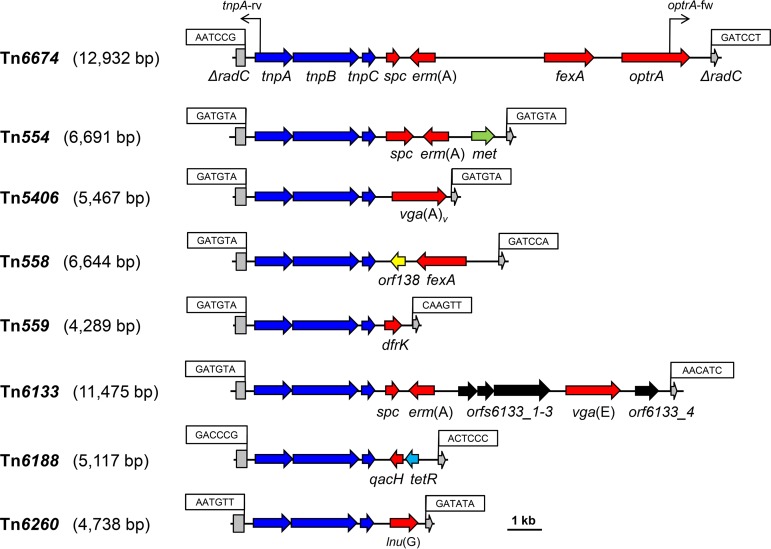
The novel 12,932-bp nonconjugative multiresistance transposon Tn6674 was identified in the chromosomal DNA of a porcine Enterococcus faecalis strain. Tn6674 belongs to the Tn554 family of transposons. It shares the same arrangement of the transposase genes tnpA, tnpB, and tnpC with Tn554. However, in addition to the Tn554-associated resistance genes spc and erm(A), Tn6674 harbored the resistance genes fexA and optrA.
KEYWORDS: Enterococcus faecalis, optrA, oxazolidinones, phenicols, transposon
ABSTRACT
The novel 12,932-bp nonconjugative multiresistance transposon Tn6674 was identified in the chromosomal DNA of a porcine Enterococcus faecalis strain. Tn6674 belongs to the Tn554 family of transposons. It shares the same arrangement of the transposase genes tnpA, tnpB, and tnpC with Tn554. However, in addition to the Tn554-associated resistance genes spc and erm(A), Tn6674 harbored the resistance genes fexA and optrA. Circular forms of Tn6674 were detected and suggest the functional activity of this transposon.
INTRODUCTION
Oxazolidinones, such as linezolid and tedizolid, are often used for the treatment of infections caused by methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant enterococci (VRE) in humans (1). Phenicols, such as florfenicol, are approved for the control of respiratory tract infections in food-producing animals (2). The gene optrA, which confers combined resistance to oxazolidinones and phenicols, has emerged during recent years (3) and poses a serious therapeutic challenge to both human and veterinary medicine. Although the optrA gene has been originally described to encode an ABC transporter, a recent study confirmed that OptrA does not mediate the efflux of oxazolidinones and phenicols (4). Meanwhile, OptrA is considered to be an ABC-F protein which acts by protecting the bacterial ribosome (5).
Since the first description of the optrA gene in Enterococcus faecalis and Enterococcus faecium (3), this gene has been identified on various plasmids of different sizes or in the chromosomal DNA of enterococci from humans and food-producing animals (1). In some cases, the optrA gene was also present in combination with cfr, cfr(B), or poxtA on the same strain or plasmid (6–8). In addition, it was also found in Staphylococcus sciuri, together with cfr and other resistance genes on the same plasmid, and in Streptococcus suis and Streptococcus gallolyticus. In S. suis, it was shown to be part of integrative and conjugative elements (ICEs) (9–11). In the present study, we identified a novel chromosome-borne transposon of the Tn554 family, designated Tn6674, which carried the optrA gene together with the resistance genes spc, erm(A), and fexA in an E. faecalis strain.
During a routine survey on antimicrobial resistance in bacteria from pigs, the florfenicol- and linezolid-resistant E. faecalis strain E1731 was isolated from a fecal sample of a pig in Henan Province, China, in 2018. Antimicrobial susceptibility testing (AST) was performed by broth microdilution according to the recommendations given in document M100 (28th ed) of the Clinical and Laboratory Standards Institute (CLSI) (12). S. aureus ATCC 29213 served as the quality control strain in AST. The AST results are shown in Table 1. Multilocus sequence typing (MLST) revealed that it belonged to sequence type 691 (ST691). The presence of the optrA gene was detected by PCR using the primers optrA-fw (5′-GCACCAGACCAATACGATACAA-3′) and optrA-rev (5′-TCCTTCTTAACCTTCTCCTTCTCA-3′), with an annealing temperature of 59°C and amplicon size of 794 bp.
