Pseudomonas aeruginosa is a major bacterial pathogen associated with a rising prevalence of antibiotic resistance. We evaluated the resistance mechanisms of P. aeruginosa against POL7080, a species-specific, first-in-class antibiotic in clinical trials that targets the lipopolysaccharide transport protein LptD. We isolated a series of POL7080-resistant strains with mutations in the two-component sensor gene pmrB.
KEYWORDS: murepavadin, POL7001, POL7080, Pseudomonas, antibiotic resistance, colistin, multidrug resistance, pmrB, polymyxins
ABSTRACT
Pseudomonas aeruginosa is a major bacterial pathogen associated with a rising prevalence of antibiotic resistance. We evaluated the resistance mechanisms of P. aeruginosa against POL7080, a species-specific, first-in-class antibiotic in clinical trials that targets the lipopolysaccharide transport protein LptD. We isolated a series of POL7080-resistant strains with mutations in the two-component sensor gene pmrB. Transcriptomic and confocal microscopy studies support a resistance mechanism shared with colistin, involving lipopolysaccharide modifications that mitigate antibiotic cell surface binding.
TEXT
Pseudomonas aeruginosa and other Gram-negative bacteria are becoming increasingly resistant to current antibiotics and pose a major threat to patients with hospital-acquired infections, compromised immune systems, or chronic pulmonary infections (1–4). Unfortunately, the discovery of new agents targeting Gram-negative bacteria is especially challenging due to an impermeable, lipopolysaccharide (LPS)-laden, outer membrane, in addition to porin mutations and efflux pumps limiting intracellular drug accumulation (5, 6). Among the last-resort antibiotics currently being used to treat severe multidrug-resistant pseudomonal infections is the polymyxin class of cationic antimicrobial peptides, including polymyxin B and colistin (polymyxin E). Recently, the first-in-class antibiotic POL7080, which is currently in phase 3 clinical trials, was reported to have species-specific activity against P. aeruginosa by inhibiting the LPS transport protein LptD (7–9). The discovery of POL7080 (and its analogue POL7001) emerged from extensive chemical modifications of the cationic antimicrobial peptide protegrin-1 (PG-1), in which a beta-hairpin was introduced to create cyclized peptidomimetic analogues (10, 11). The mechanism of action of POL7080 and its analogues differs from that of other cationic antimicrobial peptides in several key ways. Polymyxins and PG-1 interact with LPS and exhibit broad-spectrum antimicrobial activity through self-promoted uptake across the outer membrane, followed by cell lysis through poorly defined mechanisms (12–14). The LptD inhibitors POL7080 and POL7001, however, have been reported to exhibit a nonlytic mechanism of action through LptD inhibition in P. aeruginosa exclusively (7–9).
To investigate the mechanisms of resistance to POL7080 and its analogues, we selected for spontaneously resistant P. aeruginosa PA14 mutants by plating mid-log cultures on lysogeny broth (LB) agar containing 1.6 μg/ml POL7001 (∼4 times the MIC on LB agar; detailed experimental protocols are outlined in the supplemental material). We isolated 6 independent mutants and confirmed their resistance to POL7080, POL7001, and PG-1 by adapting the broth microdilution method described previously (15). All clones were highly resistant to POL7080, with 4-fold to 32-fold MIC shifts relative to PA14 (Table 1), comparable to 4 POL7080-resistant clinical isolates reported previously (16). Because resistant clones were selected in LB, MIC assays were also performed in LB, yielding results comparable to those obtained in Mueller-Hinton broth (MHB) (see Table S1 in the supplemental material). All strains remained susceptible to conventional antipseudomonal antibiotics (Table S2). Whole-genome sequencing revealed 1 or 2 single-nucleotide polymorphisms (SNPs) in each mutant, relative to the wild-type (WT) parent PA14, and all 6 isolates carried a mutation in the common gene pmrB (Table 1). PmrB is a histidine kinase and the membrane-bound sensor in the PmrA-PmrB two-component regulatory system in Gram-negative bacteria. In response to low Mg2+ levels and periplasmic antimicrobial peptides, PmrB undergoes conformational changes in its histidine kinase and methyl-accepting protein (HAMP) domain, leading to autophosphorylation, phosphoryl group transfer to its cognate response regulator PmrA, and downstream activation of transcriptional programs regulating LPS modification (17). Interestingly, mutations in the PmrA-PmrB system have been implicated in polymyxin resistance through upregulation of the lipid A deacylase PagL and the arnBCADTEF-ugd operon, resulting in LPS modifications that reduce polymyxin binding to the cell surface (18–29). Notably, the mutant PA14-pmrBG188S contained an amino acid substitution at the same site in the HAMP domain as reported previously for the colistin-resistant clinical strain PA1571-pmrBG188D, which was isolated from a cystic fibrosis patient and was found to have the PmrB substitution G188D (22). Therefore, we measured colistin activity against all 6 resistant mutants and found cross-resistance, with MIC shifts ranging from 4-fold to 32-fold (Table 1). We also tested the colistin-resistant strain PA1571-pmrBG188D with POL7080 and found that it had 8-fold cross-resistance, relative to PA14 (Table 1).
