Abstract
Cyclic di-GMP (c-di-GMP) is an important second messenger in bacteria, and its regulatory network has been extensively studied. However, information regarding the activation mechanisms of its receptors remains limited. In this study, we characterized the two-component regulator DevR as a new c-di-GMP receptor and further uncovered a novel co-activation mechanism for effective regulation of DevR in mycobacteria. We show that high c-di-GMP levels induce the expression of the devR operon in Mycobacterium smegmatis and increase mycobacterial survival under oxidative stress. The deletion of either DevR or its two-component kinase DevS significantly weakened the stimulating effect of c-di-GMP on oxidative-stress tolerance of mycobacteria. We also found that DevR senses the c-di-GMP signal through its C-terminal structure and that c-di-GMP alone does not directly affect the DNA-binding activity of DevR. Strikingly, c-di-GMP stimulated DevR phosphorylation by the kinase DevS, thereby activating DevR's DNA-binding affinity. In summary, our results indicated that c-di-GMP triggers a phosphorylation-dependent mechanism that co-activates DevR's transcriptional activity. Our findings suggest a novel paradigm for the cross-talk between c-di-GMP signaling and two-component regulatory systems that activates transcription of stress-response genes in bacteria.
Keywords: cyclic di-GMP (c-di-GMP), transcription regulation, histidine kinase, Mycobacterium smegmatis, oxidative stress, antioxidant
Introduction
Cyclic-di-GMP (c-di-GMP)3 is one of the signaling molecules utilized by bacteria to rapidly respond to environmental signals. As a global signaling molecule, c-di-GMP regulates bacterial physiological processes and environmental adaptation through its downstream receptors (1–5). Alternatively, upon sensing environmental signals, the two-component system utilizes its sensory kinase in order to phosphorylate its regulator to facilitate bacterial growth and survival under stress (6, 7). However, information regarding the activation mechanisms of c-di-GMP receptors remains limited. To date, only few examples of interplay between c-di-GMP signaling and two-component systems are available (8–11).
Oxidative stress is a common extracellular and intracellular environmental signal for bacteria. Mycobacteria belong to a type of slow-growing actinomycetes and possess a unique antioxidant capacity. For example, Mycobacterium tuberculosis is the causative agent of tuberculosis and can thrive in oxidative environments and survive under stress (12, 13). Rv3133c/Rv3132c as one of the two-component systems of M. tuberculosis was named DevR/DevS because they are differentially expressed in virulent (Dev) strains compared with avirulent strains (14, 15). The Rv3133c was also named DosR for its regulation function in dormancy survival (Dos) (16), and its partner kinase DevS was also named DosS (17). Rv2027c, the homologous kinase of DevS, was named DosT as a partner kinase of DosR (17). DevR has recently been suggested to be involved in the regulation of mycobacterial oxidative adaptation because it is up-regulated under H2O2 stress (18–20). DevR is a conserved two-component regulator and plays an important role in the adaptation of mycobacteria to hostile environments. Unphosphorylated DevR has poor DNA-binding activity in vitro. DevR phosphorylation by DevS substantially enhances its DNA-binding affinity (21). The knockout of both DosS and DosT eliminated the positive regulation of DevR (17). On the basis of the crystal structure, Wisedchaisri et al. (22, 23) found that the C-terminal DNA-binding domain of DevR (DevRC) interacts with the N-terminal domain (DevRN) of DevR and covers the phosphorylation site, Asp-54, which inhibits DevR activation by DevS. However, intermolecular inhibition is usually in a dynamic equilibrium, and DevR can be activated by a phosphorylation-dependent mechanism. When DevRC leaves the DevRN, Asp-54 is exposed and phosphorylated by DevS. Therefore, DevRC can subsequently interact with the DevRC of another DevR molecule to form a dimer, which then effectively binds with a cognate DNA (23). However, the association between oxidative stress and the phosphorylation-dependent activation of DevR in mycobacteria remains unclear. The signaling molecule for the activation that directly interacts with the two-component system DevRS has to be characterized.
Very recently, HpoR was characterized as a novel c-di-GMP receptor transcription factor, which links the c-di-GMP signal to bacterial antioxidant regulation (5). Both c-di-GMP signaling and the two-component systems DevR/DevS are involved in the regulation of adaptation to oxidative stress, thereby suggesting a potential interaction between these two systems in mycobacteria. However, the cross-talk between c-di-GMP and the two-component system and its correlation with the ability of bacteria to adapt to oxidative stress remain to be addressed. In this study, we characterized DevR as a new c-di-GMP receptor in Mycobacterium smegmatis (Msm), and we further uncovered a novel mechanism on the c-di-GMP–triggered and phosphorylation-dependent co-activation of DevR. This finding represented a novel paradigm for the cross-talk between c-di-GMP signal and two-component systems.
Results
devR operon responds to c-di-GMP signal and considerably contributes to the mycobacterial tolerance to H2O2 stress
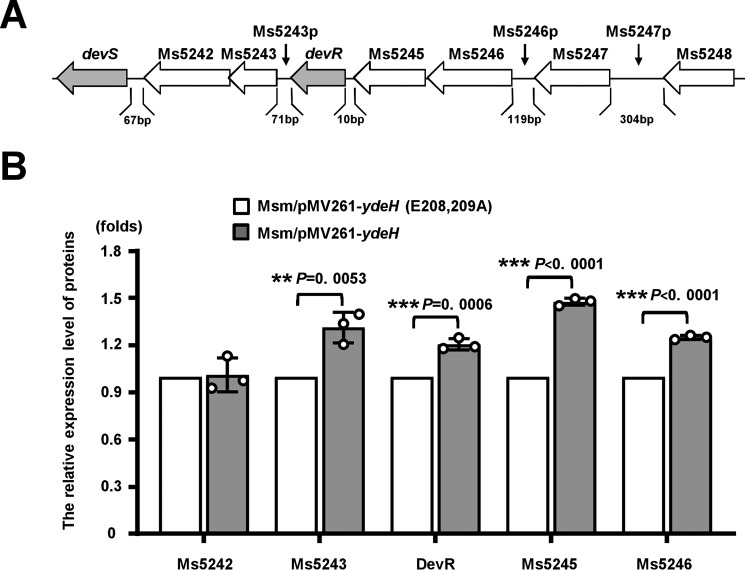
Our finding on the relationship between c-di-GMP and DevR/DevS originated from the results of a proteomic assay. Using a previously reported strategy (24), we overexpressed an Escherichia coli-derived diguanylate cyclase YdeH (25), which has a high activity of c-di-GMP synthesis in M. smegmatis, and constructed a mycobacterial strain (Msm/pMV261-ydeH) with a high level of c-di-GMP (24). The control strain (Msm/pMV261-ydeH (E208A,E209A)) expresses the mutant gene ydeH (E208A,E209A), which lost the diguanylate cyclase activity. Through a quantitative proteomic iTRAQ assay, we subsequently compared the differentially induced genes in Msm/pMV261-ydeH with Msm/pMV261-ydeH (E208, 209A). As shown in Fig. 1, most Ms5241–5246 cluster genes (Fig. 1A) designated as the devR operon (containing devR and devS genes) were induced (Fig. 1B), suggesting that the expression of the devR operon responded to the c-di-GMP signal in Msm.
Figure 1.
Quantitative proteomic assays for the effect of c-di-GMP on the gene expression of M. smegmatis. A, schematic of the devR operon (Ms5241–Ms5246) and its regulatory regions. Several neighboring noncoding regions (Ms5243p, Ms5246p, and Ms5247p), which were potentially recognized by DevR, are indicated by black arrows. B, schematic of the quantitative protein expression difference of the operon genes between Msm/pMV261-ydeH and Msm/pMV261-ydeH (E208A,E209A) (25). All error bars in the figure represented the standard deviation (S.D.) of the data derived from three biological replicates. The p values of the relative expression data were calculated by unpaired two-tailed Student's t test using GraphPad Prism 5. Asterisks denote the significant difference between two groups (**, p ≤ 0.01; ***, p ≤ 0.001).
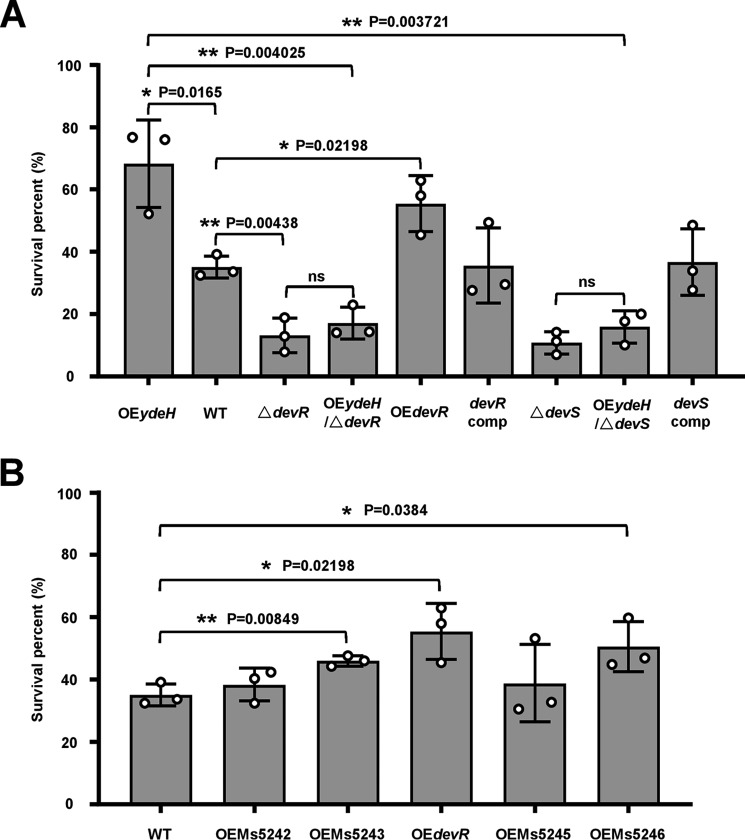
We further confirmed that devR and its operon genes significantly contributed to mycobacterial tolerance to H2O2 stress. A devR-deletion strain (ΔdevR) (Fig. S1), a devR-overexpression strain (OEdevR), and a complementation strain (devR comp) were constructed to compare their survival under H2O2 stress with the WT strain (WT). As shown in Fig. 2A, devR knockout resulted in an ∼2.7-fold decrease in mycobacterial survival rate. By contrast, devR overexpression resulted in ∼1.6-fold increase in mycobacterial survival rate under H2O2 stress. When expressing the devR gene through a pMindD-derived vector in the devR-deleted strain, the complementation strain (devR comp) re-obtained a similar survival compared with the WT strain. These results suggest that devR can play an important role for mycobacterial adaptation to H2O2 stress. Several other genes of this operon also contributed to the mycobacterial survival because their overexpression also significantly improved the mycobacterial cell counts survived under H2O2 stress (Fig. 2B) compared with the WT strain. Thus, the devR operon expression responded to the c-di-GMP signal and contributed to mycobacterial tolerance to H2O2 stress.