TABLE 1.
Resistance phenotype and genotype of E. faecalis E1731
| Antimicrobial agent | MIC (mg/liter) | Associated resistance gene(s) or mutation(s) |
|---|---|---|
| Florfenicol | 128 | optrA, fexA |
| Linezolid | 8 | optrA |
| Erythromycin | 512 | erm(A), erm(B) |
| Gentamicin | >512 | aac(A)-aph(D) |
| Lincomycin | 512 | erm(B) |
| Ciprofloxacin | 64 | GyrA S83R, ParC S80I |
| Tetracycline | 64 | tet(M), tet(L) |
| Spectinomycin | 64 | spc |
| Vancomycin | 2 | None |
To investigate the transferability of the optrA gene, conjugation and transformation experiments were performed as previously described using E. faecalis E1731 as the donor and E. faecalis JH2-2 (rifampin resistant [Rifr]) as the recipient (3, 8). For the screening of the transconjugants and transformants, brain heart infusion (BHI) agar was supplemented with 10 mg/liter florfenicol and 50 mg/liter rifampin. However, the conjugation experiments repeatedly failed. The plasmids were then extracted from the donor E. faecalis E1731, and transfer into E. faecalis JH2-2 by electrotransformation was attempted. The transformation experiments also repeatedly failed.
To gain information about the genetic environment of the optrA gene, the whole-genomic DNA of E. faecalis E1731 was sequenced by the PacBio RS II and Illumina MiSeq platforms (Shanghai Personal Biotechnology Co., Ltd., China). The sequences from the Illumina MiSeq platform were assembled using Newbler version 2.8 (454 Life Sciences, Branford, CT, USA), and PacBio sequencing reads were de novo assembled. The prediction of open reading frames (ORFs) and their annotations were performed using Glimmer 3.0. The analysis of regions flanking the optrA gene revealed the presence of a Tn554 family transposon-like element of 12,932 bp, designated Tn6674 according to the nomenclature of transposons (https://transposon.lstmed.ac.uk/). Tn6674 consisted of seven ORFs of more than 150 amino acids (aa). The first three ORFs were indistinguishable from the genes tnpA, tnpB, and tnpC, whose products are involved in the transposition of Staphylococcus aureus transposon Tn554 (13). However, the remaining four ORFs represented the resistance genes spc (resistance to spectinomycin), erm(A) (resistance to macrolides, lincosamides, and streptogramin B antibiotics), fexA (resistance to phenicols), and optrA and replaced its counterpart in Tn554, consisting of only spc and erm(A) (Fig. 1). Sequence analysis also identified several antimicrobial resistance genes or mutations related to the respective phenotypes observed in E. faecalis E1731 (Table 1).
FIG 1.
Structural comparison of E. faecalis transposon Tn6674 with other related Tn554 family transposons. Accession numbers are as follows: Tn554, X03216; Tn5406, AF186237; Tn558, AJ715531; Tn559, FN677369; Tn6133, FR772051; Tn6188, HF565366; Tn6260, KX470419; and Tn6674, MK737778. A distance scale in kilobases is given. The transposase genes (tnpA, tnpB, and tnpC), antimicrobial and disinfectant resistance genes, and genes that code for other functions are shown in blue, red, and black, respectively. The 6-bp nucleotide sequences at the transposon junctions are shown in boxes. The disrupted radC genes are indicated by asterisks. The positions of primers circ-fw/circ-rev used to detect the presence of the circular intermediates are shown by black arrows.
An NCBI BLASTN search with the Tn6674 sequence as a query sequence showed that it displayed 99% nucleotide sequence identity (query cover, 100%) with sequences deposited in GenBank (E. faecalis A101, accession no. MH018572.1; E. faecalis TZ2, accession no. MH225421.1) (14), 99% nucleotide sequence identity (query cover, 93%) with that deposited in GenBank (E. faecalis 743142, accession no. MF443377.1) (15), and 99% nucleotide sequence identity (query cover, 100%) with those of whole-genome shotgun contigs (E. faecalis EF294, accession no. QDDM01000007.1; E. faecalis 33710, accession no. QNHF01000012.1) (16, 17). In addition, a similar structure was also described in an optrA-carrying ST16 E. faecalis strains (18).