TABLE 1.
Summary of resistant mutants sequenced after selection with POL7001
| Strain | PA14 no. | Gene | SNP(s) | Protein change(s) | Function | MIC (μg/ml) in MHB (fold change)a |
|||
|---|---|---|---|---|---|---|---|---|---|
| POL7001 | POL7080 | PG-1 | Colistin | ||||||
| Wild-type PA14 | 0.050 | 0.050 | 1.3 | 0.44 | |||||
| PA14-pmrBL172del | 63160 | pmrB | CGCT506C | L172delb | Two-component system | 1.6 (32) | 1.6 (32) | 43 (32) | 14 (32) |
| PA14-pmrBG188S | 63160 | pmrB | G562A | G188S | Two-component system | 0.40 (8) | 0.20 (4) | 11 (8) | 1.8 (4) |
| 21890 | hypoc | T932G | V311G | Putative oxidoreductase | |||||
| PA14-pmrBV136L | 63160 | pmrB | G406C | V136L | Two-component system | 0.80 (16) | 0.40 (8) | >43 (>32) | 3.5 (8) |
| 43080 | vgrG14 | C1325A + C1330G | A442E + H444D | Type VI secretion | |||||
| PA14-pmrBT132P | 63160 | pmrB | A394C | T132P | Two-component system | 0.40 (8) | 0.40 (8) | >43 (>32) | 3.5 (8) |
| PA14-pmrBR155H | 63160 | pmrB | G464A | R155H | Two-component system | 1.6 (32) | 1.6 (32) | >43 (>32) | 7.0 (16) |
| PA14-pmrBA330P | 63160 | pmrB | G988C | A330P | Two-component system | 0.80 (16) | 0.80 (16) | >43 (>32) | 3.5 (8) |
| PA1571-pmrBG188D | 63160 | pmrB | G563A | G188D | Two-component system | 0.20 (4) | 0.40 (8) | 10.8 (8) | >56 (>64) |
The fold change in MIC, relative to PA14, is shown in parentheses.
In-frame deletion of L172.
Hypothetical protein.
To confirm that alterations in pmrB account for the observed resistance to POL7080 and colistin in PA14, we introduced a copy of the wild-type allele pmrBWT and the resistant alleles pmrBL172del and pmrBG188S, under the control of an arabinose promoter, into PA14 at the neutral attTn7 chromosomal site using the pUC18-derived mini-Tn7 integration system (30). We also introduced the allele pmrBG188D, which was reported previously to confer colistin resistance (22), into the wild-type background. MIC assays in the presence of 0.25% (vol/vol) arabinose demonstrated that all three mutated pmrB alleles, but not the wild-type allele, conferred POL7080 and colistin resistance (Table 2). Because the PA14-pmrBG188S mutant was only modestly resistant to POL7080 in MHB, we also determined MICs in LB, the medium in which the mutants were selected, which confirmed that all mutant alleles conferred POL7080 and colistin resistance (Table S3). Conversely, expression of the pmrBWT allele in the resistant PA14-pmrBL172del background did not restore POL7080 susceptibility, suggesting that resistant pmrB alleles were largely dominant over the wild-type allele.
TABLE 2.
