Inhaled aztreonam is increasingly used for chronic Pseudomonas aeruginosa suppression in patients with cystic fibrosis (CF), but the potential for that organism to evolve aztreonam resistance remains incompletely explored. Here, we performed genomic analysis of clonally related pre- and posttreatment CF clinical isolate pairs to identify genes that are under positive selection during aztreonam therapy in vivo.
KEYWORDS: aztreonam, Pseudomonas aeruginosa, ampC, antibiotic resistance, β-lactamase, cystic fibrosis, genome analysis, selection
ABSTRACT
Inhaled aztreonam is increasingly used for chronic Pseudomonas aeruginosa suppression in patients with cystic fibrosis (CF), but the potential for that organism to evolve aztreonam resistance remains incompletely explored. Here, we performed genomic analysis of clonally related pre- and posttreatment CF clinical isolate pairs to identify genes that are under positive selection during aztreonam therapy in vivo. We identified 16 frequently mutated genes associated with aztreonam resistance, the most prevalent being ftsI and ampC, and 13 of which increased aztreonam resistance when introduced as single gene transposon mutants. Several previously implicated aztreonam resistance genes were found to be under positive selection in clinical isolates even in the absence of inhaled aztreonam exposure, indicating that other selective pressures in the cystic fibrosis airway can promote aztreonam resistance. Given its potential to confer plasmid-mediated resistance, we further characterized mutant ampC alleles and performed artificial evolution of ampC for maximal activity against aztreonam. We found that naturally occurring ampC mutants conferred variably increased resistance to aztreonam (2- to 64-fold) and other β-lactam agents but that its maximal evolutionary capacity for hydrolyzing aztreonam was considerably higher (512- to 1,024-fold increases) and was achieved while maintaining or increasing resistance to other drugs. These studies implicate novel chromosomal aztreonam resistance determinants while highlighting that different mutations are favored during selection in vivo and in vitro, show that ampC has a high maximal potential to hydrolyze aztreonam, and provide an approach to disambiguate mutations promoting specific resistance phenotypes from those more generally increasing bacterial fitness in vivo.
INTRODUCTION
Chronic lung infections remain a serious source of morbidity and mortality for patients with cystic fibrosis (CF), and significant efforts have focused on controlling the incipient pulmonary bacterial populations present in affected individuals (1, 2). Aztreonam lysine, a fully synthetic β-lactam antibiotic, is one of several inhaled drugs that have been developed for the suppression of chronic Pseudomonas aeruginosa infections in CF patients. Like other inhaled antibiotic formulations, aztreonam lysine is able to reach high concentrations within the patient airway with only minimal systemic absorption (3, 4). Since its approval in 2010 (5), inhaled aztreonam has become widely used as therapy for CF patients residing in the United States, with nearly half of eligible individuals currently being prescribed that drug in monthly treatment cycles (6), often alternating with inhaled tobramycin (7, 8).
Although inhaled aztreonam is widely administered, the potential for P. aeruginosa to develop and maintain resistance to that antibiotic over the course of treatment is still incompletely explored. Several clinical trials have reported that P. aeruginosa can evolve modest, dose-dependent, and seemingly transient increases in aztreonam resistance following inhaled therapy (9–11), suggesting that the organism has inherent potential to combat the drug (12). Accordingly, chromosomal changes affecting aztreonam resistance in P. aeruginosa have been previously described, including those that promote active drug efflux through overexpression of the mexAB-OprM system (13–15), alteration of ftsI (PBP3) and consequent disruption of drug binding (16), and mutational overexpression (17) or coding sequence alteration (12, 18) of the chromosomal ampC β-lactamase. To more comprehensively explore the scope of aztreonam resistance mutations possible in P. aeruginosa, we recently employed in vitro selection to identify multiple known and previously unknown genes associated with increased drug resistance (12). Genes recurrently altered during aztreonam selection in vitro were also identified in a small cohort of clinical isolates from patients treated with the antibiotic (12). Nevertheless, mutations in several genes recurrently altered during artificial selection were not observed in clinical isolates, suggesting that they may incur unacceptable fitness costs in vivo, and conversely, several clinical isolates lacked mutations in any candidate genes identified during selection, indicating the existence of additional relevant pathways (12).
Here, we sought to better understand the factors most relevant to P. aeruginosa aztreonam resistance in vivo by cataloging genes under positive selection during inhaled aztreonam therapy in CF patients. We performed whole-genome sequencing and analysis of clonally related sequentially collected clinical isolate pairs obtained from the sputa of CF patients who were either treated with inhaled aztreonam or were not exposed to the drug. This approach identifies genes known to be involved in aztreonam resistance, including ftsI and ampC, while newly implicating factors with resistance contributions that we subsequently confirmed experimentally. We also observed differences between mutations recovered during in vivo and in vitro selection, likely reflecting differential fitness costs present under those conditions. As a secondary aim, given its strong signature of positive selection and its potential to be mobilized to plasmids (19, 20), we characterized resistance capabilities of mutant ampC alleles from aztreonam-resistant isolates and performed in vitro evolution of the gene to more fully investigate its existing and maximal evolutionary potential to hydrolyze aztreonam.
RESULTS
Clonal P. aeruginosa isolates demonstrate various levels of aztreonam resistance after inhaled antibiotic therapy in vivo.
We identified 64 pairs of clonally related P. aeruginosa isolates for genomic analysis that were obtained through the AIR-CF5 clinical trial, a 5-year observational study designed to monitor P. aeruginosa aztreonam susceptibility in CF patients treated with inhaled drug (21). We selected isolate pairs exhibiting at least a 4-fold difference in aztreonam MIC, regardless of whether they originated from the aztreonam-exposure or nonexposure group, leaving a final count of 60 isolate pairs (Table 1). Each pair originated from a different patient, 48 of whom underwent inhaled aztreonam therapy during the study and 12 who never received any formulation of that drug.
TABLE 1.
Paired isolate summary
| Isolate pair identifier | No. of yrs between collections | Aztreonam MIC (μg/ml) |
Fold increase in MIC | Aztreonam resistance stratification group | |
|---|---|---|---|---|---|
| Sensitive isolates | Resistant isolates | ||||
| P_1329 | 1.88 | 8 | 512 | 64 | No exposure |
| P_1266 | 1.07 | 8 | 128 | 16 | No exposure |
| P_1268 | 0.86 | 1 | 64 | 64 | No exposure |
| P_1273 | 1.04 | 8 | 64 | 8 | No exposure |
| P_1279 | 0.98 | 1 | 64 | 64 | No exposure |
| P_1455 | 1.92 | 1 | 64 | 64 | No exposure |
| P_1078 | 1.95 | 4 | 32 | 8 | No exposure |
| P_1468 | 0.90 | 4 | 32 | 8 | No exposure |
| P_1011 | 1.00 | 1 | 16 | 16 | No exposure |
| P_1085 | 1.15 | 1 | 16 | 16 | No exposure |
| P_1099 | 1.11 | 2 | 16 | 8 | No exposure |
| P_1274 | 1.01 | 1 | 16 | 16 | No exposure |
| P_1010 | 2.09 | 2 | 32 | 16 | Low resistance |
| P_1237 | 1.15 | 1 | 32 | 32 | Low resistance |
| P_1324 | 0.96 | 2 | 32 | 16 | Low resistance |
| P_1335 | 1.22 | 4 | 32 | 8 | Low resistance |
| P_1384 | 1.80 | 2 | 32 | 16 | Low resistance |
| P_1451 | 1.98 | 1 | 32 | 32 | Low resistance |
| P_1072 | 1.18 | 4 | 16 | 4 | Low resistance |
| P_1146 | 1.05 | 1 | 16 | 16 | Low resistance |
| P_1215 | 0.83 | 1 | 16 | 16 | Low resistance |
| P_1261 | 0.92 | 1 | 16 | 16 | Low resistance |
| P_1305 | 1.92 | 1 | 16 | 16 | Low resistance |
| P_1390 | 1.05 | 1 | 16 | 16 | Low resistance |
| P_1351 | 1.08 | 1 | 256 | 256 | Medium resistance |
| P_1429 | 1.01 | 2 | 256 | 128 | Medium resistance |
| P_1494 | 1.94 | 8 | 256 | 32 | Medium resistance |
| P_1505 | 0.75 | 1 | 256 | 256 | Medium resistance |
| P_1019 | 1.04 | 1 | 128 | 128 | Medium resistance |
| P_1022 | 1.19 | 2 | 128 | 64 | Medium resistance |
| P_1119 | 2.22 | 1 | 128 | 128 | Medium resistance |
| P_1144 | 0.99 | 1 | 128 | 128 | Medium resistance |
| P_1257 | 1.87 | 1 | 128 | 128 | Medium resistance |
| P_1259 | 1.95 | 2 | 128 | 64 | Medium resistance |
| P_1316 | 2.04 | 1 | 128 | 128 | Medium resistance |
| P_1323 | 1.00 | 8 | 128 | 16 | Medium resistance |
| P_1431 | 1.97 | 1 | 128 | 128 | Medium resistance |
| P_1432 | 0.91 | 2 | 128 | 64 | Medium resistance |
| P_1440 | 1.97 | 4 | 128 | 32 | Medium resistance |
| P_1499 | 1.07 | 4 | 128 | 32 | Medium resistance |
| P_1137 | 2.04 | 1 | 64 | 64 | Medium resistance |
| P_1197 | 1.86 | 1 | 64 | 64 | Medium resistance |
| P_1230 | 1.11 | 16 | 64 | 4 | Medium resistance |
| P_1334 | 2.03 | 8 | 64 | 8 | Medium resistance |
| P_1342 | 1.02 | 4 | 64 | 16 | Medium resistance |
| P_1397 | 1.82 | 2 | 64 | 32 | Medium resistance |
| P_1403 | 0.90 | 1 | 64 | 64 | Medium resistance |
| P_1437 | 0.99 | 8 | 64 | 8 | Medium resistance |
| P_1475 | 1.15 | 1 | 64 | 64 | Medium resistance |
| P_1018 | 1.11 | 2 | >2,048 | >1,024 | High resistance |
| P_1210 | 0.94 | 1 | >2,048 | >2,048 | High resistance |
| P_1411 | 0.80 | 1 | 2,048 | 2,048 | High resistance |
| P_1439 | 0.86 | 4 | 2,048 | 512 | High resistance |
| P_1472 | 1.78 | 1 | 2,048 | 2,048 | High resistance |
| P_1004 | 0.96 | 8 | 1,024 | 128 | High resistance |
| P_1115 | 0.86 | 1 | 1,024 | 1,024 | High resistance |
| P_1379 | 1.06 | 8 | 1,024 | 128 | High resistance |
| P_1038 | 1.04 | 16 | 512 | 32 | High resistance |
| P_1308 | 2.03 | 2 | 512 | 256 | High resistance |
| P_1407 | 0.81 | 64 | 512 | 8 | High resistance |
The average time between collection of paired isolates was 1.3 years (range, 0.75 to 2.22 years), and whole-genome sequencing subsequently revealed pairs to be distinguished by a range of 14 to 2,379 genomic variants (single nucleotide polymorphisms and indels). Two paired isolates from patients with no previous aztreonam exposure and three paired isolates from the aztreonam exposure group (7.8% of total) carried mutS frameshift variants that were consistent with a hypermutator phenotype (22) (see Data Set S1 in the supplemental material). Isolates from patients treated with inhaled aztreonam had an average fold difference in MIC (239.5-fold) and absolute aztreonam MIC (381 μg/ml) that were significantly (P = 0.007 and P = 0.004, respectively, 2-tailed t test) greater than seen for the group which was not exposed to aztreonam (average 29.3-fold difference, 85.3 μg/ml MIC). Half of isolate pairs from patients without inhaled aztreonam exposure had aztreonam MICs greater than 32 μg/ml, despite lacking specific exposure to that drug.
