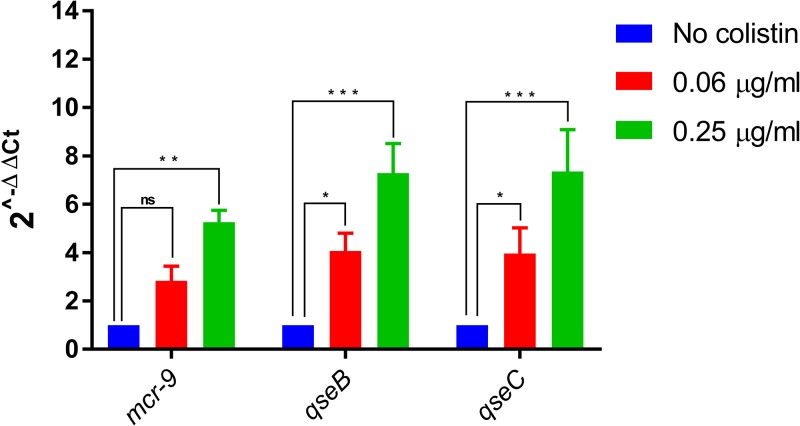
Induction experiments then were performed using recombinant strains E. coli TOP10(pNK1-mcr-9) (mcr-9 gene alone) and E. coli TOP10(pNK1-Tot) (mcr-9 together with downstream-located genes wbuC, qseC, and qseB), corresponding to low-copy-number plasmids not depending on arabinose inducibility. Although no induction of the mcr-9 expression was observed for E. coli TOP10(pNK1-mcr-9) (data not shown), a significant increase was observed for E. coli TOP10(pNK1-Tot) in the presence of subinhibitory concentrations of colistin (Fig. 5). Moreover, higher expression levels of the qseC and qseB genes were concomitantly observed in the presence of colistin (Fig. 5). These data strongly suggest that overexpression of mcr-9 observed in the presence of colistin is related to that of the QseC-QseB two-component system. Interestingly, higher expression levels were obtained in the presence of 0.5 μg/ml colistin than with 0.06 μg/ml colistin, highlighting a concentration-based modulating effect.
FIG 5.
Expression of three selected genes cloned in E. coli TOP10(pNK1-Tot) with and without colistin exposure. The genes are mcr-9, qseB, and qseC. Fold changes in mRNA levels were determined by RT-qPCR. The data have been normalized to values for reference 16S rRNA genes. Three independent replicates were performed; the data shown represent the means, and the error bars represent standard deviations. The asterisks indicate statistical significance at different levels by ANOVA: *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001. The different colistin concentrations used in our assays are shown by color, with blue indicating no colistin, red indicating 0.06 μg/ml colistin, and green indicating 0.25 μg/ml colistin.
Origin of the mcr-9 gene.
In silico analysis using the GenBank databases showed that mcr-9-like genes were present in the chromosomes of Buttiauxella species strains, including Buttiauxella ferragutiae, Buttiauxella izardii, Buttiauxella noackiae, Buttiauxella warmboldiae, Buttiauxella brennerae, Buttiauxella agrestis, and Buttiauxella gaviniae. The most closely related gene, called mcr-BG, was identified in Buttiauxella gaviniae strain CIP106356T (GenBank accession no. NZ_LXEP00000000.1). The MCR-BG protein shared 84% identity with MCR-9 and 30 to 60% with the other plasmid-encoded MCR-type enzymes (MCR-1 to -8) (GenBank accession no. WP_064511805.1). Looking at the sequences located downstream of mcr-BG, a gene homologous to wbuC was found, but neither qseC nor qseB homologues were identified. This suggested that acquisition of the mcr-9 gene from Buttiauxella spp. occurred together with wbuC as a whole fragment but that the qseC-qseB tandem originated from another source.
In fact, by performing an in silico BLAST analysis over the GenBank databases with those two genes, it appeared that they both shared 87% nucleotide identity with similar genes from Klebsiella aerogenes and Salmonella enterica, respectively. Induction experiments were performed using B. gaviniae strain CIP106356T with subinhibitory concentrations of colistin, but no overexpression of mcr-BG was observed (data not shown).
