Antibiotic tolerance, the ability to temporarily sustain viability in the presence of bactericidal antibiotics, constitutes an understudied and yet potentially widespread cause of antibiotic treatment failure. We have previously shown that the Gram-negative pathogen Vibrio cholerae can tolerate exposure to the typically bactericidal β-lactam antibiotics by assuming a spherical morphotype devoid of detectable cell wall material. However, it is unclear how widespread β-lactam tolerance is.
KEYWORDS: L-form, antibiotic, carbapenem, carbapenemase, meropenem, tolerance
ABSTRACT
Antibiotic tolerance, the ability to temporarily sustain viability in the presence of bactericidal antibiotics, constitutes an understudied and yet potentially widespread cause of antibiotic treatment failure. We have previously shown that the Gram-negative pathogen Vibrio cholerae can tolerate exposure to the typically bactericidal β-lactam antibiotics by assuming a spherical morphotype devoid of detectable cell wall material. However, it is unclear how widespread β-lactam tolerance is. Here, we tested a panel of clinically significant Gram-negative pathogens for their response to the potent, broad-spectrum carbapenem antibiotic meropenem. We show that clinical isolates of Enterobacter cloacae, Klebsiella aerogenes, and Klebsiella pneumoniae, but not Escherichia coli, exhibited moderate to high levels of tolerance of meropenem, both in laboratory growth medium and in human serum. Importantly, tolerance was mediated by cell wall-deficient spheroplasts, which readily recovered wild-type morphology and growth upon removal of antibiotic. Our results suggest that carbapenem tolerance is prevalent in clinically significant bacterial species, and we suggest that this could contribute to treatment failure associated with these organisms.
INTRODUCTION
Antibiotics are often differentiated by their ability either to inhibit bacterial growth (bacteriostatic) or to kill bacteria (bactericidal). The exact differentiation of antibiotics into these broad categories likely depends on the species and on the specific growth environment in which antibiotic susceptibility is tested (1, 2). To optimize therapy, it is essential to gain a comprehensive understanding of the various factors that modulate bacterial susceptibility to antibiotics. For example, the β-lactams (penicillins, cephalosporins, cephamycins, carbapenems, and the monobactam aztreonam), which are among the most powerful agents in our antibiotic armamentarium, prevent and/or corrupt proper cell wall (peptidoglycan) assembly (3, 4) by inhibiting the transpeptidation reaction catalyzed by penicillin-binding proteins (PBPs). Consequently, these agents typically induce cell death and lysis in susceptible bacteria, at least during rapid growth in vitro (3, 4). However, failure to eradicate infections due to organisms that are susceptible (i.e., nonresistant) to β-lactams with these agents has been described previously (5–7). This paradox can, in part, be explained by the presence of dormant persister cells, a small subpopulation that resists killing by antibiotics that require cellular activity for their lethal action (8–10). However, specimens obtained from patients treated with β-lactam antibiotics have been reported to contain spheroplasts, i.e., bacterial cells that lack a cell wall (11), and clinical isolates are often highly tolerant of β-lactam antibiotics at frequencies that cannot be explained solely by invoking the presence of rare persister cells (4, 5, 12). Spheroplast formation suggests that, in these bacteria, the antibiotic is effective in inhibiting cell wall synthesis, demonstrating that some bacteria survive antibiotic exposure in forms that are neither dormant nor resistant. We and others have previously shown that two important Gram-negative pathogens, Vibrio cholerae and Pseudomonas aeruginosa, form viable, nondividing spheroplasts under conditions of exposure to inhibitors of cell wall synthesis (12–14). Spheroplasts readily revert to wild-type rod shape and growth upon removal of antibiotic, suggesting that these cells might promote reinfection upon discontinuation of antibiotic therapy. Successful recovery of V. cholerae spheroplasts requires the cell wall stress-sensing two-component system VxrAB (also known as WigKR [15, 16]), cell wall synthesis functions, and the general cell envelope stress-sensing alternative sigma factor RpoE (14).
Spheroplast formation is reminiscent of that seen with so-called Gram-positive “L-forms,” which are irregularly dividing, cell wall-deficient cells surrounded only by their cytoplasmic membranes (17, 18). However, in striking contrast to L-forms, Gram-negative spheroplasts do not divide in the presence of antibiotic (12, 13). Division through an L-form-like mechanism is likely prevented by the presence of their strong outer membrane (OM), which exhibits almost cell wall-like mechanical properties (19). Indeed, dividing L-forms of the model Gram-negative organism Escherichia coli can be generated by inhibiting cell wall synthesis in osmostabilized growth medium, which causes the cytoplasm to “escape” its OM “shell” (20).
While the development of L-forms has been described previously as a mechanism of antibiotic resistance in Gram-positive bacteria (17, 18), it is unclear whether spheroplast formation represents a general strategy elicited by Gram-negative bacteria to tolerate cell wall synthesis inhibitors such as the β-lactams. Here, we tested a collection of well-characterized American Type Culture Collection (ATCC) and clinical Gram-negative isolates (Table 1) for their ability to tolerate exposure to the carbapenem antibiotic meropenem. We found that all of the isolates, with the notable exception of E. coli, formed cell wall-deficient spheroplasts upon exposure to meropenem and that these spheroplasts were able to fully recover to rod shape and growth upon removal of meropenem, both in a laboratory medium and in human serum. Our data suggest that spheroplast-mediated carbapenem tolerance was prevalent in clinically significant Gram-negative pathogens but was rare or absent in the E. coli isolates tested herein. Our results suggest that measurement of antibiotic susceptibility and, ultimately, treatment outcome could include more-nuanced responses, such as tolerance, in Gram-negative pathogens.
TABLE 1.
Bacterial isolates evaluated in this studya
| Isolate | Carbapenemase | Specimen source |
MHA MIC value (μg/ml) |
MHA MIC value interpretatione |
BHI+ agar MIC value (μg/ml) |
BHI+ agar MIC value interpretationf |
|---|---|---|---|---|---|---|
| E. cloacae complex ARB0008b | NA | NA | 1 | SUS | 1.5 | NA |
| E. cloacae complex ATCC 13047 | NA | NA | 0.064 | SUS | 0.094 | NA |
| E. cloacae complex WCM0001 | NA | Blood | 0.023 | SUS | 0.023 | NA |
| E. coli WCM0001 | NA | Urine | 0.016 | SUS | 0.016 | NA |
| E. coli TUV93-0 | NA | NA | 0.023 | SUS | 0.023 | NA |
| K. aerogenes ARB0007 | NA | NA | 0.064 | SUS | 0.047 | NA |
| K. aerogenes WCM0001 | NA | Respiratory/sinus | 0.032 | SUS | 0.032 | NA |
| K. pneumoniae WCM0001 | NA | Blood | 0.047 | SUS | 0.032 | NA |
| K. pneumoniae WCM0002 | NA | Respiratory/sinus | 0.032 | SUS | 0.064 | NA |
| P. aeruginosa PA14 | NA | NA | 0.25 | SUS | 0.25 | NA |
| V. cholerae N16961 | NA | NA | 0.125 | SUS | 0.125 | NA |
| E. cloacae complex 41952 | KPC | NA | >32 | RES | >32 | NA |
| E. coli 52862 | KPC | NA | >32 | RES | 32 | NA |
| K. aerogenes 28944 | KPC | NA | >32 | RES | >32 | NA |
| K. pneumoniae ARB0120c | KPC | NA | 16 | RES | 16 | NA |
| P. aeruginosa ARB0090d | KPC | NA | >32 | RES | >32 | NA |
ARB, Centers for Disease Control and Prevention and U.S. Food and Drug Administration Antibiotic Resistance Isolate Bank; ATCC, American Type Culture Collection; BHI+, brain heart infusion agar with supplements; MHA, Mueller-Hinton agar; NA, not applicable; RES, resistant; SUS, susceptible; WCM, Weill Cornell Medicine.
