Quinacrine hydrochloride is a small-molecule, orally bioavailable drug that has been used clinically as an antimalarial and for many other applications. A machine learning model trained on Ebola virus (EBOV) screening data identified quinacrine as a potent (nanomolar) in vitro inhibitor.
KEYWORDS: Ebola virus, Ebola virus disease, antiviral, quinacrine
ABSTRACT
Quinacrine hydrochloride is a small-molecule, orally bioavailable drug that has been used clinically as an antimalarial and for many other applications. A machine learning model trained on Ebola virus (EBOV) screening data identified quinacrine as a potent (nanomolar) in vitro inhibitor. In the current study, quinacrine 25 mg/kg was shown to protect 70% of mice (statistically significant) from a lethal challenge with mouse-adapted EBOV with once-daily intraperitoneal dosing for 8 days.
INTRODUCTION
In the last 5 years, we witnessed two major Ebola virus (EBOV) outbreaks in Africa. These outbreaks point to the urgent need for an effective antiviral drug that can be used to treat patients and provide protection for health care workers. In June 2018, an ethics committee in the Democratic Republic of the Congo approved the compassionate use of the five investigational therapeutics for treatment of Ebola virus disease: the antivirals favipiravir (1) and GS-5734 (remdesivir) (2) and the antibody therapeutics mAb114 (3), Zmapp (4), and REGN3470-3471-3479 (5). None of these are FDA-approved; therefore, there is still a need for rapid development of new antivirals, which points to repurposing of existing drugs (6, 7). We used in vitro screening data for EBOV to derive computational machine learning models (8). These identified quinacrine, pyronaridine, and tilorone with sub-μM 50% effective concentration [EC50] values in vitro. We address the in vivo efficacy of the antimalarial quinacrine in a mouse model (8, 9). Quinacrine hydrochloride [6‐chloro-9‐(4‐diethylamino‐1‐methylbutylamino)‐2-methoxyacridine dihydrochloride dihydrate] (Fig. 1) has a long history of clinical use as an antiprotozoal drug and a treatment for systemic lupus erythematosus (see Text S1 in the supplemental material). The current work describes the in vitro absorption, distribution, metabolism, and excretion (ADME) evaluation and the in vivo anti-EBOV efficacy of quinacrine.
FIG 1.
Quinacrine molecular structure.
Quinacrine has activity in HeLa cells but not Vero cells.
Quinacrine hydrochloride and tilorone dihydrochloride were purchased from BOC Sciences (Shirley, NY). Kolliphor HS 15 (Solutol) and sterile water for injection were purchased from Sigma-Aldrich (St. Louis, MO). Quinacrine hydrochloride was tested (using the National Institute of Allergy and Infectious Diseases [NIAID] Division of Microbiology and Infectious Diseases services) against representatives of the Herpesviridae, Togaviridae, Flaviviridae, Picornaviridae, and Poxviridae families; Bunyavirales; hepatic viruses; respiratory viruses; and other viruses.
Quinacrine was tested in vitro for its anti-EBOV activity in the Vero 76 cell line (10, 11), and the antiviral activity (EC50, >12.3 μM) was similar to the 50% cytotoxicity concentration (CC50, 12.3 μM) (see Table S1 in the supplemental material). This is in contrast to prior data in HeLa cells showing selectivity, with EC50 values of 0.35 to 0.98 μM (8, 9) and a CC50 value of 6.2 μM (8). Quinacrine demonstrated no apparent selectivity (50% selectivity index [SI50], >10) for selected additional viruses (Table S1), and the majority were tested in Vero 76 cells. This is similar to our previous observations with tilorone (12).
Quinacrine has generally favorable in vitro ADME properties.
