Abstract
Well-regulated differentiation of fibroblasts into myofibroblasts (MF) is critical for skin wound healing. Neoexpression of α-smooth muscle actin (α-SMA), an established marker for MF differentiation, is driven by TGFβ receptor (TGFβR)–mediated signaling. Hyaluronan (HA) and its receptor CD44 may also participate in this process. To further understand this process, primary mouse skin fibroblasts were isolated and treated in vitro with recombinant TGF-β1 (rTGF-β1) to induce α-SMA expression. CD44 expression was also increased. Paradoxically, CD44 knockdown by RNA interference (RNAi) led to increased α-SMA expression and α-SMA–containing stress fibers. Removal of extracellular HA or inhibition of HA synthesis had no effect on α-SMA levels, suggesting a dispensable role for HA. Exploration of mechanisms linking CD44 knockdown to α-SMA induction, using RNAi and chemical inhibitors, revealed a requirement for noncanonical TGFβR signaling through p38MAPK. Decreased monomeric G-actin but increased filamentous F-actin following CD44 RNAi suggested a possible role for myocardin-related transcription factor (MRTF), a known regulator of α-SMA transcription and itself regulated by G-actin binding. CD44 RNAi promoted nuclear accumulation of MRTF and the binding to its transcriptional cofactor SRF. MRTF knockdown abrogated the increased α-SMA expression caused by CD44 RNAi, suggesting that MRTF is required for CD44-mediated regulation of α-SMA. Finally, chemical inhibition of p38MAPK reversed nuclear MRTF accumulation after rTGF-β1 addition or CD44 RNAi, revealing a central involvement of p38MAPK in both cases. We concluded that CD44 regulates α-SMA gene expression through cooperation between two intersecting signaling pathways, one mediated by G-actin/MRTF and the other via TGFβR/p38MAPK.
Keywords: myofibroblast, CD44, actin, myocardin, p38MAPK, transforming growth factor beta (TGF-B), cytoskeleton, TGFβR, smooth muscle actin, MRTF
Introduction
Fibrosis is a common process that involves differentiation of fibroblasts into myofibroblasts (MF),2 which are cells that that mediate cellular contraction and extracellular matrix production. Fibrosis can be a normal and necessary process during tissue repair (e.g. scar formation after cutaneous wounding) (1, 2). However, dysregulated fibrosis gives rise to many pathological conditions including keloids (3), scleroderma (4), hepatic cirrhosis (5), and idiopathic pulmonary fibrosis (6). Nascent and cumulative expression of α-smooth muscle actin (α-SMA) is one of the most well-established hallmarks for MF differentiation (7). Once generated, α-SMA can incorporate into actin stress fibers and produce contractile forces which play a critical role in wound closure and scar formation during wound healing (8). Multiple transcriptional regulatory proteins have been identified that respond to biomechanical and metabolic signals generated after skin injury; together with specific binding partners, these proteins bind to the α-SMA promoter and regulate α-SMA gene expression (9). One of these is serum response factor (SRF), a mammalian transcription factor that binds to a consensus sequence CArG box (CC(A/T)6GG). This serum response element (SRE) is found in smooth muscle cell–specific contractile genes (9). Myocardin-related transcription factor A (MRTF-A), a co-activator of SRF, is normally sequestered in the cytoplasm as a result of its binding to monomeric G-actin (globular actin). In response to growth factor stimulation, or after activation of Rho/ROCK signaling and subsequent polymerization of G-actin to F-actin (filamentous actin), the amount of G-actin is decreased and MRTF is freed from sequestration, allowing it to translocate into the nucleus and bind to SRF, hence triggering serum response element–dependent α-SMA gene expression (10, 11).
TGF-β and its downstream effectors constitute one of the most potent regulatory cascades for α-SMA gene expression and MF differentiation. Among the three TGF-β ligands (TGF-β1, 2, 3), TGF-β1 is the predominant isoform and also the most studied in the context of MF differentiation (12, 13). Binding of TGF-β1 to TGFβ Receptor II (TβRII) facilitates the formation of a heterotetrameric complex between the TβRII homodimer and the TGFβ Receptor I (TβRI) homodimer. This results in the phosphorylation of the GS domain of TβRI and the activation of TβRI receptor kinase activity, leading to recruitment and phosphorylation of receptor-regulated Smads (R-Smads: Smad2 and Smad3). Phosphorylated Smad2 or Smad3 rapidly dissociate from TβRI and form a complex with Smad4, which translocates to the nucleus and activates α-SMA gene transcription (14). Although the Smad-dependent signaling pathway is widely accepted as the canonical pathway induced by TGF-β1, accumulating evidence has shown that TGF-β1 can also activate a number of other Smad-independent signaling pathways that involve mitogen-activated protein kinases (MAPKs) such as ERK1/2, JNK, p38MAPK, PI3K/AKT, and RhoA (15). Many of these molecules are also involved in α-SMA gene regulation in a tissue and cell context–dependent manner (15). In particular, studies from different groups have shown a role for p38MAPK signaling during fibrotic processes within tissues and organs as diverse as the kidney (16), eye (17), and cardiac muscle (18).
CD44 is a type I transmembrane proteoglycan with multiple isoforms that arise from alternative splicing of 10 variable exons; the most widely expressed isoform is the standard form (CD44s) (19). CD44 not only mediates cell-cell and cell-matrix interactions, but also binds growth factors and interacts with a number of growth factor receptors within the cell membrane, thereby exerting a complex role in the regulation of cell growth, survival, and differentiation (19–21). CD44 is also the principal receptor for hyaluronan (HA), a major constituent of the extracellular matrix and a mediator of some aspects of CD44 signaling (22). HA is a negatively charged, unbranched, nonsulfated glycosaminoglycan consisting of alternating disaccharide units of d-glucuronic acid and GlcNAc. HA is synthesized by HA-synthase enzymes (Has1, Has2, Has3) and degraded by hyaluronidases (Hyal1 and Hyal2) (23). Importantly, HA has been demonstrated to be involved in profibrotic signaling and myofibroblast differentiation in human lung fibroblasts (24, 25) and in the differentiation of epithelial cells and tissues as well (26, 27).
When considering CD44 and HA and their role in regulating TGF-β–mediated profibrotic signaling and MF differentiation, many issues remain unresolved because different groups have generated contradictory results. In some experimental systems, both HA and CD44 appear to be supportive and stimulatory for TGF-β–driven MF differentiation. For example, Simpson et al. (28, 29) showed that in human dermal fibroblasts, CD44 gene silencing inhibited TGF-β–induced α-SMA expression. They further demonstrated that TGF-β1–mediated MF differentiation required an interaction between EGFR, CD44, and HA in the plasma membrane which could be disrupted by treatment with hyaluronidase or 4-MU. As another example of a stimulatory role of HA and CD44 upon fibrosis and α-SMA expression, Li et al. (30) demonstrated that targeted overexpression of HAS2 in myofibroblasts generated an aggressive profibrotic phenotype in a bleomycin-induced model of lung fibrosis that could be inhibited by CD44 depletion or by blockade with a CD44 blocking antibody. On the other hand, evidence from other groups showed that CD44 can have an inhibitory effect on TGF-β receptor signaling and fibrosis in some tissues. Ansorge et al. (31) found that in a tendon-healing model, loss of CD44 (CD44 knockout mice) led to significant increases in the gene expression of TGF-β1, TGF-β3, and COL3A1, as well as in parameters indicative of good tissue repair such as material strength and tissue organization. Porsch et al. (32) showed that CD44 interacts physically with TGFβR (and also platelet-derived growth factor receptor) in the plasma membrane of primary human dermal fibroblasts. Depletion of CD44 promoted TGF-β receptor signaling activity, implying that CD44 is a negative regulator of TGF-β signaling in these fibroblasts. In a third example, Velasco et al. (33) showed that knockout of CD44 stimulated a robust activation of TGF-β receptor II and collagen type III expression and improved dermal regeneration and wound closure in Adamts5−/− mice. These contrasting examples suggest that the mechanistic relationship between CD44- and TGF-β–mediated regulation of fibrotic responses remains unclear.