For ARB0008, the meropenem MIC value determined by the ARB is 2 μg/ml (intermediate). The MIC value obtained using gradient diffusion on BHI+ agar is consistent with an interpretation of intermediate resistance.
For ARB0120, the meropenem MIC value determined by the ARB is >8 μg/ml (resistant).
For ARB0090, the meropenem MIC value determined by the ARB is >8 μg/ml (resistant).
Data were interpreted using the Clinical and Laboratories Standards Institute M100 and M45 documents.
There are no interpretative criteria for antibiotic susceptibility testing performed on BHI+ agar. Nonetheless, the essential agreement (where MIC values are ±1 doubling dilution) between the results of testing performed on MHA and BHI+ agar was 100% for all non-carbapenemase-producing isolates.
RESULTS
Tolerance of meropenem varies across Gram-negative clinical isolates.
Spheroplast-mediated β-lactam tolerance might represent an underappreciated menace in the clinical setting. To test how widespread the ability to tolerate cell wall-acting antibiotics is in clinical isolates, we assayed a panel of isolates recovered from clinical specimens representative of significant Gram-negative pathogens of the family Enterobacteriaceae: E. coli, including enterohemorrhagic E. coli (EHEC); Enterobacter cloacae; Klebsiella aerogenes (formerly Enterobacter aerogenes); and Klebsiella pneumoniae. We also tested organisms known to form spheroplasts under some conditions, namely, V. cholerae and P. aeruginosa. We used the carbapenem meropenem as a representative β-lactam. We chose meropenem due to its importance as a potent, broad-spectrum agent (21, 22) and also because in clinical practice, especially in the setting of multidrug resistance, it is often used against members of our isolate panel (23).
We conducted time-dependent killing experiments measuring both CFU (in counts per milliliter) and optical density at 600 nm (OD600). Killing experiments for all isolates were conducted in supplemented brain heart infusion (BHI+) broth under high-inoculum/slow-growth conditions (see Materials and Methods for details) to emulate the slow-growth behavior that is likely to occur during an infection (24). We chose a meropenem concentration (10 μg/ml) that is above the meropenem resistance breakpoint for Enterobacteriaceae (≥4 μg/ml), P. aeruginosa (≥8 μg/ml), and V. cholerae (≥4 μg/ml) (25, 26) and is between 6.7 × and 625 × higher than the MIC observed for each susceptible/nonresistant, non-carbapenemase-producing isolate on either Mueller-Hinton agar (MHA) or BHI+ medium (Table 1). Crucially, antibiotic susceptibility testing (AST) revealed that the meropenem MIC values did not differ significantly between media, e.g., MHA, recommended for AST by the Clinical and Laboratories Standards Institute (CLSI) (25, 26), and BHI+ agar. The essential agreement (EA) (27) between meropenem MIC values on both media was 100% for all non-carbapenemase-producing isolates included in the study (Table 1), suggesting that AST performed with BHI+ agar is comparable with standardized methods. Furthermore, the EA for meropenem quality control AST with E. coli ATCC 25922 on both MHA and BHI+ agar was 100% and within range (0.008 to 0.064 μg/ml). For comparison to the susceptible/nonresistant strains, we included a panel of conspecific clinical isolates that are carbapenem resistant due to their possession of KPC (Klebsiella pneumoniae carbapenemase).
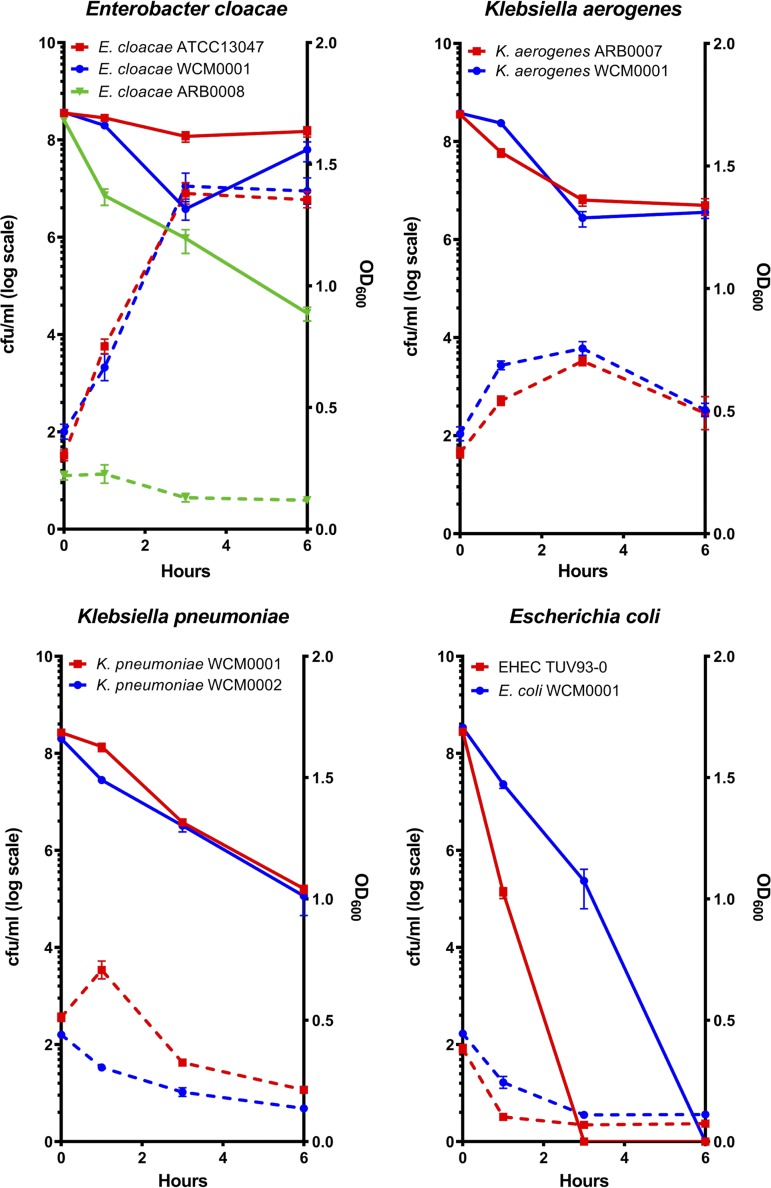
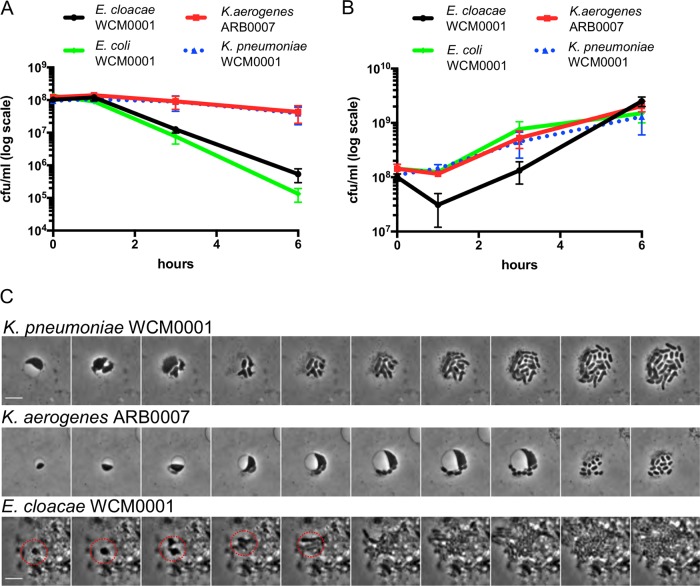
Among the susceptible/nonresistant, non-carbapenemase-producing isolates, killing and optical density dynamics data ranged widely between species and even between isolates within the same species (e.g., E. cloacae WCM0001 versus E. cloacae ARB0008) (Fig. 1). Interestingly, E. coli was considerably less tolerant than all other tested organisms with respect to both lysis behavior and survival (Fig. 1). While killing efficiency after 6 h of meropenem exposure generally ranged from ∼5-fold to ∼10-fold killing (V. cholerae N16961, P. aeruginosa PA14, E. cloacae WCM0001, E. cloacae ATCC 13047) to ∼5,000-fold killing (K. aerogenes WCM0001, E. cloacae