To facilitate the identification of genes having differing contributions to resistance levels and those occurring at different temporal stages as resistance emerged (12), the isolates from aztreonam-exposed patients were stratified based on their quantitative resistance phenotypes: 12 with low-level MICs (≤32 μg/ml), 25 with medium-level MICs between 64 μg/ml and 256 μg/ml, and 11 with highly resistant phenotypes (MICs ≥ 512 μg/ml).
Specific P. aeruginosa genes are under positive selection during inhaled aztreonam exposure.
We identified de novo mutations arising in the antibiotic-resistant member of each isolate pair in order to assess frequently mutated genes that exhibited positive selection associated with aztreonam exposure. We identified 16 candidate resistance genes (Table 2) in one or more groups of isolates as stratified by aztreonam resistance level and for which nonsynonymous mutations were specifically enriched in bacteria that were exposed to aztreonam (P ≤ 0.038). Multiple alignments were subsequently created for each candidate gene to explore the distribution of amino acid changes observed.
TABLE 2.
Genes positively selected during aztreonam exposure
| Locus name | Common gene name | Predicted gene function | % isolates with nonsynonymous mutations |
dN/dS ratio (decimal) |
Significance of dN/dS relative to no exposure group (P value) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No exposure group (n = 12) | Low resistance group (n = 12) | Medium resistance group (n = 25) | High resistance group (n = 11) | All isolates exposed to aztreonam (n = 48) | No exposure group (n = 12) | Low resistance group (n = 12) | Medium resistance group (n = 25) | High resistance group (n = 11) | All isolates exposed to aztreonam (n = 48) | ||||
| PA4110 | ampC | β-Lactamase precursor | 0.00 | 0.00 | 28.00 | 63.64 | 29.17 | 0/0 (UNDa ) | 0/0 (UND) | 7/0 (UND) | 13/0 (UND) | 20/0 (UND) | 6.20 × 10−5 |
| ERW19588 | Hypothetical protein | 0.00 | 0.00 | 24.00 | 18.18 | 16.67 | 0/0 (UND) | 0/1 (0) | 6/0 (UND) | 2/0 (UND) | 8/1 (8) | 1.80 × 10−2 | |
| PA4418 | ftsI | Penicillin binding protein 3 | 8.33 | 16.67 | 48.00 | 72.73 | 45.83 | 1/0 (UND) | 2/2 (1) | 18/0 (UND) | 15/1 (15) | 35/3 (11.67) | 9.68 × 10−3 |
| PA3168 | gyrA | DNA gyrase subunit A | 8.33 | 50.00 | 32.00 | 54.55 | 41.67 | 1/0 (UND) | 9/1 (9) | 9/2 (4.5) | 12/1 (12) | 30/4 (7.5) | 1.98 × 10−2 |
| PA4569 | ispB | Octaprenyl diphosphate synthase | 0.00 | 16.67 | 4.00 | 36.36 | 14.58 | 0/0 (UND) | 2/0 (UND) | 1/0 (UND) | 4/0 (UND) | 7/0 (UND) | 6.78 × 10−3 |
| PA0425 | mexA | Multidrug resistance protein MexA | 0.00 | 16.67 | 16.00 | 45.45 | 22.92 | 0/0 (UND) | 4/0 (UND) | 4/0 (UND) | 6/0 (UND) | 14/0 (UND) | 5.02 × 10−4 |
| PA2420 | opdJ | Outer membrane porin OpdJ | 0.00 | 8.33 | 24.00 | 9.09 | 16.67 | 0/0 (UND) | 1/1 (1) | 7/0 (UND) | 1/0 (UND) | 9/1 (9) | 6.78 × 10−3 |
| PA0958 | oprD | Outer membrane porin OprD | 8.33 | 58.33 | 32.00 | 27.27 | 37.50 | 1/0 (UND) | 9/0 (UND) | 10/1 (10) | 3/0 (UND) | 22/1 (22) | 1.88 × 10−2 |
| PA0847 | Diguanylate cyclase | 0.00 | 8.33 | 24.00 | 0.00 | 14.58 | 0/0 (UND) | 1/0 (UND) | 9/0 (UND) | 0/1 (0) | 10/1 (10) | 3.24 × 10−2 | |
| PA1866 | Hypothetical protein | 0.00 | 8.33 | 28.00 | 9.09 | 18.75 | 0/1 (0) | 1/0 (UND) | 8/2 (4) | 2/1 (2) | 11/3 (3.67) | 3.82 × 10−2 | |
| PA2480 | Probable two-component sensor | 0.00 | 0.00 | 16.00 | 36.36 | 16.67 | 0/0 (UND) | 0/0 (UND) | 4/1 (4) | 4/0 (UND) | 8/1 (8) | 6.78 × 10−3 | |
| PA2557 | Probable AMP-binding enzyme | 0.00 | 8.33 | 20.00 | 36.36 | 20.83 | 0/0 (UND) | 1/0 (UND) | 7/0 (UND) | 4/0 (UND) | 12/0 (UND) | 9.79 × 10−4 | |
| PA4681 | Hypothetical protein | 0.00 | 0.00 | 24.00 | 18.18 | 16.67 | 0/0 (UND) | 0/0 (UND) | 7/0 (UND) | 2/0 (UND) | 9/0 (UND) | 3.59 × 10−3 | |
| PA1798 | parS | Two component sensor ParS | 0.00 | 8.33 | 24.00 | 27.27 | 20.83 | 0/1 (0) | 1/2 (0.5) | 7/0 (UND) | 3/0 (UND) | 11/2 (5.5) | 1.60 × 10−2 |
| PA2426 | pvdS | Sigma factor PvdS | 0.00 | 16.67 | 28.00 | 45.45 | 29.17 | 0/0 (UND) | 2/0 (UND) | 9/0 (UND) | 6/0 (UND) | 17/0 (UND) | 6.20 × 10−5 |
| PA4282 | sbcC | Probable exonuclease | 0.00 | 8.33 | 24.00 | 18.18 | 18.75 | 0/0 (UND) | 1/0 (UND) | 8/1 (8) | 2/0 (UND) | 11/1 (11) | 9.89 × 10−3 |
UND, undefined, due to an absence of synonymous mutations.
Three genes, ftsI, ampC, and gyrA, carried recurrent missense mutations, consistent with gain-of-function changes.
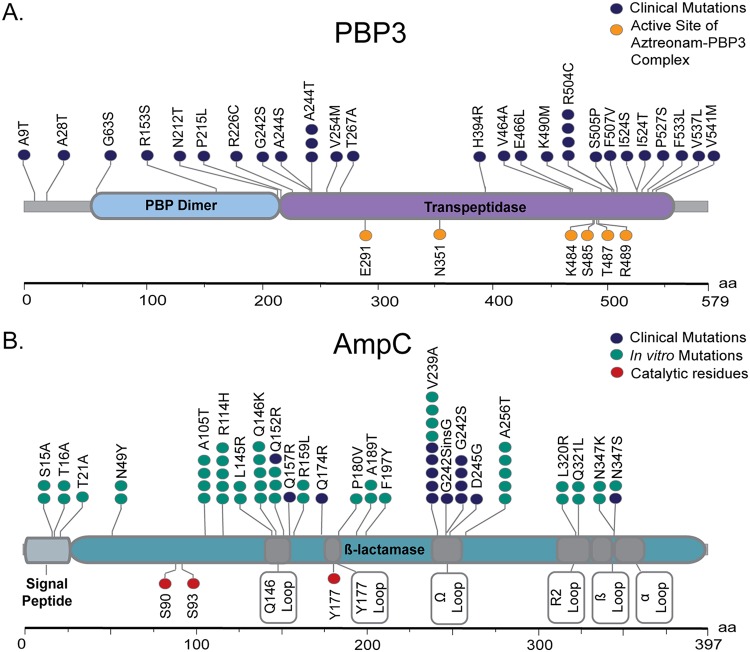
Penicillin binding protein 3 (PBP3; encoded by ftsI), which is the target of aztreonam, was the most frequently mutated target in our study. Mutations were identified in 23/60 (38.33%) isolate pairs and were most prevalent in the high-resistance group (8/11 isolates [72.73%]). We observed 25 unique missense mutations in ftsI (Fig. 1A), with affected isolates carrying a range of 1 to 4 mutations each. Two variants (A244T and R504C) were recurrently identified in more than one isolate. Five variants (A244T, R504C, P527S, H394R, and F533L) have been reported in other studies of CF clinical isolates (22, 23), three of which (A244T, R504C, and P527S) also resulted from our previous in vitro selection for aztreonam resistance (12). The majority of mutations (18/24) fell within the protein’s transpeptidase domain (residues 225 to 579) (24).