MICs in MHB with 0.25% arabinose after introduction of second pmrB alleles into PA14 and pmrBL172del backgrounds
| Background strain | attTn7 allele | MIC (μg/ml) (fold change)a |
|
|---|---|---|---|
| POL7080 | Colistin MIC | ||
| PA14 | 0.05 | 0.44 | |
| PA14 | pmrBWT | 0.10 (2) | 0.44 (1) |
| PA14 | pmrBL172del | 0.80 (16) | 1.8 (4) |
| PA14 | pmrBG188S | 0.10 (2) | 0.88 (2) |
| PA14 | pmrBG188D | 0.40 (8) | 1.8 (4) |
| PA14-pmrBL172del | 1.6 | 7.0 | |
| PA14-pmrBL172del | pmrBWT | 0.8 (0.5) | 3.5 (0.5) |
The fold change in MIC, relative to the background strain, is indicated in parentheses.
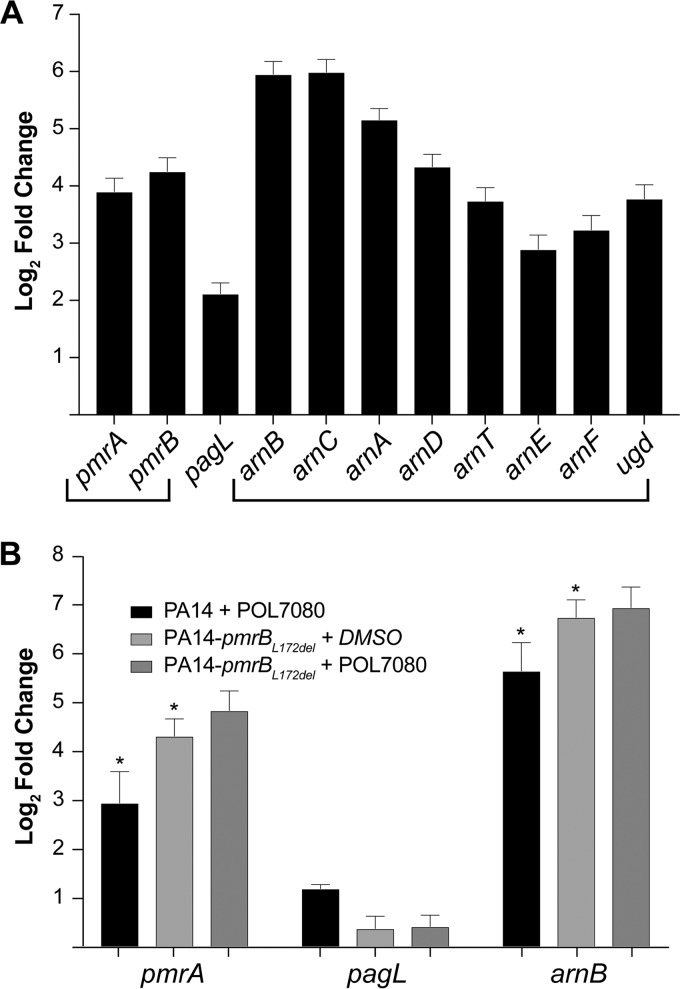
We next performed whole transcriptome sequencing to investigate the role of pmrA-pmrB in response to LptD inhibitors. After extracting total RNA from mid-log PA14 cells treated with 0.2 μg/ml POL7001 (2 times the MIC), we prepared RNA-seq libraries using the RNA TagSeq protocol (31), sequenced samples on an Illumina NextSeq instrument, and analyzed the data using Burrows-Wheeler Aligner (32) for alignment and DESeq2 (33) to determine differential gene expression. We found that LPS modification genes, including pmrA-pmrB, the lipid A deacylase gene pagL, and the entire arnBCADTEF-ugd operon, which is responsible for adding 4-amino-4-deoxy-l-arabinose (l-Ara4N) to lipid A, were significantly upregulated in response to POL7001 (Fig. 1A). The aminotransferase gene arnB, coding for the protein that catalyzes the final step in l-Ara4N addition, was among the most highly upregulated genes in the entire data set. We confirmed these findings with quantitative reverse transcription-PCR (qRT-PCR) after treating mid-log PA14 or resistant PA14-pmrBL172del cells with either POL7080 or DMSO (control) (Fig. 1B). Relative to the control, POL7080 significantly induced pmrA, arnB, and pagL expression in PA14 cells. Notably, arnB and pmrA transcript levels in untreated PA14-pmrBL172del cells exceeded those in POL7080-treated PA14 cells. Together, these data reveal that a signature transcriptional program involving key LPS modification genes is constitutively upregulated in the resistant mutant PA14-pmrBL172del. These results reveal a cellular response to LptD inhibitors that mirrors the previously reported response to polymyxins, and they suggest a shared mechanism by which pmrB mutations confer cross-resistance between POL7080 and colistin.