ARB0008, K. pneumoniae WCM0001, and WCM0002), both E. coli isolates tested were almost completely eradicated by meropenem (∼108-fold killing) (Fig. 1; see also Fig. S1 and S2 in the supplemental material). In contrast, almost all isolates grew well in the absence of meropenem, except for both isolates of P. aeruginosa, which exhibited slower growth in BHI+ than the other isolates (Fig. S3). Interestingly, E. cloacae ARB0008 had a higher meropenem MIC value than the other E. cloacae isolates but was among the susceptible/nonresistant, non-carbapenemase-producing isolates that were efficiently killed. This observation suggests that levels of susceptibility (i.e., differences in MIC values) might not necessarily correlate with tolerance, i.e., the degree of killing over time.
FIG 1.
Clinical Gram-negative pathogens exhibit various degrees of killing after exposure to meropenem. Overnight cultures of the indicated isolates were subcultured 1:10 (final volume, 5 ml) into prewarmed BHI+ liquid medium supplemented with 10 μg/ml meropenem. The optical density at 600 nm (OD600; dotted lines) and viable cell counts (CFU per milliliter; solid lines) were determined at the indicated time points. Error bars represent standard errors of the means of results from at least six biological replicates.
Similar to the results seen with V. cholerae and P. aeruginosa (Fig. S1 and S2) (13), survival of meropenem-treated cells often coincided with a substantial increase in OD600 (while the CFU count per milliliter stayed the same or decreased) (Fig. 1 and Fig. S3), demonstrating that the surviving cells were not dormant but continued to increase in mass as a population. However, since the CFU count per milliliter stayed the same or decreased, these cells did not divide in the presence of antibiotic. No increases in OD600 were observed in either isolate of E. coli, where a rapid decrease in culture turbidity indicated lysis during meropenem exposure. Thus, an appreciable level of meropenem tolerance (and, potentially, β-lactam tolerance in general) was seen in clinical isolates of some Enterobacteriaceae isolates but not (at least under conditions tested here) in E. coli. In comparison to the susceptible isolates, and as expected, both OD600 levels and CFU counts per milliliter increased in all KPC-positive isolates during meropenem exposure (Fig. S4).
Meropenem-tolerant survivors are cell wall-deficient spheroplasts.
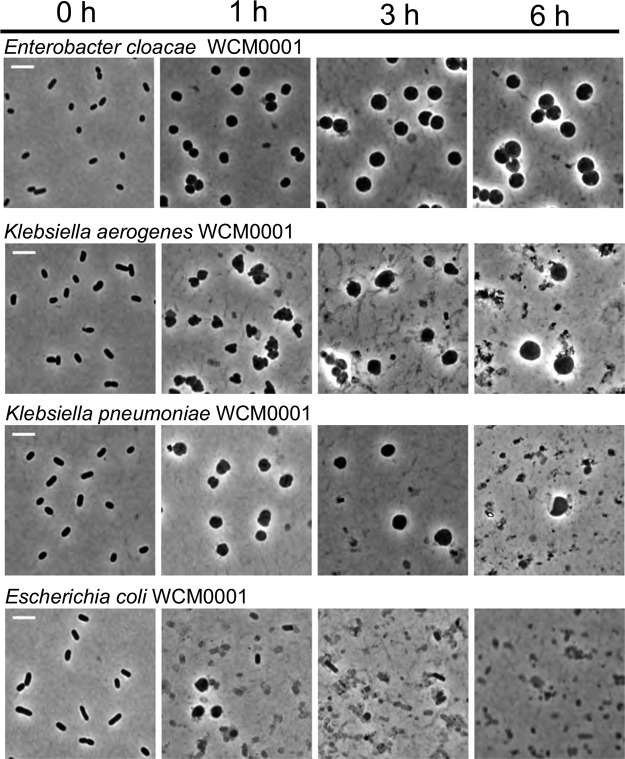
In V. cholerae and P. aeruginosa, β-lactam-tolerant cells are cell wall-deficient, metabolically active spheroplasts. In principle, the moderate to high level of tolerance that we observed in our experiments could also have been a consequence of unusually high levels of dormant persister cells in these clinical isolates due to a prolonged lag phase (28) upon emergence from stationary phase. To distinguish between these two possibilities, we withdrew samples at various time points following exposure to meropenem and imaged them. Dormant persister cells remain rod shaped in the presence of cell wall-acting antibiotics, since these cells prevent antibiotic damage through their lack of growth (29, 30). Visual examination revealed that, comparable to previous observations in V. cholerae and P. aeruginosa (12, 13), the tolerant populations of almost all isolates consisted exclusively of spherical cells (Fig. 2; see also Fig. S1, S2, and S5 to S8). The notable exceptions were the two E. coli isolates; while some spherical cells could be observed after short periods of antibiotic exposure in both tested isolates, only cell debris was observed after 6 h of exposure (Fig. 2; see also Fig. S9). In contrast to those cultures treated with meropenem, untreated bacteria retained their rod shape in BHI+ medium (Fig. S10), similar to the conspecific KPC-positive isolates with or without meropenem treatment (Fig. S11).
FIG 2.
Meropenem exposure induces spheroplast formation in Gram-negative pathogens. Overnight cultures of the indicated isolates were subcultured 1:10 (final volume, 5 ml) into prewarmed BHI+ liquid medium containing 10 μg/ml meropenem and imaged at the indicated time points. Scale bar, 5 μm.