FIG 1.
De novo mutations in proteins associated with aztreonam resistance. Mutations are displayed as lolliplots for PBP3 (A) and AmpC (B). Amino acid position is indicated along the x axis, and protein domains are shown as labeled, colored regions. Subregions in AmpC are indicated by dark gray shading, with labels shown at the bottom. The location and abundance of specific amino acid mutations are indicated by colored circles.
Mutations in β-lactamase ampC were strongly associated with highly drug-resistant phenotypes, occurring only in isolates exposed to aztreonam having MICs >64 μg/ml. A total of eight unique de novo ampC mutations (Q174R, Q152R, Q157R, V239A, G242S, G242_P243insG, D245G, and N347S) were cataloged across 11 different alleles, with a range of 1 to 4 mutations per isolate (Fig. 1B). Three mutations (V239A, G242S, and N347S) occurred in multiple isolates. Only one mutation, Q157R, was previously associated with altered β-lactam substrate specificity in P. aeruginosa (25), with the remaining seven being newly reported here.
Recurrent variants in DNA gyrase gene gyrA were also common in the isolate collection; however, all of the variants that were identified (T83I, T83A, and D87N) have been shown to cause fluoroquinolone resistance in P. aeruginosa (26). This suggests that the association of gyrA with aztreonam in our study is likely spurious and reflects uncontrolled differences in other antibiotic exposures that have occurred between patient groups.
Mutations in the remaining 13 genes were sporadic (mexA, oprD, opdJ, pvdS, sbcC, ispB, PA1866, PA2480, PA2557, and PA4681) and/or nonsense (oprD, PA4681, and ERW19588), making them more likely to represent loss-of-function changes.
Transposon mutagenesis functionally validates genes associated with aztreonam resistance in vivo.
Because the majority of genes implicated by our study were likely to be affected by loss-of-function mutations, we initially determined whether a disruption of candidate genes could lead to increased aztreonam resistance. We evaluated aztreonam sensitivities of available transposon mutants in 14 candidate genes and included bidirectional pairs of transposon disruptions where possible (27). Two genes, DNA gyrase subunit gyrA and one encoding a hypothetical protein (Q034_02961), were not available as transposon mutants, due to essentiality and absence from the MPAO1 genome, respectively. Due to variability in the measured susceptibilities of several mutants, MIC testing was performed in replicates and expressed as the average from at least three separate experiments.
All mutants demonstrated statistically significant (Student’s 2-tailed t test, P ≤ 0.013) changes in MIC relative to those for the parental MPAO1 strain and a physiologically neutral transposon control (12) (Table 3). Transposon mutants in 13 of the 14 genes showed increases in MICs from 1.5- to 5.9-fold, while transposon mutants for mexA were hypersensitive to aztreonam. Two genes, sbcC and parS, resulted in measurably dissimilar MICs among two different bidirectional transposon mutants, potentially reflecting polar effects.
TABLE 3.
Transposon mutant MICs
| Strain | Disrupted locus | Common name of disrupted gene | MIC (μg/ml) |
Fold increase over transposon control | Significance of MIC difference relative to transposon control (P value) | |
|---|---|---|---|---|---|---|
| Average | SEM | |||||
| MPAO1 (parental strain) | 1.31 | 0.13 | 0.96 | 7.85 × 10−1 | ||
| PW3303 (transposon control) | PA1274 | 1.36 | 0.15 | |||
| PW5294 | PA2557 | 4.00 | 0.00 | 2.93 | 8.66 × 10−9 | |
| PW5293 | PA2557 | 3.33 | 0.42 | 2.44 | 4.04 × 10−3 | |
| PW5085 | PA2426 | pvdS | 4.00 | 0.00 | 2.93 | 8.66 × 10−9 |
| PW2543 | PA0847 | 3.80 | 0.55 | 2.79 | 1.58 × 10−3 | |
| PW2544 | PA0847 | 4.00 | 0.00 | 2.93 | 8.66 × 10−9 | |
| PW8212 | PA4282 | sbcC | 3.82 | 0.18 | 2.80 | 2.42 × 10−9 |
| PW8213 | PA4282 | sbcC | 2.00 | 0.00 | 1.47 | 1.88 × 10−3 |
| PW5076 | PA2420 | opdJ | 4.00 | 0.00 | 2.93 | 8.66 × 10−9 |
| PW8445 | PA4418 | ftsI | 3.14 | 0.40 | 2.30 | 3.60 × 10−3 |
| PW4274 | PA1866 | 4.00 | 0.47 | 2.93 | 1.64 × 10−4 | |
| PW4275 | PA1866 | 4.36 | 0.36 | 3.20 | 3.20 × 10−6 | |
| PW5165 | PA2480 | 3.43 | 0.37 | 2.51 | 8.23 × 10−4 | |
| PW5164 | PA2480 | 4.00 | 0.00 | 2.93 | 8.66 × 10−9 | |
| PW2742 | PA0958 | oprD | 4.00 | 0.00 | 2.93 | 8.66 × 10−9 |
| PW4165 | PA1798 | parS | 4.00 | 0.47 | 2.93 | 1.64 × 10−4 |
| PW4164 | PA1798 | parS | 8.00 | 0.00 | 5.87 | 9.62 × 10−13 |
| PW7954 | PA4110 | ampC | 3.14 | 0.40 | 2.30 | 3.60 × 10−3 |
| PW7953 | PA4110 | ampC | 4.00 | 0.00 | 2.93 | 8.66 × 10−9 |
| PW1778 | PA0425 | mexA | ≤0.25 | 0.00 | 0.18 | 2.54 × 10−5 |
| PW1779 | PA0425 | mexA | ≤0.25 | 0.00 | 0.18 | 2.54 × 10−5 |
| PW8689 | PA4569 | ispB | 2.00 | 0.00 | 1.47 | 1.88 × 10−3 |
| PW8881 | PA4681 | 4.33 | 0.80 | 3.18 | 1.33 × 10−2 | |
| PW8880 | PA4681 | 4.00 | 0.00 | 2.93 | 8.66 × 10−9 | |
Different aztreonam resistance genes are under selection in vivo and in vitro.
We previously identified 15 aztreonam resistance genes that were recurrently mutated in isolates passaged for antibiotic resistance in vitro and which were subsequently experimentally validated (12), of which only two (ampC and ftsI) were also recovered through our present analysis of in vivo isolate pairs. Examination of the 13 disparate genes revealed three different reasons for their not being identified as significant in this study (see Table S1 and Data Set S1). mexR and mexT, relevant to mexAB-oprM and mexEF-oprN efflux pump function, respectively, were under positive selection in vivo in isolates from aztreonam-exposed pateints, but de novo mutations did not occur in a large enough fraction of clinical isolates to pass our filtering criteria. Eight other genes (nalD, mexF, mpl, clpA, pgi, dacB, pepA, and PA3206) were under positive selection in CF clinical isolates regardless of aztreonam exposure, and their mutation was therefore not specific to aztreonam resistance. The last seven genes (phoQ, aroB, nalC, clpS, atpA, atpD, and orfN) were not under positive selection in clinical isolates from aztreonam-exposed patients.
The differences in resistance genes that were mutated during selection in vitro and in vivo could reflect disparities in bacterial fitness incurred under those different conditions. To more fully explore whether separate aztreonam resistance mutations are favored in vitro and in vivo, we performed transposon sequencing (Tn-Seq) (28) of a comprehensive P. aeruginosa mutant library that we selected for increased antibiotic resistance. Likely owing to the small changes in aztreonam MIC conferred by individual transposon mutants (12), only 12 mutants were found to be significantly enriched on aztreonam-containing medium relative to the unselected transposon pool (see Table S2). Five of the genes (mexT, mexR, nalD, dacB, and pepA) were recurrently mutated during in vitro passaging for aztreonam resistance in our previous study (12), whereas no genes identified as under positive selection in clinical isolates were recovered as significant by Tn-Seq. These findings are consistent with gene inactivation events conferring the greatest aztreonam resistance in vitro having a fitness cost or otherwise being unfavorable in vivo.
Chromosomal ampC mutations promote aztreonam resistance.
To assess the activity of mutant ampC alleles identified in aztreonam-selected P. aeruginosa, each was cloned into an expression shuttle vector and transformed into Escherichia coli. MICs were assessed using aztreonam and other structurally distinct β-lactam antibiotics (Table 4). Ten alleles conferred 2- to 64-fold increases in aztreonam resistance, while one (allele 8) had an MIC equivalent to that for the empty plasmid vector. The alleles conferred variable levels of resistance to the other β-lactam antibiotics tested. None provided increased resistance to meropenem, consistent with the low activity of wild-type P. aeruginosa ampC with that substrate (29). One allele (allele 3) conferred a 2-fold increase in imipenem resistance. Five alleles showing increased resistance to aztreonam resulted in decreases in ampicillin resistance, and nine provided attendant gains in ceftazidime resistance. Remarkably, the allele providing the highest levels of aztreonam resistance (allele 6) also conferred the highest levels of resistance to ampicillin, ceftazidime, and cefpirome.
TABLE 4.