Figure 2.
Assays for the survival of WT and recombinant M. smegmatis under H2O2 stress. The WT and recombinant mycobacterial strains were cultured to A600 = 0.5 and treated with 9 mm H2O2 for 3 h. Cells were harvested and plated on 7H10 plates at 37 °C for 3–4 days for counting cfu. The survival percentages were calculated by cfu (with H2O2)/cfu (without H2O2) for each strain. Error bars represented the variant range of the data derived from three biological replicates. A, assays for the effects of devR, devS, and c-di-GMP on the mycobacterial survival under the stress of 9 mm H2O2. B, assays for the effects of the devR operon genes on the mycobacterial survival under 9 mm H2O2. The p values of the relative survival rate were calculated by two-tailed Student's t test using GraphPad Prism 5. Asterisks represent significant difference between two groups (*, p ≤ 0.05; **, p ≤ 0.01). WT represents the wildtype mycobacteria strain; OEydeH represents ydeH-overexpressing strain; ΔdevR represents devR-deleted strain; OEdevR represents devR-overexpressing strain; OEydeH/ΔdevR represents ydeH-overexpressing in ΔdevR strain; devR comp represents devR-complementation strain; ΔdevS represents devS-deleted strain; OEydeH/ΔdevS represents ydeH-overexpressing in ΔdevS strain; devS comp represents devS-complementation strain; OEMs5242 represents Ms5242-overexpressing strain; OEMs5243 represents Ms5243-overexpressing strain; OEMs5245 represents Ms5245-overexpressing strain; OEMs5246 represents Ms5246-overexpressing strain.
We further determined and compared the growth of several recombinant mycobacterial strains under H2O2 stress. As shown in Fig. 2A, the survival rate of ydeH-overexpressed Msm (OEydeH) increased ∼1.95-fold compared with the WT strain (WT). This result indicates that c-di-GMP enhances the survival of Msm under H2O2 stress. We also found that the knockout of either DevR or DevS in the ydeH-overexpression Msm eliminated the stimulating effect of c-di-GMP on survival. When expressing the devS gene in the devS-deleted strain, the complementation strain (devS comp) re-obtained a similar survival compared with the WT strain. Consistently, the devR complementation strain also re-obtained a similar survival percentage to the WT strain (Fig. 2A). These results indicated that both DevR and DevS were required for the enhancing effect of c-di-GMP on Msm survival under H2O2 stress.
DevR specifically binds with two regulatory sequences of the devR operon and positively regulates its expression
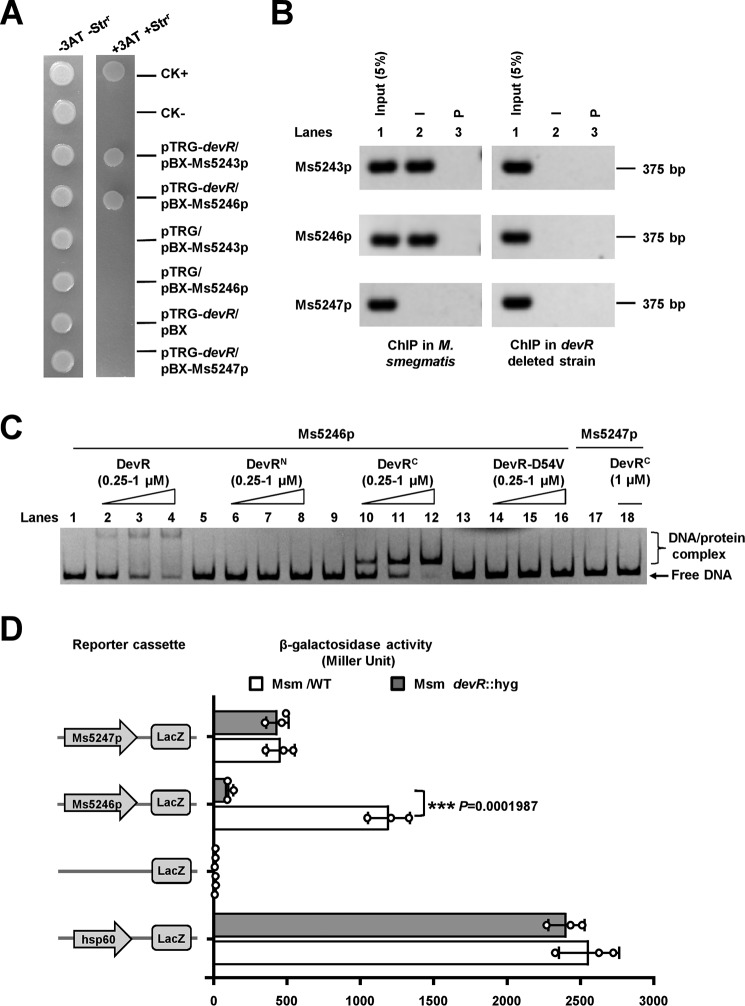
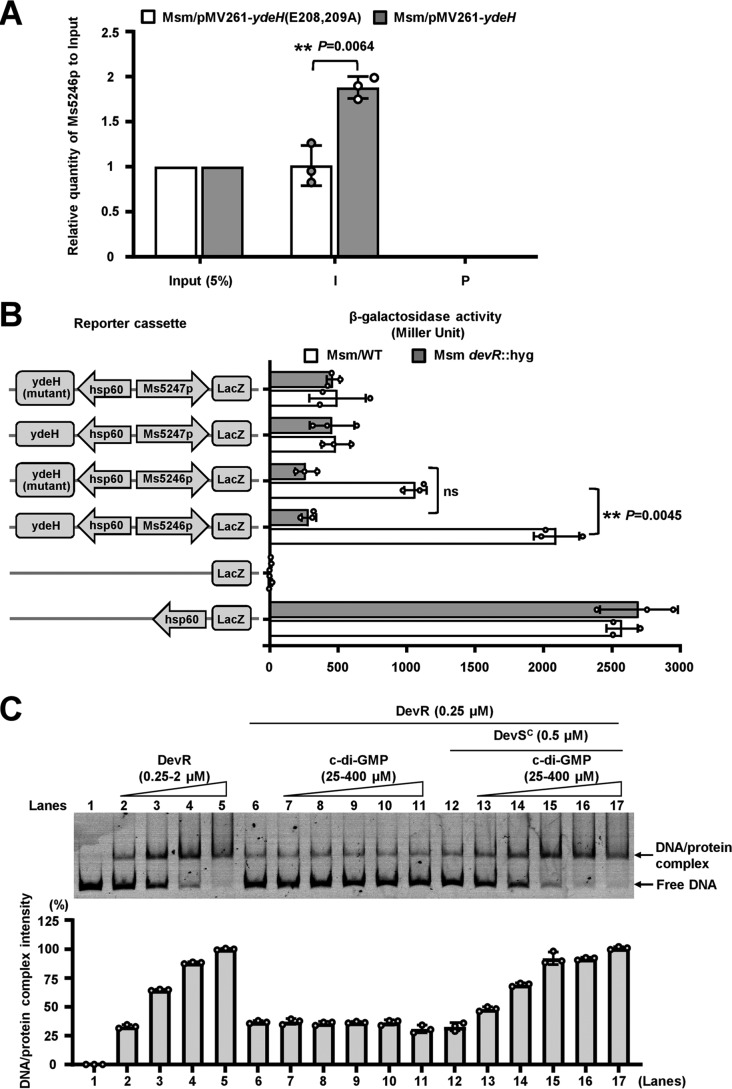
We further determined whether DevR directly regulates the operon by binding to its regulatory regions. Using bacterial one-hybrid assays (26), promoters of the M. smegmatis genes were cloned into the reporter plasmid pBXcmT and co-transformed with pTRG-devR into reporter strains to detect whether DevR can specifically bind to the potential regulatory sequences of the operon. As shown in Fig. 3A, reporter strain containing devR and target DNA Ms5243p or Ms5246p grew well, but the self-activation control strain containing either devR or DNA alone did not grow under similar conditions. This result indicated that DevR specifically interacted with the two regulatory regions. ChIP assays can further confirm that the two target DNA fragments can be specifically recovered by DevR antibody (Fig. 3B). A neighbor promoter, i.e. Ms5247p, used as a negative control was not recovered by the antisera. The specificity of DevR antibodies can be confirmed using devR deletion strains (Fig. 3B, right panel).
