There is significant interest in the development of mass spectrometry (MS) methods for antimicrobial resistance protein detection, given the ability of these methods to confirm protein expression. In this work, we studied the performance of a liquid chromatography, tandem MS multiple-reaction monitoring (LC-MS/MS MRM) method for the direct detection of the New Delhi metallo-β-lactamase (NDM) carbapenemase in clinical isolates.
KEYWORDS: New Delhi metallo-β-lactamase, mass spectrometry, multiple-reaction monitoring, tryptic peptide
ABSTRACT
There is significant interest in the development of mass spectrometry (MS) methods for antimicrobial resistance protein detection, given the ability of these methods to confirm protein expression. In this work, we studied the performance of a liquid chromatography, tandem MS multiple-reaction monitoring (LC-MS/MS MRM) method for the direct detection of the New Delhi metallo-β-lactamase (NDM) carbapenemase in clinical isolates. Using a genoproteomic approach, we selected three unique peptides (SLGNLGDADTEHYAASAR, AFGAAFPK, and ASMIVMSHSAPDSR) specific to NDM that were efficiently ionized and spectrally well-defined. These three peptides were used to build an assay with turnaround time of 90 min. In a blind set, the assay detected 21/24 blaNDM-containing isolates and 76/76 isolates with negative results, corresponding to a sensitivity value of 87.5% (95% confidence interval [CI], 67.6% to 97.3%) and a specificity value of 100% (95% CI, 95.3% to 100%). One of the missed identifications was determined by protein fractionation to be due to low (∼0.1 fm/μg) NDM protein expression (below the assay limit of detection). Parallel disk diffusion susceptibility testing demonstrated this isolate to be meropenem susceptible, consistent with low NDM expression. Total proteomic analysis of the other two missed identifications did not detect NDM peptides but detected other proteins expressed from the blaNDM-containing plasmids, confirming that the plasmids were not lost. The measurement of relative NDM concentrations over the entire isolate test set demonstrated variability spanning 4 orders of magnitude, further confirming the remarkable range that may be seen in levels of NDM expression. This report highlights the sensitivity of LC-MS/MS to variations in NDM protein expression, with implications for how this technology may be used.
INTRODUCTION
The global spread of carbapenemase-producing organisms is an urgent public health concern (1). Initially reported in 2009 in India, the New Delhi-metallo-β-lactamase (NDM) has since been detected in most countries in the world (1–3). Current methods for the rapid detection of NDM include phenotypic and PCR-based tests (4, 5). Matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) has also been used to detect carbapenemase-containing isolates that include NDM through identification of mass peaks consistent with meropenem hydrolysis degradation products. Limitations of this method are that it is relatively time-consuming (2.5 h) compared with automated cartridge-based PCR approaches and that it is unable to distinguish between carbapenemases (6). A similar method has been developed using liquid chromatography-tandem mass spectrometry (LC-MS/MS) that detects intact and hydrolyzed carbapenem products (7). Mass spectrometry approaches that detect resistance proteins or derivative peptides directly may overcome these limitations and have the advantage of providing direct evidence of NDM protein expression.
We have previously validated a rapid LC-MS/MS method for the direct detection of unique tryptic peptides of the Klebsiella pneumoniae carbapenemase (KPC) in clinical bacterial isolates with an isolation-to-result time of less than 90 min (8). This method combines theoretical analysis of dominant allelic protein sequences and experimental LC-MS/MS to select unique discriminatory peptides with robust spectral characteristics for assay development. We now apply this approach to select unique tryptic peptides of the NDM protein for assay development and examine the sensitivity of the assay to variations in NDM protein expression. NDM core peptides present in all 15 NDM allelic variants were first identified by in silico analysis. Optimal core peptides that were efficiently ionized and robustly detectable (also referred to as “high-responding peptides”) (9) were detected by data-dependent acquisition (DDA) LC-MS/MS (10, 11). The final targeted proteomic approach used a multiple-reaction monitoring assay (MRM) for analysis of three peptides highly specific to NDM. An accuracy assessment of the final method was performed using a blind sample set that included 24 blaNDM-containing isolates and 76 negative controls.
RESULTS
Prediction of theoretical core peptides for NDM.
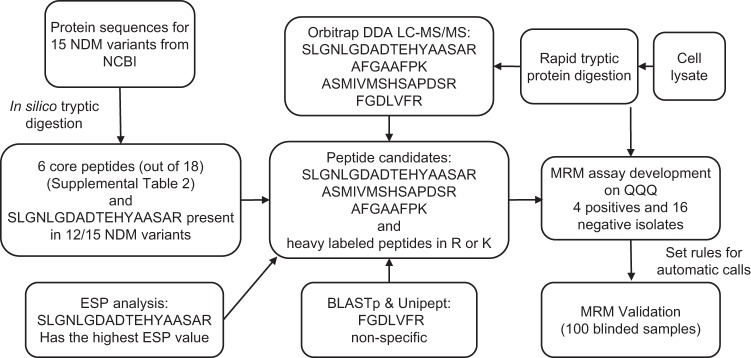
Table 1 lists the protein names, NCBI accession numbers, and amino acid substitutions for 15 NDM allelic variants analyzed in this study. Using peptidomic analysis, 6 core tryptic peptides were found within the 15 NDM variants. Figure 1 summarizes the workflow for peptide selection for NDM detection by MRM LC-MS (see also Table S2 in the supplemental material). FGDLVFR was found to be nonspecific to NDM proteins by protein blast and tryptic peptide analyses (https://unipept.ugent.be/search/single) and therefore was excluded from further study.
TABLE 1.