MICs of ampC alleles from clinical isolates
| ampC allele | AmpC amino acid substitutionsa | MIC in μg/ml (fold change relative to WT AmpC) of: |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
E. coli DH5α |
ΔampC MPAO1 |
||||||||||||
| Aztreonam | Ampicillin | Ceftazidime | Cefpirome | Meropenem | Imipenem | Aztreonam | Ampicillin | Ceftazidime | Cefpirome | Meropenem | Imipenem | ||
| None | Empty vector | 0.0625 | 2 | 0.125 | 0.0078125 | 0.0625 | 1 | 2 | 64 | 1 | 1 | 4 | 1 |
| Wild type | None | 0.25 | 64 | 0.5 | 0.015625 | 0.0625 | 1 | 16 | 1,024 | 16 | 8 | 4 | 4 |
| 1 | P7S, T105A, G242S, N347S | 1 (4) | 32 (0.5) | 2 (4) | 0.015625 (1) | 0.0625 (1) | 1 (1) | 128 (8) | 2,048 (2) | 128 (8) | 16 (2) | 4 (1) | 2 (0.5) |
| 2 | G27D, R79Q, T105A, G242S | 0.5 (2) | 32 (0.5) | 0.5 (1) | 0.0078125 (0.5) | 0.0625 (1) | 1 (1) | 64 (4) | 2,048 (2) | 64 (4) | 16 (2) | 4 (1) | 2 (0.5) |
| 3 | T105A, G242S | 0.5 (2) | 16 (0.25) | 1 (2) | 0.015625 (1) | 0.0625 (1) | 2 (2) | 32 (2) | 2,048 (2) | 64 (4) | 16 (2) | 4 (1) | 2 (0.5) |
| 4 | R79Q, T105A, Q152R, Q157R, V239A, N347S | 4 (16) | 32 (0.5) | 4 (8) | 0.015625 (1) | 0.0625 (1) | 0.5 (0.5) | 256 (16) | 2,048 (2) | 256 (16) | 32 (4) | 4 (1) | 4 (1) |
| 5 | T105A, G242_P243insG | 0.5 (2) | 32 (0.5) | 2 (4) | 0.015625 (1) | 0.0625 (1) | 1 (1) | 32 (2) | 2,048 (2) | 64 (4) | 32 (4) | 4 (1) | 2 (0.5) |
| 6 | R79Q, T105A, V239A, D245G | 16 (64) | 256 (4) | 16 (32) | 0.0625 (4) | 0.0625 (1) | 1 (1) | 256 (16) | 2,048 (2) | 256 (16) | 32 (4) | 4 (1) | 2 (0.5) |
| 7 | T105A, L176R, V239A | 1 (4) | 64 (1) | 2 (4) | 0.015625 (1) | 0.0625 (1) | 1 (1) | 32 (2) | 2,048 (2) | 64 (4) | 16 (2) | 4 (1) | 2 (0.5) |
| 8 | T21A, T105A, Q174R, G391A | 0.0625 (0.25) | 2 (0.03125) | 0.125 (0.25) | 0.0078125 (0.5) | 0.0625 (1) | 1 (1) | 2 (0.125) | 32 (0.03125) | 2 (0.125) | 1 (0.125) | 4 (1) | 2 (0.5) |
| 9 | R79Q, T105A, V239A, P274L | 0.5 (2) | 64 (1) | 1 (2) | 0.015625 (1) | 0.0625 (1) | 1 (1) | 32 (2) | 2,048 (2) | 128 (8) | 16 (2) | 4 (1) | 4 (1) |
| 10 | T105A, V239A | 1 (4) | 128 (2) | 1 (2) | 0.015625 (1) | 0.0625 (1) | 1 (1) | 64 (4) | 2,048 (2) | 64 (4) | 16 (2) | 4 (1) | 4 (1) |
| 11 | R79Q, T105A, G242S, V356I | 0.5 (2) | 64 (1) | 1 (2) | 0.015625 (1) | 0.0625 (1) | 1 (1) | 32 (2) | 1,024 (1) | 64 (4) | 8 (1) | 4 (1) | 4 (1) |
Boldface font indicates de novo mutation.
Similar resistance patterns were observed after expression shuttle vectors were transformed into a P. aeruginosa background from which chromosomal ampC had been ablated by transposon mutagenesis (27). Resistance levels for all drugs tested were higher than seen in E. coli, indicating greater basal resistance levels in P. aeruginosa. Consistent with its performance in E. coli, allele 8 conferred no increase in resistance to any of the antibiotics tested. Six alleles provided equivalent or enhanced resistance to ampicillin, ceftazidime, and cefpirome, which was not observed in E. coli. These findings indicate that higher resistance levels across multiple drugs can be achieved when expressing mutant P. aeruginosa ampC alleles in their species of origin. Somewhat contrarily, relative increases in resistance which were apparent in an E. coli background were not seen after transfer to P. aeruginosa; however, this may reflect limited ability of 2-fold serial dilution MIC testing to resolve subtle differences between resistance phenotypes occurring at high antibiotic concentrations. All alleles tested exhibited equal or reduced resistance to imipenem relative to the wild type when expressed in P. aeruginosa.
Although genomic analysis did not identify regulators of ampC expression as being under positive selection during aztreonam therapy, focused analysis of relevant genes (dacB, ampD, ampR, and mpl) identified nonsynonymous changes for multiple isolates in both the aztreonam exposure and control treatment groups (Data Set S1). As such, mutational upregulation of ampC may contribute to aztreonam resistance in a subset of cases, albeit by processes which are not specifically selected during aztreonam exposure.
Artificial evolution of ampC reveals its evolutionary potential for elevated aztreonam hydrolysis.
To more fully evaluate the evolutionary potential for ampC to confer aztreonam resistance, we lastly performed artificial evolution of that gene using cycles of mutagenic PCR, library cloning in E. coli, and selection of resultant populations against increasing concentrations of aztreonam. Bacterial growth at the highest drug concentration was harvested for plasmid and used as the template for the next round of mutagenesis. Three separate evolutionary replicates were generated in parallel. Seven to nine rounds of selection were performed before aztreonam resistance levels plateaued, after which multiple individual colonies were isolated and subjected to formal MIC analysis. Isolates from the same replicates with different aztreonam resistance phenotypes were considered to potentially harbor different alleles, and mutations were catalogued by sequencing.
Evolved populations proved largely homogeneous, with a total of five different alleles carrying a total of 19 different mutations identified. Artificially evolved alleles carried a range of 7 to 11 point mutations (Fig. 1B) and conferred between 512- to 1,024-fold increases in aztreonam resistance relative to the wild-type gene (Table 5). Moreover, all alleles either maintained original resistance levels or demonstrated increased resistance to ampicillin, ceftazidime, imipenem, and/or cefpirome. Alleles 2A and 3A uniquely showed modest 2-fold increases in meropenem resistance.
TABLE 5.
MICs of artificially evolved ampC alleles
| Selection replicate | Allele | AmpC mutations | MIC in μg/ml (fold change from WT control) of: |
|||||
|---|---|---|---|---|---|---|---|---|
| Aztreonam | Ampicillin | Ceftazidime | Cefpirome | Meropenem | Imipenem | |||
| None | Wild type | 0.125 | 32 | 0.25 | 0.03125 | 0.0625 | 1 | |
| 1 | A | S15A, T16A, N49Y, R114H, Q146K, Q152R, F197Y, V239A, A256T, Q321L, N347K | 64 (512) | 256 (8) | 256 (1,024) | 0.125 (4) | 0.0625 (1) | 1 (1) |
| 1 | B | S15A, T16A, N49Y, A105T, R114H, L145R, Q146K, Q152R, V239A, A256T, Q321L, N347K | 64 (512) | 32 (1) | 8 (32) | 0.0625 (2) | 0.0625 (1) | 1 (1) |
| 2 | A | A105T, R114H, Q146K, Q152R, R159L, V239A, A256T, L320R, Q321L, N347K | 64 (512) | 128 (4) | 32 (125) | 0.03125 (1) | 0.125 (2) | 1 (1) |
| 2 | B | T21A, A105T, R114H, Q146K, R159L, A189T, V239A, A256T, L320R | 128 (1,024) | 512 (9) | 64 (256) | 0.0625 (2) | 0.0625 (1) | 1 (1) |
| 3 | A | A105T, R114H, L145R, Q146K, P180V, A256T, N347S | 64 (512) | 64 (2) | 16 (64) | 0.03125 (1) | 0.125 (2) | 1 (1) |
DISCUSSION
It is challenging to study bacterial adaptation to antibiotics in vivo during chronic infection, because the invading organism concurrently accumulates numerous other changes which promote its survival in the host environment (30–32). Moreover, alterations to metabolic networks, including loss of nonessential metabolic functions (30), may result from selection for increased pathogenic fitness but incidentally impact antibiotic susceptibilities (33). Here, we have attempted to disambiguate de novo genomic changes in P. aeruginosa that are selected in response to inhaled aztreonam therapy from those more generally promoting fitness in the CF patient airway by identifying genes that are specifically under positive selection in patients treated with the drug. This strategy has revealed both known and previously unappreciated genes which promote aztreonam resistance in P. aeruginosa.
Sixteen genes, only three of which (ftsI, ampC, and mexA) were previously associated with aztreonam resistance (13–18), were implicated by our study (Table 2). Thirteen of these subsequently tested using transposon mutants to model gene inactivation events were functionally validated as conferring modest but statistically significant elevations in aztreonam resistance (Table 3). These resistance phenotypes were qualitatively consistent with our prior study, where the slight gains in resistance accompanying mutation of individual genes exerted additive effects when occurring multiply in the same strain (12).
Mutations in ftsI were most common, reinforcing the idea that altered aztreonam target binding is a frequent mechanism underlying drug resistance (12, 34). ftsI encodes the only essential penicillin binding protein in P. aeruginosa, and deleterious mutations in the gene result in growth defects (35). Though an increase in aztreonam resistance was seen for an ftsI transposon mutant (Table 3), which contained an insertion at the end of the transpeptidase domain and therefore likely imparts a partial loss of function (36), point mutations may confer enhanced resistance while less dramatically impacting cell physiology. Specific, recurrent ftsI missense mutations were identified from aztreonam-resistant P. aeruginosa isolates in this study (Fig. 1A) and our prior work, suggesting that particular regions of PBP3 affect the resistance phenotype. PBP3 contains two functional domains, the N-terminal (residues 1 to 221) and the C-terminal transpeptidase domains (residues 222 to 579) (36). The vast majority (19 of 25) of de novo mutations identified in vivo, and all 11 variants recovered from our in vitro evolution experiments, occurred within the transpeptidase domain (12), with 9 of them being localized to the enzyme binding pocket surrounding the PBP3 active site (24, 37). The R504C binding pocket mutation was most commonly identified in our study and has been reported in CF isolates with reduced aztreonam susceptibility (12, 22, 23). Variants at that residue have also been linked with resistance to carbapenems (38) and ceftazidime (39). Two additional binding pocket variants (P527T and F533L) have also been observed in CF clinical isolates with reduced aztreonam susceptibility (12, 22, 23). All variants within this domain are likely to influence binding pocket conformation and disrupt aztreonam binding. The newly identified binding pocket mutations (K490M, S505P, F507V, I524S, and I524T) likely affect resistance by the same mechanism. Mutations elsewhere in the transpeptidase domain have been observed in CF isolates, including two specific changes found by our study (A244T and H394R) (23), and may indirectly influence the structure of the binding pocket. The remaining PBP3 mutations (A9T, A28T, G63S, R153S, N212T, and P215L) localized to the N-terminal domain. The N-terminal domain is thought to play an accessory role in folding and stability of the transpeptidase (37), and so mutations within it could similarly influence conformation of the enzyme binding pocket.