FIG 1.
LPS modification genes upregulated in response to POL7001 and POL7080 treatment and constitutively expressed in the resistant PA14-pmrBL172del strain. (A) RNA-seq data show log2(fold change) in sequencing reads for PA14 after treatment at 37°C for 100 min with POL7001, relative to the vehicle control. Bracketed genes are located within the same operon. Upregulated genes include the pmrA-pmrB two-component regulatory system genes, the lipid A deacylase gene pagL, and the arnBCADTEF-ugd operon. (B) After treatment of PA14 and resistant PA14-pmrBL172del cells with POL7080 or dimethyl sulfoxide (DMSO) (control) at 37°C for 100 min, qRT-PCR data show log2(fold change) in LPS modification gene transcript levels (normalized to rpoD expression), relative to vehicle-treated PA14 cells. In all experiments, error bars represent standard errors of the mean of 3 biological replicates (n = 3). The coefficient of variance of raw triplicate measurements ranged from 0.5 to 8.4%. Asterisks indicate paired t test P values of <0.03 for POL7080-treated PA14 cells versus vehicle-treated PA14-pmrBL172del cells.
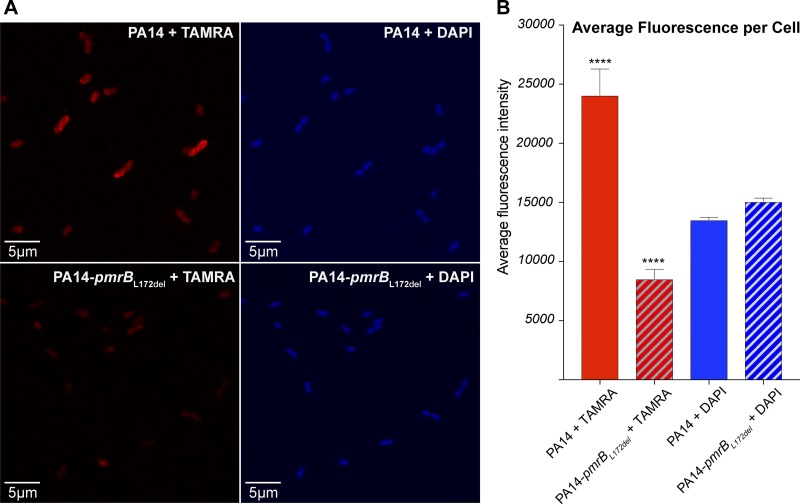
Mutations in pmrB are known to drive polymyxin resistance by l-Ara4N addition to LPS, thereby reducing drug binding to the cell surface (18–29). Therefore, we investigated whether pmrB mutations might mitigate POL7080 binding to the cell surface. We synthesized tetramethylrhodamine (TAMRA)-L27-11 (Fig. S1), a red fluorescent analogue of POL7080 with retained inhibitory activity (Table S4), to probe for differential uptake in PA14-pmrBL172del cells, relative to PA14 cells, using confocal microscopy. After treatment of mid-log PA14 or PA14-pmrBL172del cells with 1.4 μg/ml TAMRA-L27-11, the cells were washed, fixed with 4% paraformaldehyde, and stained with 4′,6-diamidino-2-phenylindole (DAPI) for nucleic acid visualization. Red-field and blue-field confocal microscopy showed comparable DAPI staining but >3-fold reduction in TAMRA-L27-11 uptake in PA14-pmrBL172del cells, relative to PA14 cells (Fig. 2), indicating less efficient drug binding at the cell surface of the resistant mutant.
FIG 2.