In this study, we sought to obtain a better understanding of how CD44 regulates TGF-β signaling and expression of α-SMA in primary murine dermal fibroblasts. We studied the impact of inhibiting CD44 gene expression by RNAi and observed an up-regulation of α-SMA. This was accompanied by changes in the cytoskeleton including an increase in F-actin polymerization, in parallel with an observed increase in nuclear accumulation of MRTF. Interestingly, CD44-mediated effects appeared to be entirely independent of HA, and although TGF-β signaling was also involved, p38MAPK rather than Smad2 turned out to be the relevant downstream effector.
Results
CD44s is the predominant CD44 isoform in primary murine dermal fibroblasts and is inhibitory to α-SMA gene expression
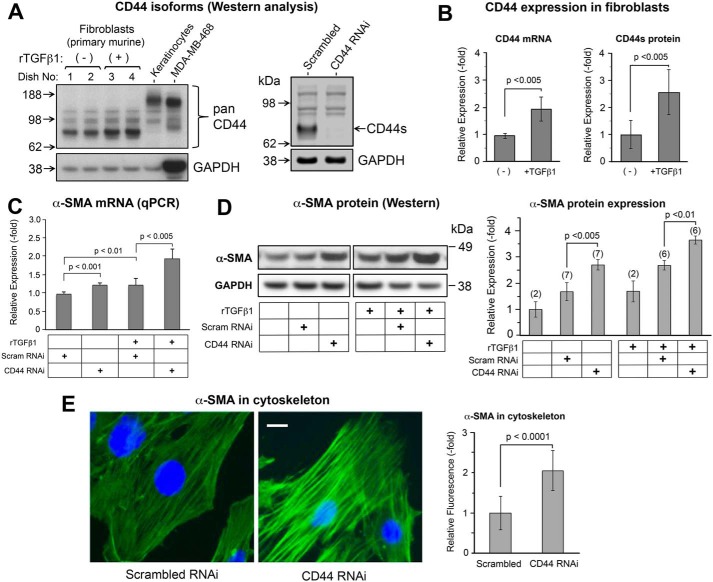
The various isoforms of CD44, including the standard form (CD44s) and other variants, are expressed in a tissue and cell context–dependent manner (19). We examined the expression of CD44 in murine dermal fibroblasts, and in primary murine keratinocytes and epithelial cancer cells for comparison, by Western blotting using a polyclonal pan-CD44 antibody. As shown in Fig. 1A, left, CD44s was the only detectable form of CD44 in fibroblasts, as confirmed by a specific knockdown experiment in which the other bands are not responsive to CD44 RNAi and therefore are nonspecific (Fig. 1A, right), and in contrast to primary keratinocytes and MDA-MB-468 breast cancer cells that express CD44E (34) and CD44v6 (35), respectively. In primary murine fibroblasts, CD44 was shown to be up-regulated at both the protein and mRNA level in response to treatment with rTGF-β1 (Fig. 1B). Because CD44 has been shown to facilitate TGF-β–driven myofibroblast differentiation in human lung fibroblasts (28, 36), we anticipated that increased CD44 gene expression after TGF-β treatment might contribute to the observed up-regulation of α-SMA. We tested this hypothesis by knocking down CD44 via RNAi. To our surprise, the expression of α-SMA was further increased in response to CD44 RNAi, at both the mRNA (Fig. 1C) and protein levels (Fig. 1D). In addition, the formation of α-SMA–positive stress fibers was significantly enhanced in fibroblasts treated with CD44 RNAi, as analyzed by immunofluorescent staining (Fig. 1E).
Figure 1.
CD44s is the predominant isoform that controls α-SMA gene expression in primary murine skin fibroblasts. A, Western blotting of CD44 isoforms in fibroblasts, at baseline and 48 h after treatment ± 2 ng/ml of rTGF-β1 (left panel). Primary mouse keratinocytes and MDA-MB-468 breast cancer cells were positive controls for large CD44 isoforms (CD44E and CD44v6, respectively). The band responsive to rTGF-β1 is CD44s. The other bands are nonspecific as shown by the knockdown experiment using CD44 RNAi (right panel). GAPDH, loading control. B, expression of CD44 mRNA by qPCR (left panel, n = 5 cultures/bar) and standard form of CD44 protein by Western blotting (right panel, n = 6 cultures/bar) in the absence or presence of rTGF-β1. Error bar, mean ± S.D. C, α-SMA mRNA levels by qPCR in cells transfected with control (scram RNAi) or CD44 siRNA (CD44 RNAi), ± 2 ng/ml of rTGF-β1 treatment for 48 h, mean ± S.D. of four independent experiments. D, α-SMA protein levels in cells treated similarly to (C); graph, mean ± S.D. of Western blot experiments (number of pooled experiments shown above each bar). E, α-SMA immunofluorescence in fibroblasts (green); nuclei counterstained with DAPI (blue). Scale bar, 20 μm. Graph, quantitation of α-SMA fluorescent staining intensity, mean ± S.D., 10 images per condition pooled from two experiments.
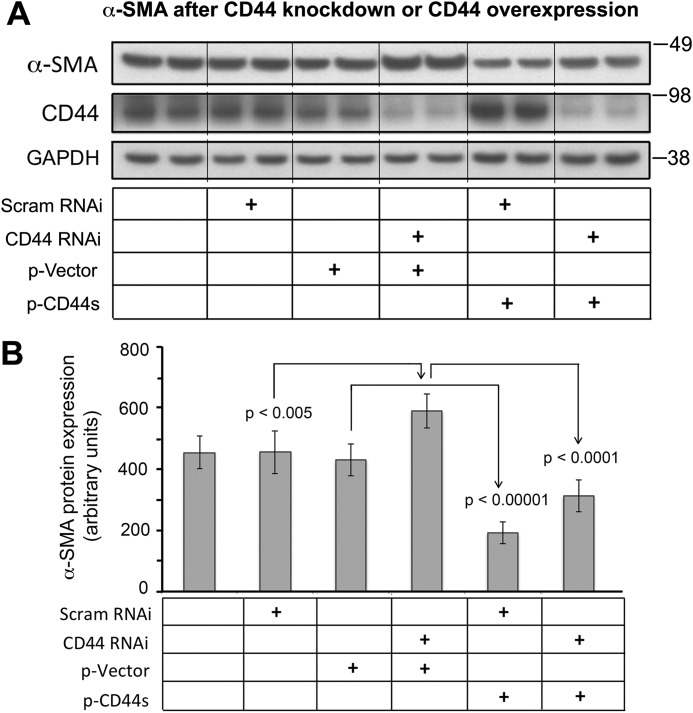
To further clarify the role of CD44s in regulating α-SMA gene expression, we forcibly expressed a CD44s plasmid vector (37) in fibroblasts and observed down-regulation of α-SMA expression on Western blots (Fig. 2, A and B). Moreover, cotransfection of CD44s plasmid and CD44 RNAi effectively attenuated the increase in α-SMA levels induced by CD44 RNAi alone (Fig. 2, A and B), indicating that forced expression of the standard form of CD44 is inhibitory to α-SMA gene expression.