In vitro ADME studies present preliminary information that can determine whether a compound is worth pursuing in in vivo studies. All in vitro ADME studies were performed by BioDuro (San Diego, CA) and were previously described in detail (9, 12). Test compounds were analyzed by reverse-phase high-performance liquid chromatography (HPLC) with a Kinetex 2.6-μm C18 100A column (3.0 by 50 mm; Phenomenex, Torrance, CA) using the Shimadzu LC-20AD system (Columbia, MD). The mobile phase consisted of solvent A (water with 0.1% formic acid) and solvent B (acetonitrile with 0.1% formic acid). The mass spectrometry (MS) detection was performed by using an API 4000 Q Trap system. The amount of parent compound was determined on the basis of the peak area ratio (compound area to internal standard [IS] area). The 50% inhibitory concentration (IC50) was calculated using GraphPad Prism 5 software. The final dimethyl sulfoxide (DMSO) percentage was 1%. For kinetic solubility determination, 396 μl of universal aqueous buffer (pH 7.4) was added to 4 μl of a 50-mM DMSO stock solution of tilorone. Wells were agitated for 4 h at 20°C and then filtered. The compound was then diluted to serial concentrations with DMSO, followed by serial dilutions with acetonitrile:H2O (1:1) before LC-MS analysis. The calculated concentration (micromoles) of soluble quinacrine was determined in reference to a standard curve. For Caco-2 permeability assessment, Caco-2 cells were grown on 24-well (pore size, 0.4 μm) polycarbonate filters. The monolayers were preincubated with prewarmed Hanks’ balanced salt solution containing 2.5% HEPES buffer (pH 7.4) for 0.5 h at 37°C. After preincubation, the buffer was removed, and quinacrine was added to reach a final concentration of 10 μM. Bovine serum albumin 2% was added to the receiver buffer for the study. The total volume was 400 μl for the apical (A) side and 1,200 μl for the basolateral (B) side. For the apical-to-basolateral transport study (A-B), 100 μl of each was collected from both sides for sample analysis at the start of the assay, and then 200 μl was collected from the apical side at 90 min (end of the study). The same time points and amounts were used for the basolateral-to-apical transport study (B-A). The apparent permeability coefficient (Papp) was calculated from the following equation: Papp (cm/s) = (V · δC/δt)/A · C, where V is the volume of the receiver cell, A is the exposed surface area (0.64 cm2), C is the initial donor concentration, and δC/δt is the change in receiver concentration over time.
For cytochrome P450 (CYP) inhibition assessment, human liver microsome solution (0.2 mg/ml), along with substrate, was aliquoted into 0.05 phosphate buffer (pH 7.4) in 1.1-ml tubes. Study samples (containing either control inhibitor or quinacrine) were added into the tubes, vortexed gently, and preincubated for 5 min at 37°C. A total of 20 μl of NADPH solution was aliquoted into all tubes, then vortexed to start the reaction and to ensure adequate mixture of the NADPH. After mixing, the tubes were incubated for 20 min at 37°C in a shaking water bath and then quenched in 300 ml formic acid/acetonitrile solution. After quenching, the samples were vortexed vigorously for 1 min and centrifuged at 4,000 rpm for 15 min (4°C). A total of 100 μl of supernatant was transferred to 0.65-ml tubes for liquid chromatography MS (LC-MS) analysis by the bioanalytical method described earlier. The CYP450 substrates and control inhibitors for each enzyme were as follows: 1A2, phenacetin, naphthoflavone; 2C9, diclofenac, sulfaphenazole; 2C19, omeprazole, tranylcypromine; 2D6, dextromethorphan, quinidine; and 3A4, midazolam, ketoconazole. Mouse liver microsome stability assessed using mouse liver microsome solution (197.5 μl, 1 mg/ml protein concentration) was aliquoted into 1.1-ml tubes, to which 2.5 μl of positive-control and quinacrine stock solutions (100 μM in DMSO) were added. The tubes were vortexed gently and preincubated for 5 min at 37°C, after which 50 μl of 5 mM NADPH or LM buffer (no NADPH buffer) was added into the tubes. For analysis, an aliquot of 15 μl was removed from each tube at 0, 5, 15, 30, and 60 min (without-NADPH reaction: 0, 30, and 60 min) and quenched with 300 μl of 25 ng/ml propranolol in acetonitrile. Samples were vigorously vortexed for 1 min and then centrifuged at 4,000 rpm for 15 min at 4°C. A total of 100 μl of supernatant from each sample was transferred to 0.65-ml tubes for LC-MS analysis. The amount of parent compound was determined on the basis of the peak area ratio (compound area to IS area) for each time point. Clearance rates were calculated by the equation CLint (μl/min/mg protein) = ln(2) · 1,000/t1/2/protein concentration, where CLint is intrinsic clearance and t1/2 is half-time.