Loss-of-function mutations in two membrane porins, OprD and OpdJ, were newly identified as mechanisms promoting aztreonam resistance, likely by decreasing outer membrane permeability to the antibiotic (40). OprD has long been recognized as a porin thought to be exclusive for the entry of carbapenem antibiotics, and loss of OprD expression is accordingly correlated with increased resistance to drugs from that class without reportedly impacting susceptibilities to other β-lactam drugs (13, 40–43). Our findings contrarily show that OprD loss promotes increased resistance to aztreonam (Table 3), indicating an additional role in aztreonam uptake. OpdJ has been identified as a probable specific outer membrane protein based on weak homology to OprD (44, 45), but the protein has been studied far less extensively and its function has not yet been experimentally elucidated. We have found that OpdJ disruption results in an aztreonam resistance phenotype equivalent to that seen with OprD loss, consistent with the two proteins having similar functional roles.
PvdS is an alternative sigma factor that serves as the major iron starvation factor of P. aeruginosa and regulates the expression of at least 26 genes or operons, including virulence factors and genes otherwise unrelated to iron metabolism (46–49). Although our study identifies a link between loss of PvdS activity and elevated aztreonam resistance, disruption of that transcriptional regulator could impart profound changes to metabolism and phenotype, making it difficult to ascertain the specific mechanism by which this phenotype is affected.
The role of MexAB-OprM in the active efflux of aztreonam has been well described (13–15), and we previously observed overexpression of this system in isolates selected for aztreonam resistance in vitro secondary to inactivation of negative regulators NalD, MexR, and NalC (12). Although these negative regulators did not harbor de novo variants and were not under significant positive selection in clinical isolates from aztreonam-exposed patients, we found that the multidrug efflux membrane subunit mexA was. Although loss of MexA resulted in aztreonam hypersensitivity (Table 3), observed mutations were not recurrent or spatially restricted to specific regions of the protein, making them less likely to fit the profile of gain-of-function changes. Though these results appear paradoxical, they are consistent with prior analyses of CF clinical isolates which have shown frequent missense and loss-of-function mutations in MexA (22, 50). Given the correlation of MexAB-OprM overproduction with increased virulence (12), it is possible that inactivating mexA mutations reflect selection for attenuation during aztreonam therapy (51) rather than increased resistance to aztreonam itself. Alternatively, it is known that loss of MexA promotes expression of the MexXY-OprM efflux system (22, 52), which has distinct antibiotic specificities and may therefore be selected in response to other antibiotics administered to these patients.
Four genes likely impacting gene regulation were under positive selection during aztreonam exposure. Two separate two-component sensors, parS and PA2480, were identified. parS encodes the sensor kinase of the P. aeruginosa ParRS regulatory system. It is reported that induction or mutational activation of this system increases resistance to multiple drugs by affecting oprD repression, efflux system activation, and lipopolysaccharide modification (53, 54). Our study indicates that loss of parS, rather than its activation, can increase resistance to aztreonam, arguing that it has additional functions in regulating alternative aztreonam resistance pathways. PA2480 is a putative two-component sensor based on protein sequence homology, but its role and regulatory targets are currently undescribed. The remaining two proteins, PA2557, a hypothetical AMP-binding enzyme, and PA0847, a diguanylate cyclase, also have possible roles in regulating gene expression (55, 56). However, without experimentally testing the function of these genes, it is difficult to hypothesize through which pathways they promote aztreonam resistance.
Two genes, PA1866 and PA4681, encode hypothetical proteins without notable homology to better-described factors in Pseudomonas or other organisms. As such, the role that these genes may play in antibiotic resistance is opaque but they will serve as interesting targets for future studies. A third gene, sbcC, has been well studied in Escherichia coli, where it is believed to serve as in the exonuclease cleavage of hairpin DNA (57), which also has an unclear relationship to aztreonam resistance.
We placed particular emphasis on the potential for the final gene, the ampC β-lactamase, to confer aztreonam resistance. Although aztreonam is known to be poorly hydrolyzed by P. aeruginosa chromosomal ampC (58), mutations which increase the enzyme’s activity against aztreonam are concerning, because resistance-causing alleles could be mobilized to plasmids and rapidly disseminated through a population (59). Unexpectedly, but consistent with earlier findings(12), two separate transposon mutants ablating ampC function resulted in minor gains in aztreonam resistance, possibly by reducing the metabolic burden of its expression (58). Regardless, more than one-quarter of aztreonam-exposed strains (14 of 48) carried de novo mutations in ampC, which were entirely absent in P. aeruginosa from patients without aztreonam exposure. Multiple recurrent ampC mutations were identified in this and our prior study(12), suggesting gain-of-function effects which could impart aztreonam resistance either through increased gene expression (17, 60) or by modifying the structure and function of the enzyme itself (12, 18).
Mutant alleles from clinical isolates harbored 8 unique de novo mutations (Fig. 1B), only two of which (Q157R and Q174R) were previously reported (25). It is probable that these variants affect enzyme substrate specificity or hydrolytic activity given their placement relative to the functional domains of ampC (61). The catalytic residues of ampC comprise Ser90-Ser93-Tyr177 (62), with boundaries of the larger active site defined by the Gln146 loop (residues 143 to 154), Tyr177 loop (residues 176 to 179), Ω-loop (residues 238 to 252), R2-loop (residues 315 to 333), β11 (residues 338 to 346), and α11 (residues 373 to 390) (Fig. 1B) (61). Five of the eight de novo mutations (Q152R, V239A, G242S, G242_P243insG, and D245G) mapped within the active site and could directly impact catalytic activity. Prior work has shown that mutations of the Ω-loop, in particular, can impart greater catalytic abilities for specific drugs (61, 63), and half of the observed mutations reside in this domain. The other three mutations (Q157R, Q174R, and N347S) were immediately adjacent to the active site and could plausibly affect its conformation.
In vitro evolution revealed the evolutionary potential of ampC to confer high levels of aztreonam resistance while maintaining or increasing activity against other β-lactam antibiotics. Artificially evolved alleles conferred greater levels of aztreonam resistance (512- to 1,024-fold increases) than those from clinical isolates (2- to 12-fold increases) (Table 4 and 5), although the total mutational burden per evolved allele was also correspondingly higher (range of 7 to 11 mutations per allele, versus a maximum of 3 mutations) (Fig. 1B). Three artificially evolved mutations (T21A, V239A, and N347S) were also identified in clinical alleles, suggesting advantages in vivo and in vitro. Six of the 19 mutations recovered (L145R, Q146K, Q152R, V239A, L320R, and Q321L) occurred within the AmpC active site and likely altered the enzyme’s specificity, while four others (R159L, P180V, N347K, and N347S) were immediately adjacent and could exert similar effects. The functional impact of the remaining nine mutations, if any, is presently unclear. Interestingly, three changes (R114H, Q146K, and A256T) that were universally present among the evolved alleles were not seen in clinical isolates, and only one of these (Q146K) mapped to an active site domain.
Surprisingly, relatively few genes identified as under positive selection for aztreonam resistance in vivo were previously implicated through in vitro selections for resistance to that drug (12), with only ampC and ftsI being identified across both studies (see Table S1 in the supplemental material). Focused analysis of the resistance genes identified from in vitro studies in the paired clinical strains revealed three separate reasons for this discrepancy. First, two genes known to affect resistance by efflux pump regulation were identified as under positive selection in aztreonam exposed isolates in vivo, but too few isolates carried mutations in those regulators to meet our selection criteria. Second, some genes that promote aztreonam resistance in vitro were not under positive selection in in vivo isolates. It is likely that such genes incur unacceptable fitness costs to mutant bacteria within CF patient lungs. Thus, the spectrum of possible aztreonam resistance mutations arising in clinical practice is likely to be more constrained than the constellation of possible mutations recovered during growth in rich media in vitro. Although the changes in aztreonam MIC conferred by individual transposon mutants has proven generally small (12), this conclusion is supported by our Tn-seq experiments (Table S2), which exclusively overlap the output of in vitro selection. Lastly, some resistance genes identified from in vitro passaging were under positive selection in clinical isolates, but this selection was equivalent whether isolates were exposed to aztreonam or not. This indicates that such genes are generally advantageous to P. aeruginosa during CF infection and shows that multiple mutations arising spontaneously during pathogenesis can promote aztreonam resistance even in the absence of that drug. Indeed, up to 64-fold differences in aztreonam resistance were observed in clinical isolate pairs from individuals who were not exposed to inhaled aztreonam therapy, and several genes identified from our study are reported as being highly mutated in isolates from CF patients (50) or to generally contribute to the P. aeruginosa resistome (22). Taken together, these findings highlight the importance of examining clinical isolates from antibiotic-treated patients when identifying resistance genes that are relevant in vivo.
In summary, leveraging signatures of positive selection specific to bacterial isolates exposed to inhaled aztreonam therapy in vivo, this work has ascertained novel resistance determinants underlying that polygenic trait. We identify ftsI and ampC mutations as being of particular importance and have demonstrated the potential for ampC to evolve high-level activity against aztreonam without sacrificing conferred resistance to other β-lactam agents. Nevertheless, we acknowledge several limitations of our study design. The patient groups used for analysis in this study were not perfectly controlled, either with respect to size or clinical characteristics, and systemic antibiotic exposures were not recorded, likely resulting in some spurious associations. For example, the gyrA mutations identified in this work are likely to be artifactual, reflecting differences in administration of fluoroquinolone agents between study populations instead of being related to aztreonam exposure. Additionally, the number or courses of aztreonam inhalation therapy in the exposed population were variable, and the timing of isolate collection in relation to drug exposure is unknown, both of which may influence resistance selection pressures at the time of isolate collection. Dedicated investigation of genes found to be mutated in posttherapy strains will be required to characterize their roles in aztreonam resistance. Finally, because we have focused on identifying the most commonly mutated aztreonam resistance factors, there are likely to be additional chromosomal resistance genes that currently remain uncharacterized.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and MIC testing.