Protein sequences of NDM variants used for peptidomic analysis
| Protein name | NCBI accession no.a | Amino acid variant(s) |
|---|---|---|
| NDM-1 | AMQ12492.1 | |
| NDM-2 | AEZ35976.1 | P28A |
| NDM-3 | AFK80349.1 | D95N |
| NDM-4 | AKN35302.1 | M153L |
| NDM-5 | AQY75714.1 | V88L; M155L |
| NDM-6 | WP_032495384.1 | A233V |
| NDM-7 | AFQ31613.1 | D130N; M154L |
| NDM-8 | BAM84089.1 | D130G; M154L |
| NDM-9 | WP_032495672.1 | E152K |
| NDM-10 | AGT37351.1 | R32S; G36D; G69S; A74T; G200R |
| NDM-11 | AJE61444.1 | M154V |
| NDM-12 | BAO79439.1 | M154L; G222D |
| NDM-13 | BAQ02518.1 | D95N; M154L |
| NDM-14 | WP_063860857.1 | D130G |
| NDM-15 | AKF43458.1 | M154L; A233V |
As of 7 May 2018.
FIG 1.
Workflow diagram of peptide selection for NDM detection by MRM LC-MS. Abbreviations used: ESP, enhanced signature peptide (predictor); DDA, data-dependent acquisition; BLASTp, protein blast; Unipept, Unipept Peptidome Analysis (Web tool).
Experimental detection of theoretically determined tryptic peptide markers.
In order to find efficiently ionized and readily detectable peptide markers (highly responsive peptides) by LC-MS/MS, we studied ATCC BAA-2146, a blaNDM-containing Klebsiella pneumoniae isolate that was sequenced previously (12). A bottom-up data-dependent acquisition proteomic analysis detected 236 proteins with 468 high-confidence peptides. These were defined as peptides that produced the highest ion current responses and that had a false-discovery rate (FDR) value of <0.01 per the results of analysis using Proteome Discoverer 1.4 software. Two core peptides (AFGAAFPK and ASMIVMSHSAPDSR) were detected. Incomplete digests and M-oxidation were observed for ASMIVMSHSAPDSR, but this peptide was retained as candidate. Three core peptides (AAITHTAR, MELPNIMHPVAK, and QEINLPVALAVVTHAHQDK) were not detected by LC-MS/MS and were not considered for further study. A high-abundance peptide, SLGNLGDADTEHYAASAR, was found to be highly specific to NDM (present in 12 out of 15 allelic variants). This peptide contains amino acid sequence variants (highlighted with italics and boldface) in NDM-12 (SLGNLDDADTEHYAASAR), NDM-6, and NDM-15 (SLGNLGDADTEHYAASVR). The enhanced signature peptide (ESP) predictor (9) value for SLGNLGDADTEHYAASAR was 0.67, which was among the highest values determined for the peptides studied (Table S2). Thus, three peptides (SLGNLGDADTEHYAASAR, AFGAAFPK, and ASMIVMSHSAPDSR) were selected for further study.
MRM assay development.
Table 2 details the peptides, transitions, and collision energy used in the MRM assay (Agilent Chip Cube triple quadrupole [QQQ]). To establish the rules for positive identification of NDM, we constructed a blind 20-sample set consisting of 4 blaNDM-containing isolates and 16 negative controls. The data were processed with Skyline 3.7 (or later versions) (13). rdotp and R ratio values measured by Skyline are shown in Table 3. rdotp data represent the normalized dot products of the light transition peak areas with the heavy transition peak areas. The R ratio data represent the ratios of light transition peak areas with the heavy transition peak areas for quantitative calculation. Figure 2 shows representative LC-MS chromatograms of three NDM peptides for two isolates used in the 20-sample assay development. On the basis of the rdotp and R ratio values determined for the 4 blaNDM-containing isolates and the 16 negative-control isolates summarized in Table 3, we set the following formal rules for analyses of results. (i) rdotp values of ≥0.95 and R ratio values of ≥0.5 were automatically called positive. (ii) rdotp values of ≤0.85 and R ratio values of ≤0.1 were automatically called negative. (iii) Manual review was required for any peptide not meeting the automatically classified positive or negative criteria as defined above. (iv) Overall positive identification of NDM required that two or more peptides scored positive by either automatic or manual review. During manual review, removal of one transition was permitted if it was judged to represent an interfering peak. The operator examined all factors for native and labeled peptides, including the retention time, the order of transition ranks, and possible interference and background noise when the signal intensity was low. When carryover was present in the intervening blank following a previous positive sample, then a rerun of the sample following the blank was permitted to eliminate the possibility of false-positive calls due to carryover.
TABLE 2.
Precursor ions, transitions, and collision energies used in MRM assaya
| Peptide | Charge | Precursor | T1 | T2 | T3 | T4 |
|---|---|---|---|---|---|---|
| SLGNLGDADTEHYAASAR | 3+ | 616.6221 | y13++ (23) | y14++ (23) | y16++ (19.9) | y11++ (23) |
| ASMIVMSHSAPDSR | 3+ | 496.9026 | y11++ (16) | y10++ (15.6) | y2+ (16) | y12++ (13) |
| AFGAAFPK | 2+ | 404.7212 | y7++ (17) | y2+ (17) | y6+ (17) |
Collision energy (eV) data are shown in parentheses.
TABLE 3.
