Abstract
The sterol regulatory element-binding proteins (SREBPs) are a family of transcription factors best known for stimulating the expression of genes encoding key lipogenic enzymes. However, SREBP functions beyond lipid metabolism are less understood. Here, we show that hepcidin antimicrobial peptide (Hamp), encoding the hormone hepcidin essential for iron homeostasis and regulated by dietary iron and inflammation, is a target gene of the two SREBP isoforms SREBP-1a/c. We found that in tissue culture, mature, active, and nuclear forms of the SREBP-1a/c proteins induce endogenous Hamp gene expression and increase the Hamp promoter activity primarily via three regulatory sequences, including an E-box. Moreover, ChIP experiments revealed that SREBP-1a binds to the Hamp gene promoter. Overexpression of nuclear SREBP-1a under the control of the phosphoenolpyruvate carboxylase-1 (Pck1) promoter in mice increased hepatic Hamp mRNA and blood hepcidin levels, and as expected, caused fatty liver. Consistent with the known effects of Hamp up-regulation, SREBP-1a–overexpressing mice displayed signs of dysregulation in iron metabolism, including reduced serum iron and increased hepatic and splenic iron storage. Conversely, liver-specific depletion of the nuclear forms of SREBPs, as in SREBP cleavage-activating protein knockout mice, impaired lipopolysaccharide-induced up-regulation of hepatic Hamp. Together, these results indicate that the SREBP-1a/c transcription regulators activate hepcidin expression and thereby contribute to the control of mammalian iron metabolism.
Keywords: DNA transcription, transcription factor, iron metabolism, transcription coregulator, inflammation, lipogenesis, Hamp, hepcidin, LPS, Mediator, SREBP
Introduction
Nonalcoholic fatty liver disease (NAFLD)3 affects nearly one-third of the adults in the United States (1). Although steatosis alone is benign, it can progress to steatohepatitis, fibrosis, cirrhosis, and hepatocellular carcinoma (2, 3). When energy is excessive, hepatic lipid accumulation is largely due to defects in key pathways of lipid metabolism, including de novo lipogenesis, fatty acid β-oxidation/esterification, and very-low density lipoprotein production/secretion. Among these pathways, de novo lipogenesis is increased in human patients with NAFLD (4–7) and animal models (8). Meanwhile, a significant number of NAFLD patients also display abnormality in iron status with increased iron accumulation in macrophages (9). However, it is unclear whether there is a cross-talk between lipogenesis and iron metabolism.
Regulation of de novo lipogenesis is a complex process that depends upon the hormone states, availability of substrates, allosteric regulation of key enzymatic activity steps, and the transcriptional control of lipogenic gene expression through key transcription factors, including sterol regulatory element-binding proteins (SREBP), carbohydrate responsive element-binding proteins (ChREBP), and liver X receptors (LXR) (10, 11). The SREBP transcription factors have three major isoforms (i.e. SREBP-1a, -1c and -2), encoded by two different genes Srebf1 and Srebf2 (12). The Srebf1 gene produces SREBP-1a or SREBP-1c isoforms (collectively called SREBP-1), of which SREBP-1a is a few amino acids longer than SREBP-1c at the N terminus and is transcriptionally more active (12). All SREBP transcription factors are translated as precursors residing in the endoplasmic reticulum membrane (13). Under conditions that favor lipid biosynthesis, SREBP precursors are processed to generate mature/active/nuclear forms of SREBP transcription factors through a proteolytic cleavage process, in which SREBP cleavage-activating protein (SCAP) is essentially required (13–16). In the nucleus, the transcriptional activities of nuclear SREBPs are regulated by a number of transcriptional cofactors, including CBP/p300 (17) and the Mediator complex (18, 19). The best-described function of SREBPs is to regulate lipid metabolism, as they are able to activate the transcription of genes encoding key enzymes for the biosynthesis of fatty acids by SREBP-1a/c or cholesterol by SREBP-2 (13–16). Thus, SREBP transcription factors play a role in the development of NAFLD, but their functions beyond lipid metabolism have been less studied.
Iron is an essential cofactor for numerous cellular proteins involved in key cellular processes, including heme biosynthesis, oxygen transport, mitochondrial respiration, DNA synthesis, and posttranslational modifications of various proteins (20–23). According to the World Health Organization (WHO) data, iron deficiency is a common condition affecting millions of people worldwide. Iron deficiency is known to cause anemia, but iron overload leads to over-production of oxygen-free radicals, causing oxidative stress that may damage organs, particularly liver and heart (20, 24). Therefore, iron homeostasis is tightly regulated under normal physiological conditions. The liver plays a pivotal role in the maintenance of iron homeostasis, as hepcidin, the only known hormone that is essential in the regulation of iron status, is primarily produced by hepatocytes (20, 24). Hepcidin regulates iron homeostasis mainly through inhibiting intestinal iron absorption and blocking iron export from macrophage (20, 24). Hepcidin is encoded by the Hamp gene, and blood hepcidin levels are primarily regulated at the transcription level in response to changes in iron levels in the body, inflammation or stress (20, 24).
In this study, we show that SREBP-1 transcription factors activate hepatic Hamp gene expression, resulting in changes of iron status. Our data suggest that the lipogenic activators SREBP-1a/c may also play a role in the regulation of iron metabolism, especially under certain pathophysiological conditions where the mature/active/nuclear forms of SREBP-1a/c proteins are up-regulated in the liver.
Results
SREBP-1 activates the Hamp gene expression in vitro
Previous studies have demonstrated that SREBP-1 transcription factors activate the target gene promoters through sterol response element (SRE) sites as well as E-boxes, where SREBP-1 proteins are in complex with upstream stimulatory factors (USF-1/2) (25–28). There are two conserved E-boxes in the Hamp gene promoter, and it has been shown that USF-1/2 transcription factors are critically required to activate the Hamp promoter and regulate iron homeostasis in mouse models (29, 30). Thus, we hypothesize that Hamp can be activated by SREBP-1 transcription factors.
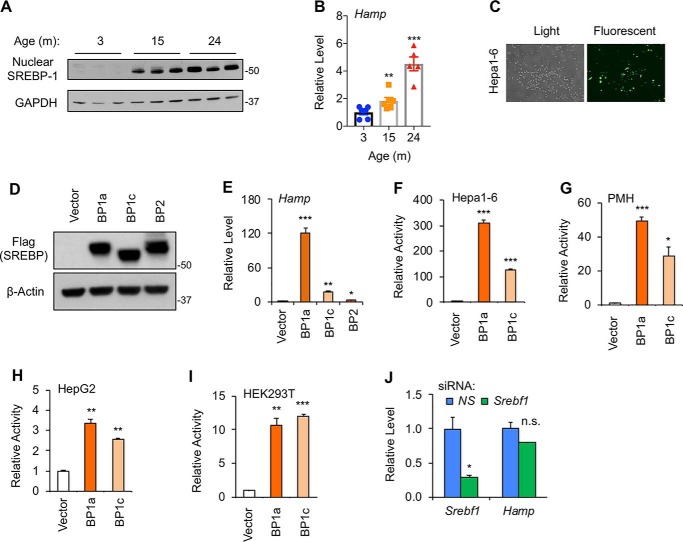
To test our hypothesis, we first asked whether hepatic Hamp expression is correlated with SREBP-1 protein levels under any physiological and/or pathophysiological conditions. For that purpose, we compared hepatic SREBP-1 proteins between young and old C57BL/6J mice that were fasted overnight. Although the nuclear forms of SREBP-1 proteins were barely detectable at the age of 3 months, they were significantly accumulated at the age of 15 or 24 months (Fig. 1A). This result confirmed our previous report showing that nuclear SREBP-1 proteins are increased during aging (31). The mRNA levels of hepatic Hamp were also elevated during aging (Fig. 1B), supporting the possibility that SREBP-1 may activate hepatic Hamp expression.
Figure 1.