Clinical P. aeruginosa isolates from patient sputa collected during the AIR-CF5 clinical trial (21) (ClinicalTrials identifier NCT01375036) were provided by Gilead Sciences, Inc., as was associated information on antibiotic sensitivities. Per the clinical trial protocol, patients were classified as being exposed to inhaled aztreonam if they received one or more courses of therapy within 12 months prior to respiratory cultures. All patients in the nonexposure group had never been exposed to inhaled aztreonam. A total of 60 paired isolates (120 total isolates) which exhibited 4-fold or greater changes in aztreonam MIC were ultimately selected for analysis.
All individual P. aeruginosa transposon mutants (36) were previously described. Electrocompetent E. coli DH5α and 10-β were purchased from New England BioLabs. E. coli λ-pir (64) was a generous gift from Pradeep Singh at the University of Washington.
All strains were maintained at 37°C in Luria-Bertani (LB) broth under selection with the appropriate antibiotic, if required, for plasmid maintenance.
Liquid MIC determination was performed according to CLSI guidelines (65), except that LB broth was used and plasmid-bearing strains were induced with 3 mM IPTG (isopropyl-β-d-thiogalactopyranoside; Thermo Scientific), where appropriate.
Microbial sequencing.
DNA was extracted using a DNeasy UltraClean Microbial DNA Isolation kit (Qiagen), and sequencing libraries were prepared as described previously (66, 67). Sequencing was performed using Illumina NextSeq 500 and MiSeq platforms with 150-bp paired-end chemistries.
Transposon sequence analysis.
Transposon mutant pool generation and sequence analysis (Tn-seq) was performed as described previously (28, 36) using aztreonam-containing medium for selection. Briefly, a pool of >110,000 unique ISlacZ-hah-tc transposon insertion mutants in the MPAO1 genome were grown on rich medium and cryopreserved in dimethyl sulfoxide (DMSO). The mutant pool was washed in Dulbecco's phosphate-buffered saline (DPBS) and recovered for 90 min in LB prior to testing on aztreonam. Mutants with enhanced aztreonam resistance were selected on 1, 2, and 4 μg/ml aztreonam-LB plates. For each concentration tested, 100 μl of the recovered transposon pool was diluted to approximately 6 × 106 CFU per 10-cm plate and incubated for 15 h. Isolates were then harvested and libraries prepared using the Tn-seq C-tailing method (28) with primers specific for transposon T8. Transposon insertions per gene were normalized based on average gene size, total number of mapped reads per sequencing run, and the read density of the Tn insertion region. After normalization, genes with a >1.5-fold increase in transposon insertion events detected on each concentration of aztreonam-containing medium relative to the parental control were considered significant.
Identification and analysis of candidate aztreonam resistance genes.
To assess the degree of positive selection for each gene, de novo genome assemblies of each aztreonam-sensitive isolate were first constructed using ABySS (68) to be used as a reference genome for each clonally related isolate pair. This measure maximized similarity in genome content between isolate pairs. Draft genomes were annotated using PROKKA (69) to provide uniform gene prediction annotation across sequenced strains. Antibiotic-resistant and antibiotic-sensitive isolates of each pair were independently aligned to the de novo assembly genome using bwa-mem (v0.7.12) (70), with single nucleotide polymorphisms and indels identified using SAMtools (v1.1) (71) as described elsewhere (66, 67). Variants were annotated with SNPEff (72). Structurally similar gene homologs with 80% or greater identity were grouped using CD-Hit (73), enabling comparison of mutations across patient isolates.
Isolates from the patients in the aztreonam treatment group were stratified according to their level of drug resistance (low resistance, ≤32 μg/ml; medium resistance, 64 μg/ml to 256 μg/ml; high resistance, ≥ 512 μg/ml). To be considered candidate aztreonam resistance determinants, genes needed to satisfy three different requirements. (i) Genes were under positive selection in one or more stratified groups of isolates from aztreonam-exposed patients, defined as the ratio of nonsynonymous to synonymous mutations (dN/dS ratio) of greater than one. (ii) The relative burden of nonsynonymous mutations was significantly higher in aztreonam-treated groups relative to isolates from the nonexposure group. Testing was performed using a Student’s 2-tailed t test for samples with unequal variance, expressing the relative number of nonsynonymous mutations (the number of nonsynonymous mutations minus the number of synonymous mutations divided by the total number of mutations) to enable testing of comparisons having zero counts for nonsynonymous or synonymous changes. (iii) Genes were mutated in at least 20% of isolates in one or more of the aztreonam drug resistance level stratification groups.
P. aeruginosa de novo gene predictions were assigned names by performing DIAMOND BLAST-p (74) searches using representative sequences from the grouped homologous gene clusters. Multiple sequence alignment of mutated proteins to assess for recurrent mutations was performed using Clustal Omega (75).
Variant detection in specific targeted genes (see Data Set S1 in the supplemental material) was separately performed against the P. aeruginosa PAO1 reference genome (GenBank accession AE004091.2) as previously described (12).
ampC cloning.
We replaced the ampR gene of E. coli-Pseudomonas shuttle vector pMMB190 (ATCC) with the gentamicin resistance cassette of pex18GM (76) to avoid possible interference in subsequent aztreonam resistance assays. The gentamicin resistance cassette was amplified using primers F_Gibson_pMMB190_GM (5′-CCGGGGATCCATTTACCG-3′, all oligonucleotides synthesized by IDT) and R_Gibson_pMMB190_GM (5′-AGACGTCAGGTGGCACTTTTC-3′) and was introduced into pMMB190 by Gibson assembly (77) to generate vector pMMB190_GM.
Wild-type or mutant ampC genes were PCR amplified from appropriate templates using primers F_Gibson_ampC_GM_pMMB190 (5′-GCTCCCGGGGCGGTTTCT-3′) and R_Gibson_ampC_GM_pMMB190 (5′-CATAGCCAGGACCGGCGTC-3′) and then inserted downstream of the lac promoter of pMMB190_GM by Gibson assembly. Plasmids were transformed into E. coli DH5α by electroporation or into P. aeruginosa as described elsewhere (64).
In vitro evolution and selection of ampC mutants.
To support the construction of high-diversity mutant libraries, we constructed a small high-efficiency cloning vector derived from pUC19 (NEB) and pMMB190_GM. The pUC19 origin of replication was amplified using primers pUC19_ori_expression_F (5′-GCGGTATCATTGCAGCACTGG-3′) and pUC19_ori_expression_R (5′-TGAGCAAAAGGCCAGCAAAAG-3′). The lacZ promoter, multiple cloning site, and gentamicin resistance cassette of pMMB190_GM were PCR amplified using primers pMMB_MCS_F (5′-GCCGACATCATAACGGTTC-3′) and pMMB_MCS_R (5′-TTTAAAAGACGTCAGGTGG-3′). The two products were then Gibson assembled to produce vector pUC_MM.
Mutagenic PCR of ampC was performed using primer sets for wild-type or mutant ampC amplification (F_Gibson_ampC_GM_pMMB190 and R_Gibson_ampC_GM_pMMB190) with the Diversify PCR Random Mutagenesis kit (Clontech) under conditions to target an average of 4.6 point-mutations per kb (3.9 mutations per gene copy). PCR products were digested with BamHI and EcoRI and then ligated overnight into appropriately digested and calf-intestinal alkaline phosphatase (CIP; NEB)-dephosphorylated pUC_MM. Ligations were purified and transformed into electrocompetent E. coli 10-β. After a 1-h recovery in SOC medium (NEB), 5 h of outgrowth was performed in 100 ml LB containing 10 μg/ml gentamicin, with libraries plated before and after growth to allow estimation of effective library size. Selections were performed using one million transformants added to 4 ml LB-aztreonam medium containing 2-fold serial dilutions of aztreonam. After 24 to 36 h of aerobic incubation, cells from the highest concentration of antibiotic with visible growth were harvested. Cryostocks were prepared from a fraction of the culture, while plasmid was extracted from the remainder and used as the template for the subsequent round of mutagenic PCR. This process was continued until resistance levels plateaued over three consecutive rounds. Three replicates of artificial selection were performed independently, in parallel.
Sanger sequencing of cloned ampC alleles was performed using primers F_Gibson_ampC_GM_pMMB190, R_Gibson_ampC_GM_pMMB190, ampC_SeqNested_F (5′-AGAAGGACCAGGCACAGATC-3′), and ampC_SeqNested_R (5′-GAACACTTGCTGCTCCATGA-3′).
Data availability.
All sequence data from this project are available from the NCBI Sequence Read Archive (SRA; https://www.ncbi.nlm.nih.gov/sra) under accession number PRJNA534096.
Supplementary Material
ACKNOWLEDGMENTS
This project was supported by Gilead Sciences, Inc.
We thank D. R. Long for helpful conversations.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00866-19.