The mcr-BG gene was cloned in plasmid pBAD and expressed in E. coli TOP10. A minor impact of the mcr-BG gene on susceptibility to polymyxins was observed once cloned and produced in E. coli, similar to what was observed with mcr-9 expressed in E. coli TOP10. After induction with arabinose, an 8-fold increase in the MIC of colistin was noticed for E. coli TOP10 producing MCR-BG (0.125 μg/ml) or producing MCR-9 (0.25 μg/ml) compared to that of E. coli TOP10 (0.015 μg/ml), while the MIC of colistin for the MCR-1-producing E. coli TOP10 recombinant strain was 4 μg/ml. Analysis of the LPS pattern of the MCR-BG-producing E. coli confirmed that a PEtN group was added to the lipid A component of the LPS of E. coli, while no specific peak was observed when analyzing the B. gaviniae strain (data not shown).
Distribution of the mcr-9 gene.
We performed an in silico analysis using the GenBank databases to see whether the mcr-9 gene was already present in the available bacterial genome sequences. Surprisingly, a 100% match at the nucleotide level was found in ca. 50 genomes, all corresponding to enterobacterial isolates, recovered from worldwide sources, either from humans or animals (swine). Among the different enterobacterial species in which mcr-9 was present were Klebsiella spp., Enterobacter spp., Salmonella spp., Leclercia spp., Citrobacter spp., Raoultella spp., Phytobacter ursingii, and Cronobacter sakazakii. Information about the colistin susceptibility status of all these strains unfortunately is not available. Diversity was observed by examining the genetic environment of the mcr-9 gene in all these genomes, with three main structures identified (Fig. 3A to C). The IS903B element was always identified upstream of mcr-9, and the wbuC gene also was systematically present downstream of it. However, different IS could be identified on the right-hand extremity, being either IS15DII, IS1R, or IS26-like. It is noteworthy that some structures did not encompass the qseC-qseB regulatory genes.
DISCUSSION
We report here the identification of an MCR determinant, detected in a colistin-resistant E. coli isolate recovered from a patient in France. Interestingly, in contrast to other MCR determinants identified so far, MCR-9 confers only reduced susceptibility, not resistance, to colistin under normal conditions in an E. coli wild-type background. However, this gene showed increased expression once induced with subinhibitory colistin concentrations, leading to resistance in E. coli according to EUCAST breakpoints (www.eucast.org).
The fact that acquisition of mcr-9 does not confer clinical resistance per se suggests a silent spread of that gene, with elevated MICs of colistin observed for the corresponding producers still categorized as susceptible. It is worth noting that we showed that the expression of mcr-9 was inducible by subinhibitory concentrations of colistin, leading to resistance in E. coli. This inducible expression was shown to be related to the presence of the qseC and qseB genes downstream of mcr-9. Those two genes encode a putative sensor kinase (QseC) and its cognate partner (QseB), with the former likely mediating dephosphorylation of the latter (30). This tandem likely has been acquired through an independent mobilization event with respect to mcr-9 acquisition. This is suggested by the lack of those genes downstream of the mcr-9-like gene in its original progenitor, B. gaviniae.
A so-called inducibility of mcr-1 gene expression was recently reported; however, the phenomenon observed was quite different. In one case, it was shown that the mcr-1 gene was truncated by IS1294b, leading to colistin susceptibility in E. coli (32). Upon colistin-based selective pressure, excision of the IS was observed, leading to restoration of the intact mcr-1 gene and subsequently to acquired resistance. In the other case, two copies of a 22-bp DNA insertion were identified in mcr-1 in Shigella sonnei, leading to colistin susceptibility (33). Upon loss of one of those two copies, restoration of the mcr-1 open reading frame was observed, leading to colistin resistance. Here, we verified that the overexpression of mcr-9 was not related to modifications in the sequences located in its upstream vicinity (data not shown).
Hence, we speculate that MCR-9 production is a source of colistin resistance upon selective pressure with that antibiotic, playing a significant clinical role in strain selection. The inducibility property observed with mcr-9 was shown to be related to the two-component system present downstream of mcr-9. Further work will be conducted to decipher the exact interactions between those plasmid-borne two-component regulatory systems and the chromosomally encoded proteins involved in lipopolysaccharide biosynthesis, particularly those modifying lipid A.