Accumulation of nuclear SREBP-1 induces Hamp gene expression. Immunoblots of the indicated proteins (A) and qRT-PCR analyses of Hamp mRNA levels (B) in livers of male C57BL/6J mice at the indicated ages. C, representative images of Hepa1–6 cells upon transfection of GFP-expressing plasmids. Immunoblots of the indicated nuclear SREBP isoforms (D), which were transiently overexpressed in Hepa1–6 cells, and qRT-PCR analyses of Hamp mRNA levels (E) upon nuclear SREBP overexpression. Luciferase reporter assays for the effects of SREBP-1a or -1c on a 3-kb human Hamp promoter in Hepa1–6 cells (F), PMH (G), HepG2 cells (H), and HEK293T cells (I). J, qRT-PCR analyses of the indicated genes in PMH upon depletion of SREBP-1 by siRNA. *, p < 0.05; **, p < 0.01; and ***, p < 0.001 versus Control (vector alone for transfection, n = 3 with independent repeats; or 3 months of age for mouse livers, n = 6. NS, nonspecific; n.s., not significant).
To determine whether the SREBP transcription factors indeed act as activators of the Hamp promoter, we overexpressed FLAG-tagged nuclear forms of SREBP-1a, SREBP-1c, or SREBP-2 isoforms by transient transfection in Hepa1–6 cells, in which transfection efficiency was relatively high (Fig. 1C). Three FLAG-tagged SREBP isoforms were overexpressed at a similar level as determined by immunoblotting using anti-FLAG antibody (Fig. 1D). As shown in Fig. 1E, overexpression of nuclear SREBP-1a or SREBP-1c robustly increased endogenous Hamp mRNA levels, and the fold of induction was consistent with the fact that SREBP-1a is transcriptionally more active than SREBP-1c. In contrast, overexpression of nuclear SREBP-2 barely induced the Hamp gene expression (Fig. 1E). These data suggest that SREBP-1 transcription factors are able to stimulate the Hamp gene expression in hepatocyte cell lines.
To determine whether SREBP-1a/c could activate the Hamp gene promoter, we performed luciferase reporter assays using a 3-kb human Hamp promoter, which has been previously defined (32, 33). As shown in Fig. 1F, overexpression of SREBP-1a or SREBP-1c robustly activated this Hamp promoter in Hepa1–6 cells. Similarly, SREBP-1a and SREBP-1c were also able to activate the Hamp promoter in primary mouse hepatocytes (PMH) (Fig. 1G) and the human hepatocellular carcinoma cell line HepG2 (Fig. 1H). Interestingly, overexpression of SREBP-1a or SREBP-1c also activated the Hamp promoter in HEK293T cells (Fig. 1I), suggesting that liver-specific factor(s) may not be required for SREBP-1a/c–induced activation of the Hamp gene promoter. Surprisingly, depletion of SREBP-1 by specific small-interfering RNA (siRNA) in PMH did not cause a significant change of Hamp (Fig. 1J). Together, these data suggest that low basal levels of nuclear SREBP-1 may contribute little to Hamp expression, but when accumulated, as in NAFLD or aging, nuclear SREBP-1a/c may activate Hamp gene transcription.
The Mediator complex regulates Hamp expression
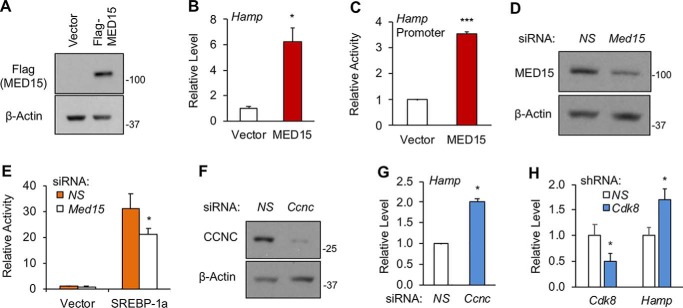
The Mediator complex is a conserved transcription cofactor that critically regulates activator-dependent transcription (34–37). In mammalian cells, the Mediator complex is comprised of about 30 subunits (34–37). We have previously shown that SREBP transcriptional activities require MED15, a subunit of the Mediator complex (18). If SREBP-1a/c can activate Hamp expression, MED15 may also regulate Hamp. To explore this possibility, we overexpressed MED15 in Hepa1–6 cells by transient transfection (Fig. 2A), and examined the effects of MED15 on the endogenous Hamp gene expression by qRT-PCR. As shown in Fig. 2B, MED15 overexpression increased the Hamp mRNA levels by 6-fold. Moreover, MED15 overexpression stimulated the Hamp promoter as detected by luciferase reporter assays (Fig. 2C). Conversely, we depleted MED15 in Hepa1–6 cells using specific siRNA (Fig. 2D). Although MED15 knockdown alone had little effects, loss of MED15 significantly reduced SREBP-1a–induced activation of the Hamp promoter (Fig. 2E).
Figure 2.
The Mediator complex subunits MED15, CCNC, and CDK8 regulate Hamp gene expression. A, immunoblots of the indicated proteins; B, qRT-PCR analyses of Hamp mRNA levels; and C, luciferase reporter assays for the activity of a 3-kb human Hamp promoter in Hepa1–6 cells after overexpression of FLAG-tagged MED15 by transient transfection. D, immunoblots of the indicated proteins and E, luciferase reporter assays for the Hamp promoter activity in Hepa1–6 cells upon depletion of MED15 by siRNA. F, immunoblots of the indicated proteins and G, qRT-PCR analyses of Hamp mRNA levels in FAO cells upon depletion of CCNC by siRNA. H, qRT-PCR analyses of the indicated genes in HepG2 cells upon depletion of CDK8 by shRNA. *, p < 0.05 and ***, p < 0.001 versus Control (NS-siRNA or shRNA, or Vector alone, n = 3 with independent repeats; NS, nonspecific).
Previously, we have showed that the Mediator subunit CDK8 together with its activating partner CCNC negatively regulates the protein stability of nuclear SREBP-1a/c through phosphorylation (19). To determine whether CCNC and CDK8 also regulate Hamp, we depleted CCNC in FAO cells using specific siRNA (Fig. 2F), and observed an up-regulation of Hamp (Fig. 2G). Similarly, when CDK8 was depleted in HepG2 cells using small hairpin RNA (shRNA) (19), we detected an increase of Hamp mRNA (Fig. 2H). Thus, MED15 activates Hamp expression, while CCNC and CDK8 suppress Hamp expression, consistent with the roles of these Mediator subunits in the regulation of nuclear SREBP-1a/c activities/levels.
SREBP-1 binds to the Hamp promoter
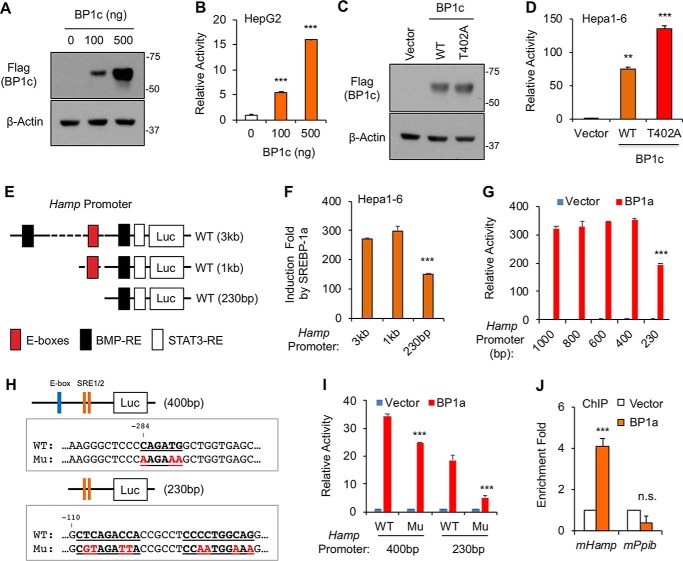
Further supporting a positive correlation between nuclear SREBP-1 protein levels and Hamp gene expression, overexpression of nuclear SREBP-1c increased the Hamp promoter activity in a dose-dependent manner in HepG2 cells (Fig. 3, A and B). Moreover, the mutant SREBP-1c/T402A, which is more stable than wildtype (WT) due to the defect of regulation by CDK8-CCNC (19), also more potently activated the Hamp promoter when the same amounts of plasmids were transfected into Hepa1–6 cells (Fig. 3, C and D). Thus, our data support that SREBP-1 transcription factors are activators of the Hamp gene promoter.