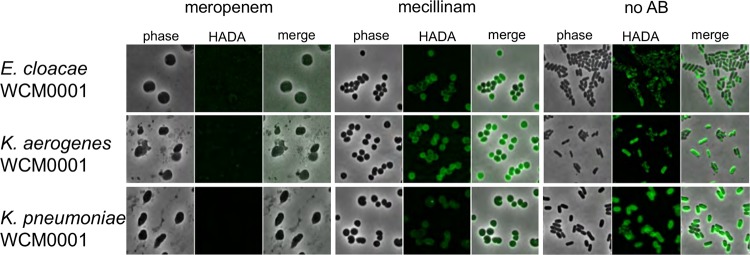
Next, we used the cell wall stain 7-hydroxycoumarin-amino-d-alanine (HADA) (31) to test whether the observed spheroplasts were able to survive meropenem exposure by synthesizing cell wall material in a meropenem-insensitive manner or by sustaining structural integrity in the absence of the cell wall. Indeed, osmotically stable, cell wall-containing spherical cells that resemble spheroplasts can be observed in E. coli when elongation-specific class B PBP2 is inhibited (32). Addition of HADA revealed little to no detectable cell wall material in meropenem-treated cells but, consistent with published data from E. coli, did result in strong staining of PBP2-inhibited (i.e., mecillinam [amdinocillin]-treated) cells (Fig. 3). The lack of detectable cell wall material in meropenem-treated cells, combined with their rapid loss of cell shape, suggests that these spheroplasts maintain structural integrity on the basis of their outer membrane rather than a reorganized cell wall. This is in line with the recent realization that the Gram-negative OM has a higher mechanical load capacity than previously appreciated (19). Finally, loss of the cell wall is typically associated with inhibition of multiple PBPs, including class A PBPs (33). Meropenem has a high affinity for PBP2 (34, 35) but also inhibits a number of other PBPs (PBP1a/b and PBP3) in E. coli and P. aeruginosa (35, 36), and our data suggest, as indicated by the absence of HADA-stained meropenem-treated cells, that multiple PBPs were inactivated at the concentration of meropenem used in our experiments.
FIG 3.
Meropenem-treated spheroplasts have no detectable cell wall material. The indicated isolates were grown in the presence of HADA and treated with vehicle (no antibiotic [no AB]) or meropenem (10 μg/ml) or mecillinam (20 μg/ml). After 6 h, cells were washed and imaged using fluorescence microscopy.
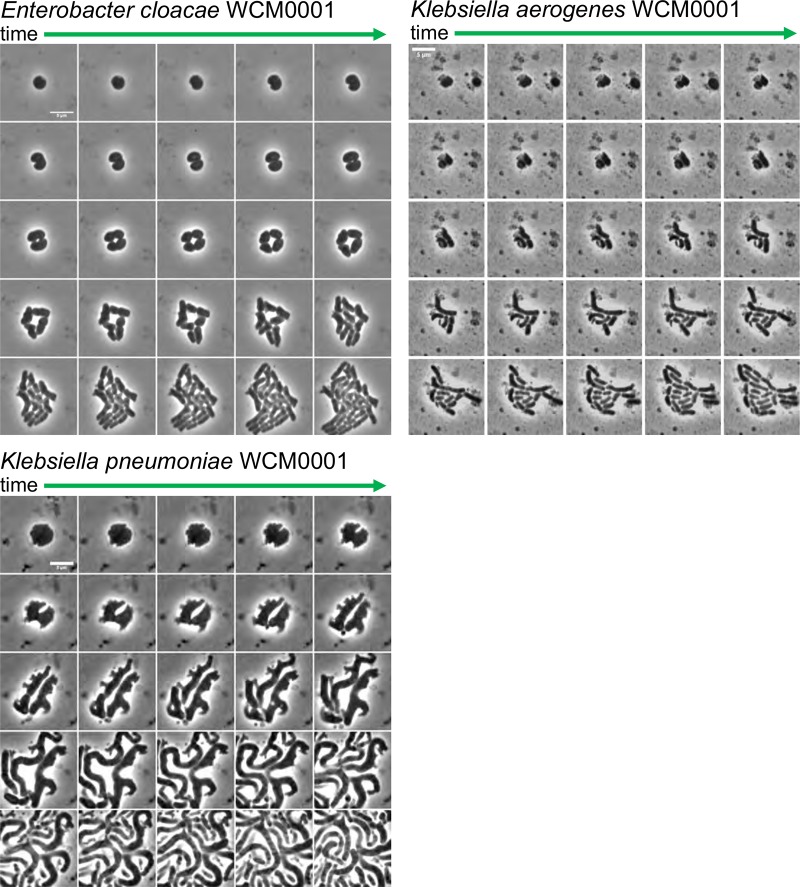
If the observed spheroplasts are truly tolerant, they should readily revert to rod shape (i.e., wild-type shape) and growth upon removal of antibiotic. To test this, we withdrew samples after 6 h of meropenem exposure, removed meropenem by addition of purified New Delhi metallo-β-lactamase-1 (NDM-1) carbapenemase, and imaged these cells using time-lapse microscopy. With various frequencies roughly reflecting the different survival rates, at least some spheroplasts from all isolates were able to recover to rod shape (Fig. 4; see also Fig. S1, S2, and S5 to S8 and Movies S1 to S9), albeit with different dynamics (cf. E. cloacae WCM0001 versus K. aerogenes WCM0001). The recovery process often included rapid division (e.g., 25 min after removing the antibiotic in K. pneumoniae WCM0001) as spherical cells, resulting in two half-spheroplasts that then increasingly approximated rod shape during subsequent division events. Taken together, our results suggest that the high tolerance levels observed for the Gram-negative pathogens tested here were not mediated by dormancy, or by a prolonged lag phase after the stationary phase, but rather by the ability to survive for extended time periods without a structurally sound cell wall.
FIG 4.
Meropenem-induced spheroplasts can recover to form an exponentially growing population. A time-lapse montage of spheroplasts imaged upon removal of meropenem after 6 h of treatment is shown. The antibiotic was removed by addition of purified NDM-1 carbapenemase, followed by time-lapse microscopy on BHI+ agarose pads (0.8% [wt/vol] agarose). Images were then acquired 5 min apart for another 2 h. Both E. coli isolates were omitted since no spheroplasts were observed after 6 h of meropenem treatment. Scale bars, 5 μm.
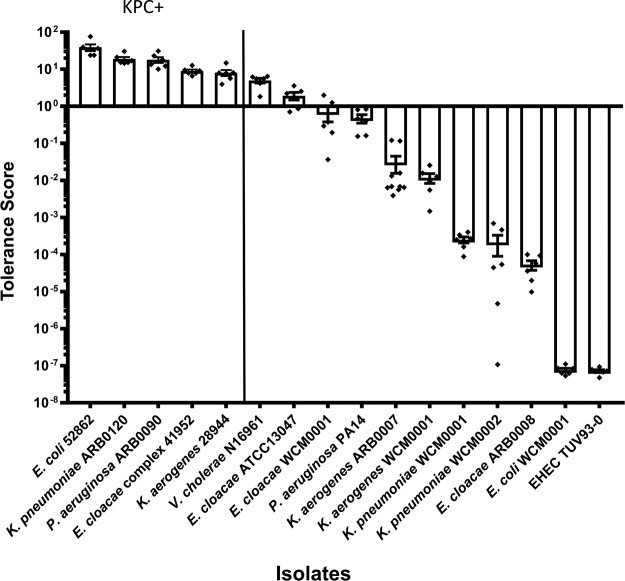
Quantification of Gram-negative tolerance: the tolerance score.