Protein binding in plasma was determined when the donor side of dialysis inserts was filled with 200 μl plasma containing 5 μM quinacrine and 0.5% DMSO and the receiver side of the dialysis inserts was filled with 350 μl of phosphate-buffered saline (100 mM, pH 7.4). The prepared dialysis apparatus was placed in a shaker (37°C, 100 rpm) for 5 h. Two tubes with plasma containing 5 μM quinacrine were prepared for stability testing; one tube was placed in the freezer (4°C) for 5 h, and the other tube was placed in a shaker (3°C, 100 rpm) for 5 h. Samples were collected from the donor and receiver sides of each dialysis insert. The same volume of blank plasma was added to buffer samples and blank buffer to plasma samples to make sure all sample mixtures contained 50% plasma and 50% buffer. A total of 50 μl of each sample was mixed with 300 μl of acetonitrile containing 25 ng/ml IS (propranolol). All samples were vortexed for 1 min and then centrifuged at 4,000 rpm (4°C, 15 min). A total of 100 μl of the supernatant was transferred to a 0.65-ml tube for LC-MS analysis. The amount of quinacrine was determined on the basis of the peak area ratio (compound area to IS area) for the two sides, and protein binding was determined using the following equation: %bound = 100 × ([area ratio of donor] 5 h · 5 − [area ratio of receiver] 5 h)/([area ratio of donor] 5 h · 5). The percentage remaining at 37°C after 5 h was calculated on the basis of the amount measured at 0°C after 5 h.
Kinetic solubility (13), CYP inhibition (14), metabolic stability (15), Caco-2 permeability (16), and plasma protein binding (in mice and humans) (17) were evaluated in vitro (Table 1). Quinacrine showed fair mouse metabolic stability, good solubility, and high absorption and is unlikely to be a P-glycoprotein substrate. It inhibits CYP450 1A2, 2C19, and 3A4 in the low μM range, and it is a potent inhibitor of CYP2D6 (IC50, 13 nM) (Table 1).
TABLE 1.
In vitro ADME properties for quinacrine
| ADME property | Data |
|---|---|
| Solubility (μM) | 80.5 at pH 7.4 |
| CYP inhibition (μM) | |
| 1A2 | 6.66 |
| 2C9 | >50 |
| 2C19 | 8.66 |
| 3A4 | 3.96 |
| 2D6 | 0.013 |
| Mouse liver microsomes | |
| t1/2 (min) | 22.5 |
| CLint (μl/min/mg protein) | 30.8 |
| Mouse plasma protein binding (%) | Binding, 91.8; stability at 5 h, 84.1 |
| Human plasma protein binding (%) | Binding 89.9; stability at 5 h, 106 |
| Caco-2 | |
| Papp A-B (cm/s) | 30.6 × E-6 |
| Papp B-A (cm/s) | 19.0 × E-6 |
| Efflux ratio | 0.92 |
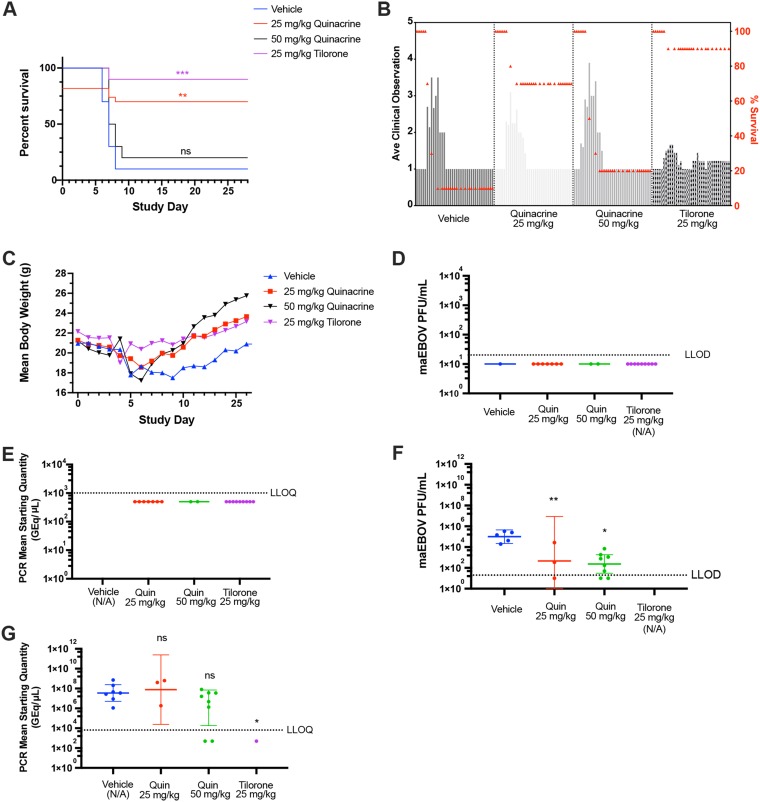
Quinacrine demonstrates in vivo efficacy in the mouse-adapted Ebola virus model.