Figure 3.
SREBP-1 binds to the Hamp gene promoter. A, immunoblots of the indicated proteins and B, luciferase reporter assays with the human Hamp promoter (3 kb) for the effects of SREBP-1c in HepG2 cells. C, immunoblots of the indicated proteins and D, luciferase reporter assays with the Hamp promoter for the effects of WT or T402A mutant SREBP-1c in Hepa1–6 cells. E, locations of some regulatory elements in the Hamp promoter. F and G, luciferase reporter assays for effects of SREBP-1a on truncated Hamp promoters in Hepa1–6 cells. H, generation of point mutations in Hamp promoter fragments. I, luciferase reporter assays for effects of SREBP-1a on WT or mutant (Mu) Hamp promoters in Hepa1–6 cells. J, ChIP followed by real-time PCR analyses for binding of overexpressed HA-tagged SREBP-1a to the indicated promoters in Hepa1–6 cells. *, p < 0.05; **, p < 0.01; and ***, p < 0.001 versus Control (vector alone or 3 kb (F), 1,000 bp (G), or WT (I) Hamp promoter, n = 3 with independent repeats).
The two conserved E-boxes on the human or mouse Hamp promoter are located between −1 kb and −230 bp (Fig. 3E). To determine whether these E-boxes are required for SREBP-dependent activation, we analyzed the activities of truncated Hamp promoters by luciferase reporter assays. As shown in Fig. 3F, deletion of the 2-kb fragment from the 5′-end of human Hamp promoter did not affect SREBP-1a–induced activation, but further deletion to −230 bp significantly reduced the Hamp promoter activity by ∼50%, suggesting that the conserved E-boxes may be involved in SREBP-dependent activation of Hamp. Further mapping suggests that the first E-box between −1 kb and −400 bp is not required, but the second E-box between −400 and −230 bp may be required (Fig. 3G). To examine this possibility, we destroyed this E-box by mutagenesis (Fig. 3H, upper panel). Indeed, the E-box–mutated Hamp promoter was significantly less induced than WT upon overexpression of nuclear SREBP-1a in luciferase reporter assays (Fig. 3I).
As shown in Fig. 3, F and G, our data also suggest that SREBP-1a activates the Hamp promoter through additional DNA sequences within the −230 bp region. By sequence analyses, we identified two sequences that look similar to the SRE in the Fasn promoter (Fig. 3H, bottom panel). To examine whether these elements are functional, we destroyed them by mutagenesis (Fig. 3H, bottom panel). As shown in Fig. 3I, luciferase reporter assays showed that the two elements are critically required for SREBP-1a–induced activation of the Hamp promoter. Together, our data indicate that SREBP-1 activates the Hamp promoter primarily via three regulatory elements, including one E-box.
To determine whether SREBP-1 proteins binds to the Hamp promoter, we performed chromatin immunoprecipitation (ChIP) assays. To efficiently pulldown nuclear SREBP-1, we transfected HA-tagged nuclear SREBP-1a into Hepa1–6 cells. HA-SREBP-1a proteins were immunoprecipitated using anti-HA antibody after cross-linking, and SREBP-1a–associated DNA was detected by real-time PCR. As shown in Fig. 3J, SREBP-1a proteins were significantly enriched on the endogenous Hamp gene promoter, but not on the promoters of non-SREBP target genes, such as Ppib. Thus, these results suggest that SREBP-1 may directly activate the Hamp promoter.
Overexpression of nuclear SREBP-1a under control of the Pck1 promoter increases hepatic Hamp gene expression in mice
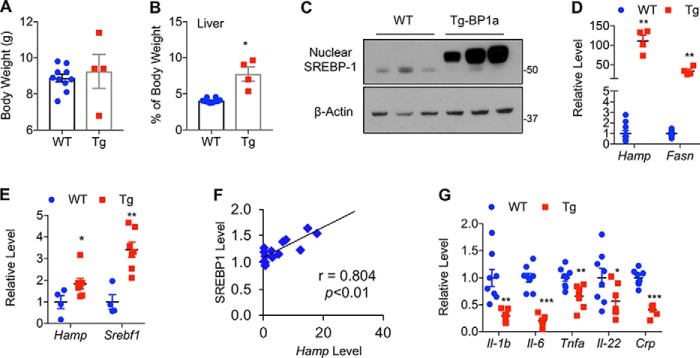
To understand the physiological significance of SREBP-1 in the regulation of Hamp gene expression, we examined SREBP-1a transgenic (Tg) mice, in which a nuclear/active form of human SREBP-1a is overexpressed under control of the Pck1 promoter (38). Although the Pck1 promoter is also active in some nonhepatocytes, it is highly active in hepatocytes during fasting, and thus it is relatively liver-specific. At the age of 3 weeks, body weights of SREBP-1a Tg and WT control mice were similar (Fig. 4A). As previously reported (38), SREBP-1a Tg mice had significantly larger livers with steatosis at the age of 3 weeks (Fig. 4B). Immunoblotting confirmed the overexpression of nuclear SREBP-1a in transgenic mouse livers (Fig. 4C). As expected, Tg mice displayed an increased expression of lipogenic genes, such as Fasn (Fig. 4D). Consistent with our tissue culture data, mRNA levels of the Hamp gene were about 100-fold higher in livers of SREBP-1a Tg mice than those of WT mice at the age of 3 weeks (Fig. 4D). Due to the increase of Hamp expression in WT mice during development through unknown mechanisms, the difference of Hamp gene expression between SREBP-1a Tg and WT mice appeared smaller at the age of 3 months, but the mRNA levels of Hamp as well as SREBP-1a target genes, such as Srebf1, were still significantly higher in livers of SREBP-1a Tg mice (Fig. 4E). Moreover, the Hamp mRNA levels detected by qRT-PCR were positively correlated with nuclear SREBP-1 protein levels measured by immunoblotting using anti-SREBP-1 antibody (Fig. 4F). These results indicate that overexpression of SREBP-1a stimulates Hamp gene expression in vivo.
Figure 4.
Liver-specific overexpression of nuclear SREBP-1a increases hepatic Hamp gene expression under normal chow. A, body weight, and B, relative liver weight of WT and SREBP-1a Tg mice. C, immunoblots of the indicated proteins and D, qRT-PCR analyses of the indicated genes in livers of WT and Tg mice at the age of 3 weeks, or 3 months (E–G). E, correlation analyses between nuclear SREBP-1 protein levels detected by immunoblotting and Hamp mRNA levels measured by qRT-PCR in livers of Tg mice. *, p < 0.05; **, p < 0.01; and ***, p < 0.001 versus WT (n = 7–13).
Because fatty liver is often accompanied by inflammation, which is a strong inducer of Hamp gene expression (39–42), we examined the expression of some inflammatory genes. However, we found that the key inflammatory cytokine genes, such as Il-1b, Il-6, and Tnfa, were expressed at lower levels in livers of SREBP-1a Tg mice at the age of 3 months (Fig. 4G). It appears that Hamp was up-regulated prior to the presence of inflammation in SREBP-1a Tg mice. Although not conclusive, our results suggest that inflammation is less likely a cause for the up-regulation of hepatic Hamp in SREBP-1a Tg mice.