Interestingly, in silico analysis identified the mcr-9 gene in isolates recovered in different parts of the world and in a variety of enterobacterial species. It would be interesting to learn about the colistin susceptibility of those mcr-9-positive isolates, since that information is not available. Broad epidemiological studies will be necessary to evaluate the extent of dissemination of that gene worldwide among human and animal isolates. Detection of the mcr-9 gene can be performed by using primers MCR-9F (5′-GGT AGT TAT TCC GCT GG-3′) and MCR-9R (5′-TCG CGG TCA GGA TTA AC-3′), which were actually validated during our study (data not shown). Looking at those genomes available in silico, it appeared that different genetic structures surrounding the mcr-9 gene could be identified, with some of them lacking the qseC-qseB regulatory genes. We speculate that the expression of mcr-9 is not inducible in strains possessing the latter structure.
Isolate 68A belongs to the phylogroup D-ST69 and is of serotype O15:H6. The phylogroup D-ST69 lineage includes the clonal group A strains first described by Johnson et al. (34) as ExPEC strains responsible for human urinary tract infections and resistant to trimethoprim-sulfamethoxazole. These strains have a typical virulence factor profile and exhibit specific O antigens (O11, O17, O73, and O77) always associated with the H18 type. However, isolate 68A was susceptible to trimethoprim-sulfamethoxazole, and the presence of the pS88-like virulence plasmid (35) and of the O15:H6 serotype has not been reported in ST69. A search of ca. 12,000 E. coli genomes in the NCBI database identified only 3 genomes corresponding to phylogroup D-ST69-O15:H6, all being of human origin (one recovered from blood in the United States, one from an intra-abdominal abscess in China, and one from feces in France), but none of these strains possessed the mcr-9 gene.
On the other hand, by searching the PLSD plasmid database (36), a total of 26 similar IncHI2 plasmids were identified from E. coli, K. pneumoniae, and Salmonella enterica, but none of them possessed the mcr-9 gene. However, 16 out of those 29 plasmid sequences contained the mcr-1.1 gene, further suggesting that such a plasmid backbone originates from strains present in animal reservoirs subjected to colistin selective pressure.
Isolate 68A was found to be resistant to colistin in susceptibility testing by broth microdilution (BMD), as recommended (EUCAST). However, it is noteworthy that the expression of mcr-9 was actually induced in the presence of colistin when performing BMD. Therefore, the MIC value actually reflects the impact of MCR-9 once the corresponding gene is induced.
Analysis of the LPS profile of isolate 68A as well as MCR-9-producing recombinant strains showed that the MCR-9 enzyme confers colistin resistance the same way as MCR-1 and MCR-3 enzymes, by adding a phosphoethanolamine group to the lipid A, although the different enzymes shared significant sequence diversities. The fact that MCR-1, -2, and -3 share similar functions was previously hypothesized through an in silico protein structure analysis (5). Here, we showed that MCR-9, along with MCR-BG and MCR-3, exhibited a very similar three-dimensional structure.
Analysis of the genetic context of the mcr-9 gene did not allow us to explain its acquisition by the IncHI2 plasmid. The putative involvement of IS903B or IS15DII in the original mobilization of the mcr-9 gene from Buttiauxella spp. remains elusive. It is noteworthy that this plasmid did not possess any other antibiotic resistance determinant; therefore, its acquisition through a coselection pressure is unlikely.
MATERIALS AND METHODS
Bacterial isolates and susceptibility testing.
The isolates were initially tested for colistin resistance using agar dilution methods. All colonies growing on plates supplemented with >2 μg/ml colistin were confirmed by the commercialized rapid polymyxin NP test (ELITech Microbiology, France) (37, 38), and MICs were determined by the BMD method using cation-adjusted Mueller-Hinton (MH) broth (38). Antimicrobial susceptibility testing for other antibiotic families was performed according to the standard disk diffusion method on MH agar plates by following CLSI recommendations (39).
WGS and bioinformatic analysis.
Whole genomic DNA of isolate 68A was sequenced as previously described (22) using Illumina NextSeq 2× 150-bp technology. The sequences were analyzed with an in-house bioinformatic pipeline named PETANC, for plasmid exploration typing assembly N′ contig ordering (22), including the PlaScope tool to assess the plasmid origin of the sequence (24).
Plasmid analysis.