Figure 2.
Effect of CD44s overexpression on α-SMA protein abundance at baseline or after CD44 knockdown. A, Western blotting of α-SMA and CD44 in fibroblasts transfected with CD44 RNAi and/or CD44s plasmid. Scram RNAi, scrambled siRNA; CD44 RNAi, transfection using CD44 siRNA; p-Vector, empty vector contro; p-CD44s, forced expression using a CD44S plasmid. B, Quantitative analysis of band intensities of α-SMA western blots, normalized to GAPDH. Mean ± S.D. of 3 experiments.
We then sought to investigate the impact of changes in CD44 expression levels on cell migration. Confluent monolayers of primary murine skin fibroblasts, transfected with either CD44-targeted siRNA or a CD44s-expressing plasmid, were subjected to an in vitro scratch wound healing assay. Wound closure rates were measured at different time points and compared among different treatment groups. As shown in Fig. S1A, CD44 RNAi inhibited migration of the fibroblasts. On the other hand, forced expression of the standard form of CD44 promoted migration of the fibroblasts (Fig. S1B).
The up-regulation of α-SMA gene expression induced by CD44 knockdown is not mediated by hyaluronan or Has2
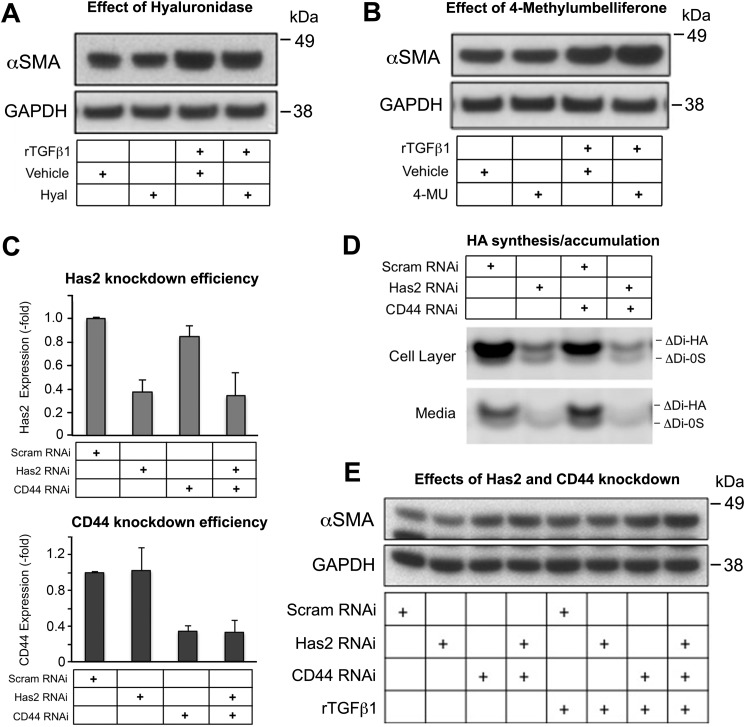
Because CD44 is the principal receptor for HA on plasma membranes of most cell types, including fibroblasts, and because HA is a critical regulator of profibrotic signaling and MF differentiation in some tissues (see the Introduction), we investigated whether HA is involved in regulating the expression of α-SMA in primary skin fibroblasts during manipulation of CD44 and/or TGF-β levels. Extracellular HA was removed by treating fibroblast cultures with hyaluronidase, and the efficient removal of extracellular HA was verified by immunofluorescent staining using HA-binding proteins as described previously (data not shown) (38). Cultures were subsequently treated with or without rTGF-β1 for 48 h. Western blot analysis showed that α-SMA gene expression was not altered by hyaluronidase treatment, whether in the absence or presence of rTGF-β1 treatment (Fig. 3A). Addition of 4-MU, a compound known to inhibit HA synthesis (38), also failed to alter α-SMA gene expression (Fig. 3B). A third manipulation was to knock down Has2, the functionally predominant Has enzyme in murine skin fibroblasts (38). Knockdown efficacies of Has2 and CD44 were verified by qPCR analyses (Fig. 3C). An examination of HA levels by fluorophore-assisted carbohydrate electrophoresis (FACE) analysis (Fig. 3D) showed that Has2 RNAi alone or together with CD44 RNAi significantly reduced the quantity of HA, both in the cell layer and in the conditioned media. However, despite marked reduction in HA levels because of Has2 knockdown, no effect on α-SMA expression was observed (Fig. 3E). CD44 knockdown on the other hand, either alone or in combination with rTGF-β1 treatment, led to consistent increases in α-SMA expression (Fig. 3E), as observed previously.
Figure 3.
Inhibition of HA synthesis by 4-methylumbelliferone (4-MU) or Has2 RNAi, or removal of extracellular HA with hyaluronidase (Hyal), has little effect upon α-SMA gene expression in murine skin fibroblasts. A, SMA protein abundance of cells treated with Hyal followed by treatment with rTGF-β1; mean ± S.D. of three experiments. Vehicle, control. B, α-SMA protein abundance in cells treated with 4-MU followed by rTGF-β1, mean ± S.D. of three experiments. GAPDH, loading control. C, knockdown efficiency with RNAi for Has2 or CD44 was validated by real-time qPCR; mean ± S.D. of three experiments. D, a representative FACE gel, showing HA disaccharide bands (Δ di-HA) from fibroblasts transfected with Has2 RNAi and/or CD44 RNAi, or control (scram RNAi). Bands from the fibroblast cell layer and from the culture media are shown. E, Western blotting of α-SMA in fibroblasts transfected with Has2 RNAi and/or CD44 RNAi, followed by rTGF-β1 treatment; results are representative of two experiments.
The regulatory effect of CD44 knockdown on α-SMA gene expression depends upon noncanonical TGF-β signaling that is partially mediated by p38MAPK
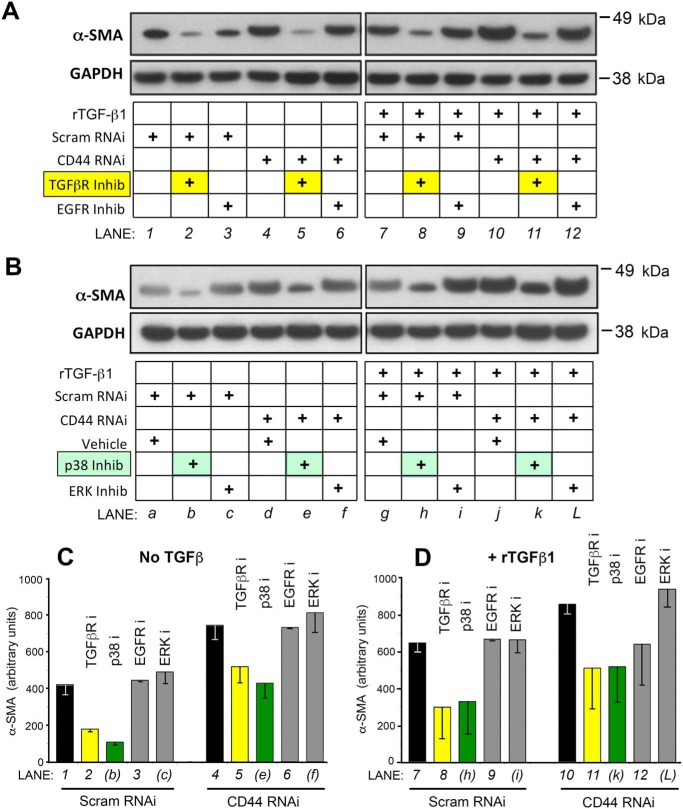
Given previous strong evidence for an important role for TGF-β signaling in regulating α-SMA gene expression, and for the involvement of CD44 (either positive or negative) upon TGFβR-regulated signaling (see the Introduction), we sought to determine which components of the TGF-β signaling cascade might be involved in the up-regulation of α-SMA caused by CD44 knockdown. Fibroblasts were pretreated with individual chemical inhibitors for membrane receptors (TGFβR or EGFR) or with RNAi or chemical inhibitors for midlevel signaling molecules (SMADs, ERK, and p38), followed by transfection with CD44 RNAi and addition of rTGF-β1. As shown in Fig. 4, A, C, and D, inhibition of TGFβR but not inhibition of EGFR suppressed α-SMA gene expression at baseline and abrogated the increased α-SMA gene expression induced by either CD44 RNAi or rTGF-β1 treatment.