Liver-specific overexpression of SREBP-1a disturbs iron status
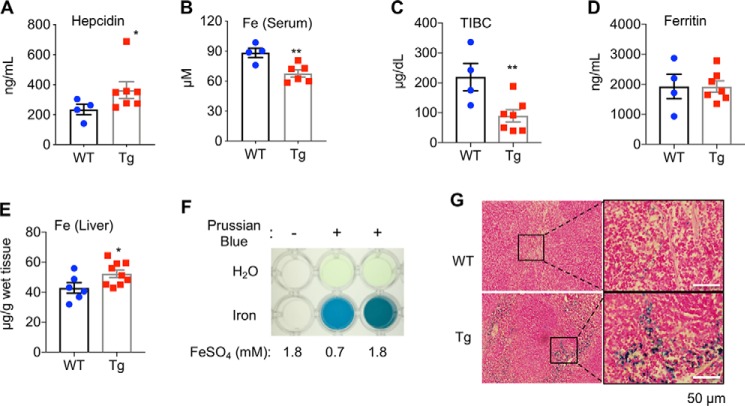
Consistent with the gene expression data, SREBP-1a Tg mice displayed an increase of hepcidin levels in blood at the age of 3 months, when fed with a normal chow diet (Fig. 5A). Because the key function of hepcidin is to regulate iron homeostasis, we then examined the iron status of SREBP-1a Tg mice. As shown in Fig. 5B, Tg mice had less non-heme iron in serum. The transferrin iron binding capacity (TIBC) was significantly lower in Tg mice (Fig. 5C), although the blood levels of ferritin were sustained (Fig. 5D). These results suggest mild dysregulation of iron metabolism in Tg mice. Moreover, Tg mice displayed an increase of non-heme iron accumulation in the liver by quantitative measurements (Fig. 5E) and spleen by Prussian blue staining (Fig. 5, F and G). Together, our data indicate that liver-specific overexpression of SREBP-1a affects iron status in vivo.
Figure 5.
Iron homeostasis is dysregulated in SREBP-1a transgenic mice. Quantitative measurements of blood: A, hepcidin levels; B, non-heme iron levels; C, TIBC; and D, ferritin levels; E, hepatic non-heme iron levels in WT and SREBP-1a Tg mice. F, positive and negative controls for Prussian blue staining. G, representative images of Prussian blue staining of spleens from WT and Tg mice. *, p < 0.05 and **, p < 0.01 versus WT (n = 7–9).
Liver-specific overexpression of SREBP-1a worsens iron homeostasis under iron-deficient states
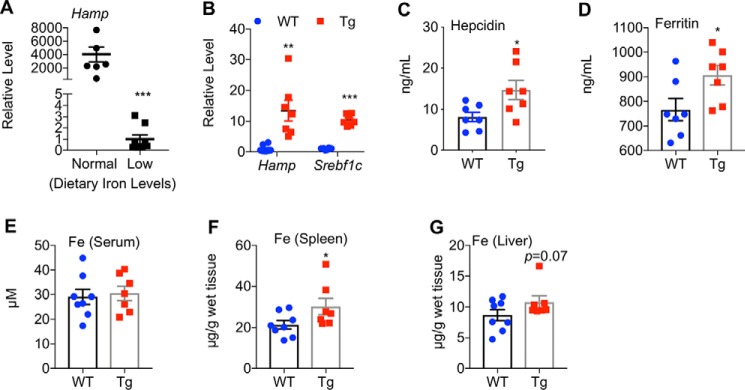
Because iron levels in diets potently affect hepatic Hamp mRNA and blood hepcidin levels (39–42), we wondered whether liver-specific overexpression of nuclear SREBP-1a also affects iron status under the iron-deficient conditions. For that purpose, we treated both SREBP-1a Tg mice and WT mice with an iron-deficient diet for 6 weeks. As expected, hepatic Hamp mRNA levels were dramatically reduced (Fig. 6A). Supporting a role of SREBP-1a in the activation of the Hamp promoter, SREBP-1a Tg mice displayed 5–30–fold higher levels of hepatic Hamp mRNA (Fig. 6B) and 2-fold higher levels of blood hepcidin when compared with WT controls (Fig. 6C). Thus, SREBP-1a seems to activate Hamp gene expression in vivo independent of dietary iron levels.
Figure 6.
SREBP-1a induces hepatic Hamp gene expression in iron-deficient states. A, effect of an iron-deficient diet on hepatic Hamp mRNA levels in WT mice. B, qRT-PCR analyses of the indicated genes in mouse livers under an iron-deficient diet. Quantitative measurements of hepcidin (C) and ferritin (D) levels in blood, and non-heme iron levels in blood (E), spleen (F), and liver (G) at the age of 4.5 months. *, p < 0.05; **, p < 0.01; and ***, p < 0.001 versus WT (n = 7–10).
Upon iron deficiency, SREBP-1a Tg mice also displayed higher levels of ferritin (Fig. 6D), but there was no difference for blood iron levels, which were already much lower than normal (Fig. 6E). Moreover, SREBP-1a Tg mice displayed a slight increase of iron accumulation in spleen (Fig. 6F) and liver (Fig. 6G) by quantitative measurements. Together, these results are consistent with increased iron deficiency in SREBP-1a Tg mice, and support a role of SREBP-1a in iron metabolism.
SREBP-1 activates Hamp expression independent of inflammation
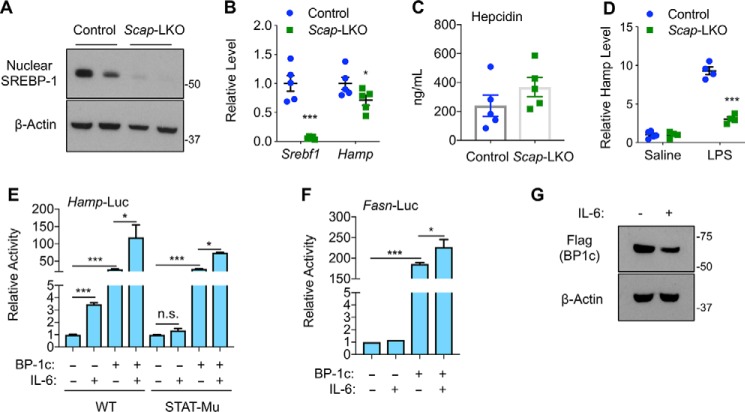
To understand the physiological role of SREBP-1 on Hamp gene expression, we examined liver-specific SCAP knockout mice (Scap-LKO), which were generated by crossing Scapflox/flox mice (43) with Albumin-Cre transgenic mice. Because SCAP is required for generation of nuclear forms of SREBP transcription factors (43), Scap-LKO mice expectedly displayed a loss of nuclear SREBP-1 proteins (Fig. 7A). As a result, SREBP-target genes, such as Srebf1, were expressed at significantly lower levels in livers of Scap-LKO mice, when fed with a normal chow diet. Although hepatic Hamp expression was also reduced in Scap-LKO mice, the difference between knockout and control mice was relatively small (Fig. 7B), and blood hepcidin levels were not significantly different (Fig. 7C). Together with SREBP-1 knockdown data from Fig. 1J, our results suggest that SREBP-1 may only have a minor effect on hepatic Hamp expression under normal physiological conditions.
Figure 7.
Nuclear SREBP-1 regulates Hamp gene expression independent of inflammation. A, immunoblots of the indicated hepatic proteins; B, qRT-PCR analyses of the indicated hepatic genes; and C, blood hepcidin levels in Scapflox/flox (Control) and liver-specific SCAP knockout (Scap-LKO) mice at the age of ∼3 months. D, effects of intraperitoneal injection of LPS (1 mg/kg body weight for 6 h) on hepatic Hamp mRNA levels in 3-month-old Control and Scap-LKO mice. Effects of IL-6 on nuclear SREBP-1c–induced activation of WT and STAT3-response element mutant (STAT-mu) Hamp promoters (E) and Fasn promoter (F), and overexpressed nuclear SREBP-1c protein levels (G). *, p < 0.05 and ***, p < 0.001 versus control (n = 5 for mice, n = 3 for tissue culture, n.s., not significant).
Because lipopolysaccharides (LPS) are known to activate Hamp expression through the IL-6/STAT3 pathway (44), we wondered whether loss of nuclear SREBP-1 affects LPS-induced Hamp expression. As shown in Fig. 7D, LPS injection robustly induced hepatic Hamp gene expression in control mice by nearly 10-fold. Surprisingly, LPS-induced up-regulation of Hamp gene expression was reduced to only 3-fold in Scap-LKO mice (Fig. 7D). Thus, although the mechanism(s) are unclear, nuclear SREBP proteins are required for LPS-induced Hamp expression in mice.