Differential uptake of TAMRA-L27-11 by PA14 cells versus PA14-pmrBL172del cells. (A) Red-field (left) and blue-field (right) confocal microscopy images of PA14 cells (top) and PA14-pmrBL172del cells (bottom) show reduced TAMRA-L27-11 uptake in PA14-pmrBL172del cells, relative to PA14 cells. All cells were DAPI stained after treatment with 1.4 μg/ml TAMRA-L27-11 for 120 min. (B) Average fluorescence intensities of TAMRA (red bars) and DAPI (blue bars) were calculated for PA14 cells (solid bars) (n = 22 cells) and resistant PA14-pmrBL172del cells (hatched bars) (n = 21 cells) using ImageJ software. The figure depicts a representative replicate from 3 independent experiments. Error bars represent standard errors of the mean, and asterisks indicate unpaired t test P values of <0.0001 for PA14 cells versus PA14-pmrBL172del cells after TAMRA-L27-11 treatment.
In summary, we report a series of pmrB mutations that confer high-level resistance to POL7080 and moderate cross-resistance to colistin. Expression analysis and confocal microscopy data support a resistance mechanism in which pmrB mutations upregulate the arnBCADTEF-ugd operon. These data align well with known mechanisms of resistance to polymyxins, in which upregulation of the arn operon has been shown to result in LPS modification with l-Ara4N reducing drug binding to the cell surface (21–23). Altogether, our findings suggest that preexisting colistin resistance may limit the utility of POL7080 in a subset of highly resistant P. aeruginosa cases and exposure to POL7080, if it is successfully developed, could inadvertently drive cross-resistance to colistin and other polymyxins.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a generous gift from Anita and Josh Bekenstein and NIH grant 1R01AI117043-04 (D.T.H.). Imaging was performed in the Microscopy Core of the Program in Membrane Biology, which is partially supported by a Centre for the Study of Inflammatory Bowel Disease grant (grant DK043351) and a Boston Area Diabetes and Endocrinology Research Center award (grant DK057521). The Zeiss confocal system is supported by NIH grant 1S10OD021577-01.
We thank Samuel Moskowitz and Lael Yonker for generously providing the clinical strain PA1571. We also thank Jonathan Livny, Nirmalya Bandyopadhyay, James Gomez, and Roby Bhattacharyya for their helpful discussions. We thank Anilkumar Nair for assistance with confocal microscopy imaging.
We declare that we have no conflicts of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00511-19.
REFERENCES
- 1.Magill SS, Edwards JR, Bamberg W, Beldavs ZG, Dumyati G, Kainer MA, Lynfield R, Maloney M, McAllister-Hollod L, Nadle J, Ray SM, Thompson DL, Wilson LE, Fridkin SK. 2014. Multistate point-prevalence survey of health care-associated infections. N Engl J Med 370:1198–1208. doi: 10.1056/NEJMoa1306801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Willyard C. 2017. The drug-resistant bacteria that pose the greatest health threats. Nature 543:15. doi: 10.1038/nature.2017.21550. [DOI] [PubMed] [Google Scholar]
- 3.Breidenstein EBM, de la Fuente-Núñez C, Hancock REW. 2011. Pseudomonas aeruginosa: all roads lead to resistance. Trends Microbiol 19:419–426. doi: 10.1016/j.tim.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 4.Lister PD, Wolter DJ, Hanson ND. 2009. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Microbiol Rev 22:582–610. doi: 10.1128/CMR.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hancock RE. 1997. The bacterial outer membrane as a drug barrier. Trends Microbiol 5:37–42. doi: 10.1016/S0966-842X(97)81773-8. [DOI] [PubMed] [Google Scholar]