Figure 4.
Chemical inhibitors reveal a role for TGF-β signaling in mediating the effects of CD44 RNAi upon α-SMA protein expression in fibroblasts. A, Fibroblasts were pretreated for 4 h with TGF-β receptor inhibitor SB431542 (TGFβR Inhib), EGF receptor inhibitor AG1478 (EGFR Inhib), or vehicle only, then transfected with CD44 RNAi or scrambled RNAi (Scram RNAi) (36-h incubation), followed by another 48 h in fresh media ± 2 ng/ml of rTGF-β1. Following these treatments, cells were harvested for Western blot analysis of α-SMA and GAPDH (loading control). Results shown are representative of four experiments. B, fibroblasts were pretreated with p38MAPK inhibitor SB202190 (p38 inhib), ERK1/2 inhibitor U0126 (ERK inhib), or vehicle only for 4 h, then transfected with CD44 RNAi or scram RNAi for 36 h, followed by ± rTGF-β1 treatment for 48 h. Results representative of four experiments. C and D, densitometric quantification of α-SMA protein bands from Western blotting (± CD44 RNAi, ± TGF-β1, and ± inhibitors) that were pooled from at least two, but more typically from three or four, experiments; mean ± S.D.
To investigate how CD44 knockdown might affect SMAD-dependent TGF-β signaling, fibroblasts were transfected with either nontargeted or CD44 RNAi for 48 h, then synchronized by serum starvation for another 24 h, followed by addition of rTGF-β1 in the media. As shown in Fig. S2A, exposure to rTGF-β1 initiated a rapid increase in phosphorylated Smad2 and phosphorylated Smad3. However, CD44 RNAi had no effect on these phosphorylations relative to scrambled RNAi controls, as analyzed by Western blotting (Fig. S2A). Furthermore, Smad2 RNAi failed to alter α-SMA gene expression in murine skin fibroblasts either at baseline or in response to rTGF-β1 treatment, nor alter the induction caused by CD44 RNAi (Fig. S3), suggesting that SMAD-mediated TGF-β signaling is dispensable for the regulation of α-SMA expression in murine skin fibroblasts. CD44 RNAi also did not alter the overall pattern of rTGF-β1–induced phosphorylation of ERK1/2 (Fig. S2B). Knockdown of CD44 exerted a mild effect on p38, with a shift in the kinetics of rTGF-β1–induced phosphorylation (peak shifted from 30 min in control cells to 6 h in CD44 knocked down cells, with both returning to baseline by 12 h) (Fig. S2C).
Although no involvement of SMAD signaling in the TGF-β–mediated regulation of α-SMA was observed in these fibroblasts, one of the Smads was found to be involved in regulating CD44 expression. Knockdown of Smad3 but not Smad 2 (Fig. S4A) blocked the TGF-β–mediated up-regulation of CD44 that we had observed (Fig. S4B).
Although CD44 did not show any major effect on p38 phosphorylation (Fig. S2C), inhibition of p38 suppressed α-SMA expression in the absence or presence of rTGF-β1, partially abrogating the increased α-SMA gene expression caused by CD44 knockdown (Fig. 4, B–D). Inhibition of the TGF-β receptor had a similar effect (Fig. 4, A, C, and D). This suggests that the noncanonical pathway (involving TGFβR and p38 activity) is essential for baseline α-SMA expression and accounts for at least some (∼50%) of the up-regulation of α-SMA expression induced by rTGF-β1. However, TGFβR and p38 are not fully responsible for induction of α-SMA expression when CD44 is knocked down (because the same proportional inhibition by TGFβR inhibitor or p38 inhibitor is maintained in the presence of CD44 RNAi). Inhibition of EGFR or ERK1/2 had no apparent effect at all on α-SMA expression (Fig. 4, A and B, respectively).
Interestingly, chemical inhibition of p38MAPK appeared to promote the induction effect of rTGF-β1 on CD44 expression and resulted in a further increased CD44 expression in primary murine dermal fibroblasts (Fig. S4C). The discrete effects on CD44 expression from inhibition of Smad3 or p38MAPK (data in Fig. S4) exemplify the complexity of TGFβR signaling in regulating target gene expression.
CD44 inhibition alters the ratio between G- and F-actins
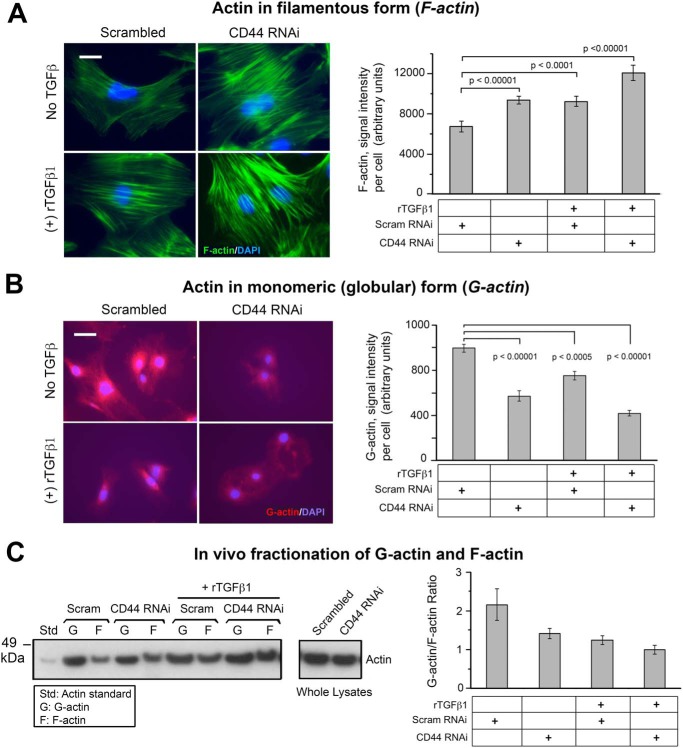
Myofibroblast differentiation is accompanied by alteration of actin cytoskeleton architecture, including the incorporation of α-SMA into stress fibers (8). After observing that CD44 RNAi increases α-SMA gene expression in mouse skin fibroblasts, we wanted to know how the actin cytoskeleton might respond in cells receiving CD44 RNAi treatment. F-actin or G-actin was labeled and visualized with fluorescence-conjugated phalloidin or DNase I, respectively. As compared with control cells, either CD44 RNAi or rTGF-β1 treatment enhanced the formation of F-actin–positive stress fibers, and a combination of these two treatments yielded the best outcome (Fig. 5A). On the other hand, these same treatments had the opposite effect on G-actin, causing a significant decrease in immunofluorescent staining intensity (Fig. 5B). When G-actin was separated from F-actin by fractionation using an in vivo G-/F-actin assay kit, examination of both fractions by Western blot analysis demonstrated a decrease in G-actin and an increase in F-actin in cells treated with either CD44 RNAi or rTGF-β1, relative to control cells that received nontargeted siRNA (Fig. 5C). Consistent with the earlier findings using immunofluorescence (Fig. 5, A and B), the changes in Fig. 5C were most pronounced when CD44 RNAi and rTGF-β1 were administered together.