To determine whether the STAT3-binding elements are involved in SREBP-1–induced activation of Hamp gene promoter, we compared WT and STAT3-binding element mutant Hamp promoters by luciferase reporter assays. As shown in Fig. 7E, IL-6 significantly induced WT, but not mutant Hamp promoter. However, nuclear forms of SREBP-1c could similarly activate both WT and mutant Hamp promoters (Fig. 7E), suggesting that SREBP-1c activation of Hamp is not through STAT3-binding elements. Interestingly, IL-6 treatment further stimulated SREBP-1c–induced activation of the Hamp promoter independent of STAT3-binding elements (Fig. 7E). IL-6 treatment similarly stimulated SREBP-1c–induced activation of the Fasn promoter (Fig. 7F). Immunoblotting analyses showed that IL-6 slightly reduced nuclear SREBP-1c (Fig. 7G), suggesting that IL-6 may increase the transcriptional activities of nuclear SREBP-1c. Nevertheless, our data suggest that SREBP-1 transcription factors may directly activate Hamp expression, and they likely act downstream of inflammatory signals.
Discussion
In this study, we identified Hamp as a novel target gene of SREBP-1 transcription factors. Nuclear forms of SREBP-1a/c could activate the Hamp promoter and increase the endogenous Hamp gene expression in vitro. In mouse models, liver-specific overexpression of nuclear SREBP-1a resulted in an increase of hepatic Hamp gene expression and elevation of blood hepcidin, which is encoded by Hamp. Consistent with the increase of blood hepcidin, iron status of SREBP-1a Tg mice was altered with a reduction of blood iron and an increase of hepatic and splenic non-heme iron. Nuclear SREBP-1 loss of function has little effects on Hamp gene expression in normal physiological states, but impaired LPS-induced Hamp expression. Because nuclear forms of SREBP-1 are often elevated or activated under certain pathophysiological conditions, such as NAFLD and aging, SREBP-1 activation of Hamp may play a role in those pathophysiological states in the regulation of iron homeostasis.
The role of SREBP-1 transcription factors in lipid metabolism is well-defined, as they can activate the transcription of a series of genes that encode the key enzymes for fatty acid biosynthesis (13–16). For this reason, SREBP-1 transcription factors are believed to play a role in human diseases relevant to dysregulation of lipid metabolism, including NAFLD. Besides lipogenic enzymes, other genes that are regulated by SREBP-1 have been less explored so far. The Hamp gene promoter contains two conserved E-boxes, which can be recognized by USF-1/2 transcription factors (45). It has been shown that USF-2 is critically required to activate the Hamp promoter and regulate iron homeostasis in vivo (29). In addition to SREs, previous studies by promoter mapping and ChIP-sequencing analyses have demonstrated that SREBP-1 transcription factors frequently bind to E-boxes likely through interaction with USF-1/2 (25–28). By ChIP assays, we show that SREBP-1a is able to specifically bind to the Hamp promoter (Fig. 3J). Our promoter mapping and mutagenesis data demonstrate that the second E-box in the Hamp promoter is involved in SREBP-1-induced activation (Fig. 3, H and I). However, SREBP-1a also activates the Hamp promoter through two DNA sequences within the −230 bp of Hamp promoter (Fig. 3, H and I). Moreover, SREBP-1 transcription factors can activate the Hamp promoter in nonhepatocytes (Fig. 1I). These data suggest that SREBP-1 may directly activate the Hamp gene promoter.
Hepcidin is an essential hormone in the regulation of iron homeostasis, and hepatocytes are the major source of hepcidin (20, 24). The hepcidin-encoding Hamp gene is primarily regulated by iron levels in the body and inflammation at the transcription level through BMP/SMAD (33, 46) and IL-6/STAT3 (32, 47, 48) pathways, respectively. To a lesser extent, the Hamp promoter is also regulated by other transcription factors, such as C/EBPα (49), CREBH (50), and USF-1/2 (29, 30). Although SREBP-1 transcription factors could robustly induce the Hamp mRNA in Hepa1–6 cells (Fig. 1E), the fold-induction in adult SREBP-1a Tg mice appeared less than that in tissue culture (Fig. 4E). This is likely due to the different levels of other factors that regulate the Hamp gene. For unknown reasons, the Hamp mRNA levels in Hepa1–6 cells are much lower than those in WT mouse livers. Thus, our data suggest that SREBP-1 activation of the Hamp promoter may be shadowed when other regulators of Hamp are present. Moreover, we cannot exclude the involvement of SREBP-1–independent regulation of Hamp by the Mediator subunits MED15, CDK8, and CCNC (Fig. 2), as the Mediator complex can also regulate the transcriptional activity of other transcription factors.
Liver-specific overexpression of SREBP-1a under the Pck1 promoter causes massive fatty liver (38). Although fatty liver usually leads to an increase of inflammation, endoplasmic reticulum stress, and oxidative stress (51), SREBP-1a Tg mice displayed less inflammation than control mice (Fig. 4G). The underlying mechanisms are unclear, but one possibility is that our experiments were done at the early stages of fatty liver. In fact, SREBP-1a Tg mice displayed much more robust Hamp gene expression when examined at 3 weeks of age (Fig. 4D) or when the mice were fed with an iron-deficient diet (Fig. 6B). Moreover, SREBP-1 activation of the Hamp promoter does not require STAT3-binding elements (Fig. 7E). Interestingly, liver-specific depletion of nuclear SREBP protein in Scap-LKO mice severely impaired LPS-induced Hamp gene expression (Fig. 7D). Thus, our data suggest that SREBP-1 induces Hamp likely independent of both iron levels and inflammation. Instead, nuclear SREBP-1 may act downstream of inflammatory signals in activation of Hamp gene expression. Future studies are necessary to elucidate the underlying molecular mechanisms.
Hepcidin regulates iron homeostasis mainly through inhibiting intestinal iron absorption and blocking iron export from macrophage (20, 24). Consistent with the increase of Hamp gene expression, blood hepcidin levels were elevated in SREBP-1a Tg mice, but the fold-change was less than that of mRNA (Figs. 5A and 6C). One possibility is that nuclear SREBP-1a may also affect hepcidin secretion and/or clearance. Another possibility is that the Pck1 promoter used for overexpressing nuclear SREBP-1a may also be active in other tissues. Nevertheless, increased hepcidin in SREBP-1a Tg mice caused a few changes in iron status, including reduced blood non-heme iron and increased accumulation of iron in splenic macrophages. These results support a role of SREBP-1 transcription factors in iron metabolism.
In summary, this study demonstrates that besides their classical roles in lipid metabolism, SREBP-1 transcription factors also regulate iron homeostasis by directly activating Hamp gene transcription. Although SREBP-1 regulation of Hamp may be less critical under normal physiological conditions because of the low abundance of nuclear/active SREBP-1 proteins (Fig. 1A), this novel regulation has pathophysiological significance, as nuclear/active SREBP-1 proteins in the liver are accumulated during aging (Fig. 1B) and in NAFLD (31). Therefore, our data provide a novel molecular insight into dysregulation of iron homeostasis in those pathophysiological states and open a new window to develop strategies to simultaneously target dysregulation of both lipid and iron metabolism.
Experimental procedures
Tissue culture
Hepa1–6, HepG2, FAO, and HEK293T cells were purchased from ATCC and cultured in Dulbecco's modified Eagle's medium (Gibco) supplemented with 10% heat-inactivated fetal bovine serum (HyClone), 2 mm glutamine (Gibco), 100 units/ml of penicillin (Gibco), and 100 μg/ml of streptomycin (Gibco) at 37 °C under humidified air containing 5% CO2. Primary mouse hepatocytes from 10- to 12-week-old male C57BL/6J mice were isolated and cultured as previously described (19).
Animal studies
All mouse experiments conformed to the protocols approved by the Animal Care and Use Committees of the Albert Einstein College of Medicine in accordance with National Institutes of Health guidelines. Pck1-nSREBP-1a transgenic mice (number 002840), Scap-flox mice (number 004162), and Albumin-Cre mice(number 003574) were purchased from the Jackson Laboratory. SREBP-1a transgenic mice were backcrossed with C57BL/6J mice for five generations to possess at least 95% of the C57BL/6J genetic background. All animals were maintained under a 12-h dark cycle with free access to water and a normal chow diet (number 5053, LabDiet; 185 ppm iron). For the establishment of iron deficiency, mice were fed an iron-deficient diet (TD.80396, Envigo; 2–6 ppm iron) for 6 weeks starting at the age of 12 weeks. Scapflox/flox mice were crossed with Albumin-Cre mice to generate liver-specific Scap-knockout (Scap-LKO) mice. Scap-LKO mice and their littermate (Scapflox/flox) control mice were treated with 1 mg/kg of LPS for 6 h before harvesting the livers.