- 6.Fernandez L, Hancock RE. 2012. Adaptive and mutational resistance: role of porins and efflux pumps in drug resistance. Clin Microbiol Rev 25:661–681. doi: 10.1128/CMR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Srinivas N, Jetter P, Ueberbacher BJ, Werneburg M, Zerbe K, Steinmann J, Van der Meijden B, Bernardini F, Lederer A, Dias RL, Misson PE, Henze H, Zumbrunn J, Gombert FO, Obrecht D, Hunziker P, Schauer S, Ziegler U, Kach A, Eberl L, Riedel K, DeMarco SJ, Robinson JA. 2010. Peptidomimetic antibiotics target outer-membrane biogenesis in Pseudomonas aeruginosa. Science 327:1010–1013. doi: 10.1126/science.1182749. [DOI] [PubMed] [Google Scholar]
- 8.Andolina G, Bencze LC, Zerbe K, Muller M, Steinmann J, Kocherla H, Mondal M, Sobek J, Moehle K, Malojcic G, Wollscheid B, Robinson JA. 2018. A peptidomimetic antibiotic interacts with the periplasmic domain of LptD from Pseudomonas aeruginosa. ACS Chem Biol 13:666–675. doi: 10.1021/acschembio.7b00822. [DOI] [PubMed] [Google Scholar]
- 9.Werneburg M, Zerbe K, Juhas M, Bigler L, Stalder U, Kaech A, Ziegler U, Obrecht D, Eberl L, Robinson JA. 2012. Inhibition of lipopolysaccharide transport to the outer membrane in Pseudomonas aeruginosa by peptidomimetic antibiotics. Chembiochem 13:1767–1775. doi: 10.1002/cbic.201200276. [DOI] [PubMed] [Google Scholar]
- 10.Shankaramma SC, Moehle K, James S, Vrijbloed JW, Obrecht D, Robinson JA. 2003. A family of macrocyclic antibiotics with a mixed peptide-peptoid β-hairpin backbone conformation. Chem Commun (Camb) 1842–1843. doi: 10.1039/B304310J. [DOI] [PubMed] [Google Scholar]
- 11.Robinson JA, Shankaramma SC, Jetter P, Kienzl U, Schwendener RA, Vrijbloed JW, Obrecht D. 2005. Properties and structure-activity studies of cyclic β-hairpin peptidomimetics based on the cationic antimicrobial peptide protegrin I. Bioorg Med Chem 13:2055–2064. doi: 10.1016/j.bmc.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 12.Deris ZZ, Swarbrick JD, Roberts KD, Azad MA, Akter J, Horne AS, Nation RL, Rogers KL, Thompson PE, Velkov T, Li J. 2014. Probing the penetration of antimicrobial polymyxin lipopeptides into Gram-negative bacteria. Bioconjug Chem 25:750–760. doi: 10.1021/bc500094d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Steinberg DA, Hurst MA, Fujii CA, Kung AH, Ho JF, Cheng FC, Loury DJ, Fiddes JC. 1997. Protegrin-1: a broad-spectrum, rapidly microbicidal peptide with in vivo activity. Antimicrob Agents Chemother 41:1738–1742. doi: 10.1128/AAC.41.8.1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang L, Dhillon P, Yan H, Farmer S, Hancock RE. 2000. Interactions of bacterial cationic peptide antibiotics with outer and cytoplasmic membranes of Pseudomonas aeruginosa. Antimicrob Agents Chemother 44:3317–3321. doi: 10.1128/aac.44.12.3317-3321.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wiegand I, Hilpert K, Hancock RE. 2008. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc 3:163–175. doi: 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
- 16.Sader HS, Dale GE, Rhomberg PR, Flamm RK. 2018. Antimicrobial activity of murepavadin tested against clinical isolates of Pseudomonas aeruginosa from the United States, Europe, and China. Antimicrob Agents Chemother 62:e00311-18. doi: 10.1128/AAC.00311-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen HD, Groisman EA. 2013. The biology of the PmrA/PmrB two-component system: the major regulator of lipopolysaccharide modifications. Annu Rev Microbiol 67:83–112. doi: 10.1146/annurev-micro-092412-155751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee JY, Na IY, Park YK, Ko KS. 2014. Genomic variations between colistin-susceptible and resistant Pseudomonas aeruginosa clinical isolates and their effects on colistin resistance. J Antimicrob Chemother 69:1248–1256. doi: 10.1093/jac/dkt531. [DOI] [PubMed] [Google Scholar]