Figure 5.
The effect of CD44 RNAi on the G-actin/F-actin ratio in fibroblasts. A, images of F-actin stained with AlexaFluor 488-conjugated phalloidin, in cells transfected with CD44 RNAi or scrambled RNAi ± 2 ng/ml of rTGF-β1 (48 h) (left). Scale bar, 50 μm. Quantitative analysis of the phalloidin signal using IPLab image analysis software; each bar, mean ± S.D. of 10 high-power fields, 3 cells/field examined per condition, pooled from two experiments (right). B, images of G-actin stained with AlexaFluor 594-conjugated DNase I, in cells transfected as in (A) (left). Scale bar, 50 μm. Quantitative analysis of fluorescent intensity; each bar, mean ± S.D. of 10 high-power fields/condition from two experiments (right). C, representative Western blotting (left) and quantitative densitometric analysis (right) of G- and F-actin in fibroblasts transfected with scrambled or CD44 siRNA ± rTGF-β1 treatment for 48 h. F- and G-actin were isolated using the in vivo G-/F-actin assay (see “Experimental procedures”). Results are expressed as G-actin/F-actin ratio; each bar, mean ± half-range of two independent experiments.
MRTF-A mediates the α-SMA induction effect of CD44 RNAi
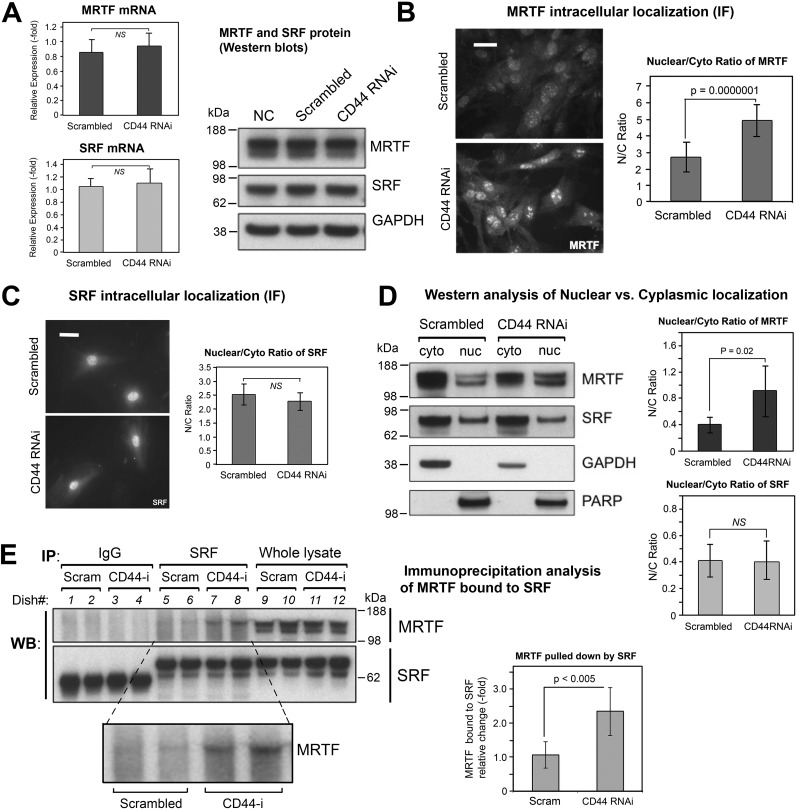
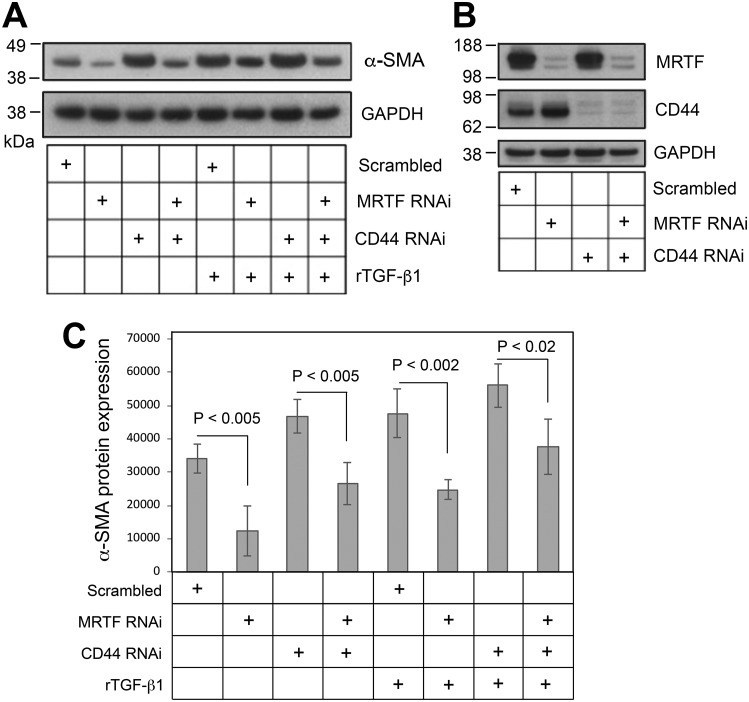
It has been demonstrated by various groups that nuclear localization of MRTF and its binding to SRF play a crucial role in regulating the transcription of contractile genes including α-SMA (11, 39, 40). Among the three homologous members of the myocardin family (myocardin, MRTF-A, MRTF-B), MRTF-A is expressed in nearly all adult tissues (41) and has a predominant role in regulating profibrotic signaling in skin tissue (42, 43). We therefore focused on the role of MRTF-A (hereafter simply called MRTF). As shown in Fig. 6A, expression levels of SRF and MRTF at both the protein and mRNA level were not affected by CD44 RNAi. However, CD44 knockdown did promote nuclear accumulation of MRTF as determined both by immunofluorescence assay (Fig. 6B) and Western blotting of MRTF in cytoplasmic and nuclear fractions (Fig. 6D). SRF subcellular localization, on the other hand, was unaffected by CD44 RNAi (Fig. 6, C and D). Furthermore, CD44 knockdown resulted in increased binding between MRTF and SRF as determined by immunoprecipitation (Fig. 6E). Lastly, knockdown of MRTF by RNAi (knockdown efficacy was verified by Western blot analysis as shown in Fig. 7B) significantly suppressed the expression α-SMA in the absence or presence of rTGF-β1 and effectively abrogated CD44 RNAi's ability to induce α-SMA gene expression (Fig. 7, A and C).
Figure 6.