Detected rdotp and R ratio values for assay development set using characterized isolates
| Sample | rdotp value/R ratio valuea
|
blaNDM | ||
|---|---|---|---|---|
| SLGNLGDADTEHYAASAR | ASMIVMSHSAPDSR | AFGAAFPK | ||
| S001 | 1.00/9.36 | 0.99/8.49 | 1.00/4.26 | Positive |
| S002 | 0.92/0.04 | 0.86/0.03 | 0.70/0.02 | Negative |
| S003 | 0.78/0.01 | 0.80/0.01 | 0.79/0.05 | Negative |
| S004 | 0.86/0.04 | 0.85/0.08 | 0.61/0.22 | Negative |
| S005 | 0.78/0.01 | 0.67/0.03 | 0.80/0.03 | Negative |
| S006 | 1.00/4.64 | 0.99/6.89 | 1.00/2.39 | Positive |
| S007 | 0.94/0.01 | 0.90/0.08 | 0.80/0.06 | Negative |
| S008 | 0.73/0.04 | 0.48/0.21 | 0.80/0.02 | Negative |
| S009 | 0.70/0.01 | 0.82/0.08 | 0.82/0.09 | Negative |
| S010 | 0.78/0.03 | 0.69/0.07 | 0.88/0.31 | Negative |
| S011 | 0.46/0.01 | 0.81/0.06 | 0.78/0.11 | Negative |
| S012 | 1.00/2.00 | 0.99/2.05 | 1.00/0.82 | Positive |
| S013 | 0.49/0.01 | 0.81/0.04 | 0.78/0.01 | Negative |
| S014 | 1.00/4.72 | 0.98/5.51 | 1.00/1.96 | Positive |
| S015 | 0.86/0.02 | 0.83/0.07 | 0.94/0.13 | Negative |
| S016 | 0.85/0.02 | 0.81/0.07 | 0.82/0.14 | Negative |
| S017 | 0.21/0.14 | 0.87/0.02 | 0.61/0.01 | Negative |
| S018 | 0.86/0.02 | 0.49/0.36 | 0.79/0.15 | Negative |
| S019 | 0.84/0.03 | 0.65/0.06 | 0.65/0.1 | Negative |
| S020 | 0.86/0.18 | 0.83/0.26 | 0.68/0.09 | Negative |
Values are given as rdotp/R ratio. Positive and negative R ratio value ranges for SLGNLGDADTEHYAASAR, 2.00 to 9.36 and 0.01 to 0.18, respectively; positive and negative R ratio value ranges for ASMIVMSHSAPDSR, 2.05 to 8.49 and 0.01 to 0.26, respectively; positive and negative R ratio value ranges for AFGAAFPK, 0.82 to 4.26 and 0.01 to 0.31, respectively.
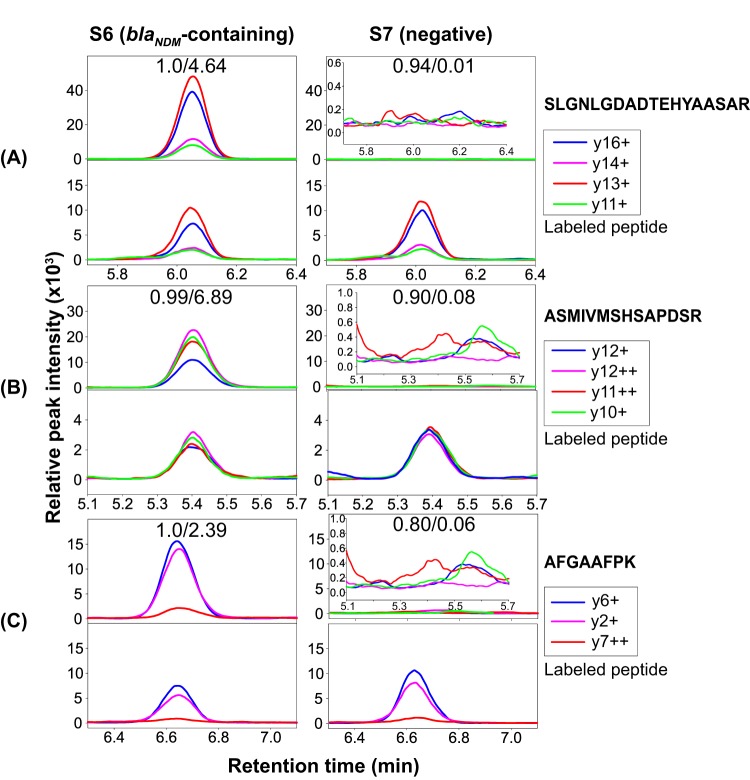
FIG 2.
Representative LC-MS chromatograms of three NDM peptides for two isolates used in 20-sample assay development. S6 is a blaNDM-containing isolate and S7 was used as a negative control. (A) SLGNLGDADTEHYAASAR. (B) ASMIVMSHSAPDSR. (C) AFGAAFPK. rdotp/R-ratio values are shown for each peptide. For S7, the signals are shown as insertions to show the details.
Performance assessment.
To test the performance of the three-peptide assay, we constructed a blind set of 100 deidentified clinical isolates consisting of 24 blaNDM-containing isolates and 76 negative-control isolates (Table S1). All 100 runs were treated independently, with blaNDM-containing isolates randomly interspersed among negative controls. Collection and analysis of LC-MS/MS data were performed by a single expert operator who was blind to the identity of the samples. The operator submitted the full list of determinations for 100 measurements prior to unblinding, and the list was matched to the result key by an independent second evaluator.
Blinded test set performance.
Automatic call rules were applied to the 3 peptides in each of 100 samples (300 peptides total). A total of 97/300 peptides were determined to be negative by the automatic call rules, and 55/300 peptides (or 89% of all 62 positively identified peptides) were called positive by the automatic rule. The remainder of the peptides (148/300) qualified for manual expert review. The majority of these manually inspected peaks were classified either as noise, represented by a higher rdotp but lower R ratio value of <=0.05, or as single-transition interference, represented by a lower rdotp value but a higher R ratio value. Automatic call rules correctly identified 19 blaNDM-containing isolates among a total of 24. An additional two isolates (L075 and L100) were manually identified as NDM positive, but with lower peak intensity (the rdotp/R ratio values for SLGNLGDADTEHYAASAR were 1.00/0.28 and 1.00/0.11 for L075 and L100, respectively). No peptide markers were detected for three blaNDM-containing isolates (L017, L097, and L099). No false-positive calls were made for the 76 negative controls, yielding overall performance levels of 87.5% sensitivity (95% confidence interval [CI], 67.6% to 97.3%) and 100% specificity (95% CI, 95.3% to 100%) for detection of NDM protein in the blind test set. Table 4 shows the rdotp values and R ratios for the 21 isolates that were identified as NDM positive. The MRM spectra for L075 and L100, the two isolates that were manually identified as NDM positive, are shown in Fig. S3 in the supplemental material. A clear match of MRM spectra was observed for SLGNLGDADTEHYAASAR for the two isolates. However, spectral interference was observed for other two peptide markers.