Since our observational data suggested variations in overall tolerance of meropenem among isolates, including differing propensities to form and recover from spheroplasts and different rates of survival, we sought to quantify the ability of this group of Gram-negative pathogens to survive and maintain cellular structural integrity during exposure to meropenem. Other assays designed to determine tolerance levels based on bacterial enumeration or growth on solid media have been developed previously (37, 38); however, given that meropenem treatment also caused an increase in OD600 in our experiments, we chose to incorporate both CFU counts and OD600 measurements in a so-called “tolerance score.” The tolerance score of a given isolate was determined by multiplying the fraction of surviving cells (CFU/ml after 6 h of treatment over initial CFU/ml) by the fraction corresponding to the OD600 measurements (OD600 measurement after 6 h of treatment over initial OD600 measurement) (see Materials and Methods for details). According to this score, V. cholerae N16961, P. aeruginosa PA14, and E. cloacae isolates WCM0001 and ATCC 13047 emerged as the most tolerant of the susceptible/nonresistant, non-carbapenemase-producing organisms, while the K. pneumoniae and K. aerogenes isolates exhibited intermediate tolerance and E. coli ranked the lowest (Fig. 5). Consistent with their ability to grow in the presence of meropenem, the KPC-producing isolates scored the highest.
FIG 5.
Meropenem tolerance scores determined for Gram-negative pathogens. Relative levels of tolerance of meropenem were quantified using a tolerance score equation based on viable cell counts and optical density data determined for broth cultures exposed to 10 μg/ml meropenem for 6 h. The tolerance score was calculated using the following equation: [(ODt6/ODt0) × (CFU/mlt6/CFU/mlt0)]. The resultant values are arranged in descending order (indicating decreasing levels of tolerance of meropenem). Error bars show standards errors of the means of results from at least six biological replicates. Raw data points are also shown.
Spheroplast formation in human serum.
To evaluate tolerance in an environment more reminiscent of growth in the human host, we performed killing experiments in human serum. CFU counts (serum growth medium is incompatible with OD measurements) were determined after 6 h of incubation with or without meropenem (Fig. 6), and cells were observed directly for spheroplast formation. All isolates grew in serum growth medium (Fig. 6B), but compared to the results seen with BHI+ medium, killing by meropenem was reduced for some isolates. Klebsiella aerogenes ARB0007 and K. pneumoniae WCM0001 were almost completely tolerant, with only an ∼5-fold decrease in viability over the 6-h period compared to the 10-fold to 100-fold killing that occurred in BHI+ medium with these isolates. In contrast, E. cloacae WCM0001 was killed at a higher rate in human serum than in BHI+ medium. However, spheroplasts were observed for all isolates, except for E. coli, and recovery to wild-type rod shape morphology and growth upon removal of meropenem by addition of NDM-1 was efficient (Fig. 6C). These data suggest that the degree of tolerance is growth medium and isolate specific (though E. coli was still the least tolerant) but that spheroplast formation as a means of tolerating exposure to meropenem was conserved across the bacteria and conditions tested here.
FIG 6.
Spheroplast formation and recovery in human serum. (A and B) The indicated isolates were grown overnight in serum growth medium (SGM), diluted 1:10 (final volume, 500 μl) into fresh SGM, and incubated in the presence (A) or absence (B) of 10 μg/ml meropenem. Cells were plated (CFU per milliliter) at the indicated time points. (C) After 6 h of incubation, purified NDM-1 carbapenemase was added to remove meropenem, followed by time-lapse microscopy on agarose pads containing 40% (vol/vol) human serum. All values represent means of results from three biological replicates; error bars represent standard errors of the means. The red circle indicates the initial growth steps of a recovering spheroplast within cell debris.
DISCUSSION
In contrast to antibiotic resistance (the ability to grow in the presence of antibiotics), the phenomenon of antibiotic tolerance (the ability to resist killing by bactericidal antibiotics for extended time periods) remains understudied. While much effort has been directed at understanding persister cells (9) (multidrug-tolerant, dormant, or near-dormant phenotypic variants produced as a small fraction of bacterial populations), it is unclear what other strategies exist among bacteria to resist killing by ordinarily bactericidal antibiotics. We and others have previously described a tolerance mechanism (via formation of stable spheroplasts) by which Gram-negative bacteria are able to survive the normally lethal event of cell wall degradation that is caused by exposure to β-lactam antibiotics and other inhibitors of cell wall synthesis (12–15). Cell wall-less Gram-negative spheroplasts are formed by the majority population, are not dormant (i.e., not persisters), and presumably rely on their strong OM to maintain structural integrity (14). Here, we show that this phenomenon is more widespread than previously recognized and is prevalent in members of the family Enterobacteriaceae, a clinically important group of bacterial organisms. Gram-negative clinical isolates that are susceptible/nonresistant to meropenem as determined using conventional MIC-based methods failed to be eradicated at appreciable levels of antibiotic, and surviving cells were spheroplasts devoid of detectable cell wall material. Our data raise the possibility that rapid death and lysis of majority populations after β-lactam therapy might be the exception, not the norm, in clinical practice. Therefore, in addition to persister formation (8–10, 39), heteroresistance (40), and overt resistance, these data highlight a fourth potential mechanism for β-lactam therapy treatment failure.
Importantly, spheroplast-like structures have been isolated from patients infected with Gram-negative pathogens and treated with β-lactam antibiotics (11), implying that these cells are able to survive without a cell wall in the human host. Consistent with this idea, we observed spheroplast formation in our collection of Enterobacteriaceae during exposure to meropenem in human serum growth medium. Therefore, we consider it likely that spheroplasts, similarly to persisters, can be responsible for recalcitrant infections. Indeed, we present the first evidence that carbapenem-induced spheroplasts in clinically significant members of the family Enterobacteriaceae can readily revert to wild-type rod shape and growth upon removal of carbapenem. While Gram-negative spheroplast formation has been noted in previous studies (11, 41), evidence of spheroplast recovery (i.e., reversion to wild-type, rod-shaped, growing cells) was lacking. As such, spheroplast formation has not been incorporated into models of antibiotic susceptibility, particularly in the human host. Future experiments will determine if spheroplasts that are able to revert to a growing population are also observable during infections in patients treated with β-lactam antibiotics and if their formation correlates with treatment outcomes.
In addition to providing a reservoir for a large number of cells that can repopulate an infection after antibiotic therapy is discontinued, a majority population of damaged but viable cells, such as spheroplasts post-antibiotic treatment, poses other health risks. β-Lactam antibiotics have been suggested to induce the generation of reactive oxygen species as well as the SOS DNA damage response and could have mutagenic potential (42, 43). A large reservoir of damaged cells might thus increase the possibility of developing broad resistance to other antibiotics (44, 45). Furthermore, though we did not test this directly, spheroplasts could in principle continue to produce virulence factors (toxins, proteases, and other tissue-damaging enzymes), and thus disease, during antibiotic therapy.