Test article preparation for in vivo studies required that the dose formulations were aseptically prepared under yellow light by mixing the appropriate amount of test article in melted Kolliphor HS 15 (Solutol) (20% of final volume) using a vortex mixer for 30 s. The remaining sterile water (Sigma-Aldrich) was added, and the formulations were mixed using a vortex mixer for 30 s and sonicating for 15 min. The final 20% Kolliphor HS 15 dose formulations were clear red solutions. All work with mouse-adapted EBOV (maEBOV)-challenged mice was approved by the University of Texas Medical Branch’s (UTMB) institutional animal care and use committee and was done in accordance with all applicable sections of the Final Rules of the Animal Welfare Act regulations (9 CFR parts 1, 2, and 3) and the Guide for the Care and Use of Laboratory Animals (18). This work was conducted in UTMB’s American Association for Accreditation of Laboratory Animal Care-accredited GNL BSL4 laboratory. BALB/c mice (Envigo Laboratories) aged 6 to 8 weeks at delivery with a weight range of 16.4 to 27.1 g on study day (SD) were acclimated to study housing for 7 days before the virus challenge. Animals were monitored daily by visual examination. Clinical scoring and health assessments were performed and documented at each observation using a quantitative assessment of pain and distress scoring system. Animals were scored based on the following observations: 1, healthy; 2, mild signs of lethargy, some fur ruffling, no hunched posture; 3, fur ruffling, hunched posture, mild signs of lethargy; 4, ruffled, hunched posture, increased lethargy, and limited mobility; 5, moribund (ruffled, hunched posture with reduced or minimal mobility consistent with inability to reach food or water or a ±20% weight loss). Once animals reached a clinical score of 3, they were observed twice daily with 6 to 8 h between observations. Animals with advanced disease (score of 4) were observed a third time. The third observation occurred 4 to 6 h after the afternoon observation. All surviving animals were humanely euthanized on SD28. Mice were weighed daily through SD7. After this period and for the remainder of the study, animals were weighed every 3 days and monitored at least once per day for the development of clinical signs. Ebola virus Mus musculus/COD/1976/Mayinga-CDC-808012 (maEBOV) stock virus from clarified supernatant from maEBOV-infected Vero E6 cell monolayers in growth medium/20% fetal bovine serum/0.01 M Tris was diluted from the 1.05 × 107 PFU/ml stock to a target challenge dose concentration of 1,000 PFU/ml such that animals received a target of 100 PFU per 100-μl dose. Virus administration was performed via intraperitoneal (i.p.) injection, and the viral dose administered was verified through plaque assay analysis of the prepared virus suspension. Dosing was performed once daily beginning 1 h ± 15 min postchallenge for 8 days (8 total doses). Viral titer was determined by taking the average of three randomly selected vials determined via a plaque assay on Vero E6 cells. Analysis of sterility, mycoplasma, and endotoxin found that all were negative. Viral load in the serum was determined by reverse transcription-quantitative PCR (qRT-PCR) and standard plaque assay as described previously (12). Briefly, serum samples were removed from frozen storage, thawed, and serially diluted in filtered sterilized dilution medium (minimal essential medium/1% heat-inactivated fetal bovine serum/1% penicillin-streptomycin) for analysis. Plaque assays were conducted on serum dilutions (12). Because of volume limitations, neat serum was not analyzed. For qRT-PCR, samples were processed for RNA extraction and purification using the Zymo Direct-zol RNA mini prep kit for viral RNA extraction and purification and were analyzed via quantitative RT-PCR in conjunction with real-time qRT-PCR of filoviruses using TaqMan chemistry and the Bio-Rad CFX96 RT-PCR detection system (12). For quantification purposes, an HPLC-purified synthetic EBOV RNA standard derived from the conserved EBOV glycoprotein gene was used.
Only 10% of challenged vehicle control mice survived until SD7, as previously published (12), with one negative-control animal surviving until the end of the study (Fig. 2A). Ninety percent of the mice in the positive-control tilorone-treated group (25 mg/kg) lived to the end of the study, as previously published (12). The highest-dose group of quinacrine (50 mg/kg) was associated with only 20% survival, whereas the 25-mg/kg group was associated with 70% survival, a statistically significant increase over the vehicle-treated group (Fig. 2A). The average clinical scores of the quinacrine-treated groups followed a similar pattern to that of the vehicle group, with an increase in the scores starting at SD4 and returning to a prechallenge score by SD10. Interestingly, the average clinical score in the tilorone positive-control group had a mitigated increase in this time frame (Fig. 2B). The mean body weights of the quinacrine-25-mg/kg- and tilorone-treated groups returned to prechallenge values by the end of the study (Fig. 2C). The weights in the quinacrine-50-mg/kg-treated group appeared to be higher than those of the quinacrine-25-mg/kg- and tilorone-treated groups, but this was not statistically significant and was likely an artifact of the lower survival rate in this group. There was no detectable viremia in the serum of any of the mice sacrificed at the end of the study according to plaque assay or qRT-PCR (Fig. 2D and E). There was a statistically significant reduction in the geometric mean of viable EBOV virus in serum samples of both quinacrine groups compared with those of the vehicle group as quantified via plaque assay (Dunnett’s multiple-comparison test; quinacrine 25 and 50 mg/kg with adjusted P values of 0.093 and 0.0016, respectively) (Fig. 2F). However, comparison of the viral loads of mice sacrificed because of clinical score using qRT-PCR (Fig. 2G) did not show a statistically significant reduction in the quinacrine-treated groups (Dunnett’s multiple-comparison test) (Fig. 2G).