TABLE 4.
rdotp values and R ratios for 24 blaNDM-containing isolatesa
| Sample no. | CDC/FDA AR bank or NIH isolate |
SLGNLGDADTEHYAASAR rdotp/R value |
SLGNLGDADTEHYAASAR single-peptide call |
ASMIVMSHSAPDSR rdotp/R value |
ASMIVMSHSAPDSR single-peptide call |
AFGAAFPK rdotp/R value |
AFGAAFPK single-peptide call |
Final call | Concn (fm/μg) |
|---|---|---|---|---|---|---|---|---|---|
| L002 | CDC_148 | 1/6.87 | Positive | 0.99/6.21 | Positive | 1/3.05 | Positive | Positive | 85.9 |
| L004 | CDC_150 | 1.0/17.0 | Positive | 0.98/18.7 | Positive | 1.0/7.36 | Positive | Positive | 212.5 |
| L008 | CDC_057 | 1.0/0.58 | Positive | 0.99/0.55 | Positive | 0.98/0.35 | Positivec | Positive | 7.3 |
| L016 | CDC_152 | 1.0/8.02 | Positive | 0.98/6.72 | Positive | 1.0/3.06 | Positive | Positive | 100.3 |
| L024 | CDC_038 | 1.0/9.46 | Positive | 0.98/8.82 | Positive | 1.0/3.83 | Positive | Positive | 118.3 |
| L027 | CDC_143 | 1.0/5.25 | Positive | 0.99/3.47 | Positive | 0.99/2.01 | Positive | Positive | 65.6 |
| L030 | CDC_146 | 1.0/9.71 | Positive | 0.98/6.44 | Positive | 1.0/3.55 | Positive | Positive | 121.4 |
| L032 | CDC_151 | 1.0/51.02 | Positive | 0.99/38.15 | Positive | 1.0/19.44 | Positive | Positive | 637.8 |
| L048 | NDM-33 | 1.0/40.3 | Positive | 0.98/31.52 | Positive | 1.0/15.83 | Positive | Positive | 503.8 |
| L050 | CDC_158 | 1.0/5.58 | Positive | 0.98/3.41 | Positive | 1.0/1.94 | Positive | Positive | 69.8 |
| L051 | CDC_127 | 1.0/17.48 | Positive | 0.99/16.15 | Positive | 1.0/6.5 | Positive | Positive | 218.5 |
| L054 | CDC_162 | 1.0/48.27 | Positive | 0.99/48.01 | Positive | 1.0/19.33 | Positive | Positive | 603.4 |
| L057 | CDC_149 | 1.0/39.65 | Positive | 0.97/40.2 | Positive | 1.0/15.37 | Positive | Positive | 495.6 |
| L063 | CDC_137 | 0.95/0.01 | Negativeb | 0.98/28.08 | Positive | 1.0/12.28 | Positive | Positivee | 351.0 |
| L075 | CDC_082 | 1.0/0.28 | Positived | 0.94/0.33 | Positivec,d,e | 0.93/0.2 | Positivec ,d | Positive | 3.5 |
| L082 | CDC_145 | 1.0/5.54 | Positive | 0.98/4.21 | Positive | 0.99/2.05 | Positive | Positive | 69.3 |
| L086 | CDC_139 | 1.0/5.08 | Positive | 0.99/4.18 | Positive | 1.0/2.04 | Positive | Positive | 63.5 |
| L089 | CDC_157 | 1.0/12.3 | Positive | 0.96/9.39 | Positive | 1.0/4.94 | Positive | Positive | 153.8 |
| L094 | CDC_119 | 1.0/1.87 | Positive | 0.98/1.68 | Positive | 1.0/0.81 | positve | Positive | 23.4 |
| L098 | CDC_138 | 1.0/86.58 | Positive | 0.98/64.12 | Positive | 1.0/30.76 | Positive | Positive | 1,082.3 |
| L100 | CDC_037 | 1.0/0.11 | Positived | 0.93/0.08 | positivec,d,e | 0.94/0.11 | Possible positive | Positive | 1.4 |
| L017 | NDM-34 | 0.99/0.05 | Negative | 0.96/0.04 | Negative | 0.95/0.23c | Negative | Negative | Not detectable |
| L017g | NDM-34 | 0.94/0.04 | Negative | 0.94/0.04 | Negative | 0.93/0.10c | Negative | Negativeh | Not detectable |
| L092 | CDC_118 | 0.94/0.02 | Negativeb | 0.79/0.03 | negative | 0.85/0.05 | Negativec | Negative | Not detectable |
| L092g | CDC_118 | 1.0/17.9 | Positive | 1.0/32.5 | Positive | 1.0/7.8 | Positive | Positiveh | 223.8 |
| L099 | CDC_128 | 0.99/0.05 | Negativef | 0.99/0.05 | Negativec | 0.94/0.04 | Negativeb ,c | Negative | Not detectable |
| L099g | CDC_128 | 1.0/26.2 | Positive | 1.0/50.6 | Positive | 1.0/11.2 | Positive | Positiveh | 327.5 |
Three repeated tests for L017, L092, and L099 were also included in the analysis; those data were not included in the calculations of sensitivity and specificity.
Noise.
Interference.
Weak signal.
Removal of one transition.
Carryover.
A new preparation was tested after 100-sample validation was completed. The values reported represent averages of results from three replicates.
These data were not included in the calculation of assay sensitivity and specificity.
Discordant analyses.