Figure 3.
Assays for interactions between DevR and its target DNAs. A, bacterial one-hybrid assays for the interaction between DevR and Ms5246p or Ms5243p. Co-transformants containing pBX-Ms5246p/pTRG-devR or pBX-Ms5243p/pTRG-devR plasmids grew well on the screening medium. Positive control strain (CK+) containing pBX-Rv2031/pTRG-Rv3133c grew well on the screening medium, but the negative control strain (CK−) containing the empty vectors pBX/pTRG, pBX-Ms5243p/pTRG, pBX-Ms5246p/pTRG, pBX/pTRG-devR, and pBX-Ms5247p/pTRG-devR plasmids did not. B, ChIP assays. ChIP using preimmune (P) or immune sera (I) raised against DevR. The promoter Ms5247p was used as the negative control. The devR-deleted strain (right panel) was used for the negative control strain. C, EMSAs. The specific DNA-binding activity of DevR or the C-terminal DNA-binding domain of DevR (DevRC) on the Ms5246p promoter DNA can be observed on the gel when the Ms5246p DNA substrate was co-incubated with increasing amounts of protein. By contrast, no binding activity was observed for the N-terminal domain of DevR (DevRN) (lanes 6–8) and the phosphorylation site mutant protein DevR-D54V (lanes 14–16). Additionally, the negative control promoter Ms5247p was not be bound by DevRC (lane 18). D, β-gal activity assays. Left column: the schematic representation of plasmids is used to generate reporter strains. The hsp60–lacZ was used as the positive control. Null promoter lacZ and Ms5247p–lacZ were used as negative controls. β-Galactosidase activity was examined and presented as Miller units (right panel) both in WT and devR-deleted strains. Error bars represent the variant range of the data derived from three biological replicates. The p values of the relative expression data were calculated by unpaired two-tailed Student's t test using GraphPad Prism 5. The p values of the results (p ≤ 0.001) are indicated by *** on the right of the column.
An electrophoretic mobility-shift assay (EMSA) was performed to further confirm the interaction between DevR and target DNA (Fig. 3C). With increasing amounts of DevR protein (0.25–1 μm) in the reactions (Fig. 3C, lanes 2–4), a stepwise increase in the amount of shifted DNA was clearly observed. DevRC demonstrated a slightly improved DNA-binding ability (Fig. 3C, lanes 10–12). By contrast, no DNA-binding activity can be clearly observed for DevRN (Fig. 3C, lanes 6–8) and DevR-D54V (lanes 14–16) under the same experimental conditions. The negative control promoter Ms5247p was not bound by DevRC (Fig. 3C, lane 18). Additionally, the unlabeled Ms5246p could competitively inhibit the binding of FITC-labeled Ms5246p to DevR (Fig. S2, lanes 6 and 7); by contrast, the unlabeled Ms5247p did not compete for the binding of DevR to the labeled Ms5246p. These results suggested that DevR specifically bound the regulatory sequence of the devR operon through DevRC.
We subsequently constructed a series of promoter lacZ co-expression plasmids to examine the regulatory effect of DevR on the gene expression by β-gal activity assays. As shown in Fig. 3D, the strong promoter hsp60 substantially promoted lacZ expression in both WT and devR-deleted Msm strains compared with the nonpromoter lacZ plasmid. These results indicated that the report system worked well. When Ms5246p was used as a promoter, lacZ expression was significantly down-regulated in the devR-deleted mutant Msm strains compared with the WT strains. An insignificant difference between the WT and mutant strain was observed in lacZ expression when a negative control, i.e. Ms5247p, was used as the promoter. Therefore, DevR specifically recognized two regulatory sequences of the devR operon through its C terminus and positively regulated the operon's expression.
c-di-GMP physically interacts with DevR protein
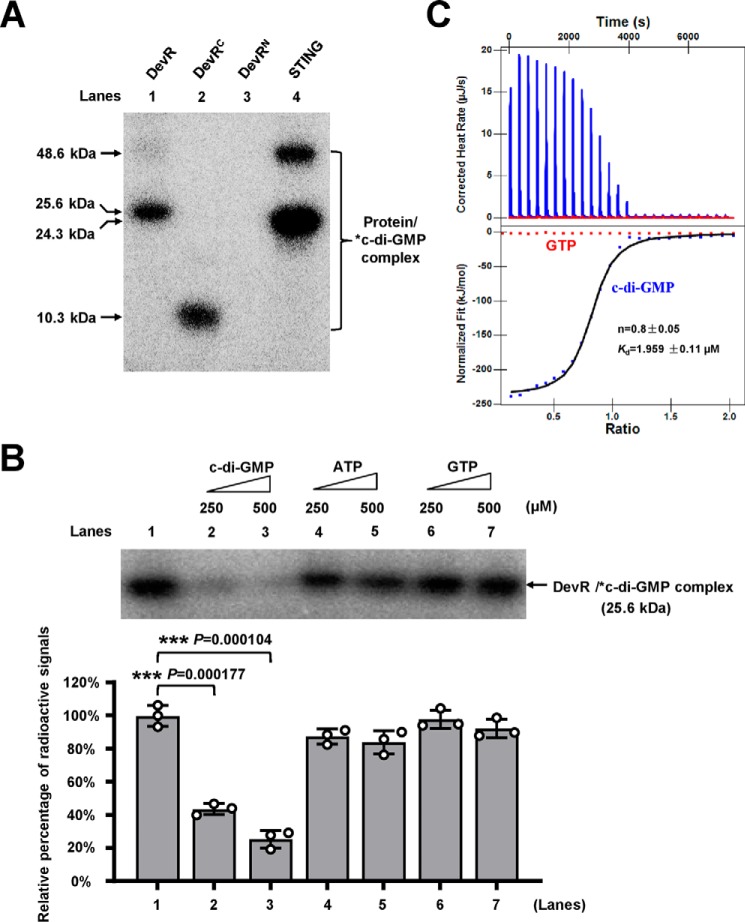
The expression of the devR operon responds to the c-di-GMP signal, and DevR is a positive regulator of the operon, suggesting that DevR would be a direct sensor of c-di-GMP signaling. A cross-linking assay confirmed this hypothesis. As shown in Fig. 4A, an autoradiograph signal corresponding to DevR was bound to radioactively labeled c-di-GMP on a PAGE (Fig. 4A, lane 1), indicating that DevR can bind with c-di-GMP. Similarly, DevRC protein also bound c-[32P]di-GMP to form a specific autoradiograph signal on the gel (Fig. 4A, lane 2). In addition, a previously reported receptor protein STING bound c-di-GMP well (Fig. 4A, lane 4) (27, 28). By contrast, no binding signal can be observed for DevRN protein (Fig. 4A, lane 3). The addition of unlabeled c-di-GMP at 50- and 100-fold excess to the reaction mixtures competitively inhibited the binding of DevR to c-[32P]di-GMP (Fig. 4B, lanes 2 and 3). This result indicated the specificity of DevR binding to c-di-GMP. A negative control nucleotide, either GTP or ATP, did not alter the binding of DevR to c-di-GMP even at a 100-fold higher concentration (500 μm) (Fig. 4B, lanes 5 and 7). These results indicated a specific interaction between DevR and c-di-GMP.
Figure 4.
Assays for the specific interaction between c-di-GMP and DevR. A, UV cross-linking assay. STING was used as the positive control (lane 4), and 12 μm DevR (lane 1), DevRC (lane 2), and DevRN (lane 3) were co-incubated with 5 μm labeled c-di-GMP for 30 min on ice, respectively. After that, reaction samples were irradiated by UV for 30 min and assayed on 12% (w/v) SDS-PAGE. Radioactive gel was exposed to a storage phosphor screen (GE Healthcare). Radioactively-labeled nucleotides are indicated by *. B, competitive experiment assays. Different amounts of unlabeled c-di-GMP, ATP, or GTP were added to the mixture with the radioactively-labeled c-di-GMP and co-incubated with 12 μm DevR for 30 min on ice, respectively. Then the assays were conducted as described in A. *c-di-GMP–DevR complex was quantified as shown in the lower panel. The error bars represent the variant range of the quantified data derived from three repeat experiments. The p values of the quantified data were calculated by unpaired two-tailed Student's t test using GraphPad Prism 5. Asterisks in the figure denote the significant difference between two groups (***, p ≤ 0.001). C, ITC assays. Original titration data and integrated heat measurements are shown in the upper and lower plots, respectively. The solid line in the bottom panel represents the best fit to a one-site binding model of the interaction of DevR with c-di-GMP. GTP was used as the negative control, and no interaction between DevR and GTP was observed.
An isothermal titration calorimetry (ITC) assay further confirmed this specific interaction. Fig. 4C (upper panel) shows the raw data for c-di-GMP titration against DevR and indicates that the binding reaction was exothermic. The integrated heat measurements are shown in the lower panel of Fig. 4C. The binding stoichiometry between DevR and c-di-GMP was ∼1:1. The binding affinity of the interaction (Kd) was 1.959 ± 0.11 μm. By contrast, no specific binding with DevR was observed when GTP was used as a negative control molecule for a similar assay. Our results showed that c-di-GMP can specifically interact with DevR, which is a novel c-di-GMP receptor transcription factor.
c-di-GMP regulates the DNA-binding activity of DevR
A direct interaction between c-di-GMP and DevR suggests that the second messenger regulates the DNA-binding activity of DevR. We first utilized ChIP assays to examine the regulation of a high level of c-di-GMP on the Ms5246p DNA-binding activity of DevR in the M. smegmatis cell. As shown in Fig. 5A, DevR can precipitate ∼1.9-fold lower than the Ms5246p DNA fragments from the ydeH (E208A,E209A)-overexpressing strain than in the ydeH-overexpressing strain (Fig. 5A), indicating that high level of c-di-GMP can obviously stimulate the intracellular DNA-binding affinity of DevR in Msm. It was further confirmed through the β-gal activity assays (Fig. 5B).