FIG 2.
Efficacy of quinacrine against maEBOV in the mouse model. (A) The survival curves for the i.p.-dosed positive-control tilorone (25 mg/kg) and quinacrine test articles (25 and 50 mg/kg) were compared with that of vehicle control. The quinacrine 25-mg/kg and tilorone 25-mg/kg survival curves were statistically significantly different from vehicle using a log-rank (Mantel-Cox) test (P = 0.0040 and 0.0004, respectively). (B) Mean clinical scoring results with overlaid percent survival. (C) Mean body weight results. (D) Plaque assay for viable EBOV in serum samples by group (mice sacrificed at the end of study). (E) qRT-PCR measurement of viral RNA in serum samples by group (mice sacrificed at the end of study). (F) Plaque assay for viable EBOV in serum samples (mice sacrificed based on clinical score). Quinacrine 25- and 50-mg/kg doses were statistically significantly decreased compared with vehicle (Dunnett’s multiple-comparison test; adjusted P = 0.093 and 0.0016, respectively). (G) qRT-PCR measurement of viral RNA in serum samples (mice sacrificed based on clinical score). Results were not statistically significantly different from vehicle (Dunnett’s multiple-comparison test).
Despite quinacrine’s early public use predating FDA approval, there are still considerable in vitro and in vivo data available, but data on in vitro ADME are limited. Prior in vivo pharmacokinetics and toxicity data tended to agree with our in vitro data (see Text S2 in the supplemental material) and provided a basis for EBOV efficacy studies in mice. The higher dose did not result in statistically significant survival, but it did reduce the viable EBOV in mice sacrificed midstudy because of high clinical scores. The low survival rate may indicate potential toxicity at higher doses when delivered by this route, pointing to the potential in future studies of lower doses or use of the oral route. Based on a 2016 publication (19), the interspecies allometric scaling for dose conversion from animal to human studies equates a 25-mg/kg mouse i.p. quinacrine dose equivalent to ∼2.025 mg/kg in humans (∼121.5 mg for an average person, which is in line with previous dosing as an antimalarial).
While quinacrine is a potent antiprotozoal, the mechanism of action has not been conclusively determined (see Text S3 in the supplemental material). Quinacrine has also been shown to have antiviral activity in vitro against the dengue virus (DENV2) and Zika virus, with EC50 values of 7.09 ± 9.18 and 2.27 ± 0.14 μM, respectively (20). In vivo data show significant inhibition of vaccinia virus in mice treated up to 8 days before challenge (21). The cell entry pathway of viruses of the Flavivirus genus (including dengue virus) (22), Zika virus (23), vaccinia virus (24), and EBOV (25, 26) involves fusion of the viral membrane with the endosomal membrane in late endosomes and lysosomes, which are highly acidic environments. Quinacrine and many other structurally similar antimalarials are lysosomotropic amines (see Text S4 in the supplemental material). These compounds can diffuse freely and rapidly across the membranes of acidic cytoplasmic organelles in their unprotonated form; however, once they enter the acidic environment, they become protonated, which halts free diffusion out, causing substantial accumulation in these organelles (27). Raising the pH in these organelles likely mitigates the enzymatic activity necessary for viral entry, suggesting a possible mechanism. Specifically, EBOV entry requires cathepsin B activity (28, 29) to prime the viral glycoprotein (30), and its activity is highly pH dependent (31).
In summary, the in vivo EBOV efficacy data for quinacrine combined with prior exposure to millions of patients demonstrate that it would make a viable candidate for repurposing and clinical testing for Ebola virus disease alongside the current drugs being evaluated under compassionate use in Africa.