Plasmid analysis was performed using the Kieser extraction method (26), followed by gel electrophoresis in order to further confirm the size of the plasmid containing the mcr-9 gene using the E. coli strain 50192 harboring four plasmids of 154, 66, 48, and 7 kb as plasmid size markers. The determination of the incompatibility group was confirmed by PBRT (27).
Analysis of the LPS modification.
The LPS of E. coli TOP10 (unmodified lipid A), of isolate E. coli 68A (mcr-9 positive) grown in the presence or absence of colistin, of the E. coli(pBAD-mcr-9) clone carrying the mcr-9 gene in the pBAD vector, grown in the presence or absence of l-arabinose, of B. gaviniae, and of the E. coli(pBAD-mcr-3.12) clone carrying the mcr-3.12 gene as a control were analyzed by mass spectrometry (29). Lipid A was extracted using an acetic acid-based procedure as previously described (40, 41). Once extracted, 0.7 μl of the concentrate was spotted on a matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) plate, followed by addition of 0.7 μl of norharmane matrix (Sigma-Aldrich, St. Louis, Missouri), and then air dried. The samples were analyzed on a Vitek MS instrument (bioMérieux, Marcy l’Étoile, France) in the negative-ion mode.
Cloning and overexpression of the mcr-9 gene.
The mcr-9 gene was cloned into the arabinose-inducible pBAD vector in order to determine the impact of the expression of the MCR-9 putative phosphoethanolamine transferase on colistin susceptibility. Primers used for cloning are indicated in Table 1. Induction of the pBAD vector was performed using MH broth supplemented with 1% l-arabinose as previously described (42). In addition, to avoid using an arabinose-inducible vector, for the purpose of colistin inducibility assays, the mcr-9 gene and the full mcr-9 cassette were cloned in low-copy-number plasmid pNK1 (42) and transformed in E. coli TOP10, giving recombinant plasmids pNK-mcr-9 and pNK-Tot.
TABLE 1.
Oligonucleotide sequences used in this work
| Primer | Sequence | Application |
|---|---|---|
| 16S | Forward, 5′-GTGCAATATTCCCCACTGCT-3′ | RT-qPCR |
| Reverse, 5′-CGATCCCTAGCTGGTCTGAG-3′ | ||
| mcr-BG | Forward, 5′-ACTACCAGGATTACGCCTCG-3′ | RT-qPCR |
| Reverse, 5′-GCGACTGGAAGGGTTCTTTG-3′ | ||
| mcr-9 | Forward, 5′-CGGTACCGCTACCGCAATAT-3′ | RT-qPCR |
| Reverse, 5′-ATAACAGCGAGACACCGGTT-3′ | ||
| mcr-1 | Forward, 5′-ACACTTATGGCACGGTCTATG-3′ | RT-qPCR |
| Reverse, 5′-GCACACCCAAACCAATGATAC-3′ | ||
| repA IncHI2 | Forward, 5′-GTAACCACTAAATACCCGGG-3′ | RT-qPCR |
| Reverse, 5′-TTCCTGGTTTCGGTTTAGCC-3′ | ||
| qseB | Forward, 5′-TGACATTGTGCTCCTGGATC-3′ | RT-qPCR |
| Reverse, 5′-AGTTCGCTGAACTCGAACGG-3′ | ||
| qseC | Forward, 5′-ATATGACAACCCTGACTCGC-3′ | RT-qPCR |
| Reverse, 5′-AATTGGGTGGGTGAAAGTCG-3′ | ||
| mcr-9 (for cloning in pBAD) | Forward, 5′-ATGCCTGTACTTTTCAGGG-3′ | Cloning |
| Reverse, 5′-AACAATCGATTAGCCACGGC-3′ | ||
| mcr-9 (for cloning in pNK1) | Forward, 5′-TTCTCTGATGAACGTTCCGC-3′ | Cloning |
| Reverse, 5′-TGTCACGTCAACTGGATGAC-3′ |
Colistin induction assays.