(i) Sample L063. As noted above, the SLGNLGDADTEHYAASAR peptide is not present in NDM-6, NDM-12, and NDM-15. The variant form, SLGNLDDADTEHYAASAR, is specific to NDM-12, and SLGNLGDADTEHYAASVR is specific to NDM-6 and NDM-15. To confirm the specific variant form of SLGNLGDADTEHYAASAR in isolate L063, a separate MRM assay consisting of three peptides (SLGNLGDADTEHYAASAR, SLGNLDDADTEHYAASAR, and SLGNLGDADTEHYAASVR) was developed with labeled peptides. A strong signal of SLGNLGDADTEHYAASVR was detected in isolate L063, and no signals corresponding to SLGNLGDADTEHYAASAR and SLGNLDDADTEHYAASAR were detected, demonstrating the ability of the assay to detect NDM-6 and NDM-15 with these peptides (Fig. S1).
(ii) Samples L017, L092, and L099. Following the completion of the performance evaluation with the blind test set, we sought to study the NDM protein in the false-negative samples using higher-sensitivity methods, to understand the mechanism of assay failure. To increase the detection sensitivity, 10 μg of the lysate for samples L017, L092, and L099 was fractionated by the use of a C18 column at high pH and their fractions were analyzed by the MRM assay using the Agilent Chip Cube QQQ as described above. All three NDM peptides were detected for sample L017, and no peptide markers were detected for samples L092 and L099. The chromatograph of the three peptide markers for sample L017 after high-pH fractionation enrichment is shown in Fig. S2. The results indicated that the level of expression of NDM in lysate L017 was low (estimated to be in the range of 0.12 fm/μg of isolate lysate based on the concentration of labeled peptides and total amount of the proteins used in fractionation). Kirby-Bauer testing for sample L017 (Proteus mirabilis), performed on two separate isolate preparations, revealed that the meropenem disk diameter (23 mm) was in the range indicating susceptibility. This finding is consistent with the low quantity of NDM present in the MRM assay in the absence of other mechanisms conferring carbapenem resistance in this isolate (Table 4).
Given the results, a second preparation of L017, L092, and L099 was tested. Single-colony subcultures for each of these three isolates were prepared, and cells were harvested from three different locations on each plate for new lysate preparations. A second blind assessment was performed in which these nine lysates were treated as independent samples and tested with two additional blaNDM-containing isolates and nine blaNDM-negative isolates. Surprisingly, these two new preparations (all six lysates) of samples L092 and L099 demonstrated high concentrations of all three NDM tryptic peptides, in contrast to the undetectable levels in the first preparation (Table S3). MRM of samples L092 and L099 (Table S4) performed in triplicate indicated that the reproducibility of the R ratio values for each peptide was within 10% among the samples. No NDM peptides were detected in any of the three new lysates from L017, as seen in the first sample preparation.
In a separate experiment, targeted LC-MS/MS using a high-mass-resolution mass spectrometer (Orbitrap Lumos) was applied to samples L017, L021 (as negative control), L024 (as positive control), L075, and L100. L017 had very low NDM expression, and the MRM assay failed to detect NDM peptide markers in this sample. L075 and L100 had low NDM abundance and were the only two samples whose spectra were manually identified as NDM positive. The LC-MS/MS chromatograms for three peptide markers in each of these five samples are shown in Fig. S4. Clearly, use of the high-resolution mass spectrometer and longer gradient reduced interference and improved detection in the isolates with low NDM abundance, especially for AFGAAFPK. For sample L017, two NDM peptide markers were correctly detected by Orbitrap Lumos.
(iii) Total proteomics of samples L092 and L099. In order to determine if plasmid loss accounted for the substantial variations in NDM protein concentrations detected in the two separate subcultures of L092 and L099, we used a total proteomic approach (detailed in the supplemental material) to study the plasmid-encoded proteins present in the original and second set of L092 and L099 samples. Highly expressed NDM protein was detected in the recultured L092 and L099 samples with 6 unique peptides for each sample whereas no NDM was detected in the original L092 and L099 samples, consistent with the results of the original and repeated MRM LC-MS/MS assays. Totals of 22 and 20 high-confidence plasmid proteins were detected for the two L092 preparations, representing products of genes carried by all three plasmids (Table S5a). Totals of 16 and 12 high-confidence plasmid proteins were detected for the two preparations of L099, representing products of genes located on all three plasmids (Table S5b). These findings indicate that loss of the plasmids containing the blaNDM gene does not explain the lack of detection of the NDM protein in the first set of isolates and demonstrate that substantial variations in the concentrations of NDM can be present in different preparations of the same isolates, grown under identical conditions.
DISCUSSION
There has been significant recent interest in the development of mass spectrometry-based methods for antimicrobial resistance protein detection. The ability of these methods to measure protein concentrations quantitatively may provide complementary functional information beyond that given by PCR-based assays limited to detecting the presence or absence of a gene. In this work, we studied the performance of an LC-MS/MS MRM method for the direct detection of NDM carbapenemase in clinical isolates. A rapid assay with a turnaround time of 90 min was developed based on three unique peptides specific to the NDM protein that were efficiently ionized and spectrally well defined. To characterize the performance of this assay, a blind isolate set containing 24 blaNDM-containing and 76 negative-control isolates was tested. The assay detected 21/24 blaNDM-containing and 76/76 negative isolates, corresponding to a sensitivity value of 87.5% (95% confidence interval [CI] 67.6% to 97.3%) and a specificity value of 100% (95% CI, 95.3% to 100%).
We undertook a detailed study of the three samples in which NDM protein was not detected by the MRM assay. One of the missed identifications (L017) was determined by protein fractionation and targeted LC-MS/MS by Orbitrap Lumos high-mass-resolution mass spectrometer to have been due to low NDM protein expression (∼0.1 fm/μg, which was below the MRM assay’s limit of detection). Interestingly, parallel disk diffusion susceptibility testing demonstrated this isolate to be meropenem susceptible, consistent with low NDM expression. Total proteomic analysis performed on the other two isolates (L092 and L099) did not detect NDM peptides but did detect other proteins expressed from the same plasmids containing the blaNDM gene, arguing against plasmid loss. Surprisingly, repeat assays of subculture preparations of these two isolates grown under identical nonselective conditions revealed high concentrations of detectable NDM peptides in the second round of testing, demonstrating the remarkable variability in expression of NDM that may occur.