The effect of CD44 knockdown on gene expression and subcellular localization of MRTF and SRF. A, mRNA levels of MRTF and of SRF by qPCR (left), mean ± S.D. of three experiments; protein levels of MRTF and SRF by Western blot analysis (right), showing no difference in response to CD44 RNAi treatment. B, MRTF-A immunofluorescent staining in fibroblasts transfected with CD44 RNAi or scram RNAi (left). Scale bar, 50 μm. Digital analysis of staining intensity to provide MRTF nuclear/cytoplasmic ratio (right); mean ± S.D., 100 cells analyzed per condition. C, SRF immunofluorescent staining in cells transfected as in (B) (left); scale bar 50 μm. Digital analysis of SRF Nuc/Cyto ratio (right). D, subcellular fractionation of MRTF in fibroblasts transfected with CD44 RNAi or scram RNAi. Left, representative Western blotting of MRTF from cytoplasmic (cyto) or nuclear (nuc) fractions, along with GAPDH and PARP as controls for purity of the cyto and nuc fractions, respectively. Right, Nuc/Cyto ratio of MRTF, determined from densitometric scanning (IPLab) of bands in two independent experiments, mean ± S.D. E, immunoprecipitation analyses to assess binding between SRF and MRTF in fibroblasts transfected with CD44 or scram RNAi. An anti-SRF antibody (SRF) or normal rabbit immunoglobulin (IgG) was added to the cell lysates to pulldown protein complexes, and the membrane probed with anti-MRTF or anti-SRF antibodies (left). Quantitative analysis of the SRF-bound MRTF band (inset) is shown in the graph (right); bars, mean ± S.D. of two experiments with duplicate samples.
Figure 7.
Knockdown of MRTF partially abrogates the α-SMA up-regulation induced by CD44 knockdown or rTGF-β1 treatment. A, α-SMA Western blotting in fibroblasts transfected with MRTF RNAi and/or CD44 RNAi, ± rTGF-β1. B, knockdown efficacy (Western blotting) of MRTF and CD44 for the experiment shown in (A). C, densitometric analyses of α-SMA Western blots using IPLab imaging software, normalized to GAPDH, pooled from four experiments; bars, mean ± range.
The regulatory effect of CD44 RNAi and TGF-β receptor signaling on the subcellular localization of MRTF depends on p38MAPK activity
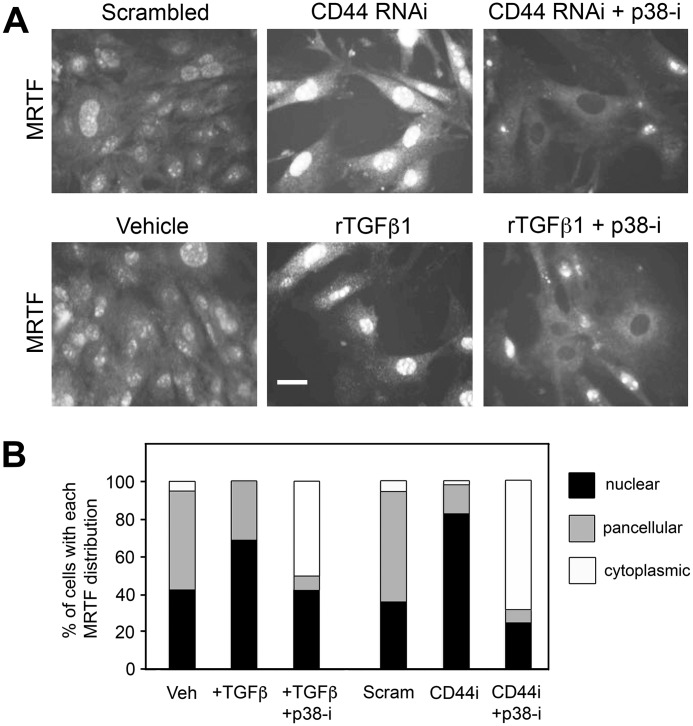
Nuclear accumulation of MRTF and binding to SRF play a central role in regulating α-SMA gene expression. As shown in Fig. 8, A and B, either CD44 RNAi or rTGF-β1 treatment was able to induce nuclear accumulation of MRTF in primary murine dermal fibroblasts. Either effect could be prevented by pretreatment of the cells with SB202190 (p38-i), a chemical inhibitor of p38MAPK. The expression level of MRTF was not altered by treatment with SB202190 as determined by Western blot analysis (data not shown). During p38MAPK inhibition, MRTF appeared completely absent from a majority of nuclei. These results indicate that the regulatory effects of both CD44 RNAi and TGF-β1 upon subcellular localization of MRTF are dependent on p38MAPK activity.
Figure 8.
Effects of CD44 RNAi and TGF-β receptor signaling on the subcellular localization of MRTF-A are dependent upon the p38MAPK activity. A, upper panel, immunofluorescently stained MRTF in fibroblasts treated with either scrambled siRNA, or CD44 RNAi in the presence or absence of SB202190, a p38MAPK inhibitor (p38-i). Lower panel, immunofluorescently stained MRTF in fibroblasts treated with either vehicle control or rTGF-β1 in the presence or absence of SB202190 (p38-i). Scale bar, 50 μm. B, analysis of subcellular MRTF, in which each cell was scored for its predominant intracellular distribution (cytoplasmic, nuclear, or pancellular). Each bar represents 100 to 300 cells analyzed in six images per treatment condition.
Discussion
The significance of the study presented here is that, to our knowledge, this report is the first description of a direct regulatory effect of CD44 on α-SMA gene expression via an actin/MRTF/SRF-dependent pathway. Our working model for α-SMA regulation in mouse fibroblasts is summarized in Fig. 9. Most previous studies that investigated CD44's role in profibrotic signaling and regulation of α-SMA expression focused on characterizing the interactions between CD44 and EGFR, or between CD44 and TGFβR in the plasma membrane, but contradictory observations were generated depending upon the cell type and experimental conditions (see the Introduction). Thus, CD44 was stimulatory for α-SMA expression in some systems and inhibitory in others. Curious about this discrepancy, we examined primary murine skin fibroblasts and found that knockdown of CD44 actually promoted α-SMA expression in these cells (notably similar to what Porsch et al. (32 had reported in human dermal fibroblasts)). We then sought to determine whether this effect is mediated by EGFR or TGFβR signaling and found that knockdown of CD44 had little to no impact upon either pathway, at least in terms of canonical signaling. Blockade of EGFR or ERK1/2 did nothing to blunt the effect of CD44 knockdown, and ERK1/2 phosphorylation was unaffected by CD44 RNAi, suggesting no involvement of EGFR signaling. Regarding TGFβR signaling, we found that CD44 knockdown did not alter the phosphorylation of Smad2/3, and had only mild effects on phosphorylation of p38MAPK, suggesting that CD44 RNAi does not directly modulate TGFβR activation. The mild effect on p38MAPK might be achieved via other pathways or could be an indirect effect. Our experimental approach of knocking down Smad2 failed to rescue the increase in α-SMA expression caused by CD44 RNAi, suggesting that the classical TGFβR/Smad pathway is dispensable in regulating α-SMA expression in primary murine skin fibroblasts. These findings motivated us to explore other pathways that might be responsible for mediating CD44's regulatory effect on α-SMA. We observed that increased α-SMA gene expression after CD44 depletion was accompanied by a pronounced shift in the actin cytoskeleton, with a significant increase in F-actin filaments and a decrease in monomeric G-actin in the cytosol, accompanied by increased nuclear accumulation of MRTF and enhanced MRTF-SRF binding. Moreover, knockdown of MRTF markedly reversed the up-regulated gene expression of α-SMA induced by CD44 RNAi, suggesting that MRTF mediates the regulatory effect of CD44 RNAi on α-SMA.