For induction assays with colistin, growth conditions were as follows. Buttiauxella gaviniae was grown in an LB agar plate, clinical isolate 68A was grown in an LB agar plate supplemented with colistin (1 μg/ml) or left unsupplemented, and E. coli TOP10 pNK1-mcr-9 and pNK1-Tot recombinant strains were plated in LB agar supplemented with kanamycin (30 μg/ml). All these strains then were grown overnight in liquid medium before the mRNA extraction assay, with kanamycin (30 μg/ml) being the selective agent of the pNK1 plasmid. A 1/10 dilution culture then was performed with or without colistin. Induction assays were performed using subinhibitory concentrations of colistin, namely, 0.12 μg/ml for B. gaviniae, E. coli TOP10(pNK1-mcr-9), and E. coli TOP10(pNK1-Tot), 2 μg/ml for clinical isolate 68A, originally recovered onto an LB agar plate without colistin, and 12 μg/ml for clinical isolate 68A, originally recovered from an LB agar plate with colistin. Primers used for cloning are indicated in Table 1.
mRNA extraction.
Cultures of isolate 68A and the recombinant E. coli TOP10 strains harboring plasmids pNK1-mcr-9 and pNK1-Tot were used for mRNA extraction. They were grown for 3 h. Total RNA was extracted by using the TRIzol Max bacterial RNA isolation kit (Thermo Fisher Scientific, Waltham, MA) as specified by the manufacturer. The RNA samples were treated with DNase to remove contaminating DNA traces by a DNA-free kit (Thermo Fisher Scientific). Finally, RNA samples without traces of DNA were cleaned with the RNeasy PowerClean Pro cleanup kit (Qiagen, Hilden, Germany) and measured with a NanoDrop 2000 spectrophotometer (Thermo Scientific, Littau, Switzerland).
RT-qPCR.
A total of 200 ng of each RNA sample was reverse transcribed using the PrimeScript RT reagent kit (TaKaRa, Saint-Germain-en-Laye, France) according to the manufacturer’s instructions. Quantitative real-time PCR (RT-qPCR) was performed using a RotorGene Q (Qiagen, Hilden, Germany). Sequences of primers used are listed in Table 1. Reactions were set up in a total volume of 25 μl with a Rotor-Gene SYBR green PCR kit (Qiagen, Hilden, Germany). The selected genes in this assay were 16S rRNA (E. coli and B. gaviniae reference gene), mcr-1, and mcr-9, together with a fragment corresponding to the iteron of the IncHI2 plasmid. Three independent replicates were performed with and without induction. The obtained threshold cycle (CT) values were analyzed by the 2−ΔΔCT method (43). Relative expression levels were calculated and compared with those of samples that were not induced. All values were corrected with values for the appropriate reference gene.
ACKNOWLEDGMENTS
This work has been funded by the University of Fribourg, the Swiss National Science Foundation (project FNS-407240_177381), and the Laboratoire Européen Associé INSERM Emerging Antibiotic Resistance in Gram-Negative Bacteria. E.D. was partially supported by the Fondation pour la Recherche Médicale (Equipe FRM 2016, grant number DEQ20161136698). G.R. was supported by the Assistance Publique–Hôpitaux de Paris (Poste d’Accueil AP-HP–CEA [Commissariat à l’Énergie Atomique et Aux Énergies Alternatives] 2016–2018). We are grateful to Daniel Haft, from NCBI/NLM/NIH/DHHS, for his contribution to the MCR nomenclature and assigning the MCR-9 name to the present enzyme. The work performed by M.P. was funded by the European Union's Horizon 2020 research and innovation program under Marie Skłodowska-Curie grant agreement no. 675412 (New Diagnostics for Infectious Diseases [ND4ID]). We are also grateful to A. Van Belkum and the bioMérieux company for supporting M.P.