- 19.Lee JY, Park YK, Chung ES, Na IY, Ko KS. 2016. Evolved resistance to colistin and its loss due to genetic reversion in Pseudomonas aeruginosa. Sci Rep 6:25543. doi: 10.1038/srep25543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller AK, Brannon MK, Stevens L, Johansen HK, Selgrade SE, Miller SI, Hoiby N, Moskowitz SM. 2011. PhoQ mutations promote lipid A modification and polymyxin resistance of Pseudomonas aeruginosa found in colistin-treated cystic fibrosis patients. Antimicrob Agents Chemother 55:5761–5769. doi: 10.1128/AAC.05391-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moskowitz SM, Ernst RK, Miller SI. 2004. PmrAB, a two-component regulatory system of Pseudomonas aeruginosa that modulates resistance to cationic antimicrobial peptides and addition of aminoarabinose to lipid A. J Bacteriol 186:575–579. doi: 10.1128/jb.186.2.575-579.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moskowitz SM, Brannon MK, Dasgupta N, Pier M, Sgambati N, Miller AK, Selgrade SE, Miller SI, Denton M, Conway SP, Johansen HK, Hoiby N. 2012. PmrB mutations promote polymyxin resistance of Pseudomonas aeruginosa isolated from colistin-treated cystic fibrosis patients. Antimicrob Agents Chemother 56:1019–1030. doi: 10.1128/AAC.05829-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han ML, Velkov T, Zhu Y, Roberts KD, Le Brun AP, Chow SH, Gutu AD, Moskowitz SM, Shen HH, Li J. 2018. Polymyxin-induced lipid A deacylation in Pseudomonas aeruginosa perturbs polymyxin penetration and confers high-level resistance. ACS Chem Biol 13:121–130. doi: 10.1021/acschembio.7b00836. [DOI] [PubMed] [Google Scholar]
- 24.Han ML, Zhu Y, Creek DJ, Lin YW, Anderson D, Shen HH, Tsuji B, Gutu AD, Moskowitz SM, Velkov T, Li J. 2018. Alterations of metabolic and lipid profiles in polymyxin-resistant Pseudomonas aeruginosa. Antimicrob Agents Chemother 62:e02656-17. doi: 10.1128/AAC.02656-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soon RL, Nation RL, Cockram S, Moffatt JH, Harper M, Adler B, Boyce JD, Larson I, Li J. 2011. Different surface charge of colistin-susceptible and resistant Acinetobacter baumannii cells measured with zeta potential as a function of growth phase and colistin treatment. J Antimicrob Chemother 66:126–133. doi: 10.1093/jac/dkq422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McPhee JB, Lewenza S, Hancock RE. 2003. Cationic antimicrobial peptides activate a two-component regulatory system, PmrA-PmrB, that regulates resistance to polymyxin B and cationic antimicrobial peptides in Pseudomonas aeruginosa. Mol Microbiol 50:205–217. doi: 10.1046/j.1365-2958.2003.03673.x. [DOI] [PubMed] [Google Scholar]
- 27.Fernández L, Alvarez-Ortega C, Wiegand I, Olivares J, Kocíncová D, Lam JS, Martínez JL, Hancock REW. 2013. Characterization of the polymyxin B resistome of Pseudomonas aeruginosa. Antimicrob Agents Chemother 57:110–119. doi: 10.1128/AAC.01583-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gunn JS, Lim KB, Krueger J, Kim K, Guo L, Hackett M, Miller SI. 1998. PmrA-PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol Microbiol 27:1171–1182. doi: 10.1046/j.1365-2958.1998.00757.x. [DOI] [PubMed] [Google Scholar]
- 29.Cannatelli A, Giani T, Aiezza N, Di Pilato V, Principe L, Luzzaro F, Galeotti CL, Rossolini GM. 2017. An allelic variant of the PmrB sensor kinase responsible for colistin resistance in an Escherichia coli strain of clinical origin. Sci Rep 7:5071. doi: 10.1038/s41598-017-05167-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choi KH, Schweizer HP. 2006. Mini-Tn7 insertion in bacteria with single attTn7 sites: example Pseudomonas aeruginosa. Nat Protoc 1:153–161. doi: 10.1038/nprot.2006.24. [DOI] [PubMed] [Google Scholar]
- 31.Shishkin AA, Giannoukos G, Kucukural A, Ciulla D, Busby M, Surka C, Chen J, Bhattacharyya RP, Rudy RF, Patel MM, Novod N, Hung DT, Gnirke A, Garber M, Guttman M, Livny J. 2015. Simultaneous generation of many RNA-seq libraries in a single reaction. Nat Methods 12:323–325. doi: 10.1038/nmeth.3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.