REFERENCES
- 1.Filkins LM, O’Toole GA. 2015. Cystic fibrosis lung infections: polymicrobial, complex, and hard to treat. PLoS Pathog 11:e1005258. doi: 10.1371/journal.ppat.1005258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lyczak JB, Cannon CL, Pier GB. 2002. Lung infections associated with cystic fibrosis. Clin Microbiol Rev 15:194–222. doi: 10.1128/cmr.15.2.194-222.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Elborn JS, Henig NR. 2010. Optimal airway antimicrobial therapy for cystic fibrosis: the role of inhaled aztreonam lysine. Expert Opin Pharmacother 11:1373–1385. doi: 10.1517/14656566.2010.482102. [DOI] [PubMed] [Google Scholar]
- 4.Zeitler K, Salvas B, Stevens V, Brown J. 2012. Aztreonam lysine for inhalation: new formulation of an old antibiotic. Am J Health Syst Pharm 69:107–115. doi: 10.2146/ajhp100624. [DOI] [PubMed] [Google Scholar]
- 5.O'Sullivan BP, Yasothan U, Kirkpatrick P. 2010. Inhaled aztreonam. Nat Rev Drug Discov 9:357–358. doi: 10.1038/nrd3170. [DOI] [PubMed] [Google Scholar]
- 6.Cystic Fibrosis Foundation. 2016. Cystic Fibrosis Foundation patient registry 2016 annual data report 94. Cystic Fibrosis Foundation, Bethesda, MD. [Google Scholar]
- 7.Rojo-Molinero E, Macià MD, Rubio R, Moyà B, Cabot G, López-Causapé C, Pérez JL, Cantón R, Oliver A. 2016. Sequential treatment of biofilms with aztreonam and tobramycin is a novel strategy for combating Pseudomonas aeruginosa chronic respiratory infections. Antimicrob Agents Chemother 60:2912–2922. doi: 10.1128/AAC.00196-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flume PA, Clancy JP, Retsch-Bogart GZ, Tullis DE, Bresnik M, Derchak PA, Lewis SA, Ramsey BW. 2016. Continuous alternating inhaled antibiotics for chronic pseudomonal infection in cystic fibrosis. J Cyst Fibros 15:809–815. doi: 10.1016/j.jcf.2016.05.001. [DOI] [PubMed] [Google Scholar]
- 9.Oermann CM, McCoy KS, Retsch-Bogart GZ, Gibson RL, McKevitt M, Montgomery AB. 2011. Pseudomonas aeruginosa antibiotic susceptibility during long-term use of aztreonam for inhalation solution (AZLI). J Antimicrob Chemother 66:2398–2404. doi: 10.1093/jac/dkr303. [DOI] [PubMed] [Google Scholar]
- 10.Wainwright CE, Quittner AL, Geller DE, Nakamura C, Wooldridge JL, Gibson RL, Lewis S, Montgomery AB. 2011. Aztreonam for inhalation solution (AZLI) in patients with cystic fibrosis, mild lung impairment, and P. aeruginosa. J Cyst Fibros 10:234–242. doi: 10.1016/j.jcf.2011.02.007. [DOI] [PubMed] [Google Scholar]
- 11.McCoy KS, Quittner AL, Oermann CM, Gibson RL, Retsch-Bogart GZ, Montgomery AB. 2008. Inhaled aztreonam lysine for chronic airway Pseudomonas aeruginosa in cystic fibrosis. Am J Respir Crit Care Med 178:921–928. doi: 10.1164/rccm.200712-1804OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jorth P, McLean K, Ratjen A, Secor PR, Bautista GE, Ravishankar S, Rezayat A, Garudathri J, Harrison JJ, Harwood RA, Penewit K, Waalkes A, Singh PK, Salipante SJ. 2017. Evolved aztreonam resistance is multifactorial and can produce hypervirulence in Pseudomonas aeruginosa. mBio 8:e00517-17. doi: 10.1128/mBio.00517-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quale J, Bratu S, Gupta J, Landman D. 2006. Interplay of efflux system, ampC, and oprD expression in carbapenem resistance of Pseudomonas aeruginosa clinical isolates. Antimicrob Agents Chemother 50:1633–1641. doi: 10.1128/AAC.50.5.1633-1641.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Masuda N, Sakagawa E, Ohya S, Gotoh N, Tsujimoto H, Nishino T. 2000. Substrate specificities of MexAB-OprM, MexCD-OprJ, and MexXY-OprM efflux pumps in Pseudomonas aeruginosa. Antimicrob Agents Chemother 44:3322–3327. doi: 10.1128/aac.44.12.3322-3327.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Llanes C, Hocquet D, Vogne C, Benali-Baitich D, Neuwirth C, Plésiat P. 2004. Clinical strains of Pseudomonas aeruginosa overproducing MexAB-OprM and MexXY efflux pumps simultaneously. Antimicrob Agents Chemother 48:1797–1802. doi: 10.1128/aac.48.5.1797-1802.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liao X, Hancock RE. 1997. Susceptibility to beta-lactam antibiotics of Pseudomonas aeruginosa overproducing penicillin-binding protein 3. Antimicrob Agents Chemother 41:1158–1161. doi: 10.1128/AAC.41.5.1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lister P, Wolter D, Hanson N. 2009. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Microbiol Rev 22:582–610. doi: 10.1128/CMR.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berrazeg M, Jeannot K, Enguéné VYN, Broutin I, Loeffert S, Fournier D, Plésiat P. 2015. Mutations in β-lactamase AmpC increase resistance of Pseudomonas aeruginosa isolates to antipseudomonal cephalosporins. Antimicrob Agents Chemother 59:6248–6255. doi: 10.1128/AAC.00825-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chérif T, Saidani M, Decré D, Boutiba-Ben Boubaker I, Arlet G. 2016. Cooccurrence of multiple AmpC β-lactamases in Escherichia coli, Klebsiella pneumoniae, and Proteus mirabilis in Tunisia. Antimicrob Agents Chemother 60:44–51. doi: 10.1128/AAC.00828-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gupta G, Tak V, Mathur P. 2014. Detection of AmpC β lactamases in Gram-negative bacteria. J Lab Physicians 6:1–6. doi: 10.4103/0974-2727.129082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keating C, Zuckerman JB, Singh P, Gurtovaya O, Bresnik M, Marshall B, Saiman L. 2018. Are changes in aztreonam susceptibility associated with inhaled aztreonam exposure? Results of a prospective 5-year study of Pseudomonas aeruginosa isolates from CF patients in the United States, abstr 446. Pediatr Pulmonol 53:S317. [Google Scholar]
- 22.López-Causapé C, Sommer LM, Cabot G, Rubio R, Ocampo-Sosa AA, Johansen HK, Figuerola J, Cantón R, Kidd TJ, Molin S, Oliver A. 2017. Evolution of the Pseudomonas aeruginosa mutational resistome in an international cystic fibrosis clone. Sci Rep 7:5555. doi: 10.1038/s41598-017-05621-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clark ST, Sinha U, Zhang Y, Wang PW, Donaldson SL, Coburn B, Waters VJ, Yau YCW, Tullis DE, Guttman DS, Hwang DM. 2019. Penicillin binding protein 3 is a common adaptive target among Pseudomonas aeruginosa isolates from adult cystic fibrosis patients treated with β-lactams. Int J Antimicrob Agents 53:620–628. doi: 10.1016/j.ijantimicag.2019.01.009. [DOI] [PubMed] [Google Scholar]
- 24.Han S, Zaniewski RP, Marr ES, Lacey BM, Tomaras AP, Evdokimov A, Miller JR, Shanmugasundaram V. 2010. Structural basis for effectiveness of siderophore-conjugated monocarbams against clinically relevant strains of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 107:22002–22007. doi: 10.1073/pnas.1013092107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cabot G, Bruchmann S, Mulet X, Zamorano L, Moyà B, Juan C, Haussler S, Oliver A. 2014. Pseudomonas aeruginosa ceftolozane-tazobactam resistance development requires multiple mutations leading to overexpression and structural modification of AmpC. Antimicrob Agents Chemother 58:3091–3099. doi: 10.1128/AAC.02462-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kos VN, Déraspe M, McLaughlin RE, Whiteaker JD, Roy PH, Alm RA, Corbeil J, Gardner H. 2015. The resistome of Pseudomonas aeruginosa in relationship to phenotypic susceptibility. Antimicrob Agents Chemother 59:427–436. doi: 10.1128/AAC.03954-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Held K, Ramage E, Jacobs M, Gallagher L, Manoil C. 2012. Sequence-verified two-allele transposon mutant library for Pseudomonas aeruginosa PAO1. J Bacteriol 194:6387–6389. doi: 10.1128/JB.01479-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gallagher LA. 2019. Methods for Tn-Seq analysis in Acinetobacter baumannii, p. 115–134. In Biswas I, Rather PN (ed), Acinetobacter baumannii: methods and protocols. Springer, New York, NY. [DOI] [PubMed] [Google Scholar]
- 29.Queenan AM, Shang W, Flamm R, Bush K. 2010. Hydrolysis and inhibition profiles of β-lactamases from molecular classes A to D with doripenem, imipenem, and meropenem. Antimicrob Agents Chemother 54:565–569. doi: 10.1128/AAC.01004-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.La Rosa R, Johansen HK, Molin S. 2018. Convergent metabolic specialization through distinct evolutionary paths in Pseudomonas aeruginosa. mBio 9:e00269-18. doi: 10.1128/mBio.00269-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Winstanley C, O’Brien S, Brockhurst MA. 2016. Pseudomonas aeruginosa evolutionary adaptation and diversification in cystic fibrosis chronic lung infections. Trends Microbiol 24:327–337. doi: 10.1016/j.tim.2016.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Diaz Caballero J, Clark ST, Coburn B, Zhang Y, Wang PW, Donaldson SL, Tullis DE, Yau YCW, Waters VJ, Hwang DM, Guttman DS. 2015. Selective sweeps and parallel pathoadaptation drive Pseudomonas aeruginosa evolution in the cystic fibrosis lung. mBio 6:e00981-15. doi: 10.1128/mBio.00981-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Evans TJ. 2015. Small colony variants of Pseudomonas aeruginosa in chronic bacterial infection of the lung in cystic fibrosis. Future Microbiol 10:231–239. doi: 10.2217/fmb.14.107. [DOI] [PubMed] [Google Scholar]
- 34.Alm RA, Johnstone MR, Lahiri SD. 2015. Characterization of Escherichia coli NDM isolates with decreased susceptibility to aztreonam/avibactam: role of a novel insertion in PBP3. J Antimicrob Chemother 70:1420–1428. doi: 10.1093/jac/dku568. [DOI] [PubMed] [Google Scholar]