REFERENCES
- 1.Poirel L, Jayol A, Nordmann P. 2017. Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin Microbiol Rev 30:557–596. doi: 10.1128/CMR.00064-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sun J, Zhang H, Liu YH, Feng Y. 2018. Towards understanding MCR-like colistin resistance. Trends Microbiol 26:794–808. doi: 10.1016/j.tim.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 3.Liu Y-Y, Wang Y, Walsh TR, Yi L-X, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu L-F, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu J-H, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 4.Xavier BB, Lammens C, Ruhal R, Kumar-Singh S, Butaye P, Goossens H, Malhotra-Kumar S. 2016. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Euro Surveill 21 https://www.eurosurveillance.org/content/10.2807/1560-7917.ES.2016.21.27.30280. [DOI] [PubMed] [Google Scholar]
- 5.Yin W, Li H, Shen Y, Liu Z, Wang S, Shen Z, Zhang R, Walsh TR, Shen J, Wang Y. 2017. Novel plasmid-mediated colistin resistance gene mcr-3 in Escherichia coli. mBio 8:e1166-17. doi: 10.1128/mBio.00543-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Litrup E, Kiil K, Hammerum AM, Roer L, Nielsen EM, Torpdahl M. 2017. Plasmid-borne colistin resistance gene mcr-3 in Salmonella isolates from human infections, Denmark, 2009-17. Euro Surveill 03:22. doi: 10.2807/1560-7917.ES.2017.22.31.30587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carattoli A, Villa L, Feudi C, Curcio L, Orsini S, Luppi A, Pezzotti G, Magistrali CF. 2017. Novel plasmid-mediated colistin resistance mcr-4 gene in Salmonella and Escherichia coli, Italy 2013, Spain and Belgium, 2015 to 2016. Euro Surveill 3:30589. doi: 10.2807/1560-7917.ES.2017.22.31.30589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borowiak M, Fischer J, Hammerl JA, Hendriksen RS, Szabo I, Malorny B. 2017. Identification of a novel transposon-associated phosphoethanolamine transferase gene, mcr-5, conferring colistin resistance in d-tartrate fermenting Salmonella enterica subsp. enterica serovar Paratyphi B. J Antimicrob Chemother 72:3317–3324. doi: 10.1093/jac/dkx327. [DOI] [PubMed] [Google Scholar]
- 9.AbuOun M, Stubberfield EJ, Duggett NA, Kirchner M, Dormer L, Nunez-Garcia J, Randall LP, Lemma F, Crook DW, Teale C, Smith RP, Anjum MF. 2018. mcr-1 and mcr-2 (mcr-6.1) variant genes identified in Moraxella species isolated from pigs in Great Britain from 2014 to 2015. J Antimicrob Chemother 73:2904. doi: 10.1093/jac/dky272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang YQ, Li YX, Lei CW, Zhang AY, Wang HN. 2018. Novel plasmid-mediated colistin resistance gene mcr-7.1 in Klebsiella pneumoniae. J Antimicrob Chemother 73:1791–1795. doi: 10.1093/jac/dky111. [DOI] [PubMed] [Google Scholar]
- 11.Wang X, Wang Y, Zhou Y, Li J, Yin W, Wang S, Zhang S, Shen J, Shen Z, Wang Y. 2018. Emergence of a novel mobile colistin resistance gene, mcr-8, in NDM-producing Klebsiella pneumoniae. Emerg Microbes Infect 7:122. doi: 10.1038/s41426-018-0124-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kieffer N, Nordmann P, Poirel L. 2017. Moraxella species as potential sources of MCR-like polymyxin resistance determinants. Antimicrob Agents Chemother 61:e00129-17. doi: 10.1128/AAC.00129-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poirel L, Kieffer N, Fernandez-Garayzabal JF, Vela AI, Larpin Y, Nordmann P. 2017. MCR-2-mediated plasmid-borne polymyxin resistance most likely originates from Moraxella pluranimalium. J Antimicrob Chemother 72:2947–2949. doi: 10.1093/jac/dkx225. [DOI] [PubMed] [Google Scholar]
- 14.Ling Z, Yin W, Li H, Zhang Q, Wang X, Wang Z, Ke Y, Wang Y, Shen J. 2017. Chromosome-mediated mcr-3 variants in Aeromonas veronii from chicken meat. Antimicrob Agents Chemother 61:e01272-17. doi: 10.1128/AAC.01272-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eichhorn I, Feudi C, Wang Y, Kaspar H, Feßler AT, Lübke-Becker A, Michael GB, Shen J, Schwarz S. 2018. Identification of novel variants of the colistin resistance gene mcr-3 in Aeromonas spp. from the national resistance monitoring programme GERM-Vet and from diagnostic submissions. J Antimicrob Chemother 73:1217–1221. doi: 10.1093/jac/dkx538. [DOI] [PubMed] [Google Scholar]