The spheroplasts observed here are reminiscent of Gram-positive L-forms. However, unlike L-forms, Gram-negative spheroplasts fail to replicate in the presence of antibiotics and recover to wild-type rod shape and growth only when the antibiotic is removed. L-form cell division relies on membrane lipid overproduction and subsequent stochastic blebbing (46). We speculate that in Gram-negative spheroplasts, L-form-like proliferation is prevented due to their cytoplasmic membranes being confined by their rigid OMs, thus preventing division through membrane blebs. Interestingly, the recovery dynamics reported for the clinical isolates tested here resemble those observed for osmostabilized E. coli spheroplasts generated by lysozyme treatment (resulting in cell wall degradation) (32). This suggests that spheroplast recovery might be accomplished through a mechanism that is widely conserved among Gram-negative bacteria. It also raises the possibility that E. coli may survive β-lactam exposure in the human host through spheroplast formation under some conditions, if the specific infection environment is osmotically stabilizing.
In summary, our work demonstrates that the ability to survive the presence of bactericidal β-lactam antibiotics does not solely rely on classical resistance or dormancy but instead could also be dependent upon an intrinsic tolerance mechanism in Gram-negative bacteria that are otherwise fully susceptible to the damage induced by cell wall-acting agents. Rather than preventing harmful effects of antibiotics (as in resistance and dormancy), these tolerant spheroplasts survive by circumventing the essentiality of the antibiotic’s main target, the cell wall. Our observations underscore the necessity of studying clinical isolates and non-model organisms to gain a more complete understanding of the complex processes underlying the susceptibility to antibiotics in clinical settings.
MATERIALS AND METHODS
Chemicals, media, and growth conditions.
Meropenem (TCI Chemicals, Portland, OR) was formulated as a 10 mg/ml stock solution in distilled water and stored at −20°C. BHI medium (per liter: 17.5 g brain heart infusion from solids, 10.0 g pancreatic digest of gelatin, 2.0 g dextrose, 5.0 g sodium chloride, 2.5 g disodium phosphate) was purchased from RPI (Wilmington, NC) and prepared as a broth according to package instructions, with 15 g per liter agar added for solid media. All isolates were grown in supplemented BHI (BHI+) medium with added NAD (Sigma-Aldrich, St. Louis, MO) and hemin (Beantown Chemical, Hudson, NH) (both at a final concentration of 15 μg/ml). All isolates were grown overnight in a 37°C shaking incubator prior to initiating the experiment.
Bacterial isolates.
The bacterial isolates are summarized in Table 1. The clinical isolates were identified to the species level or (in the case of E. cloacae) complex level using matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI Biotyper, Bruker Daltonics, Inc., Billerica, MA) according to the manufacturer’s instructions. Meropenem AST (Table 1) was performed for all isolates using gradient diffusion (Etest, bioMérieux, Inc., Durham, NC) according to the manufacturer’s instructions on both MHA and BHI+ agar (i.e., solid media). Each day of testing, quality control testing of the Etest strips was performed on both MHA and BHI+ agar with E. coli ATCC 25922. In all cases, quality control testing passed. The resultant AST data obtained with MHA was interpreted using the CLSI M100 (Enterobacteriaceae and P. aeruginosa) and M45 (V. cholerae) documents (25, 26). The presence of the blaKPC gene was confirmed for the KPC-producing isolates by the use of the Xpert Carba-R assay (Cepheid, Sunnyvale, CA, USA) according to the manufacturer’s instructions.
Microscopy.
All images were taken on a Leica MDi8 microscope (Leica Microsystems, GmbH, Wetzlar, Germany) with a PECON TempController 2000-1 stage (Erbach, Germany) heated at 37°C for growth experiments or maintained at room temperature for static images. Time-lapse microscopy was performed by imaging frames 5 min apart, and data were processed in ImageJ (47). HADA-stained cells were imaged at 365-nm excitation for 1 s of exposure time. Images were minimally processed in ImageJ by subtracting background and adjusting brightness/contrast uniformly across all fluorescent images.
Time-dependent killing experiments.
Overnight cultures of each isolate were grown at 37°C in liquid BHI+ medium and diluted 1:10 the following day in fresh, prewarmed BHI+ medium containing a final concentration of 10 μg/ml meropenem. At each time point, samples were diluted 5-fold in blank medium and OD600 was measured. At the same time point, viable cell counts were also determined by 10-fold serial dilution of cells in BHI+ agar and spot-plating 10 μl of each dilution on BHI+ agar plates. Colonies were counted after 24 h of growth at 30°C. Images were taken by placing cells on BHI+ agarose pads (0.8% [wt/vol] agarose). Cells were concentrated by centrifugation (8,000 × g, 5 min) where necessary.
Tolerance score.
The tolerance score was calculated from measurements of OD600 and CFU/ml after 6 h of exposure to meropenem. The score (representing the OD fold change value multiplied by the survival fraction value) was calculated as follows: (ODt6/ODt0) × (CFU/mlt6/CFU/mlt0) (where t6 represents 6 h of exposure and t0 represents time zero).
Time-dependent killing assays in human serum.
To generate serum growth medium (SGM), human serum (Rockland Pharmaceuticals, Limerick, PA) was thawed on ice and diluted in Dulbecco’s modified Eagle’s medium (DMEM; VWR, Radnor, PA) to 40% (vol/vol). Bacteria were inoculated from frozen stocks into 300 μl of SGM in Eppendorf tubes and incubated at 37°C overnight without agitation. After incubation, cells were diluted 10-fold into 450 μl of fresh SGM, followed by addition of meropenem (10 μg/ml). Survival was measured by diluting and spot-plating for determinations of CFU counts per milliliter at the indicated times. For recovery time-lapse images, cells were concentrated 10-fold (via centrifugation at 8,000 × g for 5 min) and the antibiotic was inactivated by addition of 5 μl of purified NDM-1 (5 mg/ml). Time-lapse images were obtained at 37°C on SGM plus 0.8% (wt/vol) agarose.
HADA staining following antibiotic treatment.
Cultures were grown with shaking at 37°C in BHI+ liquid media and subcultured the next day at 1:10 (final volume, 1 ml) containing 50 μM HADA (31) (7-hydroxycoumarin-amino-d-alanine) with or without meropenem (10 μg/ml). At each time point, 100 μl of the culture was harvested and washed three times with 200 μl BHI+ medium by centrifugation (8,000 × g, 5 min) to remove antibiotic and excess HADA. After the third wash, cells were concentrated 10-fold and imaged on BHI+ agarose pads (0.8% [wt/vol] agarose). Where indicated, HADA staining and imaging were performed as described above after treatment with 20 μg/ml mecillinam (Sigma-Aldrich, St. Louis, MO). Images were analyzed in ImageJ and are minimally processed (background removal).
Purification of New Delhi metallo-β-lactamase-1 (NDM-1).
Isolate E. cloacae ATCC BAA-2468 was used as a template for the PCR amplification of the blaNDM-1 gene. SignalP 4.1 was used to predict the membrane localization signal sequence of NDM-1. PCR primers BR_83 (5′-cagcagcggcctggtgccgcgcggcagccaGTGCATGCCCGGTGAAATCCG-3′) and BR_84 (5′-cagcttcctttcgggctttgttagcagccgCATGGCTCAGCGCAGCTTGTC-3′) (lowercase indicates homology overhangs for isothermal assembly cloning) were designed to amplify the gene without the predicted signal sequence. Following PCR using Q5 DNA polymerase (New England Biolabs, Ipswich, MA) and the BR_83/BR_84 primer pair, the product was cloned into pET-15b N-terminal 6×His expression plasmid (New England Biolabs).