Figure 5.
Effect of c-di-GMP on the DNA-binding activity of DevR in vitro and in vivo in M. smegmatis. A, ChIP-qPCR assays for the effect of c-di-GMP on the intracellular DNA-binding activity of DevR in M. smegmatis. Input (5%) indicated that the supernatant of disrupted cells was diluted to 5% and used as the template for PCR. ChIP was performed using preimmune (P) or immune sera (I) raised against DevR. These experiments were quantified using qPCR. The p values of the relative expression data were calculated by two-tailed Student's t test using GraphPad Prism 5. ** represent significant difference (p ≤ 0.01) between the two groups. B, effect of the presence or absence of DevR on the c-di-GMP–triggered activities of Ms5246p. Left column, schematic representation of plasmids used to generate reporter strains. Right column, schematic representation of β-gal activity determined in both Msm/WT and Msm hyg::devR (devR-deleted) strains. Data are presented as Miller units (right panel). For statistical analysis, two-tailed Student's t tests were performed using GraphPad Prism 5. Null promoter lacZ, hsp60-lacZ, and Ms5247p were used as controls. Values presented are the averages of three independent biological experiments. The p values of the results (p ≤ 0.01) are indicated by **. C, c-di-GMP indirectly stimulated the DNA-binding activity of DevR. c-di-GMP alone did not stimulate the DNA-binding activity of DevR (lanes 7–11), but c-di-GMP obviously stimulated the activity in the presence of 0.5 μm autophosphorylated DevSC (lanes 13–17). The DevR–DNA complex was quantified, and the mean values of three independent experiments along with error bars are shown. ns, no significant difference between two groups.
EMSA was further used to determine the regulation of c-di-GMP on DevR DNA-binding activity in vitro. As shown in Fig. 5C, when increasing amounts of c-di-GMP (25–400 μm) are added into the reactions, we cannot observe a corresponding increase in the amounts of shifted DNA substrates by DevR (Fig. 5C, lanes 7–11) and DevRC (Fig. S3). DevR phosphorylation by DevS considerably enhances its DNA-binding affinity (21), thereby suggesting that DevS is involved in the c-di-GMP–triggered DNA-binding activity of DevR. We used EMSA to test this assumption. As shown in Fig. 5C, c-di-GMP could enhance the DNA-binding ability of DevR in the presence of DevS (lanes 13–17). These results indicated that c-di-GMP indirectly but not directly stimulated the DNA-binding activity of DevR. Therefore, c-di-GMP modulated the DNA-binding ability of DevR in vivo in Msm and in vitro.
Two-component kinase DevS is required to stimulate DNA-binding affinity of DevR by c-di-GMP in vivo in Msm
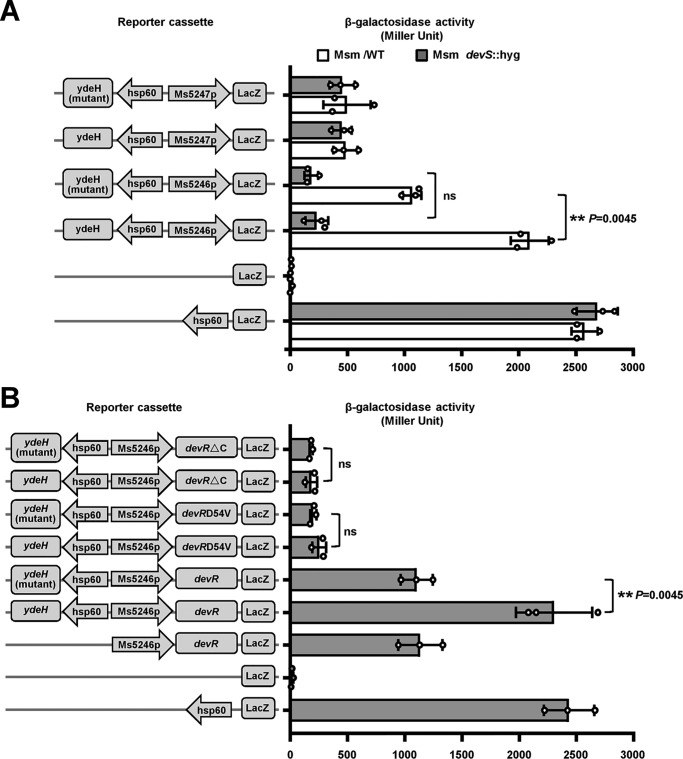
We constructed a devS-deleted Msm strain (Fig. S4) and a series of promoter lacZ co-expression plasmids, and we utilized β-gal activity assays to examine whether DevS is required to stimulate the DNA-binding ability of DevR by c-di-GMP in Msm. Overexpressing YdeH in Msm, which can improve the c-di-GMP level, significantly enhanced the expression of Ms5246p-lacZ in the WT strains but not in the devS-deleted mutant Msm strains (Fig. 6A). In comparison, we cannot observe a similar enhancement when overexpressing YdeH (E208A,E209A), which lost the activity of c-di-GMP synthesis. No significant difference between the WT and devS-deleted strains was observed in lacZ expression when a negative control, i.e. Ms5247p, was used as the promoter. The strong promoter hsp60 substantially promoted lacZ expression in both WT and devS-deleted Msm strains compared with the nonpromoter lacZ plasmid (Fig. 6A). These results suggested that DevS was required for the c-di-GMP–triggered DNA-binding activity of DevR in Msm.
Figure 6.
β-Gal activity assays for the effect of two-component kinase DevS on the c-di-GMP–triggered DNA-binding affinity of DevR in M. smegmatis. A, effect of the presence or absence of DevS on the c-di-GMP–triggered activities of Ms5246p. B, effect of phosphorylation site Asp-54 of DevR on the c-di-GMP receptor activation by DevS. Left column, schematic representation of plasmids used to generate reporter strains. Right column, schematic representation of β-gal activity determined in both WT and devS-deleted M. smegmatis strains. Data are presented as Miller units (right panel). For statistical analysis, two-tailed Student's t tests were performed using GraphPad Prism 5. Null promoter lacZ, hsp60-lacZ, and Ms5247p were used as controls. Values presented are the averages of three independent biological experiments. The p values of the results (p ≤ 0.01) are indicated by **. ns, no significant difference between two groups.
To further confirm this observation, we constructed a phosphorylation site mutant of DevR, DevR-D54V, which inhibits the activation of DevR by DevS (29), and performed a similar β-gal activity assay. WT DevR can significantly enhance the lacZ expression through Ms5246p in the ydeH-overexpressing Msm strains, but DevR-D54V cannot (Fig. 6B). The effect of DevR-D54V on the lacZ expression was very similar to that of DevR-ΔC, in which the DNA-binding domain was deleted. Hence, either deleting two-component kinase DevS or mutating the phosphorylation site of DevR eliminated the stimulation effect of c-di-GMP on the DNA-binding activity of DevR in Msm.
c-di-GMP stimulates DevR phosphorylation by DevS
To further pursue the mechanism of the c-di-GMP–triggered activation of DevR by DevS, we next utilized the purified DevSC protein to determine the effect of c-di-GMP on its kinase activity in vitro. As shown in Fig. 7A, in a time-course assay, purified DevSC protein had good DevS autophosphorylation activity (lanes 1–3). However, insignificant stimulation or inhibition on DevS autophosphorylation activity was observed in the presence of 500 μm c-di-GMP (Fig. 7A, lanes 4–6) compared with the reactions in the absence of c-di-GMP. This result indicated that c-di-GMP did not affect the autophosphorylation activity of DevS. We further determine the effect of c-di-GMP on the protein kinase activity of DevS for the phosphorylation of its partner regulator DevR (Fig. S5). As shown in Fig. 7B, with increasing amounts of c-di-GMP (0–400 μm) in the reactions (lanes 3–6), a stepwise increase in the amount of a 32P-labeled autoradiograph signal corresponding to DevR was clearly observed on a polyacrylamide gel. Meanwhile, the amount of the signal corresponding to DevSC decreased (Fig. 7B, lanes 3–6). This result suggested that c-di-GMP can stimulate the DevR-phosphorylation by its two-component kinase DevS.
Figure 7.