Plasmids
pGL4 plasmids of 3-kb WT and STAT3-mutant human Hamp promoter were kindly provided by Dr. Martina Muckenthaler (University of Heidelberg, Germany). The truncated (1 kb, 800 bp, 600 bp, 400 bp, and 230 bp) Hamp-luc plasmids were generated by PCR. The second E-box at −284 bp (CAGATG) was mutated to AAGAAA in 400 bp Hamp-luc by PCR. Similarly, two putative SREs (CTCAGACCA and CCCCTGGCAG) were mutated to CGTAGATTA and CCAATGGAAA, respectively, in 230 bp Hamp-luc by PCR.
Real-time PCR analysis
Total RNA (1 μg) was used for cDNA synthesis using the iScript cDNA synthesis kit (Bio-Rad). Real-time PCR was performed using FastStart Universal SYBR Green Master (Roche Applied Science). Sequence information for specific primers used in this study is listed in Table 1. All reactions were normalized to the Gapdh or Ppib levels.
Table 1.
List of primers for real-time RT-PCR (h, human; m, mouse)
| Gene Name | Primer sequences |
|---|---|
| hHamp | Forward: 5′-TGACCAGTGGCTCTGTTTTCC |
| Reverse: 5′-ATGGGGAAGTGGGTGTCTCG | |
| hCdk8 | Forward: 5′-AAAGTTGGCCGAGGCACTTA |
| Reverse: 5′-TGCCGACATAGAGATCCCAG | |
| hPpib | Forward: 5′-AACGCAACATGAAGGTGCTC |
| Reverse: 5′-CAAGATCACCCGGCCTACA | |
| mHamp | Forward: 5′-CTGAGCAGCACCACCTATCT |
| Reverse: 5′-GTATCGCAATGTCTGCCCTG | |
| mSrebf1 | Forward: 5′-CCAGCGGCTGCCTTCACACA |
| Reverse: 5′-CCAGCCGAAAAGCGAGGCCA | |
| mSrebf1c | Forward: 5′-GGAGCCATGGATTGCACATT |
| Reverse: 5′-GGCCCGGGAAGTCACTGT | |
| mFasn | Forward: 5′-CACTGCATTGACGGCCGGGT |
| Reverse: 5′-GGACAAGCCCAGGCTGCGAG | |
| mIl1b | Forward: 5′-CAACCAACAAGTGATATTCTCCATG |
| Reverse: 5′-GATCCACACTCTCCAGCTGCA | |
| mIl6 | Forward: 5′-ATGGATGCTACCAAACTGGAT |
| Reverse: 5′-TGAAGGACTCTGGCTTTGTCT | |
| mTnfa | Forward: 5′-TCCCAGGTTCTCTTCAAGGGA |
| Reverse: 5′-GGTGAGGAGCACGTAGTCGG | |
| mIl-22 | Forward: 5′-ATGAGTTTTTCCCTTATGGGGAC |
| Reverse: 5′-GCTGGAAGTTGGACACCTCAA | |
| mCrp | Forward: 5′-CTGCACAAGGGCTACACTGT |
| Reverse: 5′-TCTCCCACCAAAGACTGCTTT | |
| mGapdh | Forward: 5′-GGGAAGCCCATCACCATCT |
| Reverse: 5′-GCCTCACCCCATTTGATGTT | |
| mPpib | Forward: 5′-CAAGCATGTGGTTTTCGGCA |
| Reverse: 5′-CGTGTCTGTCTTGGTGCTCT |
Protein extraction and immunoblotting
Protein extraction and immunoblotting were performed as previously described (19). The following primary antibodies with a dilution of 1:1,000 were used: anti-CCNC (ab85927, Abcam), anti-β-actin (A5060, Sigma), anti-FLAG (F3165, Sigma), anti-MED15 (A302–422A, Bethyl Lab), anti-SREBP-1 (2A4, Santa Cruz Biotechnology), and anti-GAPDH (ab9483, Abcam).
Transfection and luciferase reporter assays
The procedures for transfection of plasmid DNA or siRNA and luciferase reporter assays have been described previously (19). For luciferase reporter assays, cells were seeded at a density of 1 × 105 per well in 24-well-plates. The next day, cells were co-transfected pcDNA-4TO-Flag-SREBPs or empty vector with 200 ng of various Hamp-luc plasmids and 50 ng of Renilla luciferase control plasmid using the Lipofectamine 3000 transfection reagent. About 8–20 h after transfection, cells were lysed and analyzed using the Dual-Luciferase System (Promega) according to the manufacturer's instructions. The activity of firefly luciferase was normalized by the corresponding activity of Renilla luciferase.
ChIP assay
The pBABE-HA-nSREBP1a or control pBABE-HA together with hHamp-luc (3 kb) was co-transfected with the control system (human Fasn-luc, HA-G4DBD-VP16-TAD, and 2×GBE-luc) into Hepa1–6 cells. After culture for 24 h, cells were cross-linked in 1% formaldehyde for 10 min before being re-suspended in the lysis buffer (1% SDS, 10 mm EDTA, and 50 mm Tris-HCl, pH 8.1) containing protease inhibitor mixture (1 mm PMSF and 1 μg/ml each of aprotinin and leupeptin). The sample was sonicated on ice until the cross-linked chromatin DNA was sheared to an average length of ∼500 bp. The sonicated cell supernatant was diluted 10-fold in ChIP dilution buffer (0.01% SDS, 1.0% Triton X-100, 2 mm EDTA, 150 mm NaCl, and 20 mm Tris-HCl, pH 8.1) containing protease inhibitors mixture. The sample was pre-cleaned with protein A/G-Sepharose, and then incubated with anti-HA or control IgG antibody overnight at 4 °C. The antibody complexes were recovered with protein A/G-Sepharose at 4 °C for 1 h. DNA within the complexes was released with reverse cross-linking buffer, extracted with phenol-chloroform, precipitated with ethanol, and analyzed with the SYBR Green-based real-time PCR. Cycle threshold (Ct) values were obtained for input (1% of starting chromatin), control ChIP, and anti-HA ChIP samples of the same treatment. The results were normalized by the transfection control (HA-G4DBD-VP16-TAD binding to GBE). The sequences of qPCR primers are listed in Table 2.
Table 2.
List of primers for real-Time ChIP-PCR
| Gene Name | Primer sequences |
|---|---|
| Human Fasn promoter | Forward: 5′-GTCCCCGGGAAGCTGCTAAG |
| Reverse: 5′-CGGGGTTACTGCCGGTCATC | |
| Synthetic Gal4-responsive Promoter | Forward: 5′-CCACCATGGAAGACGCCAAA |
| Reverse: 5′-AGGAACCAGGGCGTATCTCT | |
| Human Hamp promoter | Forward: 5′-AAAACACCACGTGCGGATCG |
| Reverse: 5′-TCTCACCACACGTGCATAGG | |
| Mouse Hamp promoter | Forward: 5′-CTCTGCTGGCTGTAGGTGAC |
| Reverse: 5′-CGGGAGAGATAAGCGGGAAC | |
| Mouse Ppib promoter | Forward: 5′-ACGCCGTGACAAAAGTCTGA |
| Reverse: 5′-GCAGCGTTGGGATTGGTATC |
Iron assays
Serum iron was measured using the Iron Colorimetric Assay Kit (K390-100, Biovision, Milpitas, CA) according to the manufacturer's instructions. Livers and spleens were isolated from 12- to 18-week-old male mice and stored at −80 °C before analysis. Tissue non-heme iron concentrations were determined as previously described (52). Fresh frozen tissue sections (livers and spleens, 5 μm) were stained with Prussian blue stain for iron and counterstained with nuclear fast red (Polysciences, Warrington, PA).