The analysis of the regions flanking Tn6674 revealed that Tn6674 inserted into the gene radC gene, which encodes a DNA repair protein (Fig. 1). The radC gene had been described to be a common integration site for transposons of the Tn554 family, such as Tn554, Tn5406, Tn558, Tn559, Tn6133, Tn6188, and Tn6260 in various Gram-positive bacteria (19–25). In E. faecalis E1731, Tn6674 displayed the hexanucleotide sequence 5′-AATCCG-3′ at the left-end junction and the sequence 5′-GATCCT-3′ at the right-end junction, which marked the boundaries of the integrated segment. This is similar to that in the Staphylococcus lentus transposon Tn558 (21). Transposons of the Tn554 family, including Tn6674, do not contain inverted repeats at their termini and also do not generate a duplication of the target sequence at the integration site. Instead, studies on serial transposition of Tn554 into primary and secondary target sites revealed that the sequences at the junctions of Tn554 varied with respect to the target sites, as follows: with each new transposition event, the sequence originally present in the target site is found at the left terminus of Tn554, whereas the former left-end junction is now found at the right terminus, and the former right-end junction is lost (21, 26).
Tn6674 has an overall structure similar to that of other members of the Tn554 family of transposons. However, all so-far-known Tn554-like transposons differ distinctly in their resistance gene regions. These are composed of spc and erm(A) in Tn554 from S. aureus (13), a vga(A) variant in Tn5406 from S. aureus (20), fexA in Tn558 from S. lentus (21), dfrK in Tn559 from S. aureus (22), spc, erm(A), and vga(E) in Tn6133 from S. aureus (23), lnu(G) in Tn6260 from E. faecalis (25), and spc, erm(A), fexA, and optrA in Tn6674 from E. faecalis.
Previous studies have shown that the active transposons, such as Tn5406, Tn558, and Tn559, can excise from their host DNA and form circular intermediates which precede their integration into a new target site (20–22). Thus, an inverse PCR assay was set up to detect whether these circular intermediates were present in E. faecalis E1731. For this, the pair of primers circ-fw (5′-TAGATGAACCGACAAACC-3′) and circ-rev (5′-TCAACCAACCTACGAAGT-3′), with an annealing temperature of 47°C and amplicon size of 1,445 bp, was used. Sequence analysis of this amplicon consisted of 380 bp, which included part of tnpA and its upstream region at the left end of Tn6674, whereas the remaining 1,065 bp of the amplicon represented the right end of Tn6674. The presence of circular Tn6674 forms suggested that this transposon was active in E. faecalis E1731.
On the one hand, the location of optrA as part of transposon Tn6674 suggests a novel mechanism for the dissemination of optrA in enterococci, which is different from IS1216-mediated recombination that was previously described in other studies (27, 28). On the other hand, its location as an integral part of a functionally active transposon will likely accelerate the dissemination of the optrA gene. Since the optrA gene confers combined resistance to oxazolidinones used in human medicine (1) and phenicols used in food-producing animals (3, 8, 9), the prudent use of both classes of antimicrobial agents in their respective fields is urgently needed.
Data availability.
The complete sequence of Tn6674 determined in this study has been deposited in GenBank under accession number MK737778.
ACKNOWLEDGMENTS
We thank Don B. Clewell (University of Michigan) for providing E. faecalis reference strain JH2-2.
This work was supported by grants from the National Natural Science Foundation of China (grant U1604103), the Program for Innovative Research Team (in Science and Technology) in University of Henan Province (grant 18IRTSTHN020), and the German Federal Ministry of Education and Research (BMBF) under project number 01KI1727D as part of the Research Network Zoonotic Infectious Diseases.