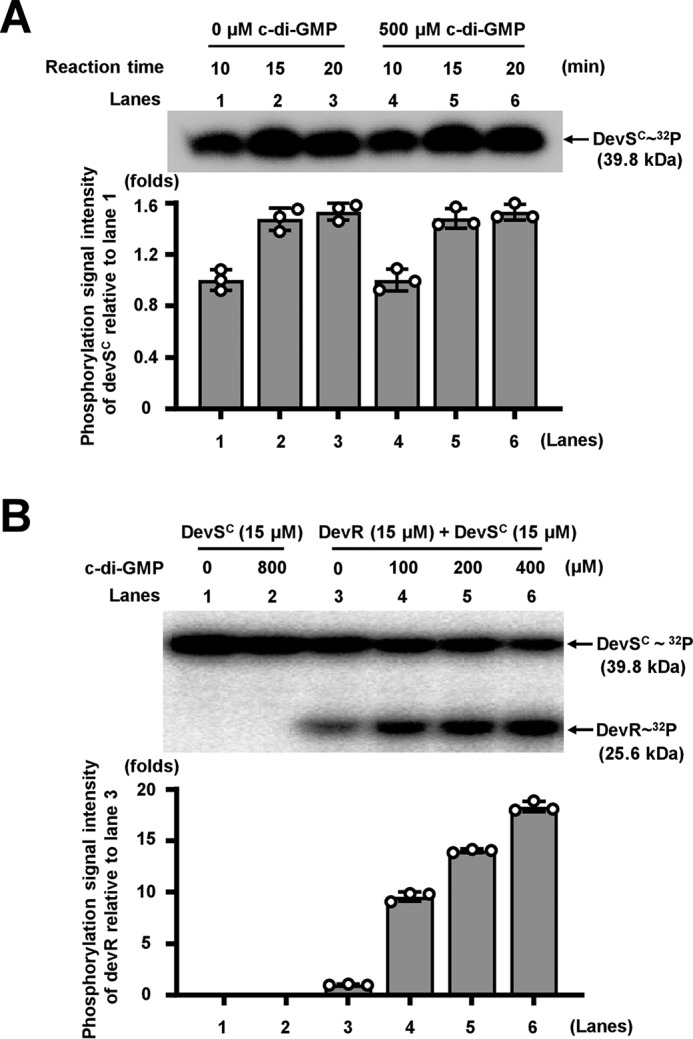
Assays for the effects of c-di-GMP on the kinase activity of DevS and their co-regulation on mycobacterial tolerance to oxidative stress. A, effect of c-di-GMP on the autophosphorylation activity of DevS. DevSC was co-incubated with [γ-32P]ATP, and 10-μl aliquots were removed from the mixture at 10, 15, and 20 min (lanes 1–3), and reaction was quenched immediately. Activity was also detected in the presence of 500 μm c-di-GMP (lanes 4–6). The phosphorylation signal intensity of DevSC was quantified and is shown in the lower panel. B, effect of c-di-GMP on the phosphorylation of DevR by DevS. A total of 15 μm DevSC was fully autophosphorylated for 60 min and mixed with 15 μm DevR for further phosphotransfer reactions in the presence of 100–400 μm c-di-GMP. The phosphorylation signal intensity of DevR was quantified and is shown in the lower panel.
Discussion
c-di-GMP is an important second messenger in bacteria, and its regulatory function has been extensively studied (2–5, 27, 30–35). However, the activation mechanisms of the signaling molecule's receptors remain largely unclear. In this study, we characterized a two-component regulator DevR as a new c-di-GMP–responsive transcription factor in mycobacteria and further uncovered a novel co-activation mechanism of DevR. Our data support that DevR positively regulates the expression of the devR operon and contributes to the mycobacterial oxidative stress tolerance. c-di-GMP directly targets DevR and activates its DNA-binding affinity by enhancing DevR phosphorylation with DevS. Therefore, although c-di-GMP acts as a global signal molecule, its cross-talk with the two-component system DevR/DevS significantly contributed to the regulation of mycobacterial tolerance to oxidative stress.
In our study, DevR senses c-di-GMP through its C-terminal DNA-binding domain. Several types of c-di-GMP–binding motifs have been identified in previous studies, which include the PilZ domain in the Pseudomonas aeruginosa PilZ protein (36), the degenerate GGDEF/EAL domain in the P. aeruginosa PelD protein (30), the GEMM (RNA effectors) domain in the Vibrio cholerae Vc1 protein (38), and the AAAδ54 interaction domain in the P. aeruginosa FleQ protein (39). However, we failed to find any of these known c-di-GMP–binding motifs within the DevRC protein. Our results imply that DevR represents a novel class of receptors/effectors in M. smegmatis.
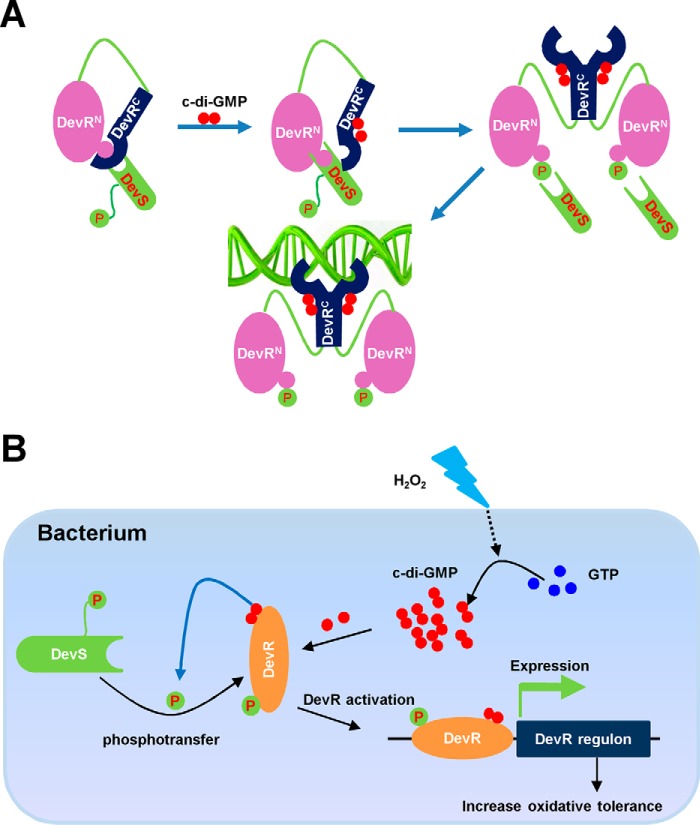
Transcription factors have been successfully characterized as c-di-GMP receptors in different bacterial species (2, 3, 24, 39–41). Generally, c-di-GMP directly stimulates or inhibits the DNA-binding activities of these receptor transcriptional factors (2, 3, 39–41). Different from most previously reported activation models of receptor transcription factors, c-di-GMP physically interacts with DevR but cannot directly affect the DNA-binding activity of DevR in this study. Alternatively, c-di-GMP stimulates the phosphorylation of DevR by its two-component kinase DevS, which finally results in the enhancement of the DNA-binding ability of DevR. Our finding is consistent with a previous structural study of DevR (23). Wisedchaisri et al. (23) solved the crystal structure and proposed that the DevRC of DevR is responsible for DNA binding, which interacts with the DevRN of DevR. The intermolecular interaction covers the phosphorylation site Asp-54, which inhibits the activation of DevR by DevS (23). In this study, we confirm that DevR sensed the c-di-GMP signal through DevRC, and the binding of c-di-GMP stimulated the DevR phosphorylation by its two-component kinase DevS, thereby further triggering the rearrangement of DevR and dimer formation, which finally activates the interaction between DevR and target DNA. Our finding supports a model shown in Fig. 8A representing a novel model for the activation of the c-di-GMP receptor regulator.
Figure 8.
Schematic representation of the mechanism on DevR activation by c-di-GMP and two-component system DevS/DevR and its regulation on mycobacterial tolerance to oxidative stress. A, DevR-activation mechanism co-triggered by c-di-GMP and DevS. DevRN dynamically interacts with DevRC and covers the phosphorylation site (Asp-54). Upon binding of c-di-GMP with the C-terminal domain of DevR (DevRC), its structure is rearranged, thereby disrupting the dynamic interaction between DevRC and DevRN. This allowed Asp-54 to be exposed. Therefore, DevS can transfer the phosphor group to Asp-54 of DevRN, which induced an interaction of DevRC with a second subunit to form an activated DevR dimer. B, model for c-di-GMP–triggered and DevS/DevR-dependent oxidative stress tolerance in mycobacteria. H2O2 induced c-di-GMP accumulation in mycobacteria, and then c-di-GMP bound to DevRC and refolded it to expose the phosphorylation site Asp-54, which enhanced the phosphotransfer between DevS and DevR and activated DevR. The activated DevR bound to the target promoter and up-regulated the expression of target genes to increase the survival rate of mycobacteria under oxidative stress.
The regulatory role of c-di-GMP has been associated with the mycobacterial adaptation to oxidative stress (5). In this study, DevR has been characterized as a new c-di-GMP–responsive receptor regulator, which further links the function of c-di-GMP to stress tolerance in mycobacteria. Recently, DevR was found to be up-regulated under H2O2 stress both in M. smegmatis and in M. tuberculosis (18–20), thereby implying that the regulator may be involved in the oxidative stress response. However, the signaling pathway and molecular mechanism remain unclear. Previously, it was found that H2O2 induces c-di-GMP accumulation in Msm (5). This current study further characterized DevR as a novel c-di-GMP receptor transcriptional factor and found that DevR positively regulated bacterial tolerance to oxidative stress in M. smegmatis. Our data together with previous findings support a model in which, under oxidative stress, c-di-GMP is induced (5) and physically interacts with DevR. Thereafter, this stimulates the phosphorylation of DevR by its two-component kinase DevS and activates the DNA-binding affinity of DevR (Fig. 8B). Then, DevR positively regulates the expression of devR regulon and triggers the mycobacterial adaptation to oxidative stress. Our finding extends the second messenger's regulatory function to stress tolerance in bacteria.
c-di-GMP signaling and two-component systems are two important mechanisms utilized by bacteria to fight against environmental stress. Findings from this study show that the cross-talk between the two systems triggers oxidative stress tolerance in mycobacteria. Several modes have been reported for the interaction between c-di-GMP signal and the two-component system. On the one hand, the two-component system directly regulates the expression or activity of diguanylate cyclase and phosphodiesterase. For example, yaiC in E. coli CFT073 (8) and RapA in Pseudomonas fluorescens are activated by phoB/phoR (9). RavR in Xanthomonas campestris is a diguanylate cyclase in the form of a nonphosphorylated state but obtains phosphodiesterase activity after it is phosphorylated by RavA (11). On the other hand, c-di-GMP physically interacted with two-component kinases. For example, c-di-GMP directly binds to the sensory kinase SgmT in Myxococcus xanthus and spatially sequesters SgmT (10). However, whether and how the c-di-GMP mechanism directly interacts with the response regulator of the two-component system remain unclear. In this study, on the basis of proteomic screening, we detect the correlation of the c-di-GMP signal with the expression of the DevR operon, which contributes to oxidative stress tolerance in Msm. Subsequently, we characterize a direct interaction between c-di-GMP and the two-component regulator DevR. Finally, we uncover a novel mechanism on the c-di-GMP–triggered and phosphorylation-dependent activation of DevR by its two-component kinase DevS. This finding represents a novel paradigm for the cross-talk between the c-di-GMP signal and the two-component systems.