Supplementary Material
ACKNOWLEDGMENTS
We thank the technical staff at UTMB for performing the high containment studies, including Terry Juelich, Birte Kalveram, Shane Massey, David Perez, Jennifer Smith, Rose Wanjala, and Lihong Zhang. We gratefully acknowledge Dan Contoit and the team at Bioduro, as well as discussions with Mary Lingerfelt, Manu Anantpadma, Robert Davey, and Joel Freundlich. Mindy Davis is gratefully acknowledged for assistance with the NIAID virus screening capabilities (task order no. B22).
This project has been funded in whole or in part by federal funds from the Division of Microbiology and Infectious Diseases, NIAID, National Institutes of Health (NIH), Department of Health and Human Services, under contract no. HHSN272201000040I, task order HHSN27200011 (UTMB primary investigator, David Beasley). We kindly acknowledge NIH funding from the National Center for Advancing Translational Sciences (R21TR001718, to S.E.).
T.R.L. is an employee at Collaborations Pharmaceuticals, Inc. S.E. is the CEO of Collaborations Pharmaceuticals, Inc.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01142-19.
REFERENCES
- 1.Sissoko D, Laouenan C, Folkesson E, M’Lebing A-B, Beavogui A-H, Baize S, Camara A-M, Maes P, Shepherd S, Danel C, Carazo S, Conde MN, Gala J-L, Colin G, Savini H, Bore JA, Le Marcis F, Koundouno FR, Petitjean F, Lamah M-C, Diederich S, Tounkara A, Poelart G, Berbain E, Dindart J-M, Duraffour S, Lefevre A, Leno T, Peyrouset O, Irenge L, Bangoura NF, Palich R, Hinzmann J, Kraus A, Barry TS, Berette S, Bongono A, Camara MS, Chanfreau Munoz V, Doumbouya L, Souley H, Kighoma PM, Koundouno FR, Réné L, Loua CM, Massala V, Moumouni K, Provost C, Samake N, Sekou C, Soumah A, Arnould I, Komano MS, Gustin L, Berutto C, Camara D, Camara FS, Colpaert J, Delamou L, Jansson L, Kourouma E, Loua M, Malme K, Manfrin E, Maomou A, Milinouno A, Ombelet S, Sidiboun AY, Verreckt I, Yombouno P, Bocquin A, Carbonnelle C, Carmoi T, Frange P, Mely S, Nguyen V-K, Pannetier D, Taburet A-M, Treluyer J-M, Kolie J, Moh R, Gonzalez MC, Kuisma E, Liedigk B, Ngabo D, Rudolf M, Thom R, Kerber R, Gabriel M, Di Caro A, Wölfel R, Badir J, Bentahir M, Deccache Y, Dumont C, Durant J-F, El Bakkouri K, Gasasira Uwamahoro M, Smits B, Toufik N, Van Cauwenberghe S, Ezzedine K, Dortenzio E, Pizarro L, Etienne A, Guedj J, Fizet A, Barte de Sainte Fare E, Murgue B, Tran-Minh T, Rapp C, Piguet P, Poncin M, Draguez B, Allaford Duverger T, Barbe S, Baret G, Defourny I, Carroll M, Raoul H, Augier A, Eholie SP, Yazdanpanah Y, Levy-Marchal C, Antierrens A, Van Herp M, Günther S, de Lamballerie X, Keïta S, Mentre F, Anglaret X, Malvy D, JIKI Study Group. 2016. Experimental treatment with favipiravir for Ebola virus disease (the JIKI trial): a historically controlled, single-arm proof-of-concept trial in Guinea. PLoS Med 13:e1001967. doi: 10.1371/journal.pmed.1001967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Warren TK, Jordan R, Lo MK, Ray AS, Mackman RL, Soloveva V, Siegel D, Perron M, Bannister R, Hui HC, Larson N, Strickley R, Wells J, Stuthman KS, Van Tongeren SA, Garza NL, Donnelly G, Shurtleff AC, Retterer CJ, Gharaibeh D, Zamani R, Kenny T, Eaton BP, Grimes E, Welch LS, Gomba L, Wilhelmsen CL, Nichols DK, Nuss JE, Nagle ER, Kugelman JR, Palacios G, Doerffler E, Neville S, Carra E, Clarke MO, Zhang L, Lew W, Ross B, Wang Q, Chun K, Wolfe L, Babusis D, Park Y, Stray KM, Trancheva I, Feng JY, Barauskas O, Xu Y, Wong P, Braun MR, Flint M, McMullan LK, Chen SS, Fearns R, Swaminathan S, Mayers DL, Spiropoulou CF, Lee WA, Nichol ST, Cihlar T, Bavari S. 2016. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature 531:381–385. doi: 10.1038/nature17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Corti D, Misasi J, Mulangu S, Stanley DA, Kanekiyo M, Wollen S, Ploquin A, Doria-Rose NA, Staupe RP, Bailey M, Shi W, Choe M, Marcus H, Thompson EA, Cagigi A, Silacci C, Fernandez-Rodriguez B, Perez L, Sallusto F, Vanzetta F, Agatic G, Cameroni E, Kisalu N, Gordon I, Ledgerwood JE, Mascola JR, Graham BS, Muyembe-Tamfun JJ, Trefry JC, Lanzavecchia A, Sullivan NJ. 2016. Protective monotherapy against lethal Ebola virus infection by a potently neutralizing antibody. Science 351:1339–1342. doi: 10.1126/science.aad5224. [DOI] [PubMed] [Google Scholar]