REFERENCES
- 1.Sadowy E. 2018. Linezolid resistance genes and genetic elements enhancing their dissemination in enterococci and streptococci. Plasmid 99:89–98. doi: 10.1016/j.plasmid.2018.09.011. [DOI] [PubMed] [Google Scholar]
- 2.Priebe S, Schwarz S. 2003. In vitro activities of florfenicol against bovine and porcine respiratory tract pathogens. Antimicrob Agents Chemother 47:2703–2705. doi: 10.1128/AAC.47.8.2703-2705.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Y, Lv Y, Cai J, Schwarz S, Cui L, Hu Z, Zhang R, Li J, Zhao Q, He T, Wang D, Wang Z, Shen Y, Li Y, Feßler AT, Wu C, Yu H, Deng X, Xia X, Shen J. 2015. A novel gene, optrA, that confers transferable resistance to oxazolidinones and phenicols and its presence in Enterococcus faecalis and Enterococcus faecium of human and animal origin. J Antimicrob Chemother 70:2182–2190. doi: 10.1093/jac/dkv116. [DOI] [PubMed] [Google Scholar]
- 4.Wang Y, Li X, Wang Y, Schwarz S, Shen J, Xia X. 2018. Intracellular accumulation of linezolid and florfenicol in optrA-producing Enterococcus faecalis and Staphylococcus aureus. Molecules 23:3195. doi: 10.3390/molecules23123195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharkey LK, Edwards TA, O'Neill AJ. 2016. ABC-F proteins mediate antibiotic resistance through ribosomal protection. mBio 7:e01975-15. doi: 10.1128/mBio.01975-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lazaris A, Coleman DC, Kearns AM, Pichon B, Kinnevey PM, Earls MR, Boyle B, O’Connell B, Brennan GI, Shore AC. 2017. Novel multiresistance cfr plasmids in linezolid-resistant methicillin-resistant Staphylococcus epidermidis and vancomycin-resistant Enterococcus faecium (VRE) from a hospital outbreak: co-location of cfr and optrA in VRE. J Antimicrob Chemother 72:3252–3257. doi: 10.1093/jac/dkx292. [DOI] [PubMed] [Google Scholar]
- 7.Kuroda M, Sekizuka T, Matsui H, Suzuki K, Seki H, Saito M, Hanaki H. 2018. Complete genome sequence and characterization of linezolid-resistant Enterococcus faecalis clinical isolate KUB3006 carrying a cfr(B)-transposon on its chromosome and optrA-plasmid. Front Microbiol 9:2576. doi: 10.3389/fmicb.2018.02576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hao W, Shan X, Li D, Schwarz S, Zhang SM, Li XS, Du XD. 2019. Analysis of a poxtA- and optrA-co-carrying conjugative multiresistance plasmid from Enterococcus faecalis. J Antimicrob Chemother 74:1771–1775. doi: 10.1093/jac/dkz109. [DOI] [PubMed] [Google Scholar]
- 9.Li D, Wang Y, Schwarz S, Cai J, Fan R, Li J, Feßler AT, Zhang R, Wu C, Shen J. 2016. Co-location of the oxazolidinone resistance genes optrA and cfr on a multiresistance plasmid from Staphylococcus sciuri. J Antimicrob Chemother 71:1474–1478. doi: 10.1093/jac/dkw040. [DOI] [PubMed] [Google Scholar]
- 10.Huang J, Chen L, Wu Z, Wang L. 2017. Retrospective analysis of genome sequences revealed the wide dissemination of optrA in Gram-positive bacteria. J Antimicrob Chemother 72:614–616. doi: 10.1093/jac/dkw488. [DOI] [PubMed] [Google Scholar]
- 11.Mendes RE, Deshpande L, Streit JM, Sader HS, Castanheira M, Hogan PA, Flamm RK. 2018. ZAAPS programme results for 2016: an activity and spectrum analysis of linezolid using clinical isolates from medical centres in 42 countries. J Antimicrob Chemother 73:1880–1887. doi: 10.1093/jac/dky099. [DOI] [PubMed] [Google Scholar]
- 12.Clinical and Laboratory Standards Institute. 2018. Performance standards for antimicrobial susceptibility testing, 28th ed. CLSI supplement M100 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 13.Murphy E, Huwyler L, de Freire Bastos MDC. 1985. Transposon Tn554: complete nucleotide sequence and isolation of transposition-defective and antibiotic-sensitive mutants. EMBO J 4:3357–3365. doi: 10.1002/j.1460-2075.1985.tb04089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cai J, Schwarz S, Chi D, Wang Z, Zhang R, Wang Y. 2019. Faecal carriage of optrA-positive enterococci in asymptomatic healthy humans in Hangzhou, China. Clin Microbiol Infect 25:630.e1–630.e6. doi: 10.1016/j.cmi.2018.07.025. [DOI] [PubMed] [Google Scholar]