Experimental procedures
Expression and purification of recombinant proteins
The genes in this study were amplified by PCR using their respective primer pairs as follows: 5′-CCCGGAATTCGCATGATCAGGGTTTTTCTGG-3′ and 5′-CTAGTCTAGACTAGTTGCGCCGGTCCAGT-3′ for devRMs; 5′-CCCGGAATTCCGATGATCAGGGTTTTTCTGG-3′ and 5′-CTAGTCTAGACTAGGAGCGTTCGGCGTCG-3′ for devRN (1–144 aa); 5′-CTAGGAATTCTTATGTCCGATCCGCTCTCGGGCCT-3′ and 5′-CTAGTCTAGACTAGTTGCGCCGGTCCAGTT-3′ for devRC (144–211 aa); and 5′-GATAAGAATTCACATGACGCGCGACATCGGC-3′ and 5′-GACGGCGAGTAGTCTAGAGGGTCACCTTCC-3′ for devSC (220–571 aa) from genomic DNA of M. smegmatis mc2 155. The mutant gene of devR-D54V was produced through site-directed mutagenesis by overlapping extension–PCR. The mutagenic PCR primers were designed as follows: 5′-ATGTCGCGGTGCTCGAAGTGCGGCTGCCCGA-3′ and 5′-TCGGGCAGCCGCACTTCGAGCACCGCGACAT-3′. The amplified DNA fragments were cloned into modified pET28a or pMV261 vectors to produce recombinant plasmids (Table S1). The expression strains of E. coli BL21 containing the recombinant plasmids were grown in 1 liter of LB medium at 37 °C up to an A600 of 0.8. Protein expression was induced with 0.6 mm isopropyl d-1-thiogalactopyranoside at 30 °C for 5 h. The cells were harvested and resuspended in binding buffer (100 mm Tris-HCl, pH 8.0, 500 mm NaCl, and 10 mm imidazole) and sonicated. The proteins were purified on a Ni2+-affinity column as described previously (42). The elution was dialyzed against the buffer (50 mm Tris-HCl, pH 7.5, 100 mm NaCl, 10% glycerol) for 2 h and stored at −80 °C.
Quantitative proteomic (iTRAQ) analysis
M. smegmatis was grown in 7H9 medium at 37 °C and cultured to A600 1.2. The cells were suspended in lysis buffer (7 m urea, 2 m thiourea, 4% CHAPS, 40 mm Tris-HCl, pH 8.5, 1 mm phenylmethylsulfonyl fluoride, 2 mm EDTA) and sonicated in ice. The proteins were reduced with 10 mm DTT at 56 °C for 1 h and alkylated by 55 mm iodoacetamide in the darkroom for 1 h. The final samples were kept at −80 °C for further analysis. The next step is iTRAQ labeling and SCX fractionation. 100 μg of proteins were digested with Trypsin Gold (Promega, Madison, WI) at 37 °C for 16 h and further processed according to the manufacturer's protocol (Applied Biosystems). SCX chromatography was performed with an LC-20AB HPLC pump system (Shimadzu, Kyoto, Japan). For LC-ESI-MS/MS analysis, each fraction was resuspended in buffer A (5% acetonitrile, 0.1% formic acid) and centrifuged at 20,000 × g for 10 min. 10 μl of supernatants were loaded, and data acquisition was performed with a triple TOF 5600 system (AB SCIEX, Concord, Ontario, Canada). Raw data files were converted into MGF files, and protein identifications were performed by using a Mascot search engine (version 2.3.02, Matrix Science, London, UK) against a database containing M. smegmatis mc2 155 protein sequences. The quantitative protein ratios were weighted and normalized by the median ratio in Mascot.
Cross-linking assays
The cross-linking assays were conducted as described previously (5) with modifications. Briefly, radioactively-labeled c-di-GMP was enzymatically produced from [α-32P]GTP by E. coli diguanylate cyclase encoded by the ydeH gene (25). 10 μm YdeH was co-incubated with 30 μm [α-32P]GTP in DGC buffer (50 mm Tris-HCl, pH 7.5, 50 mm NaCl, and 5 mm MgCl2) at 37 °C for 12 h. 12 μm DevR, DevRN, DevRC, or STING proteins were co-incubated with 5 μm radioactively-labeled c-di-GMP in DGC buffer on ice for 30 min. For competitive experiment assays, the competing cold ligands c-di-GMP, ATP, and GTP were added at the same time with radioactively-labeled c-di-GMP and co-incubated with 12 μm DevR for 30 min on ice, respectively. Then, samples were irradiated by UV for 30 min and directly subjected to 12% SDS-PAGE. Electrophoresis was performed at 120 V for 1 h. Gels were exposed to a storage phosphor screen (GE Healthcare) overnight. Images were acquired using a Typhoon Scanner (GE Healthcare).
Bacterial one-hybrid assays
Bacterial one-hybrid assays were performed as described previously (26) with modifications. devR was cloned into pTRG vector (Stratagene). Promoters of the M. smegmatis genes were cloned into pBXcmT vector. All recombinant pTRG and pBXcmT plasmids were transformed into E. coli XL1-Blue MRF′ Kan strain (Stratagene) (Table S1). Co-transformants containing the pBX-Rv2031/pTRG-Rv3133c plasmids (26) served as positive control, and co-transformants containing the empty vectors pBX and pTRG served as negative control. Positive-growth co-transformants were selected on a selective screening medium plate containing 20 mm 3-amino-1,2,4-triazole, 16 μg/ml streptomycin, 15 μg/ml tetracycline, 34 μg/ml chloramphenicol, and 50 μg/ml kanamycin.
ChIP assay
ChIP was carried out as described previously (5) with some modifications. The late-logarithmic phase cells of M. smegmatis mc2155 were fixed with 1% formaldehyde at room temperature for 20 min and stopped with 125 mm glycine. The cross-linked cells were harvested and resuspended in 1 ml of TBSTT buffer (20 mm Tris-HCl, 150 mm NaCl, 0.1% Tween 20, and 0.1% Triton X-100). The sample was sonicated, and the supernatant extract was collected by centrifugation. The sample extracts were incubated with 10 μl of antibodies against DevR or preimmune serum under rotation for 3 h at 4 °C. The complexes were immunoprecipitated with 20 μl of 50% protein A–agarose under rotation for 1 h at 4 °C. The immunocomplexes were collected by centrifugation and resuspended in 100 μl of TE (20 mm Tris-HCl, pH 7.8, 10 mm EDTA, 0.5% SDS). Then, the cross-linking was reversed for 6 h at 65 °C. The DNA samples of the input and ChIP were purified and analyzed by PCR. Three pairs of primers were used for PCR analysis: 5′-ACCGGAATTCGCAAGTCCCTGCTGGACAAC-3′ and 5′-CTAGTCTAGATCAACAGGTCCCAGCACTC-3′ for Ms5243p; 5′-ACCGGAATTCCGGTGATCGAGAAGCTCA-3′ and 5′-CTAGTCTAGAAGGAACAGCTTCAGCTCGC-3′ for Ms5246p; and 5′-ACCGGAATTCCCGTACCAGGACAAGGT-3′ and 5′-CTAGTCTAGAGGAACGGCGCTCTTCATG-3′ for Ms5247p.
Quantitative PCR analysis
qPCRs were performed as described previously (5). The 25-μl reaction mixture of PCR contained 10 μl of 2× SYBR qPCR Mix kit (Aidlab, China), 200 nm Ms5246p-specific primer pairs(5′-GGCGCAGTCGCTCGAA-3′ and 5′-TCCCACCCGCAGGTTGTC-3′), and 2 μl of the immunoprecipitated and purified DNA samples from the ChIP assays. Each reaction was performed in triplicate. Ms5246p was amplified and detected using a CFX96 instrument (Bio-Rad) with the following protocol: 95 °C for 5 min, followed by 40 cycles of 95 °C for 15 s, 60 °C for 15 s, and 72 °C for 45 s; melting curve, 55–99 °C, 0.5 °C/10 s, 25 °C for 5 min. The data were analyzed with Bio-Rad CFX Manager version 2.1. Amplification specificity was assessed using melting-curve analysis. The relative quantity of Ms5246p in ChIP was normalized to the quantity of Ms5246p in input. The degree of change in relative quantity of Ms5246p was calculated using the 2−ΔΔCt method (43). For statistical analysis, two-tailed Student's t tests were performed.