We further studied the relative concentrations of NDM in the 24 blaNDM-containing isolates (Table 4) using the R ratio value, which is based on the ratio of the concentration of SLGNLGDADTEHYAASAR (or of a corresponding variant peptide) and the concentration of its labeled peptide. The levels of NDM protein expression in these 24 isolates ranged over 4 orders of magnitude from 0.1 fm/μg to 1,000 fm/μg of total protein or peptides, showing remarkably variable NDM expression. It may be possible to increase the detection sensitivity of the MRM assay by enriching the peptides in the mixture using either fractionation or antibody pulldown of targeted peptides (14–16). While fractionation can improve the detection limit of the assay, it would increase the analysis time, which would impact its implementation in clinical practice.
A limitation of this assay is the requirement for manual interpretation of spectra that do not clearly meet positive or negative criteria. While larger sample sizes may assist in refining these criteria, some manual interpretation of spectra would be required for samples that do not lie within positive or negative boundaries. The MRM spectra for two isolates (L075 and L100) that were manually identified as NDM positive are shown in Fig. S3 in the supplemental material. A clear match of MRM spectra was observed for SLGNLGDADTEHYAASAR for the two isolates. However, interfering transitions were observed for the other two peptide markers. Thus, for analysis of isolates with low levels of NDM expression, training for manual interpretation of the MRM spectra may be required and this may limit widespread implementation of this assay in its current format for clinical purposes.
Our analysis suggests that the three core peptides that we selected for our assay represent highly specific NDM. Other investigators have demonstrated that NDM can be identified using the peptide FGDLVFR (17). FGDLVFR is a core peptide for NDM but does not appear to be specific to NDM based on BLASTp analysis and was not further evaluated in our study for this reason.
In conclusion, we describe the evaluation of an MRM assay for the direct detection of NDM in cultured clinical isolates by LC-MS/MS. The total assay time from cell lysate to LC-MS/MS assay results for one sample was less than 90 min. Our results highlight the dramatic variability that may be seen in NDM protein concentrations as well as some potential limitations of the current analytic approach at the lower limits of peptide detection. Fractionation or antibody pulldown may enhance the detection of specific peptide markers and increase the limits of detection used with this analytic approach.
MATERIALS AND METHODS
Bacterial isolates.
25 blaNDM-containing isolates were obtained from the Centers for Disease Control and Food and Drug Administration Antibiotic Resistance Isolate Bank (ARISOLATEBANK), the National Institutes of Health Clinical Center isolate collection, and the American Type Culture Collection (ATCC). An additional 92 bacterial isolates were used as negative-control samples (see Table S1 in the supplemental material). All bacterial isolates were grown on blood agar plates (Remel, Lenexa, KS) for 18 to 24 h at 35°C with 5% CO2.
Antimicrobial susceptibility testing (AST).
AST was performed by standard Kirby-Bauer disk diffusion methods using Mueller-Hinton agar plates (Remel, Lenexa, KS) and meropenem-containing disks (Becton, Dickinson and Company, Sparks, MD). Disk diameters were interpreted using CLSI M100 (28th edition) (18).
Analysis of NDM sequences.
The protein sequences of 15 NDM allelic variants (Table 1) were downloaded (https://www.ncbi.nlm.nih.gov/protein; last accessed May 2018). Core tryptic peptides were defined as those tryptic peptides present in all 15 NDM allelic variants (Table S2) using methods for sequence alignment, in silico tryptic digestion, and core peptide identification as described previously for KPC variants (8). The uniqueness of the identified tryptic peptides to NDM was analyzed using both the Unipept Peptidome Analysis Web tool (http://unipept.ugent.be/) and protein blast (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins; last accessed December 2016).
Tryptic protein digestion.
A loop of 10 μl bacterial cells was lysed with formic acid (FA) and acetonitrile (ACN) for LC-MS/MS preparation as previously described (19). Cell lysates (2 μl) were lyophilized, resuspended in 96 μl of 100 mM NH4HCO3, and digested in a water bath for 15 min at 55°C with the addition of 4 μl of 0.1μg/μl trypsin (Promega, Madison, WI). Digested samples were filtered with an Ultrafree centrifugal filter (Merck Millipore, MA) (0.5 ml, 0.22 μm pore size) before injection into the LC-MS. A Qubit 2.0 Fluorometer (Thermo Fisher, San Jose, CA) was used for total peptide concentration measurement. Concentrations over 100 μg/ml were diluted to 100 μg/ml with 100 mM NH4HCO3. For initial identification of NDM peptides, 200 μl volumes of formic acid/acetonitrile (FA/ACN) lysate were processed per a protocol described previously (8). The lysates were digested for 30 min at 55°C in a CEM Discoverer microwave system (CEM, Mathews, NC) (8). The digests were then diluted 160×, and 10 μl volumes of the diluted digests were loaded onto an Orbitrap Fusion LC-MS apparatus for protein identification.
High-performance liquid chromatography (HPLC) fractionation.
Peptide fractionation was performed on an Agilent 6540 quadrupole time of flight (QTOF) LC-MS system. The retention times for labeled peptides SLGNLGDADTEHYAASAR, ASMIVMSHSAPDSR, and AFGAAFPK were determined using a Waters Xbridge C18 column (4.6 by 100 mm; 2.5 μm pore size) under conditions of high pH (20) with 10 mM tetraethylammonium bicarbonate H2O/ACN mobile phases with a flow rate of 0.5 ml/min. The fractionation required two LC-MS runs. In the first, the three heavy (6 pmol) labeled peptides were detected as single ions (M+H)+ and eluted out at retention times of 7.8, 9.6, and 10.5 min, respectively. On the second LC-MS run, the tube connected to the MS was disconnected and reconnected to a Beckman S100 fraction collector. The tryptic digests (10 μg) of the test isolates were then loaded onto the column, and the fractions were collected between retention times of 6.8 to 11.5 min for 10 s per fraction (30 fractions). Each fraction was transferred to a glass total-recovery vial (Waters; catalog no. 186000384c). After speed dry, the digests were resuspended in a mixture consisting of 7 μl of 100 mM NH4HCO3 and 2 μl of labeled peptide mix (2.5 fm/μl). An 8 μl volume was injected into an Agilent CubeChip 6495 QQQ apparatus for MRM analysis.