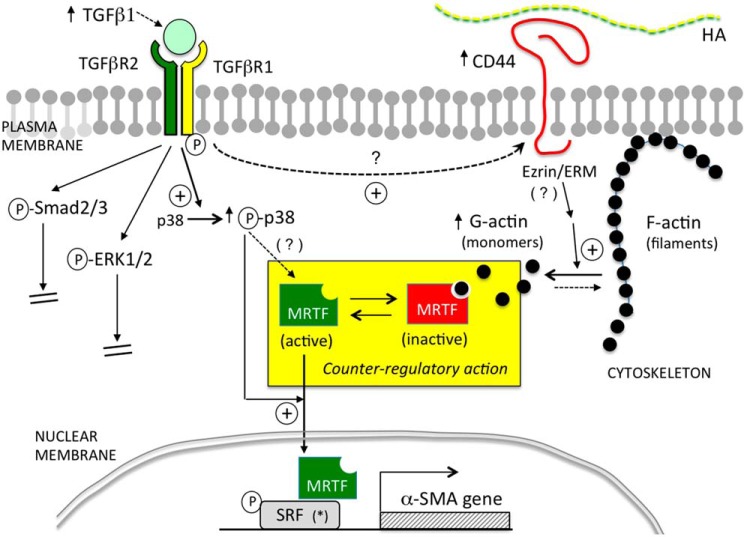
Figure 9.
Summary diagram. Cartoon illustration of the pathways examined in this study. In the context of primary murine dermal fibroblasts, TGF-β/TGFβR-mediated signaling positively regulates α-SMA expression via p38MAPK instead of via Smad2/3 or ERK1/2. CD44 negatively regulates α-SMA expression by inhibiting actin polymerization, leading to more globular G-actin in the cytoplasm which binds and sequesters MRTF, preventing MRTF from entering the nucleus and binding to SRF. Activation of p38MAPK is required for up-regulation of α-SMA expression caused by either TGFβR signaling or knockdown of CD44.
The fact that MRTF (together with its obligatory binding partner SRF) can control myofibroblast differentiation was documented previously in cells from various tissues including cardiac, lung and skin fibroblasts, and kidney and ovarian epithelial cells (44). Crider et al. (39) showed that knockdown of MRTF-A/B effectively reduced the expression of smooth muscle–specific proteins (α-SMA, SM22-α, etc.) induced by TGF-β1 treatment, whereas forced expression of a constitutively active MRTF-A construct markedly increased the expression of α-SMA and SM22-α, along with contractile function in rat embryonic fibroblasts. In skin, Velasquez et al. (40) showed that treating mice with a small molecule, isoxazole, promoted wound healing in vivo by regulating MRTF stability and activity. Our results showed that MRTF is crucial not only for α-SMA expression in primary murine dermal fibroblasts but also for mediating CD44's regulatory effect. This effect of CD44 upon the functionality of MRTF appears to be mediated through the actin cytoskeleton system, as shown by our data that demonstrate shifts in the G- to F-actin profile (Fig. 4).
The nature of the link between MRTF, actin, and α-SMA transcription is well-established from previous studies. MRTFs contain a basic region (B1) and an adjacent Glu-rich domain through which they associate with SRF (10, 45), and an N-terminal domain that contains three Arg-Pro-X-X-X-Glu-Leu (RPEL) motifs that mediate MRTF binding with G-actin. At baseline, when levels of actin polymerization are low, monomeric G-actins bind to MRTF via the RPEL motifs and sequester MRTF in the cytoplasm. Upon stimulation by growth factors and cytokines, activation of Rho GTPases (RhoA, Rac, and Cdc42) induces actin polymerization through conversion of G-actin into F-actin filaments, thus freeing MRTF to enter nuclei and bind to SRF (11). In addition to this well-established regulatory mechanism, phosphorylation on certain residues of MRTF regulates MRTF subcellular localization and function. Both ERK and p38MAPK can phosphorylate MRTF at certain sites. ERK-mediated phosphorylation of MRTF-A at serine 454 inhibits nuclear localization of MRTF induced by serum (46), and p38MAPK/MK2 can stimulate MRTF-A phosphorylation at serine 312 and serine 333 (47). MRTF phosphorylation by p38MAPK in HeLa cells or mouse embryonic fibroblasts (46, 47) did not appear to affect MRTF binding to actin nor its subcellular localization, but such effects could be cell context–dependent because regulation of MRTF nuclear accumulation and α-SMA promoter activity via activation of p38MAPK were demonstrable in porcine kidney epithelial cells (48). Here in our study, chemical inhibition of p38MAPK blocked α-SMA expression at baseline and aborted the α-SMA induction effect caused by CD44 RNAi. Although CD44 knockdown did not appear to affect p38MAPK phosphorylation status, chemical inhibition of p38MAPK led to marked cytoplasmic sequestration of MRTF, effectively abrogating the increased α-SMA gene expression induced by CD44 RNAi. This indicates that p38MAPK kinase activity is required and permissive for CD44's regulatory effect on α-SMA gene expression via MRTF. Our observation is in harmony with findings from many other groups that show p38MAPK plays a crucial role in mediating TGF-β–induced MF differentiation and tissue fibrosis (16–18).
We showed in this report that in primary murine fibroblasts, only the shortest CD44 isoform known as CD44 standard (CD44s) is detectable by Western blot analysis, as opposed to other variants that may be produced by alternative splicing in certain tissues, such as variant isoforms v6 or v7/8 that play a role in regulating cellular behavior in certain fibroblasts (49, 50). We also demonstrated that forced expression of CD44s suppressed α-SMA gene expression at baseline and abrogated the increased α-SMA gene expression caused by CD44 RNAi, further strengthening our argument that CD44s plays a predominant role in regulating α-SMA gene expression in primary murine skin fibroblasts. How CD44 is regulating the G- to F-actin cytoskeletal changes that regulate MRTF, and why such a relationship was not widely recognized before, are very interesting questions. One reason may be that CD44 manipulation (e.g. CD44 deletion or overexpression) typically yields only mild phenotypes. For example, mice with global CD44 deletion showed only minor abnormalities in leukocyte behavior (19), although injury brought out more significant differences, such as a delay in wound re-epithelialization in mice with epidermal-specific deletion of CD44 (CD44 floxed-k14Cre) (51). In wounded BALB/c mice, CD44 deficiency did not alter skin wound closure rates, but did attenuate FAP-positive fibroblast accumulation during wound healing along with increased fibrillar collagen accumulation (52).
An important unanswered question is how CD44 might be controlling the G- to F-actin cytoskeletal filament changes that regulate MRTF. A strong connection between CD44 and regulation of cytoskeletal dynamics has been demonstrated in many studies. For example, CD44 has a well-established role as an organizer of the cortical actin cytoskeleton through direct interactions with ERM (ezrin/radixin/moesin) (19, 53) or other membrane-associated cytoskeletal proteins such as ankyrin (54). In addition, CD44 may also regulate actin cytoskeleton by modulating the activity of Rho GTPases (55–63). For example, CD44 can mediate cell adhesion (64–66). Disruption of epithelial cell-cell contacts by calcium deprivation can trigger the activation of small GTPases and lead to nuclear accumulation of MRTF in a Rho-, Rac1-, or p38-dependent manner (48, 64–68). In our system, CD44 knockdown might disrupt cell-cell contacts, triggering GTPase activation and increasing nuclear MRTF. Protein kinase C, a family of serine/threonine kinases, is another set of candidates for involvement because they mediate intracellular signaling, remodel cytoskeletal microfilaments by interacting with various actin-binding proteins (69) and can regulate the expression and function of CD44 (70–73). In future follow-up studies, any of the above molecules might be considered as candidates for understanding how CD44 inhibition (or overexpression) regulates changes in the G-/F-actin ratio and subcellular relocalization of MRTF.