- 16.Carroll LM, Gaballa A, Guldimann C, Sullivan G, Henderson LO, Wiedmann M. 2019. Identification of novel mobilized colistin resistance gene mcr-9 in a multidrug-resistant, colistin-susceptible Salmonella enterica serotype Typhimurium isolate. mBio 10:e00853-19. doi: 10.1128/mBio.00853-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kempf I, Jouy E, Chauvin C. 2016. Colistin use and colistin resistance in bacteria from animals. Int J Antimicrob Agents 48:598–606. doi: 10.1016/j.ijantimicag.2016.09.016. [DOI] [PubMed] [Google Scholar]
- 18.Kieffer N, Aires-de-Sousa M, Nordmann P, Poirel L. 2017. High rate of MCR-1-producing Escherichia coli and Klebsiella pneumoniae among pigs, Portugal. Emerg Infect Dis 23:2023–2029. doi: 10.3201/eid2312.170883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poirel L, Madec JY, Lupo A, Schink AK, Kieffer N, Nordmann P, Schwarz S. 2018. Antimicrobial resistance in Escherichia coli. Microbiol Spectr 6:ARBA-0026-2017. doi: 10.1128/microbiolspec.ARBA-0026-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwarz S, Johnson AP. 2016. Transferable resistance to colistin: a new but old threat. J Antimicrob Chemother 71:2066–2070. doi: 10.1093/jac/dkw274. [DOI] [PubMed] [Google Scholar]
- 21.Zurfluh K, Stephan R, Widmer A, Poirel L, Nordmann P, Nüesch HJ, Hächler H, Nüesch-Inderbinen M. 2017. Screening for fecal carriage of MCR-producing Enterobacteriaceae in healthy humans and primary care patients. Antimicrob Resist Infect Control 6:28. doi: 10.1186/s13756-017-0186-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bourrel AS, Poirel L, Royer G, Darty M, Vuillemin X, Kieffer N, Clermont O, Denamur E, Nordmann P, Decousser JW, IAME Resistance Group. 2019. Colistin resistance in Parisian inpatient faecal Escherichia coli as the result of two distinct evolutionary pathways. J Antimicrob Chemother 74:1521–1530. doi: 10.1093/jac/dkz090. [DOI] [PubMed] [Google Scholar]
- 23.Peigne C, Bidet P, Mahjoub-Messai F, Plainvert C, Barbe V, Médigue C, Frapy E, Nassif X, Denamur E, Bingen E, Bonacorsi S. 2009. The plasmid of Escherichia coli strain S88 (O45:K1:H7) that causes neonatal meningitis is closely related to avian pathogenic E. coli plasmids and is associated with high-level bacteremia in a neonatal rat meningitis model. Infect Immun 77:2272–2284. doi: 10.1128/IAI.01333-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Royer G, Decousser JW, Branger C, Dubois M, Médigue C, Denamur E, Vallenet D. 2018. PlaScope: a targeted approach to assess the plasmidome from genome assemblies at the species level. Microb Genom 4:e000211. doi: 10.1099/mgen.0.000211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. 2005. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 63:219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 26.Kieser T. 1984. Factors affecting the isolation of CCC DNA from Streptomyces lividans and Escherichia coli. Plasmid 12:19–36. doi: 10.1016/0147-619X(84)90063-5. [DOI] [PubMed] [Google Scholar]
- 27.Liu YY, Chandler CE, Leung LM, McElheny CL, Mettus RT, Shanks RMQ, Liu JH, Goodlett DR, Ernst RK, Doi Y. 2017. Structural modification of lipopolysaccharide conferred by mcr-1 in Gram-negative ESKAPE pathogens. Antimicrob Agents Chemother 61:e00580-17. doi: 10.1128/AAC.00580-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun J, Xu Y, Gao R, Lin J, Wei W, Srinivas S, Li D, Yang RS, Li XP, Liao XP, Liu YH, Feng Y. 2017. Deciphering MCR-2 colistin resistance. mBio 8:e00625-17. doi: 10.1128/mBio.00625-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kieffer N, Nordmann P, Moreno AM, Zanolli Moreno L, Chaby R, Breton A, Tissières P, Poirel L. 2018. Genetic and functional characterization of an MCR-3-like enzyme-producing Escherichia coli isolate recovered from swine in Brazil. Antimicrob