Labeled peptides.
Peptide standards containing heavy isotopic labels in R (U-13C6 and U-15N4) or K (U-13C6 and U-15N2) C-terminal amino acids were purchased (JPT, Berlin, Germany). The characterization and concentration data were provided by the manufacturer. The labeled peptides were stored in 100 mM NH4HCO3 at 15 pm/μl or 1 pm/μl and −20°C. They were further diluted with 100 mM NH4HCO3 to reduce their concentrations as described below.
MRM assay.
The MRM assay was run on an Agilent CubeChip 6495 QQQ apparatus with a high-capacity chip (C18; Agilent catalog no. G4240-62010) (160 nl, 150 mm) as previously described (21). The mobile phases consisted of 0.1% FA, 5% ACN, and 95% H2O (buffer A) and 0.1% FA, 95% ACN, and 5% H2O (buffer B). The gradient was run from 5% to 20% buffer B over 7 min with a flow rate of 0.4 μl/min. The total assay time was 15 min. Dwell time was 20 ms for all transitions, with Q1 and Q3 mass resolution of 0.7 Da (unit). Other MS settings included the following: delta EMV+ value, 300; cell accelerator voltage, 2; gas temperature, 200°C; gas flow rate, 11 liters/min. Table 2 lists the peptides and transitions as well as collision energy for each transition. The labeled peptide mix was composed of 5 fm/μl each for SLGNLGDADTEHYAASAR, ASMIVMSHSAPDSR, and AFGAAFPK. A 4 μl volume of labeled peptide mix was added to 16 μl of digested peptide solution in a silanized vial (National C4000-S9; Thermo), and a 4 μl volume was injected into the LC-MS apparatus. A separate MRM assay was also developed to detect variant forms of SLGNLGDADTEHYAASAR and SLGNLDDADTEHYAASAR for NDM-12 and SLGNLGDADTEHYAASVR for NDM-6 and NDM-15. Details of the protocol are described in the supplemental material.
Tryptic peptide identification by LC-MS/MS.
Bottom-up protein identification was carried out using an Orbitrap Fusion or Lumos mass spectrometer (Thermo Fisher Scientific) as previously described (19). Briefly, 1 μg of tryptic digests was separated on an EasySpray column (Thermo Fisher ES803; 50 cm by 75 μm inner diameter [ID] packed with PepMap RSLC C18 2-μm-diameter particles) using a 120 min linear gradient of 5% to 35% ACN–0.1% FA at a flow rate of 300 nl/min. Mass analysis was carried out in data-dependent analysis mode, where MS1 scans at 60,000 mass resolution were carried out with the full MS range from m/z 375 to 1,500 and 10 higher-energy collisional dissociation (HCD) MS2 scans at 30,000 resolution were sequentially carried out using an Orbitrap system. LC-MS/MS data were searched against a custom FASTA database composed of Escherichia coli protein sequences (4,212 sequences downloaded from https://www.uniprot.org/ in July 2016) and 15 sequences of NDM variants by the use of Proteome Discoverer 1.4 (Thermo Fisher Scientific) and Scaffold 4 (Proteome Software Inc., Portland, OR) as previously described (19, 21). Additional total proteomic analysis for repeat extractions from new subcultures of samples L092 and L099 was performed as detailed in the supplemental material.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the CDC and FDA Antibiotic Resistance Isolate Bank (ARISOLATEBANK), from which 22 blaNDM-containing isolates (listed in Table 4) were obtained.
This work was supported in part by the Intramural Research Programs of the National Institutes of Health Clinical Center (H.W., J.R.S., S.K.D., J.-H.Y., A.F.S., and J.P.D.); the National Institute of Allergy and Infectious Diseases (J.P.D.); the National Heart, Lung, and Blood Institute (M.G. and Y.C.); and the National Institute of Diabetes and Digestive and Kidney Diseases and Johns Hopkins University (A.Z.R.). The content is solely our responsibility and does not necessarily represent the official views of the NIH or the U.S. Government. S.K.D. has been involved in a collaborative agreement with Bruker Daltonics, Inc., to develop organism databases for MALDI-TOF MS, independently of this study. Bruker Daltonics, Inc., had no role in the work published here.
We declare that we have no competing financial interests.
H.W., S.K.D., A.F.S., and J.P.D. conceived the project design. H.W., J.R.S., S.K.D., Y.C., and J.-H.Y. carried out the experiments. H.W., S.K.D., J.R.S., A.F.S., and J.P.D. performed primary analysis of the data, and Y.C., M.G., and A.Z.R. critically reviewed the primary analysis and provided LC-MS instrument support. H.W., J.R.S., S.K.D., A.F.S., and J.P.D. cowrote the manuscript. All of us critically evaluated and edited the manuscript.
We have no conflicts to disclose.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00461-19.