The plasmid was transformed by heat shock into chemically competent E. coli BL21(DE3) cells (New England Biolabs). Using the transformed cells, 1 liter LB medium cultures were grown from single colonies shaking at 37°C. At an OD600 of ∼0.3, cells were induced with 1 mM isopropyl-β-d-thiogalactopyranoside (Sigma-Aldrich, St. Louis, MO) and grown for an additional 3 h at 37°C. Cells were harvested by centrifugation (11,200 × g, 20 min), and the pellets were frozen at −80°C. After lysis by sonication, the protein was found to be insoluble (likely due to its proposed membrane anchor [48, 49]). Insoluble protein pellets were resolubilized in 3 M urea (VWR, Radnor, PA) and purified using immobilized metal affinity chromatography and nickel-nitrilotriacetic acid (Ni-NTA) resin (Qiagen, Hilden, Germany). Eluted proteins were renatured by three-step dialysis to a final buffer composition of 20 mM Tris, 150 mM NaCl, 50 μM ZnSO4, and 30% (vol/vol) glycerol. The resulting protein was quantified by Bradford assay (50) and its functionality verified in a biological assay of the ability of purified NDM-1 to restore growth of meropenem-susceptible E. coli MG1655 on agar containing meropenem (10 μg/ml).
Supplementary Material
ACKNOWLEDGMENTS
Isolates E. cloacae ARB0008, K. aerogenes ARB0007, K. pneumoniae ARB0120, and P. aeruginosa ARB0090 were obtained from the CDC and FDA Antibiotic Resistance Isolate Bank (https://www.cdc.gov/drugresistance/resistance-bank/index.html). Isolates E. cloacae complex 41952, E. coli 52862, and K. aerogenes 28944 were gifts from Barry Kreiswirth (Rutgers University, NJ). We thank Matthew K. Waldor (Harvard Medical School, MA) and Pamela V. Chang (Cornell University, NY) for the gifts of V. cholerae N16961 and E. coli TUV93-0, respectively.
Research in the Dörr laboratory is supported by NIAID grant 1R01AI143704. Research in the VanNieuwenhze laboratory is supported by NIH grant GM113172.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00756-19.
REFERENCES
- 1.Nemeth J, Oesch G, Kuster SP. 2015. Bacteriostatic versus bactericidal antibiotics for patients with serious bacterial infections: systematic review and meta-analysis. J Antimicrob Chemother 70:382–395. doi: 10.1093/jac/dku379. [DOI] [PubMed] [Google Scholar]
- 2.Balouiri M, Sadiki M, Ibnsouda SK. 2016. Methods for in vitro evaluating antimicrobial activity: a review. J Pharm Anal 6:71–79. doi: 10.1016/j.jpha.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cho H, Uehara T, Bernhardt TG. 2014. Beta-lactam antibiotics induce a lethal malfunctioning of the bacterial cell wall synthesis machinery. Cell 159:1300–1311. doi: 10.1016/j.cell.2014.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tomasz A. 1979. From penicillin-binding proteins to the lysis and death of bacteria: a 1979 view. Rev Infect Dis 1:434–467. doi: 10.1093/clinids/1.3.434. [DOI] [PubMed] [Google Scholar]
- 5.Gonzalez BE, Martinez-Aguilar G, Mason EO Jr, Kaplan SL. 2004. Azithromycin compared with beta-lactam antibiotic treatment failures in pneumococcal infections of children. Pediatr Infect Dis J 23:399–405. doi: 10.1097/01.inf.0000122605.34902.49. [DOI] [PubMed] [Google Scholar]
- 6.Lemaitre BC, Mazigh DA, Scavizzi MR. 1991. Failure of beta-lactam antibiotics and marked efficacy of fluoroquinolones in treatment of murine Yersinia pseudotuberculosis infection. Antimicrob Agents Chemother 35:1785–1790. doi: 10.1128/aac.35.9.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patel JA, Reisner B, Vizirinia N, Owen M, Chonmaitree T, Howie V. 1995. Bacteriologic failure of amoxicillin-clavulanate in treatment of acute otitis media caused by nontypeable Haemophilus influenzae. J Pediatr 126:799–806. doi: 10.1016/s0022-3476(95)70415-9. [DOI] [PubMed] [Google Scholar]
- 8.Conlon BP, Rowe SE, Lewis K. 2015. Persister cells in biofilm associated infections. Adv Exp Med Biol 831:1–9. doi: 10.1007/978-3-319-09782-4_1. [DOI] [PubMed] [Google Scholar]
- 9.Lewis K. 2010. Persister cells. Annu Rev Microbiol 64:357–372. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- 10.Brauner A, Fridman O, Gefen O, Balaban NQ. 2016. Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat Rev Microbiol 14:320–330. doi: 10.1038/nrmicro.2016.34. [DOI] [PubMed] [Google Scholar]
- 11.Roberts D, Higgs E, Rutman a, Cole P. 1984. Isolation of spheroplastic forms of Haemophilus influenzae from sputum in conventionally treated chronic bronchial sepsis using selective medium supplemented with N-acetyl-d-glucosamine: possible reservoir for re-emergence of infection. Br Med J 289:1409–1412. doi: 10.1136/bmj.289.6456.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Monahan LG, Turnbull L, Osvath SR, Birch D, Charles IG, Whitchurch CB. 2014. Rapid conversion of Pseudomonas aeruginosa to a spherical cell morphotype facilitates tolerance to carbapenems and penicillins but increases susceptibility to antimicrobial peptides. Antimicrob Agents Chemother 58:1956–1962. doi: 10.1128/AAC.01901-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dörr T, Davis BM, Waldor MK. 2015. Endopeptidase-mediated beta lactam tolerance. PLoS Pathog 11:e1004850. doi: 10.1371/journal.ppat.1004850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weaver AI, Murphy SG, Umans BD, Tallavajhala S, Onyekwere I, Wittels S, Shin JH, VanNieuwenhze M, Waldor MK, Dörr T. 2018. Genetic determinants of penicillin tolerance in Vibrio cholerae. Antimicrob Agents Chemother 62:e01326-18. doi: 10.1128/AAC.01326-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dörr T, Alvarez L, Delgado F, Davis BM, Cava F, Waldor MK. 2016. A cell wall damage response mediated by a sensor kinase/response regulator pair enables beta-lactam tolerance. Proc Natl Acad Sci U S A 113:404–409. doi: 10.1073/pnas.1520333113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng AT, Ottemann KM, Yildiz FH. 2015. Vibrio cholerae response regulator VxrB controls colonization and regulates the type VI secretion system. PLoS Pathog 11:e1004933. doi: 10.1371/journal.ppat.1004933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Errington J, Mickiewicz K, Kawai Y, Wu LJ. 2016. L-form bacteria, chronic diseases and the origins of life. Philos Trans R Soc Lond B Biol Sci 371. doi: 10.1098/rstb.2015.0494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Errington J. 2017. Cell wall-deficient, L-form bacteria in the 21st century: a personal perspective. Biochem Soc Trans 45:287–295. doi: 10.1042/BST20160435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rojas ER, Billings G, Odermatt PD, Auer GK, Zhu L, Miguel A, Chang F, Weibel DB, Theriot JA, Huang KC. 2018. The outer membrane is an essential load-bearing element in Gram-negative bacteria. Nature 559:617–621. doi: 10.1038/s41586-018-0344-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Billings G, Ouzounov N, Ursell T, Desmarais SM, Shaevitz J, Gitai Z, Huang KC. 2014. De novo morphogenesis in L-forms via geometric control of cell growth. Mol Microbiol 93:883–896. doi: 10.1111/mmi.12703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bonomo RA, Burd EM, Conly J, Limbago BM, Poirel L, Segre JA, Westblade LF. 2018. Carbapenemase-producing organisms: a global scourge. Clin Infect Dis 66:1290–1297. doi: 10.1093/cid/cix893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Breilh D, Texier-Maugein J, Allaouchiche B, Saux MC, Boselli E. 2013. Carbapenems. J Chemother 25:1–17. doi: 10.1179/1973947812Y.0000000032. [DOI] [PubMed] [Google Scholar]