Agents Chemother 62:e00278-18. doi: 10.1128/AAC.00278-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Breland EJ, Zhang EW, Bermudez T, Martinez CR III, Hadjifrangiskou M. 2017. The histidine residue of QseC is required for canonical signaling between QseB and PmrB in uropathogenic Escherichia coli. J Bacteriol 199:e00060-17. doi: 10.1128/JB.00060-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poirel L, Kieffer N, Brink A, Coetze J, Jayol A, Nordmann P. 2016. Genetic features of MCR-1-producing colistin-resistant Escherichia coli isolates in South Africa. Antimicrob Agents Chemother 60:4394–4397. doi: 10.1128/AAC.00444-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou K, Luo Q, Wang Q, Huang C, Lu H, Rossen JWA, Xiao Y, Li L. 2018. Silent transmission of an IS1294b-deactivated mcr-1 gene with inducible colistin resistance. Int J Antimicrob Agents 51:822–828. doi: 10.1016/j.ijantimicag.2018.01.004. [DOI] [PubMed] [Google Scholar]
- 33.Pham Thanh D, Thanh Tuyen H, Nguyen Thi Nguyen T, Chung The H, Wick RR, Thwaites GE, Baker S, Holt KE. 2016. Inducible colistin resistance via a disrupted plasmid-borne mcr-1 gene in a 2008 Vietnamese Shigella sonnei isolate. J Antimicrob Chemother 71:2314–2317. doi: 10.1093/jac/dkw173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson JR, Murray AC, Kuskowski MA, Schubert S, Prère MF, Picard B, Colodner R, Raz R, Trans-Global Initiative for Antimicrobial Resistance Initiative (TIARA) Investigators. 2005. Distribution and characteristics of Escherichia coli clonal group A. Emerg Infect Dis 11:141–145. doi: 10.3201/eid1101.040418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lemaître C, Mahjoub-Messai F, Dupont D, Caro V, Diancourt L, Bingen E, Bidet P, Bonacorsi S. 2013. A conserved virulence plasmidic region contributes to the virulence of the multiresistant Escherichia coli meningitis strain S286 belonging to phylogenetic group C. PLoS One 8:e74423. doi: 10.1371/journal.pone.0074423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Galata V, Fehlmann T, Backes C, Keller A. 2019. PLSDB: a resource of complete bacterial plasmids. Nucleic Acids Res 47:D195–D202. doi: 10.1093/nar/gky1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nordmann P, Jayol A, Poirel L. 2016. Rapid detection of polymyxin resistance in Enterobacteriaceae. Emerg Infect Dis 22:1038–1043. doi: 10.3201/eid2206.151840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jayol A, Kieffer N, Poirel L, Guérin F, Güneser D, Cattoir V, Nordmann P. 2018. Evaluation of the rapid polymyxin NP test and its industrial version for the detection of polymyxin-resistant Enterobacteriaceae. Diagn Microbiol Infect Dis 92:90–94. doi: 10.1016/j.diagmicrobio.2018.05.006. [DOI] [PubMed] [Google Scholar]
- 39.Clinical and Laboratory Standards Institute. 2018. Performance standards for antimicrobial susceptibility testing; 28th informational supplement (M100–S28). Clinical and Laboratory Standards Institute, Wayne (PA). [Google Scholar]
- 40.Kocsis B, Kilar A, Péter S, Dörnyel A, Sandor V, Kilar F. 2017. Mass spectrometry for profiling LOS and lipid A structures from whole-cell lysates: directly from a few bacterial colonies or from liquid broth cultures. Methods Mol Biol 1600:187–198. doi: 10.1007/978-1-4939-6958-6_17. [DOI] [PubMed] [Google Scholar]
- 41.Breton A, Novikov A, Martin R, Tissieres P, Caroff M. 2017. Structural and biological characteristics of different forms of V. filiformis lipid A: use of MS to highlight structural discrepancies. J Lipid Res 58:543–552. doi: 10.1194/jlr.M072900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poirel L, Kieffer N, Nordmann P. 2017. In vitro study of ISApl1-mediated mobilization of the colistin resistance gene mcr-1. Antimicrob Agents Chemother 61:e00127-17. doi: 10.1128/AAC.00127-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gouy M, Guindon S, Gascuel O. 2010. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol 27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]