REFERENCES
- 1.Bonomo RA, Burd EM, Conly J, Limbago BM, Poirel L, Segre JA, Westblade LF. 2018. Carbapenemase-producing organisms: a global scourge. Clin Infect Dis 66:1290–1297. doi: 10.1093/cid/cix893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khan AU, Maryam L, Zarrilli R. 2017. Structure, genetics and worldwide spread of New Delhi metallo-beta-lactamase (NDM): a threat to public health. BMC Microbiol 17:101. doi: 10.1186/s12866-017-1012-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, Walsh TR. 2009. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother 53:5046–5054. doi: 10.1128/AAC.00774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mancini S, Kieffer N, Poirel L, Nordmann P. 2017. Evaluation of the RAPIDEC® CARBA NP and beta-CARBA® tests for rapid detection of carbapenemase-producing Enterobacteriaceae. Diagn Microbiol Infect Dis 88:293–297. doi: 10.1016/j.diagmicrobio.2017.05.006. [DOI] [PubMed] [Google Scholar]
- 5.Traczewski MM, Carretto E, Canton R, Moore NM, Brovarone F, Nardini P, Visiello R, García-Castillo M, Ruiz-Garbajosa P, Tato M. 2018. Multicenter evaluation of the Xpert Carba-R assay for detection of carbapenemase genes in Gram-negative isolates. J Clin Microbiol 56:e00272-18. doi: 10.1128/JCM.00272-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hrabak J, Studentova V, Walkova R, Zemlickova H, Jakubu V, Chudackova E, Gniadkowski M, Pfeifer Y, Perry JD, Wilkinson K, Bergerova T. 2012. Detection of NDM-1, VIM-1, KPC, OXA-48, and OXA-162 carbapenemases by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J Clin Microbiol 50:2441–2443. doi: 10.1128/JCM.01002-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kulkarni MV, Zurita AN, Pyka JS, Murray TS, Hodsdon ME, Peaper DR. 2014. Use of imipenem to detect KPC, NDM, OXA, IMP, and VIM carbapenemase activity from Gram-negative rods in 75 minutes using liquid chromatography-tandem mass spectrometry. J Clin Microbiol 52:2500–2505. doi: 10.1128/JCM.00547-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang H, Drake SK, Youn JH, Rosenberg AZ, Chen Y, Gucek M, Suffredini AF, Dekker JP. 2017. Peptide markers for rapid detection of KPC carbapenemase by LC-MS/MS. Sci Rep 7:2531. doi: 10.1038/s41598-017-02749-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fusaro VA, Mani DR, Mesirov JP, Carr SA. 2009. Prediction of high-responding peptides for targeted protein assays by mass spectrometry. Nat Biotechnol 27:190–198. doi: 10.1038/nbt.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bauer M, Ahrne E, Baron AP, Glatter T, Fava LL, Santamaria A, Nigg EA, Schmidt A. 2014. Evaluation of data-dependent and -independent mass spectrometric workflows for sensitive quantification of proteins and phosphorylation sites. J Proteome Res 13:5973–5988. doi: 10.1021/pr500860c. [DOI] [PubMed] [Google Scholar]
- 11.Hu A, Noble WS, Wolf-Yadlin A. 2016. Technical advances in proteomics: new developments in data-independent acquisition. F1000Res 5:419. doi: 10.12688/f1000research.7042.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leski T, Vora GJ, Taitt CR. 2012. Multidrug resistance determinants from NDM-1-producing Klebsiella pneumoniae in the USA. Int J Antimicrob Agents 40:282–284. doi: 10.1016/j.ijantimicag.2012.05.019. [DOI] [PubMed] [Google Scholar]
- 13.MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, MacCoss MJ. 2010. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anderson NL, Jackson A, Smith D, Hardie D, Borchers C, Pearson TW. 2009. SISCAPA peptide enrichment on magnetic beads using an in-line bead trap device. Mol Cell Proteomics 8:995–1005. doi: 10.1074/mcp.M800446-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neubert H, Gale J, Muirhead D. 2010. Online high-flow peptide immunoaffinity enrichment and nanoflow LC-MS/MS: assay development for total salivary pepsin/pepsinogen. Clin Chem 56:1413–1423. doi: 10.1373/clinchem.2010.144576. [DOI] [PubMed] [Google Scholar]
- 16.Shi T, Fillmore TL, Sun X, Zhao R, Schepmoes AA, Hossain M, Xie F, Wu S, Kim JS, Jones N, Moore RJ, Pasa-Tolic L, Kagan J, Rodland KD, Liu T, Tang K, Camp DG II, Smith RD, Qian WJ. 2012. Antibody-free, targeted mass-spectrometric approach for quantification of proteins at low picogram per milliliter levels in human plasma/serum. Proc Natl Acad Sci U S A 109:15395–15400. doi: 10.1073/pnas.1204366109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cecchini T, Yoon EJ, Charretier Y, Bardet C, Beaulieu C, Lacoux X, Docquier JD, Lemoine J, Courvalin P, Grillot-Courvalin C, Charrier JP. 2018. Deciphering multifactorial resistance phenotypes in Acinetobacter baumannii by genomics and targeted label-free proteomics. Mol Cell Proteomics 17:442–456. doi: 10.1074/mcp.RA117.000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.CLSI. 2018. Performance standards for antimicrobial susceptibility testing, 28th ed CLSI supplement M100 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 19.Lau AF, Wang H, Weingarten RA, Drake SK, Suffredini AF, Garfield MK, Chen Y, Gucek M, Youn JH, Stock F, Tso H, DeLeo J, Cimino JJ, Frank KM, Dekker JP. 2014. A rapid matrix-assisted laser desorption ionization–time of flight mass spectrometry-based method for single-plasmid tracking in an outbreak of carbapenem-resistant Enterobacteriaceae. J Clin Microbiol 52:2804–2812. doi: 10.1128/JCM.00694-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang F, Shen Y, Camp DG II, Smith RD. 2012. High-pH reversed-phase chromatography with fraction concatenation for 2D proteomic analysis. Expert Rev Proteomics 9:129–134. doi: 10.1586/epr.12.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang H, Drake SK, Yong C, Gucek M, Tropea M, Rosenberg AZ, Dekker JP, Suffredini AF. 2016. A novel peptidomic approach to strain typing of clinical Acinetobacter baumannii isolates using mass spectrometry. Clin Chem 62:866–875. doi: 10.1373/clinchem.2015.253468. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.