ELISA
ELISA for mouse Hepcidin-1 (HMC-001, Intrinsic LifeSciences), mouse saturation rate of transferrin total iron binding capacity (TIBC) (E03T0359, BlueGene Biotech, Shanghai, China), and Ferritin (ab157713, Abcam) were performed according to the manufacturer's instructions.
Statistical analysis
Data were presented as mean ± S.D. and compared between two groups using a two-tailed Student's t test. Difference was considered statistically significant if p < 0.05.
Author contributions
A. M. X. resources; A. M. X., Z. S., and F. Y. data curation; A. M. X. formal analysis; A. M. X., Z. S., and F. Y. validation; A. M. X., Z. S., and F. Y. investigation; A. M. X. visualization; A. M. X. and Z. S. methodology; A. M. X. and F. Y. writing-original draft; A. M. X., Z. S., and F. Y. writing-review and editing; F. Y. conceptualization; F. Y. supervision; F. Y. funding acquisition; F. Y. project administration.
Acknowledgments
We thank Dr. Martina Muckenthaler (University of Heidelberg, Germany) for the generous gift of the Hamp luciferase reporter constructs and our colleagues for helpful comments.
This work was supported by National Institutes of Health Grants P30 DK041296 and DK020541 and R01 DK110063 and DK098439. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- NAFLD
- non-alcoholic fatty liver disease
- SREBP
- sterol regulatory element-binding protein
- SCAP
- SREBP cleavage activating protein
- C/EBPα
- CCAAT/enhancer-binding protein α
- USF
- upstream stimulatory factor
- PMH
- primary mouse hepatocyte
- Mu
- mutant
- shRNA
- small hairpin RNA
- qRT
- quantitative RT
- Tg
- transgenic
- HA
- hemagglutinin
- TIBC
- transferrin iron binding capacity
- LPS
- lipopolysaccharide
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- STAT
- signal transducers and activators of transcription
- IL
- interleukin.
References
- 1. Browning J. D., Szczepaniak L. S., Dobbins R., Nuremberg P., Horton J. D., Cohen J. C., Grundy S. M., and Hobbs H. H. (2004) Prevalence of hepatic steatosis in an urban population in the United States: impact of ethnicity. Hepatology 40, 1387–1395 10.1002/hep.20466 [DOI] [PubMed] [Google Scholar]
- 2. Brunt E. M. (2010) Pathology of nonalcoholic fatty liver disease. Nat. Rev. Gastroenterol. Hepatol. 7, 195–203 10.1038/nrgastro.2010.21 [DOI] [PubMed] [Google Scholar]
- 3. Tiniakos D. G., Vos M. B., and Brunt E. M. (2010) Nonalcoholic fatty liver disease: pathology and pathogenesis. Annu. Rev. Pathol. 5, 145–171 10.1146/annurev-pathol-121808-102132 [DOI] [PubMed] [Google Scholar]
- 4. Diraison F., Moulin P., and Beylot M. (2003) Contribution of hepatic de novo lipogenesis and reesterification of plasma non esterified fatty acids to plasma triglyceride synthesis during non-alcoholic fatty liver disease. Diabetes Metab. 29, 478–485 10.1016/S1262-3636(07)70061-7 [DOI] [PubMed] [Google Scholar]
- 5. Donnelly K. L., Smith C. I., Schwarzenberg S. J., Jessurun J., Boldt M. D., and Parks E. J. (2005) Sources of fatty acids stored in liver and secreted via lipoproteins in patients with nonalcoholic fatty liver disease. J. Clin. Invest. 115, 1343–1351 10.1172/JCI23621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lambert J. E., Ramos-Roman M. A., Browning J. D., and Parks E. J. (2014) Increased de novo lipogenesis is a distinct characteristic of individuals with nonalcoholic fatty liver disease. Gastroenterology 146, 726–735 10.1053/j.gastro.2013.11.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ameer F., Scandiuzzi L., Hasnain S., Kalbacher H., and Zaidi N. (2014) De novo lipogenesis in health and disease. Metabolism 63, 895–902 10.1016/j.metabol.2014.04.003 [DOI] [PubMed] [Google Scholar]
- 8. Yen T. T., Allan J. A., Yu P. L., Acton M. A., and Pearson D. V. (1976) Triacylglycerol contents and in vivo lipogenesis of ob/ob, db/db and Avy/a mice. Biochim. Biophys. Acta 441, 213–220 10.1016/0005-2760(76)90164-8 [DOI] [PubMed] [Google Scholar]
- 9. Aigner E., Feldman A., and Datz C. (2014) Obesity as an emerging risk factor for iron deficiency. Nutrients 6, 3587–3600 10.3390/nu6093587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wong R. H., and Sul H. S. (2010) Insulin signaling in fatty acid and fat synthesis: a transcriptional perspective. Curr. Opin. Pharmacol. 10, 684–691 10.1016/j.coph.2010.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Song Z., Xiaoli A. M., and Yang F. (2018) Regulation and metabolic significance of de novo lipogenesis in adipose tissues. Nutrients 10 10.3393/nu10101383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xiaoli, and Yang F. (2013) Mediating lipid biosynthesis: Implications for cardiovascular disease. Trends Cardiovasc. Med. 23, 269–273 10.1016/j.tcm.2013.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Osborne T. F., and Espenshade P. J. (2009) Evolutionary conservation and adaptation in the mechanism that regulates SREBP action: what a long, strange tRIP it's been. Genes Dev. 23, 2578–2591 10.1101/gad.1854309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Horton J. D. (2002) Sterol regulatory element-binding proteins: transcriptional activators of lipid synthesis. Biochem. Soc. Trans. 30, 1091–1095 10.1042/bst0301091 [DOI] [PubMed] [Google Scholar]
- 15. Jeon T. I., and Osborne T. F. (2012) SREBPs: metabolic integrators in physiology and metabolism. Trends Endocrinol. Metab. 23, 65–72 10.1016/j.tem.2011.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rui L. (2014) Energy metabolism in the liver. Compr. Physiol. 4, 177–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Giandomenico V., Simonsson M., Grönroos E., and Ericsson J. (2003) Coactivator-dependent acetylation stabilizes members of the SREBP family of transcription factors. Mol. Cell. Biol. 23, 2587–2599 10.1128/MCB.23.7.2587-2599.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yang F., Vought B. W., Satterlee J. S., Walker A. K., Jim Sun Z. Y., Watts J. L., DeBeaumont R., Saito R. M., Hyberts S. G., Yang S., Macol C., Iyer L., Tjian R., et al. (2006) An ARC/Mediator subunit required for SREBP control of cholesterol and lipid homeostasis. Nature 442, 700–704 10.1038/nature04942 [DOI] [PubMed] [Google Scholar]
- 19. Zhao X., Feng D., Wang Q., Abdulla A., Xie X. J., Zhou J., Sun Y., Yang E. S., Liu L. P., Vaitheesvaran B., Bridges L., Kurland I. J., Strich R., Ni J. Q., Wang C., et al. (2012) Regulation of lipogenesis by cyclin-dependent kinase 8-mediated control of SREBP-1. J. Clin. Investig. 122, 2417–2427 10.1172/JCI61462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ganz T., and Nemeth E. (2012) Hepcidin and iron homeostasis. Biochim. Biophys. Acta 1823, 1434–1443 10.1016/j.bbamcr.2012.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rishi G., and Subramaniam V. N. (2017) The liver in regulation of iron homeostasis. Am. J. Physiol. Gastrointest. Liver Physiol. 313, G157–G165 10.1152/ajpgi.00004.2017 [DOI] [PubMed] [Google Scholar]
- 22. Anderson G. J., and Frazer D. M. (2017) Current understanding of iron homeostasis. Am. J. Clin. Nutr. 106, 1559S–1566S 10.3945/ajcn.117.155804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Camaschella C., and Pagani A. (2018) Advances in understanding iron metabolism and its crosstalk with erythropoiesis. Br. J. Haematol. 182, 481–494 10.1111/bjh.15403 [DOI] [PubMed] [Google Scholar]