- 4.Qiu X, Wong G, Audet J, Bello A, Fernando L, Alimonti JB, Fausther-Bovendo H, Wei H, Aviles J, Hiatt E, Johnson A, Morton J, Swope K, Bohorov O, Bohorova N, Goodman C, Kim D, Pauly MH, Velasco J, Pettitt J, Olinger GG, Whaley K, Xu B, Strong JE, Zeitlin L, Kobinger GP. 2014. Reversion of advanced Ebola virus disease in nonhuman primates with ZMapp. Nature 514:47–53. doi: 10.1038/nature13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sivapalasingam S, Kamal M, Slim R, Hosain R, Shao W, Stoltz R, Yen J, Pologe LG, Cao Y, Partridge M, Sumner G, Lipsich L. 2018. Safety, pharmacokinetics, and immunogenicity of a co-formulated cocktail of three human monoclonal antibodies targeting Ebola virus glycoprotein in healthy adults: a randomised, first-in-human phase 1 study. Lancet Infect Dis 18:884–893. doi: 10.1016/S1473-3099(18)30397-9. [DOI] [PubMed] [Google Scholar]
- 6.Hernandez HW, Soeung M, Zorn KM, Ashoura N, Mottin M, Andrade CH, Caffrey CR, de Siqueira-Neto JL, Ekins S. 2018. High throughput and computational repurposing for neglected diseases. Pharm Res 36:27. doi: 10.1007/s11095-018-2558-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baker NC, Ekins S, Williams AJ, Tropsha A. 2018. A bibliometric review of drug repurposing. Drug Discov Today 23:661–672. doi: 10.1016/j.drudis.2018.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ekins S, Freundlich JS, Clark AM, Anantpadma M, Davey RA, Madrid P. 2015. Machine learning models identify molecules active against the Ebola virus in vitro. F1000Res 4:1091. doi: 10.12688/f1000research.7217.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anantpadma M, Lane T, Zorn KM, Lingerfelt MA, Clark AM, Freundlich JS, Davey RA, Madrid PB, Ekins S. 2019. Ebola virus Bayesian machine learning models enable new in vitro leads. ACS Omega 4:2353–2361. doi: 10.1021/acsomega.8b02948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Desmyter J, Melnick JL, Rawls WE. 1968. Defectiveness of interferon production and of rubella virus interference in a line of African green monkey kidney cells (Vero). J Virol 2:955–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Emeny JM, Morgan MJ. 1979. Regulation of the interferon system: evidence that Vero cells have a genetic defect in interferon production. J Gen Virol 43:247–252. doi: 10.1099/0022-1317-43-1-247. [DOI] [PubMed] [Google Scholar]
- 12.Ekins S, Lingerfelt MA, Comer JE, Freiberg AN, Mirsalis JC, O'Loughlin K, Harutyunyan A, McFarlane C, Green CE, Madrid PB. 2017. Efficacy of tilorone dihydrochloride against Ebola virus infection. Antimicrob Agents Chemother 62:e01711-17. doi: 10.1128/AAC.01711-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stoner CL, Gifford E, Stankovic C, Lepsy CS, Brodfuehrer J, Prasad JV, Surendran N. 2004. Implementation of an ADME enabling selection and visualization tool for drug discovery. J Pharm Sci 93:1131–1141. doi: 10.1002/jps.20020. [DOI] [PubMed] [Google Scholar]
- 14.Ansede JH, Thakker DR. 2004. High-throughput screening for stability and inhibitory activity of compounds toward cytochrome P450-mediated metabolism. J Pharm Sci 93:239–255. doi: 10.1002/jps.10545. [DOI] [PubMed] [Google Scholar]
- 15.Di L, Kerns EH, Hong Y, Kleintop TA, McConnell OJ, Huryn DM. 2003. Optimization of a higher throughput microsomal stability screening assay for profiling drug discovery candidates. J Biomol Screen 8:453–462. doi: 10.1177/1087057103255988. [DOI] [PubMed] [Google Scholar]