- 15.Deshpande LM, Castanheira M, Flamm RK, Mendes RE. 2018. Evolving oxazolidinone resistance mechanisms in a worldwide collection of enterococcal clinical isolates: results from the SENTRY Antimicrobial Surveillance Program. J Antimicrob Chemother 73:2314–2322. doi: 10.1093/jac/dky188. [DOI] [PubMed] [Google Scholar]
- 16.Zhou W, Gao S, Xu H, Zhang Z, Chen F, Shen H, Zhang C. 2019. Distribution of the optrA gene in Enterococcus isolates at a tertiary care hospital in China. J Glob Antimicrob Resist 17:180–186. doi: 10.1016/j.jgar.2019.01.001. [DOI] [PubMed] [Google Scholar]
- 17.Chen M, Pan H, Lou Y, Wu Z, Zhang J, Huang Y, Yu W, Qiu Y. 2018. Epidemiological characteristics and genetic structure of linezolid-resistant Enterococcus faecalis. Infect Drug Resist 11:2397–2409. doi: 10.2147/IDR.S181339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsilipounidaki K, Gerontopoulos A, Papagiannitsis C, Petinaki E. 2019. First detection of an optrA-positive, linezolid-resistant ST16 Enterococcus faecalis from human in Greece. New Microbes New Infect 29:100515. doi: 10.1016/j.nmni.2019.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murphy E, Reinheimer E, Huwyler L. 1991. Mutational analysis of att554, the target of the site-specific transposon Tn554. Plasmid 26:20–29. doi: 10.1016/0147-619X(91)90033-S. [DOI] [PubMed] [Google Scholar]
- 20.Haroche J, Allignet J, El Solh N. 2002. Tn5406, a new staphylococcal transposon conferring resistance to streptogramin A and related compounds including dalfopristin. Antimicrob Agents Chemother 46:2337–2343. doi: 10.1128/AAC.46.8.2337-2343.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kehrenberg C, Schwarz S. 2005. Florfenicol-chloramphenicol exporter gene fexA is part of the novel transposon Tn558. Antimicrob Agents Chemother 49:813–815. doi: 10.1128/AAC.49.2.813-815.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kadlec K, Schwarz S. 2010. Identification of the novel dfrK-carrying transposon Tn559 in a porcine methicillin-susceptible Staphylococcus aureus ST398 strain. Antimicrob Agents Chemother 54:3475–3477. doi: 10.1128/AAC.00464-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwendener S, Perreten V. 2011. New transposon Tn6133 in methicillin-resistant Staphylococcus aureus ST398 contains vga(E), a novel streptogramin A, pleuromutilin, and lincosamide resistance gene. Antimicrob Agents Chemother 55:4900–4904. doi: 10.1128/AAC.00528-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Müller A, Rychli K, Muhterem-Uyar M, Zaiser A, Stessl B, Guinane CM, Cotter PD, Wagner M, Schmitz-Esser S. 2013. Tn6188-a novel transposon in Listeria monocytogenes responsible for tolerance to benzalkonium chloride. PLoS One 8:e76835. doi: 10.1371/journal.pone.0076835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu XQ, Wang XM, Li H, Shang YH, Pan YS, Wu CM, Wang Y, Du XD, Shen JZ. 2017. Novel lnu(G) gene conferring resistance to lincomycin by nucleotidylation, located on Tn6260 from Enterococcus faecalis E531. J Antimicrob Chemother 72:993–997. doi: 10.1093/jac/dkw549. [DOI] [PubMed] [Google Scholar]
- 26.Murphy E, Lofdahl S. 1984. Transposition of Tn554 does not generate a target duplication. Nature 307:292–294. doi: 10.1038/307292a0. [DOI] [PubMed] [Google Scholar]
- 27.He T, Shen Y, Schwarz S, Cai J, Lv Y, Li J, Feßler AT, Zhang R, Wu C, Shen J, Wang Y. 2016. Genetic environment of the transferable oxazolidinone/phenicol resistance gene optrA in Enterococcus faecalis isolates of human and animal origin. J Antimicrob Chemother 71:1466–1473. doi: 10.1093/jac/dkw016. [DOI] [PubMed] [Google Scholar]
- 28.Morroni G, Brenciani A, Antonelli A, D'Andrea MM, Di Pilato V, Fioriti S, Mingoia M, Vignaroli C, Cirioni O, Biavasco F, Varaldo PE, Rossolini GM, Giovanetti E. 2018. Characterization of a multiresistance plasmid carrying the optrA and cfr resistance genes from an Enterococcus faecium clinical isolate. Front Microbiol 9:2189. doi: 10.3389/fmicb.2018.02189. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The complete sequence of Tn6674 determined in this study has been deposited in GenBank under accession number MK737778.