- 24. Means R. T., Jr. (2013) Hepcidin and iron regulation in health and disease. Am. J. Med. Sci. 345, 57–60 10.1097/MAJ.0b013e318253caf1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yokoyama C., Wang X., Briggs M. R., Admon A., Wu J., Hua X., Goldstein J. L., and Brown M. S. (1993) SREBP-1, a basic-helix-loop-helix-leucine zipper protein that controls transcription of the low density lipoprotein receptor gene. Cell 75, 187–197 10.1016/S0092-8674(05)80095-9 [DOI] [PubMed] [Google Scholar]
- 26. Griffin M. J., Wong R. H., Pandya N., and Sul H. S. (2007) Direct interaction between USF and SREBP-1c mediates synergistic activation of the fatty-acid synthase promoter. J. Biol. Chem. 282, 5453–5467 10.1074/jbc.M610566200 [DOI] [PubMed] [Google Scholar]
- 27. Seo Y. K., Chong H. K., Infante A. M., Im S. S., Xie X., and Osborne T. F. (2009) Genome-wide analysis of SREBP-1 binding in mouse liver chromatin reveals a preference for promoter proximal binding to a new motif. Proc. Natl. Acad. Sci. U.S.A. 106, 13765–13769 10.1073/pnas.0904246106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Reed B. D., Charos A. E., Szekely A. M., Weissman S. M., and Snyder M. (2008) Genome-wide occupancy of SREBP1 and its partners NFY and SP1 reveals novel functional roles and combinatorial regulation of distinct classes of genes. PLoS Genet. 4, e1000133 10.1371/journal.pgen.1000133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nicolas G., Bennoun M., Devaux I., Beaumont C., Grandchamp B., Kahn A., and Vaulont S. (2001) Lack of hepcidin gene expression and severe tissue iron overload in upstream stimulatory factor 2 (USF2) knockout mice. Proc. Natl. Acad. Sci. U.S.A. 98, 8780–8785 10.1073/pnas.151179498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bayele H. K., McArdle H., and Srai S. K. (2006) Cis and trans regulation of hepcidin expression by upstream stimulatory factor. Blood 108, 4237–4245 10.1182/blood-2005-07-027037 [DOI] [PubMed] [Google Scholar]
- 31. Feng D., Youn D. Y., Zhao X., Gao Y., Quinn W. J. 3rd, Xiaoli A. M., Sun Y., Birnbaum M. J., Pessin J. E., and Yang F. (2015) mTORC1 down-regulates cyclin-dependent kinase 8 (CDK8) and cyclin C (CycC). PLoS ONE 10, e0126240 10.1371/journal.pone.0126240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Verga Falzacappa M. V., Vujic Spasic M., Kessler R., Stolte J., Hentze M. W., and Muckenthaler M. U. (2007) STAT3 mediates hepatic hepcidin expression and its inflammatory stimulation. Blood 109, 353–358 10.1182/blood-2006-07-033969 [DOI] [PubMed] [Google Scholar]
- 33. Truksa J., Lee P., and Beutler E. (2009) Two BMP responsive elements, STAT, and bZIP/HNF4/COUP motifs of the hepcidin promoter are critical for BMP, SMAD1, and HJV responsiveness. Blood 113, 688–695 10.1182/blood-2008-05-160184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Malik S., and Roeder R. G. (2010) The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat. Rev. Genet. 11, 761–772 10.1038/nrg2901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Taatjes D. J. (2010) The human Mediator complex: a versatile, genome-wide regulator of transcription. Trends Biochem. Sci. 35, 315–322 10.1016/j.tibs.2010.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Conaway R. C., and Conaway J. W. (2011) Origins and activity of the Mediator complex. Semin. Cell Dev. Biol. 22, 729–734 10.1016/j.semcdb.2011.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Allen B. L., and Taatjes D. J. (2015) The Mediator complex: a central integrator of transcription. Nat. Rev. Mol. Cell Biol. 16, 155–166 10.1038/nrm3951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shimano H., Horton J. D., Hammer R. E., Shimomura I., Brown M. S., and Goldstein J. L. (1996) Overproduction of cholesterol and fatty acids causes massive liver enlargement in transgenic mice expressing truncated SREBP-1a. J. Clin. Invest. 98, 1575–1584 10.1172/JCI118951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Coffey R., and Ganz T. (2017) Iron homeostasis: an anthropocentric perspective. J. Biol. Chem. 292, 12727–12734 10.1074/jbc.R117.781823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Knutson M. D. (2017) Iron transport proteins: gateways of cellular and systemic iron homeostasis. J. Biol. Chem. 292, 12735–12743 10.1074/jbc.R117.786632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Reichert C. O., da Cunha J., Levy D., Maselli L. M. F., Bydlowski S. P., and Spada C. (2017) Hepcidin: homeostasis and diseases related to iron metabolism. Acta Haematol. 137, 220–236 10.1159/000471838 [DOI] [PubMed] [Google Scholar]
- 42. Parrow N. L., and Fleming R. E. (2014) Bone morphogenetic proteins as regulators of iron metabolism. Annu. Rev. Nutr. 34, 77–94 10.1146/annurev-nutr-071813-105646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Matsuda M., Korn B. S., Hammer R. E., Moon Y. A., Komuro R., Horton J. D., Goldstein J. L., Brown M. S., and Shimomura I. (2001) SREBP cleavage-activating protein (SCAP) is required for increased lipid synthesis in liver induced by cholesterol deprivation and insulin elevation. Genes Develop. 15, 1206–1216 10.1101/gad.891301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Huang H., Constante M., Layoun A., and Santos M. M. (2009) Contribution of STAT3 and SMAD4 pathways to the regulation of hepcidin by opposing stimuli. Blood 113, 3593–3599 10.1182/blood-2008-08-173641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Corre S., and Galibert M. D. (2005) Upstream stimulating factors: highly versatile stress-responsive transcription factors. Pigment Cell Res. 18, 337–348 10.1111/j.1600-0749.2005.00262.x [DOI] [PubMed] [Google Scholar]
- 46. Casanovas G., Mleczko-Sanecka K., Altamura S., Hentze M. W., and Muckenthaler M. U. (2009) Bone morphogenetic protein (BMP)-responsive elements located in the proximal and distal hepcidin promoter are critical for its response to HJV/BMP/SMAD. J. Mol. Med. (Berl.) 87, 471–480 10.1007/s00109-009-0447-2 [DOI] [PubMed] [Google Scholar]
- 47. Wrighting D. M., and Andrews N. C. (2006) Interleukin-6 induces hepcidin expression through STAT3. Blood 108, 3204–3209 10.1182/blood-2006-06-027631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pietrangelo A., Dierssen U., Valli L., Garuti C., Rump A., Corradini E., Ernst M., Klein C., and Trautwein C. (2007) STAT3 is required for IL-6-gp130-dependent activation of hepcidin in vivo. Gastroenterology 132, 294–300 10.1053/j.gastro.2006.10.018 [DOI] [PubMed] [Google Scholar]
- 49. Pinto J. P., Ribeiro S., Pontes H., Thowfeequ S., Tosh D., Carvalho F., and Porto G. (2008) Erythropoietin mediates hepcidin expression in hepatocytes through EPOR signaling and regulation of C/EBPα. Blood 111, 5727–5733 10.1182/blood-2007-08-106195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Vecchi C., Montosi G., Zhang K., Lamberti I., Duncan S. A., Kaufman R. J., and Pietrangelo A. (2009) ER stress controls iron metabolism through induction of hepcidin. Science 325, 877–880 10.1126/science.1176639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Engin A. (2017) Non-alcoholic fatty liver disease. Adv. Exp. Med. Biol. 960, 443–467 10.1007/978-3-319-48382-5_19 [DOI] [PubMed] [Google Scholar]
- 52. Roy C. N., Mak H. H., Akpan I., Losyev G., Zurakowski D., and Andrews N. C. (2007) Hepcidin antimicrobial peptide transgenic mice exhibit features of the anemia of inflammation. Blood 109, 4038–4044 10.1182/blood-2006-10-051755 [DOI] [PMC free article] [PubMed] [Google Scholar]