EMSA
EMSAs were carried out as described previously (24). Briefly, the DNA fragments for EMSA were amplified by PCR. A pair of primers (5′-ACCGGAATTCCGGTGATCGAGAAGCTCA-3′ and 5′-CTAGTCTAGAAGGAACAGCTTCAGCTCGC-3′) was used to amplify the upstream regulatory sequence of devR operon, named as Ms5246p (375 bp). For competition assay, the Ms5246p DNA fragment was re-amplified with the FITC-labeled reverse primer. The DNA substrates (5 ng/μl) and different amounts of proteins were co-incubated in EMSA buffer (50 mm Tris-HCl, pH 7.5, 100 mm NaCl, 5 mm MgCl2, 10% glycerol) for 30 min on ice. The reaction mixtures were then subjected to 4.5% native polyacrylamide gel. Electrophoresis was performed at 150 V at room temperature for 1.5 h in 0.5× Tris borate-EDTA buffer. Images of gels were acquired using a Typhoon scanner (GE Healthcare).
β-Gal activity assays
β-Gal activity experiments were performed in M. smegmatis by constructing promoter–lacZ fusions based on the expression vector of pMV261 (5). The elements, including promoters ydeH, ydeH (E208,209A), devR, devRD54V, and lacZ, were sequentially cloned into the pMV261 backbone as indicated in the figures. The recombinant plasmids were transformed into the devR knockout strain, devS knockout strain, or WT M. smegmatis strain. Mycobacterial strains were grown in 7H9 medium at 37 °C until late logarithmic phage. For detecting DevR expression under H2O2 stress, the logarithmic strains continued to culture with the addition of 6 mm H2O2 for 3 h. The cells were harvested and washed with phosphate-buffered saline (PBS). β-Gal measurements were performed as described previously (44).
Isothermal titration calorimetry (ITC) analysis
ITC assays were performed at 25 °C with a nano-ITC low volume isothermal calorimeter (TA Instruments, New Castle, DE) controlled by ITC-run software as described previously (28). Briefly, the DevR protein was dialyzed in the buffer (20 mm Tris base, 100 mm NaCl, 5 mm MgCl2, pH 7.5), and all buffers were degassed before use. DevR (13.5 μm) and the c-di-GMP (100 μm) were added into the sample cell (350 μl) and the syringe (50 μl), respectively. Data were recorded automatically and subsequently analyzed using the NanoAnalyze software provided by the manufacturer. In control experiments, the c-di-GMP solution was titrated into the buffer in sample cells to obtain the heat of dilution. The value of the heat of dilution was then subtracted from the experimental curve in the final analysis. All the titration curves were fitted to the independent-site binding model.
Autophosphorylation and phosphotransfer assays
The autophosphorylation and phosphotransfer assays were performed as described previously (45) with modifications. Briefly, for autophosphorylation, DevSC (15 μm), which represents the active C-terminal fragment of MSMEG_5241 encoded by 220–571 amino acids, was co-incubated with 5 μCi of [γ-32P]ATP (3000 Ci/mm) in 30 μl of reaction buffer (50 mm Tris-HCl, pH 8.0, 50 mm KCl, 25 mm MgCl2, 50 μm ATP) at room temperature. 10-μl aliquots were removed from the mixture at 10, 15, and 20 min and were stopped with the addition of 5 μl of SDS-PAGE loading buffer. For detecting the effect of c-di-GMP on phosphotransfer, DevSC was first autophosphorylated for 60 min. 15 μm DevR was incubated with different amounts of c-di-GMP on ice for 30 min. Then, the mixtures were co-incubated with autophosphorylated DevSC at room temperature for 2 min and stopped immediately with addition SDS-PAGE loading buffer. The radioactive SDS-polyacrylamide gels were exposed to a storage phosphor screen (GE Healthcare) overnight. Images were obtained using Typhoon Scanner (GE Healthcare).
Assays for survival of mycobacteria under H2O2 stress
Assays for the sensitivity of mycobacteria to H2O2 were carried out as described previously (37) with several modifications. Briefly, the WT and recombinant mycobacterial strains were cultured to mid-log phage and then diluted in fresh 7H9 medium until A600 = 0.1. Then, 9 mm H2O2 were added into the medium when the A600 of the cells reached to 0.5 and then continued to culture for 3 h. The cells were harvested and washed with PBS, then diluted with PBS, and plated onto 7H10 plates to count the colony-forming units (cfu). The survival percentages were calculated by cfu (with H2O2)/cfu (without H2O2) for each strain.
Author contributions
Q. H., W. L., and Z.-G. H. conceptualization; Q. H., J. Z., Y. C., and L. H. data curation; Q. H. formal analysis; Q. H., W. L., and Z.-G. H. investigation; Q. H. methodology; Q. H., W. L., and Z.-G. H. writing-original draft; Q. H., W. L., and Z.-G. H. project administration; Q. H., W. L., and Z.-G. H. writing-review and editing; W. L. and Z.-G. H. supervision; W. L. and Z.-G. H. funding acquisition.
Supplementary Material
This work was supported by the National Natural Science Foundation of China Grant 31730005, National Key R&D Program of China Grant 2017YFD0500300, and NSFC Grants 31670075 and 31870036. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S5 and Table S1.
The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD010623.
- c-di-GMP
- cyclic di-GMP
- iTRAQ
- isobaric tags for relative and absolute quantitation
- aa
- amino acid
- Msm
- M. smegmatis
- qPCR
- quantitative PCR
- ITC
- isothermal titration calorimetry
- cfu
- colony-forming unit
- SCX
- strong cation exchange choematography.
References
- 1. Lori C., Ozaki S., Steiner S., Böhm R., Abel S., Dubey B. N., Schirmer T., Hiller S., and Jenal U. (2015) Cyclic di-GMP acts as a cell cycle oscillator to drive chromosome replication. Nature 523, 236–239 10.1038/nature14473 [DOI] [PubMed] [Google Scholar]
- 2. Tschowri N., Schumacher M. A., Schlimpert S., Chinnam N. B., Findlay K. C., Brennan R. G., and Buttner M. J. (2014) Tetrameric c-di-GMP mediates effective transcription factor dimerization to control Streptomyces development. Cell 158, 1136–1147 10.1016/j.cell.2014.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Krasteva P. V., Fong J. C., Shikuma N. J., Beyhan S., Navarro M. V., Yildiz F. H., and Sondermann H. (2010) Vibrio cholerae VpsT regulates matrix production and motility by directly Sensing cyclic di-GMP. Science 327, 866–868 10.1126/science.1181185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boehm A., Kaiser M., Li H., Spangler C., Kasper C. A., Ackermann M., Kaever V., Sourjik V., Roth V., and Jenal U. (2010) Second messenger-mediated adjustment of bacterial swimming velocity. Cell 141, 107–116 10.1016/j.cell.2010.01.018 [DOI] [PubMed] [Google Scholar]
- 5. Li W., Li M., Hu L., Zhu J., Xie Z., Chen J., and He Z. G. (2018) HpoR, a novel c-di-GMP effective transcription factor, links the second messenger's regulatory function to the mycobacterial antioxidant defense. Nucleic Acids Res. 46, 3595–3611 10.1093/nar/gky146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Oyston P. C., Dorrell N., Williams K., Li S. R., Green M., Titball R. W., and Wren B. W. (2000) The response regulator PhoP is important for survival under conditions of macrophage-induced stress and virulence in Yersinia pestis. Infect. Immun. 68, 3419–3425 10.1128/IAI.68.6.3419-3425.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Altamirano-Silva P., Meza-Torres J., Castillo-Zeledón A., Ruiz-Villalobos N., Zuñiga-Pereira A. M., Chacón-Díaz C., Moreno E., Guzmán-Verri C., and Chaves-Olarte E. (2018) Brucella abortus senses the intracellular environment through the BvrR/BvrS two-component system, which allows B. abortus to adapt to its replicative niche. Infect. Immun. 86, e00713 10.1128/IAI.00713-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Crépin S., Porcheron G., Houle S., Harel J., and Dozois C. M. (2017) Altered regulation of the diguanylate cyclase YaiC reduces production of type 1 fimbriae in a Pst mutant of uropathogenic Escherichia coli CFT073. J. Bacteriol. 199, e00168 10.1128/JB.00168-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Monds R. D., Newell P. D., Gross R. H., and O'Toole G. A. (2007) Phosphate-dependent modulation of c-di-GMP levels regulates Pseudomonas fluorescens Pf0-1 biofilm formation by controlling secretion of the adhesin LapA. Mol. Microbiol. 63, 656–679 10.1111/j.1365-2958.2006.05539.x [DOI] [PubMed] [Google Scholar]
- 10. Petters T., Zhang X., Nesper J., Treuner-Lange A., Gomez-Santos N., Hoppert M., Jenal U., and Søgaard-Andersen L. (2012) The orphan histidine protein kinase SgmT is a c-di-GMP receptor and regulates composition of the extracellular matrix together with the orphan DNA binding response regulator DigR in Myxococcus xanthus. Mol. Microbiol. 84, 147–165 10.1111/j.1365-2958.2012.08015.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tao J., Li C., Luo C., and He C. (2014) RavA/RavR two-component system regulates Xanthomonas campestris pathogenesis and c-di-GMP turnover. FEMS Microbiol. Lett. 358, 81–90 10.1111/1574-6968.12529 [DOI] [PubMed] [Google Scholar]
- 12. Ehrt S., and Schnappinger D. (2009) Mycobacterial survival strategies in the phagosome: defence against host stresses. Cell. Microbiol. 11, 1170–1178 10.1111/j.1462-5822.2009.01335.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stokes R. W., and Waddell S. J. (2009) Adjusting to a new home: Mycobacterium tuberculosis gene expression in response to an intracellular lifestyle. Future Microbiol. 4, 1317–1335 10.2217/fmb.09.94 [DOI] [PubMed] [Google Scholar]