- 23.Baldwin CM, Lyseng-Williamson KA, Keam SJ. 2008. Meropenem: a review of its use in the treatment of serious bacterial infections. Drugs 68:803–838. doi: 10.2165/00003495-200868060-00006. [DOI] [PubMed] [Google Scholar]
- 24.Haugan MS, Charbon G, Frimodt-Møller N, Løbner-Olesen A. 2018. Chromosome replication as a measure of bacterial growth rate during Escherichia coli infection in the mouse peritonitis model. Sci Rep 8:14961. doi: 10.1038/s41598-018-33264-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.CLSI. 2019. Performance standards for antimicrobial susceptibility testing, 29th ed CLSI supplement M100 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 26.CLSI. 2015. Methods for antimicrobial dilution and disk susceptibility testing of infrequently isolated or fastidious bacteria, 3rd ed CLSI guideline M45. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 27.Patel JB, Sharp S, Novak-Weekley S. 2013. Verification of antimicrobial susceptibility testing methods: a practical approach. Clin Microbiol Newsl 35:103–109. doi: 10.1016/j.clinmicnews.2013.06.001. [DOI] [Google Scholar]
- 28.Fridman O, Goldberg A, Ronin I, Shoresh N, Balaban NQ. 2014. Optimization of lag time underlies antibiotic tolerance in evolved bacterial populations. Nature 513:418–421. doi: 10.1038/nature13469. [DOI] [PubMed] [Google Scholar]
- 29.Shah D, Zhang Z, Khodursky A, Kaldalu N, Kurg K, Lewis K. 2006. Persisters: a distinct physiological state of E. coli. BMC Microbiol 6:53. doi: 10.1186/1471-2180-6-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. 2004. Bacterial persistence as a phenotypic switch. Science 305:1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- 31.Kuru E, Hughes HV, Brown PJ, Hall E, Tekkam S, Cava F, de Pedro MA, Brun YV, VanNieuwenhze MS. 2012. In situ probing of newly synthesized peptidoglycan in live bacteria with fluorescent d-amino acids. Angew Chem Int Ed Engl 51:12519–12523. doi: 10.1002/anie.201206749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ranjit DK, Young KD. 2013. The Rcs stress response and accessory envelope proteins are required for de novo generation of cell shape in Escherichia coli. J Bacteriol 195:2452–2462. doi: 10.1128/JB.00160-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Satta G, Cornaglia G, Mazzariol A, Golini G, Valisena S, Fontana R. 1995. Target for bacteriostatic and bactericidal activities of beta-lactam antibiotics against Escherichia coli resides in different penicillin-binding proteins. Antimicrob Agents Chemother 39:812–818. doi: 10.1128/aac.39.4.812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhanel GG, Simor AE, Vercaigne L, Mandell L; Canadian Carbapenem Discussion Group. 1998. Imipenem and meropenem: Comparison of in vitro activity, pharmacokinetics, clinical trials and adverse effects. Can J Infect Dis 9:215–228. doi: 10.1155/1998/831425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sumita Y, Fukasawa M. 1995. Potent activity of meropenem against Escherichia coli arising from its simultaneous binding to penicillin-binding proteins 2 and 3. J Antimicrob Chemother 36:53–64. doi: 10.1093/jac/36.1.53. [DOI] [PubMed] [Google Scholar]
- 36.Sumita Y, Tada E, Nouda H, Okuda T, Fukasawa M. 1992. Mode of action of meropenem, a new carbapenem antibiotic. Chemotherapy 40:90–102. doi: 10.1159/000239004. [DOI] [Google Scholar]
- 37.Brauner A, Shoresh N, Fridman O, Balaban NQ. 2017. An experimental framework for quantifying bacterial tolerance. Biophys J 112:2664–2671. doi: 10.1016/j.bpj.2017.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gefen O, Chekol B, Strahilevitz J, Balaban NQ. 2017. TDtest: easy detection of bacterial tolerance and persistence in clinical isolates by a modified disk-diffusion assay. Sci Rep 7:1–9. doi: 10.1038/srep41284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lewis K. 2007. Persister cells, dormancy and infectious disease. Nat Rev Microbiol 5:48–56. doi: 10.1038/nrmicro1557. [DOI] [PubMed] [Google Scholar]
- 40.El-Halfawy OM, Valvano MA. 2015. Antimicrobial heteroresistance: an emerging field in need of clarity. Clin Microbiol Rev 28:191–207. doi: 10.1128/CMR.00058-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Laar TA, Chen T, You T, Leung KP. 2015. Sublethal concentrations of carbapenems alter cell morphology and genomic expression of Klebsiella pneumoniae biofilms. Antimicrob Agents Chemother 59:1707–1717. doi: 10.1128/AAC.04581-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miller C, Thomsen LE, Gaggero C, Mosseri R, Ingmer H, Cohen SN. 2004. SOS response induction by beta-lactams and bacterial defense against antibiotic lethality. Science 305:1629–1631. doi: 10.1126/science.1101630. [DOI] [PubMed] [Google Scholar]
- 43.Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. 2007. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130:797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- 44.Windels EM, Michiels JE, Fauvart M, Wenseleers T, Van den Bergh B, Michiels J. 2019. Bacterial persistence promotes the evolution of antibiotic resistance by increasing survival and mutation rates. ISME J 13:1239–1251. doi: 10.1038/s41396-019-0344-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Levin-Reisman I, Ronin I, Gefen O, Braniss I, Shoresh N, Balaban NQ. 2017. Antibiotic tolerance facilitates the evolution of resistance. Science 355:826–830. doi: 10.1126/science.aaj2191. [DOI] [PubMed] [Google Scholar]
- 46.Mercier R, Kawai Y, Errington J. 2013. Excess membrane synthesis drives a primitive mode of cell proliferation. Cell 152:997–1007. doi: 10.1016/j.cell.2013.01.043. [DOI] [PubMed] [Google Scholar]
- 47.Rueden CT, Schindelin J, Hiner MC, DeZonia BE, Walter AE, Arena ET, Eliceiri KW. 2017. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics 18:529. doi: 10.1186/s12859-017-1934-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.King D, Strynadka N. 2011. Crystal structure of New Delhi metallo-beta-lactamase reveals molecular basis for antibiotic resistance. Protein Sci 20:1484–1491. doi: 10.1002/pro.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gonzalez LJ, Bahr G, Nakashige TG, Nolan EM, Bonomo RA, Vila AJ. 2016. Membrane anchoring stabilizes and favors secretion of New Delhi metallo-beta-lactamase. Nat Chem Biol 12:516–522. doi: 10.1038/nchembio.2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.