- 35.Chen W, Zhang Y-M, Davies C. 2016. Penicillin-binding protein 3 is essential for growth of Pseudomonas aeruginosa. Antimicrob Agents Chemother 61:e01651-16. doi: 10.1128/AAC.01651-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jacobs MA, Alwood A, Thaipisuttikul I, Spencer D, Haugen E, Ernst S, Will O, Kaul R, Raymond C, Levy R, Chun-Rong L, Guenthner D, Bovee D, Olson MV, Manoil C. 2003. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 100:14339–14344. doi: 10.1073/pnas.2036282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sainsbury S, Bird L, Rao V, Shepherd SM, Stuart DI, Hunter WN, Owens RJ, Ren J. 2011. Crystal structures of penicillin-binding protein 3 from Pseudomonas aeruginosa: comparison of native and antibiotic-bound forms. J Mol Biol 405:173–184. doi: 10.1016/j.jmb.2010.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cabot G, Zamorano L, Moyà B, Juan C, Navas A, Blázquez J, Oliver A. 2016. Evolution of Pseudomonas aeruginosa antimicrobial resistance and fitness under low and high mutation rates. Antimicrob Agents Chemother 60:1767–1778. doi: 10.1128/AAC.02676-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cabot G, Florit-Mendoza L, Sánchez-Diener I, Zamorano L, Oliver A. 2018. Deciphering β-lactamase-independent β-lactam resistance evolution trajectories in Pseudomonas aeruginosa. J Antimicrob Chemother 73:3322–3331. doi: 10.1093/jac/dky364. [DOI] [PubMed] [Google Scholar]
- 40.Strateva T, Yordanov D. 2009. Pseudomonas aeruginosa - a phenomenon of bacterial resistance. J Med Microbiol 58:1133–1148. doi: 10.1099/jmm.0.009142-0. [DOI] [PubMed] [Google Scholar]
- 41.Huang H, Hancock RE. 1993. Genetic definition of the substrate selectivity of outer membrane porin protein OprD of Pseudomonas aeruginosa. J Bacteriol 175:7793–7800. doi: 10.1128/jb.175.24.7793-7800.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Livermore DM. 2001. Of Pseudomonas, porins, pumps and carbapenems. J Antimicrob Chemother 47:247–250. doi: 10.1093/jac/47.3.247. [DOI] [PubMed] [Google Scholar]
- 43.Tomás M, Doumith M, Warner M, Turton JF, Beceiro A, Bou G, Livermore DM, Woodford N. 2010. Efflux pumps, OprD porin, AmpC β-lactamase, and multiresistance in Pseudomonas aeruginosa isolates from cystic fibrosis patients. Antimicrob Agents Chemother 54:2219–2224. doi: 10.1128/AAC.00816-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hancock REW, Brinkman F. 2002. Function of pseudomonas porins in uptake and efflux. Annu Rev Microbiol 56:17–38. doi: 10.1146/annurev.micro.56.012302.160310. [DOI] [PubMed] [Google Scholar]
- 45.Tamber S, Ochs MM, Hancock R. 2006. Role of the novel OprD family of porins in nutrient uptake in Pseudomonas aeruginosa. J Bacteriol 188:45–54. doi: 10.1128/JB.188.1.45-54.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tiburzi F, Imperi F, Visca P. 2008. Intracellular levels and activity of PvdS, the major iron starvation sigma factor of Pseudomonas aeruginosa. Mol Microbiol 67:213–227. doi: 10.1111/j.1365-2958.2007.06051.x. [DOI] [PubMed] [Google Scholar]
- 47.Imperi F, Tiburzi F, Fimia GM, Visca P. 2010. Transcriptional control of the pvdS iron starvation sigma factor gene by the master regulator of sulfur metabolism CysB in Pseudomonas aeruginosa. Environ Microbiol 12:1630–1642. doi: 10.1111/j.1462-2920.2010.02210.x. [DOI] [PubMed] [Google Scholar]
- 48.Leoni L, Orsi N, de Lorenzo V, Visca P. 2000. Functional analysis of PvdS, an iron starvation sigma factor of Pseudomonas aeruginosa. J Bacteriol 182:1481–1491. doi: 10.1128/jb.182.6.1481-1491.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Llamas MA, Imperi F, Visca P, Lamont IL. 2014. Cell-surface signaling in Pseudomonas: stress responses, iron transport, and pathogenicity. FEMS Microbiol Rev 38:569–597. doi: 10.1111/1574-6976.12078. [DOI] [PubMed] [Google Scholar]
- 50.Greipel L, Fischer S, Klockgether J, Dorda M, Mielke S, Wiehlmann L, Cramer N, Tümmler B. 2016. Molecular epidemiology of mutations in antimicrobial resistance loci of Pseudomonas aeruginosa isolates from airways of cystic fibrosis patients. Antimicrob Agents Chemother 60:6726–6734. doi: 10.1128/AAC.00724-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lorè NI, Cigana C, De Fino I, Riva C, Juhas M, Schwager S, Eberl L, Bragonzi A. 2012. Cystic fibrosis-niche adaptation of Pseudomonas aeruginosa reduces virulence in multiple infection hosts. PLoS One 7:e35648. doi: 10.1371/journal.pone.0035648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vettoretti L, Plesiat P, Muller C, El Garch F, Phan G, Attree I, Ducruix A, Llanes C. 2009. Efflux unbalance in Pseudomonas aeruginosa isolates from cystic fibrosis patients. Antimicrob Agents Chemother 53:1987–1997. doi: 10.1128/AAC.01024-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Muller C, Plésiat P, Jeannot K. 2011. A two-component regulatory system interconnects resistance to polymyxins, aminoglycosides, fluoroquinolones, and β-lactams in Pseudomonas aeruginosa. Antimicrob Agents Chemother 55:1211–1221. doi: 10.1128/AAC.01252-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fernández L, Gooderham WJ, Bains M, McPhee JB, Wiegand I, Hancock R. 2010. Adaptive resistance to the “last hope” antibiotics polymyxin B and colistin in Pseudomonas aeruginosa is mediated by the novel two-component regulatory system ParR-ParS. Antimicrob Agents Chemother 54:3372–3382. doi: 10.1128/AAC.00242-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Botsford JL, Harman JG. 1992. Cyclic AMP in prokaryotes. Microbiol Rev 56:100–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kulasakara H, Lee V, Brencic A, Liberati N, Urbach J, Miyata S, Lee DG, Neely AN, Hyodo M, Hayakawa Y, Ausubel FM, Lory S. 2006. Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3′-5′)-cyclic-GMP in virulence. Proc Natl Acad Sci U S A 103:2839–2844. doi: 10.1073/pnas.0511090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Connelly JC, Kirkham LA, Leach DR. 1998. The SbcCD nuclease of Escherichia coli is a structural maintenance of chromosomes (SMC) family protein that cleaves hairpin DNA. Proc Natl Acad Sci U S A 95:7969–7974. doi: 10.1073/pnas.95.14.7969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sakurai Y, Yoshida Y, Saitoh K, Nemoto M, Yamaguchi A, Sawai T. 1990. Characteristics of aztreonam as a substrate, inhibitor and inducer for β-lactamases. J Antibiot (Tokyo) 43:403–410. doi: 10.7164/antibiotics.43.403. [DOI] [PubMed] [Google Scholar]
- 59.Pereira SG, Cardoso O. 2014. Mobile genetic elements of Pseudomonas aeruginosa isolates from hydrotherapy facility and respiratory infections. Clin Microbiol Infect 20:O203–O206. doi: 10.1111/1469-0691.12359. [DOI] [PubMed] [Google Scholar]
- 60.Zamorano L, Moyà B, Juan C, Mulet X, Blázquez J, Oliver A. 2014. The Pseudomonas aeruginosa CreBC two-component system plays a major role in the response to β-lactams, fitness, biofilm growth, and global regulation. Antimicrob Agents Chemother 58:5084–5095. doi: 10.1128/AAC.02556-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim JY, Jung HI, An YJ, Lee JH, Kim SJ, Jeong SH, Lee KJ, Suh P-G, Lee H-S, Lee SH, Cha S-S. 2006. Structural basis for the extended substrate spectrum of CMY-10, a plasmid-encoded class C beta-lactamase. Mol Microbiol 60:907–916. doi: 10.1111/j.1365-2958.2006.05146.x. [DOI] [PubMed] [Google Scholar]
- 62.Lahiri SD, Johnstone MR, Ross PL, McLaughlin RE, Olivier NB, Alm RA. 2014. Avibactam and class C β-lactamases: mechanism of inhibition, conservation of the binding pocket, and implications for resistance. Antimicrob Agents Chemother 58:5704–5713. doi: 10.1128/AAC.03057-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Raimondi A, Sisto F, Nikaido H. 2001. Mutation in Serratia marcescens AmpC β-lactamase producing high-level resistance to ceftazidime and cefpirome. Antimicrob Agents Chemother 45:2331–2339. doi: 10.1128/AAC.45.8.2331-2339.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hmelo LR, Borlee BR, Almblad H, Love ME, Randall TE, Tseng BS, Lin C, Irie Y, Storek KM, Yang JJ, Siehnel RJ, Howell PL, Singh PK, Tolker-Nielsen T, Parsek MR, Schweizer HP, Harrison JJ. 2015. Precision-engineering the Pseudomonas aeruginosa genome with two-step allelic exchange. Nat Protoc 10:1820–1841. doi: 10.1038/nprot.2015.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Clinical and Laboratory Standards Institute. 2018. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 11th ed. M07Ed11. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 66.Salipante SJ, SenGupta DJ, Cummings LA, Land TA, Hoogestraat DR, Cookson BT. 2015. Application of whole-genome sequencing for bacterial strain typing in molecular epidemiology. J Clin Microbiol 53:1072–1079. doi: 10.1128/JCM.03385-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Roach DJ, Burton JN, Lee C, Stackhouse B, Butler-Wu SM, Cookson BT, Shendure J, Salipante SJ. 2015. A year of infection in the intensive care unit: prospective whole genome sequencing of bacterial clinical isolates reveals cryptic transmissions and novel microbiota. PLoS Genet 11:e1005413. doi: 10.1371/journal.pgen.1005413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJM, Birol I. 2009. ABySS: a parallel assembler for short read sequence data. Genome Res 19:1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 70.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fu L, Niu B, Zhu Z, Wu S, Li W. 2012. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Buchfink B, Xie C, Huson DH. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods 12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 75.Sievers F, Higgins DG. 2014. Clustal Omega, accurate alignment of very large numbers of sequences, p 105–116. In Russell DJ. (ed), Multiple sequence alignment methods. Humana Press, Totowa, NJ. [DOI] [PubMed] [Google Scholar]
- 76.Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweize HP. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86. doi: 10.1016/s0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- 77.Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequence data from this project are available from the NCBI Sequence Read Archive (SRA; https://www.ncbi.nlm.nih.gov/sra) under accession number PRJNA534096.