- 14. Kinger A. K., and Tyagi J. S. (1993) Identification and cloning of genes differentially expressed in the virulent strain of Mycobacterium tuberculosis. Gene 131, 113–117 10.1016/0378-1119(93)90678-V [DOI] [PubMed] [Google Scholar]
- 15. Dasgupta N., Kapur V., Singh K. K., Das T. K., Sachdeva S., Jyothisri K., and Tyagi J. S. (2000) Characterization of a two-component system, devR-devS, of Mycobacterium tuberculosis. Tuber. Lung Dis. 80, 141–159 10.1054/tuld.2000.0240 [DOI] [PubMed] [Google Scholar]
- 16. Boon C., and Dick T. (2002) Mycobacterium bovis BCG response regulator essential for hypoxic dormancy. J. Bacteriol. 184, 6760–6767 10.1128/JB.184.24.6760-6767.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Roberts D. M., Liao R. P., Wisedchaisri G., Hol W. G., and Sherman D. R. (2004) Two sensor kinases contribute to the hypoxic response of Mycobacterium tuberculosis. J. Biol. Chem. 279, 23082–23087 10.1074/jbc.M401230200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kendall S. L., Movahedzadeh F., Rison S. C., Wernisch L., Parish T., Duncan K., Betts J. C., and Stoker N. G. (2004) The Mycobacterium tuberculosis dosRS two-component system is induced by multiple stresses. Tuberculosis 84, 247–255 10.1016/j.tube.2003.12.007 [DOI] [PubMed] [Google Scholar]
- 19. Nde C. W., Toghrol F., Jang H. J., and Bentley W. E. (2011) Toxicogenomic response of Mycobacterium bovis BCG to peracetic acid and a comparative analysis of the M. bovis BCG response to three oxidative disinfectants. Appl. Microbiol. Biotechnol. 90, 277–304 10.1007/s00253-010-2931-6 [DOI] [PubMed] [Google Scholar]
- 20. Li X., Wu J., Han J., Hu Y., and Mi K. (2015) Distinct responses of Mycobacterium smegmatis to exposure to low and high levels of hydrogen peroxide. PLoS One 10, e0134595 10.1371/journal.pone.0134595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chauhan S., and Tyagi J. S. (2008) Cooperative binding of phosphorylated DevR to upstream sites is necessary and sufficient for activation of the Rv3134c-devRS operon in Mycobacterium tuberculosis: implication in the induction of DevR target genes. J. Bacteriol. 190, 4301–4312 10.1128/JB.01308-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wisedchaisri G., Wu M., Rice A. E., Roberts D. M., Sherman D. R., and Hol W. G. (2005) Structures of Mycobacterium tuberculosis DosR and DosR–DNA complex involved in gene activation during adaptation to hypoxic latency. J. Mol. Biol. 354, 630–641 10.1016/j.jmb.2005.09.048 [DOI] [PubMed] [Google Scholar]
- 23. Wisedchaisri G., Wu M., Sherman D. R., and Hol W. G. (2008) Crystal structures of the response regulator DosR from Mycobacterium tuberculosis suggest a helix rearrangement mechanism for phosphorylation activation. J. Mol. Biol. 378, 227–242 10.1016/j.jmb.2008.02.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li W., and He Z. G. (2012) LtmA, a novel cyclic di-GMP-responsive activator, broadly regulates the expression of lipid transport and metabolism genes in Mycobacterium smegmatis. Nucleic Acids Res. 40, 11292–11307 10.1093/nar/gks923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zähringer F., Massa C., and Schirmer T. (2011) Efficient enzymatic production of the bacterial second messenger c-di-GMP by the diguanylate cyclase YdeH from E. coli. Appl. Biochem. Biotechnol. 163, 71–79 10.1007/s12010-010-9017-x [DOI] [PubMed] [Google Scholar]
- 26. Guo M., Feng H., Zhang J., Wang W., Wang Y., Li Y., Gao C., Chen H., Feng Y., and He Z. G. (2009) Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. Genome Res. 19, 1301–1308 10.1101/gr.086595.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Burdette D. L., Monroe K. M., Sotelo-Troha K., Iwig J. S., Eckert B., Hyodo M., Hayakawa Y., and Vance R. E. (2011) STING is a direct innate immune sensor of cyclic di-GMP. Nature 478, 515–518 10.1038/nature10429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li W., Cui T., Hu L., Wang Z., Li Z., and He Z. G. (2015) Cyclic diguanylate monophosphate directly binds to human siderocalin and inhibits its antibacterial activity. Nat. Commun. 6, 8330 10.1038/ncomms9330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Saini D. K., Malhotra V., Dey D., Pant N., Das T. K., and Tyagi J. S. (2004) DevR–DevS is a bona fide two-component system of Mycobacterium tuberculosis that is hypoxia-responsive in the absence of the DNA-binding domain of DevR. Microbiology 150, 865–875 10.1099/mic.0.26218-0 [DOI] [PubMed] [Google Scholar]
- 30. Lee V. T., Matewish J. M., Kessler J. L., Hyodo M., Hayakawa Y., and Lory S. (2007) A cyclic-di-GMP receptor required for bacterial exopolysaccharide production. Mol. Microbiol. 65, 1474–1484 10.1111/j.1365-2958.2007.05879.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wilksch J. J., Yang J., Clements A., Gabbe J. L., Short K. R., Cao H., Cavaliere R., James C. E., Whitchurch C. B., Schembri M. A., Chuah M. L., Liang Z. X., Wijburg O. L., Jenney A. W., Lithgow T., and Strugnell R. A. (2011) MrkH, a novel c-di-GMP-dependent transcriptional activator, controls Klebsiella pneumoniae biofilm formation by regulating type 3 fmbriae expression. PLoS Pathog. 7, e1002204 10.1371/journal.ppat.1002204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ryan R. P., Fouhy Y., Lucey J. F., Jiang B. L., He Y. Q., Feng J. X., Tang J. L., and Dow J. M. (2007) Cyclic di-GMP signalling in the virulence and environmental adaptation of Xanthomonas campestris. Mol. Microbiol. 63, 429–442 10.1111/j.1365-2958.2006.05531.x [DOI] [PubMed] [Google Scholar]
- 33. Duerig A., Abel S., Folcher M., Nicollier M., Schwede T., Amiot N., Giese B., and Jenal U. (2009) Second messenger-mediated spatiotemporal control of protein degradation regulates bacterial cell cycle progression. Genes Dev. 23, 93–104 10.1101/gad.502409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Parvatiyar K., Zhang Z., Teles R. M., Ouyang S., Jiang Y., Iyer S. S., Zaver S. A., Schenk M., Zeng S., Zhong W., Liu Z. J., Modlin R. L., Liu Y. J., and Cheng G. (2012) The helicase DDX41 recognizes the bacterial secondary messengers cyclic di-GMP and cyclic di-AMP to activate a type I interferon immune response. Nat. Immunol. 13, 1155–1161 10.1038/ni.2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Strempel N., Nusser M., Neidig A., Brenner-Weiss G., and Overhage J. (2017) The oxidative stress agent hypochlorite stimulates c-di-GMP synthesis and biofilm formation in Pseudomonas aeruginosa. Front. Microbiol. 8, 2311 10.3389/fmicb.2017.02311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Amikam D., and Galperin M. Y. (2006) PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics 22, 3–6 10.1093/bioinformatics/bti739 [DOI] [PubMed] [Google Scholar]
- 37. Colangeli R., Haq A., Arcus V. L., Summers E., Magliozzo R. S., McBride A., Mitra A. K., Radjainia M., Khajo A., Jacobs W. R. Jr, Salgame P., and Alland D. (2009) The multifunctional histone-like protein Lsr2 protects mycobacteria against reactive oxygen intermediates. Proc. Natl. Acad. Sci. U.S.A. 106, 4414–4418 10.1073/pnas.0810126106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kariisa A. T., Weeks K., and Tamayo R. (2016) The RNA domain Vc1 regulates downstream gene expression in response to cyclic diguanylate in Vibrio cholerae. PLoS One 11, e0148478 10.1371/journal.pone.0148478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hickman J. W., and Harwood C. S. (2008) Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol. Microbiol. 69, 376–389 10.1111/j.1365-2958.2008.06281.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fazli M., O'Connell A., Nilsson M., Niehaus K., Dow J. M., Givskov M., Ryan R. P., and Tolker-Nielsen T. (2011) The CRP/FNR family protein Bcam1349 is a c-di-GMP effector that regulates biofilm formation in the respiratory pathogen Burkholderia cenocepacia. Mol. Microbiol. 82, 327–341 10.1111/j.1365-2958.2011.07814.x [DOI] [PubMed] [Google Scholar]
- 41. Tao F., He Y. W., Wu D. H., Swarup S., and Zhang L. H. (2010) The cyclic nucleotide monophosphate domain of Xanthomonas campestris global regulator Clp defines a new class of cyclic di-GMP effectors. J. Bacteriol. 192, 1020–1029 10.1128/JB.01253-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li W., Hu L., Xie Z., Xu H., Li M., Cui T., and He Z. G. (2018) Cyclic di-GMP integrates functionally divergent transcription factors into a regulation pathway for antioxidant defense. Nucleic Acids Res. 46, 7270–7283 10.1093/nar/gky611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Livak K. J., and Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔC(T)) method. Methods 25, 402–408 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 44. Miller J. H. (1972) Experiments in Molecular Genetics, pp. 352–355, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 45. Saini D. K., Pant N., Das T. K., and Tyagi J. S. (2002) Cloning, overexpression, purification, and matrix-assisted refolding of DevS (Rv3132c) histidine protein kinase of Mycobacterium tuberculosis. Protein Expr. Purif. 25, 203–208 10.1006/prep.2002.1628 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.