- 16.Stoner CL, Cleton A, Johnson K, Oh DM, Hallak H, Brodfuehrer J, Surendran N, Han HK. 2004. Integrated oral bioavailability projection using in vitro screening data as a selection tool in drug discovery. Int J Pharm 269:241–249. doi: 10.1016/j.ijpharm.2003.09.006. [DOI] [PubMed] [Google Scholar]
- 17.Andersson TB, Bredberg E, Ericsson H, Sjoberg H. 2004. An evaluation of the in vitro metabolism data for predicting the clearance and drug-drug interaction potential of CYP2C9 substrates. Drug Metab Dispos 32:715–721. doi: 10.1124/dmd.32.7.715. [DOI] [PubMed] [Google Scholar]
- 18.National Research Council. 2011. Guide for the care and use of laboratory animals. National Academies Press, Washington, DC. doi: 10.17226/12910. [DOI] [Google Scholar]
- 19.Nair AB, Jacob S. 2016. A simple practice guide for dose conversion between animals and human. J Basic Clin Pharm 7:27–31. doi: 10.4103/0976-0105.177703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Balasubramanian A, Teramoto T, Kulkarni AA, Bhattacharjee AK, Padmanabhan R. 2017. Antiviral activities of selected antimalarials against dengue virus type 2 and Zika virus. Antiviral Res 137:141–150. doi: 10.1016/j.antiviral.2016.11.015. [DOI] [PubMed] [Google Scholar]
- 21.Glaz ET, Szolgay E, Stoger I, Talas M. 1973. Antiviral activity and induction of interferon-like substance by quinacrine and acranil. Antimicrob Agents Chemother 3:537–541. doi: 10.1128/aac.3.5.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mukhopadhyay S, Kuhn RJ, Rossmann MG. 2005. A structural perspective of the flavivirus life cycle. Nat Rev Microbiol 3:13–22. doi: 10.1038/nrmicro1067. [DOI] [PubMed] [Google Scholar]
- 23.Agrelli A, de Moura RR, Crovella S, Brandão LAC. 2019. Zika virus entry mechanisms in human cells. Infect Genet Evol 69:22–29. doi: 10.1016/j.meegid.2019.01.018. [DOI] [PubMed] [Google Scholar]
- 24.Townsley AC, Weisberg AS, Wagenaar TR, Moss B. 2006. Vaccinia virus entry into cells via a low-pH-dependent endosomal pathway. J Virol 80:8899–8908. doi: 10.1128/JVI.01053-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hunt CL, Lennemann NJ, Maury W. 2012. Filovirus entry: a novelty in the viral fusion world. Viruses 4:258–275. doi: 10.3390/v4020258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aman MJ. 2016. Chasing Ebola through the endosomal labyrinth. mBio 7:e00346. doi: 10.1128/mBio.00346-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin RE, Marchetti RV, Cowan AI, Howitt SM, Broer S, Kirk K. 2009. Chloroquine transport via the malaria parasite's chloroquine resistance transporter. Science 325:1680–1682. doi: 10.1126/science.1175667. [DOI] [PubMed] [Google Scholar]
- 28.Dube D, Schornberg KL, Shoemaker CJ, Delos SE, Stantchev TS, Clouse KA, Broder CC, White JM. 2010. Cell adhesion-dependent membrane trafficking of a binding partner for the ebolavirus glycoprotein is a determinant of viral entry. Proc Natl Acad Sci U S A 107:16637–16642. doi: 10.1073/pnas.1008509107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee JE, Fusco ML, Hessell AJ, Oswald WB, Burton DR, Saphire EO. 2008. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature 454:177–182. doi: 10.1038/nature07082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller EH, Obernosterer G, Raaben M, Herbert AS, Deffieu MS, Krishnan A, Ndungo E, Sandesara RG, Carette JE, Kuehne AI, Ruthel G, Pfeffer SR, Dye JM, Whelan SP, Brummelkamp TR, Chandran K. 2012. Ebola virus entry requires the host-programmed recognition of an intracellular receptor. EMBO J 31:1947–1960. doi: 10.1038/emboj.2012.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Werle B, Julke B, Lah T, Spiess E, Ebert W. 1997. Cathepsin B fraction active at physiological pH of 7.5 is of prognostic significance in squamous cell carcinoma of human lung. Br J Cancer 75:1137–1143. doi: 10.1038/bjc.1997.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.