Another surprising finding was the absence of any apparent role for HA in the effects of CD44 in our system. Studies from researchers at Cardiff University established both HA and CD44 as important facilitators of TGF-β–driven MF differentiation in human lung fibroblasts (see the Introduction) which was mediated by the canonical Smad2/3-dependent pathway (24, 25). Conversely, Evanko et al. (74) showed that reduction or removal of HA led to enhanced MF differentiation in human lung fibroblasts (along with increased fibronectin and type I collagen deposition) suggesting that HA has a negative effect on α-SMA gene expression and MF differentiation. In our study, we found that CD44 inhibits MF differentiation in primary murine fibroblasts, independently of HA. This is not unprecedented. For example, Schmitt and colleagues (37) showed that in human embryonic kidney cells, CD44 is a positive regulator of canonical Wnt signaling by regulating the phosphorylation and membrane localization of low-density lipoprotein receptor–related protein 6 (LRP6), and this regulation was not affected by either hyaluronidase treatment or by antibodies that block HA/CD44 binding, suggesting that CD44-mediated amplification of Wnt signaling is independent of HA. The reason for these discrepancies remains to be elucidated, but it may be that differences in species, type of tissue, and cell culture conditions are important contributing factors. Also, it is well-recognized that HA is not the only ligand for CD44. CD44 can bind to osteopontin, fibronectin, type I collagen, fibrin, P-selectin, and E-selectin, any of which might theoretically play a role in HA-independent CD44-mediated signaling (19, 20, 75).
In conclusion, the present study shows that CD44 regulates α-SMA gene expression in primary murine dermal fibroblasts by controlling actin cytoskeleton dynamics and subcellular localization of MRTF in an HA-independent manner. The subcellular localization of MRTF is also susceptible to p38MAPK activity, which is a noncanonical downstream target of TGFβR signaling. These two pathways intersect at the level of MRTF and mediate the regulatory effect of CD44 on α-SMA gene expression. These findings add new components to the complexity of how CD44 interacts with membrane receptor kinases such as TGFβR and provide novel insights about how CD44 may regulate profibrotic signaling and myofibroblast differentiation. Importantly, further experiments are needed to elucidate which ligand(s) bind to CD44 to allow regulation of α-SMA expression and which pathway(s) or molecule(s) are involved in mediating CD44's role in the dynamic changes in MRTF and the actin cytoskeleton in primary mouse skin fibroblasts.
Experimental procedures
Primary cell culture
Primary mouse dermal fibroblasts were isolated from the skin of 2- to 3-day-old pups from WT C57BL/6J mice (The Jackson Laboratory, Bar Harbor, ME) following a previously published protocol, as approved by the hospital's animal review committee (Cleveland Clinic IACUC) (38). Briefly, the entire trunk skin was removed and incubated overnight in 0.25% trypsin without EDTA, followed by mechanical separation of epidermis from dermis. To isolate fibroblasts, the dermis was finely diced and incubated with 400 units/ml of collagenase type I (Worthington Biochemical) for 30 min at 37 °C, then with 100 units/ml of DNase I (Worthington Biochemical) for 10 min at 37 °C. The suspension was then passed through a 100-μm cell strainer and centrifuged at 300 rpm for 3 min. The fibroblasts were collected and cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS), 1% penicillin/streptomycin, and 1.0 g/liter of glucose. The cells were maintained at 37 °C with 5% CO2. Recombinant human TGF-β1 was purchased from R&D Systems. For all experiments, subconfluent cells from passage 2 or 3 were used.
Inhibition of HAS2, CD44, Smad2, and MRTF gene expression by RNAi
ON-TARGETplus SMARTpool siRNA and control siRNA were purchased from Dharmacon (Lafayette, CO): CD44 (L-041132-01), Has2 (L-042589-01), Smad2 (L-040707-00), Smad3 (L-040706-00), MRTF/Mkl1 (L-054350-00), and nontargeted scrambled siRNA (D-001810-10). The siRNAs were reverse transfected into mouse skin fibroblasts using Lipofectamine RNAiMAX (Invitrogen/Thermo Scientific) as described previously (38). Briefly, for a 6-well tissue culture plate format, 30 pmol of siRNA was mixed with 8 μl of Lipofectamine RNAiMAX in 500 ml of Opti-MEM with a final duplex concentration of 100 nm. The siRNA/transfection reagent mixture was added to the plate first, followed by seeding the fibroblasts at a density of 2.2 × 105 per well. The cells were re-fed with fresh antibiotic-free medium supplemented with 10% FBS 24 h after transfection and further incubated for another 36–48 h before harvesting for assays. The knockdown efficacy of target gene expression by RNAi was confirmed using quantitative real-time PCR and Western blotting of the targeted gene product.
CD44 plasmid purification and transfection
The plasmid for the standard form of CD44 (CD44s) was a generous gift from Dr. Veronique Orian-Rousseau (37). The CD44s insert in the plasmid was confirmed by DNA sequencing. The CD44s plasmid or an empty pcDNA3.1 mammalian expression vector (Invitrogen/Thermo Fisher Scientific) was amplified using One ShotTM Top10 chemically competent Escherichia coli (catalog no. C4040-10, Invitrogen/Thermo Scientific) and purified using the Qiagen Plasmid Maxi Kit (catalog no. 12162, Qiagen, Hilden, Germany), then transfected into cells using Lipofectamine 3000 (Invitrogen/Thermo Scientific), according to the manufacturers' instructions.
Cell migration assay
Primary murine dermal fibroblasts were plated at a density of 3 × 105/well into a 6-well plate and transfected with either CD44-targeted siRNA (or scrambled siRNA as control) or CD44s expression plasmid (or an empty pcDNA3.1 vector). At 48 h after transfection, two parallel wounds were made by scraping the confluent cell culture with the tip of a 10 μl polystyrene pipette tip. The floating cells and debris were washed away and the medium was replaced with low serum (1% FBS) medium. The wound closure was monitored by an inverted microscope (Leica DFC7000 T) at five different positions along the wound, immediately and after varying time periods up to 24 h. The cell migration (wound closure) was quantified using the edge-finding function of Image J. The data were statistically analyzed by Student's t test.
RNA isolation and quantitative real-time RT-PCR
RNA was prepared from fibroblasts using TRIzol reagent (Life Technologies) per the manufacturer's instruction. cDNA was then synthesized using random primers and Superscript III reverse transcriptase (Life Technologies). For real-time PCR, TaqMan gene expression probes for Has2 (catalog no. Mm00515089_m1), CD44 (catalog no. Mm01277161_m1), α-SMA (Mm01546133_m1), MRTF-A (Mm00461840_m1), SRF (Mm00491032_m1), and 18S rRNA (endogenous control, catalog no. 4333760F) were purchased from Applied Biosystems/Life Technologies. The mRNA transcript levels of all target genes were measured in triplicate on an Applied Biosystems 7500 Real-time PCR system and calculated using the 2-ΔΔCt method (Waltham, MA). The mRNA levels were presented as -fold differences relative to untreated normal controls.
In vivo G-/F-actin assay
Soluble G-actin was separated from filamentous F-actin using the G-Actin/F-Actin In Vivo Assay Kit (catalog no. BK037, Cytoskeleton, Inc., Denver, CO) according to the manufacturer's instructions. Briefly, fibroblasts were harvested by scraping into lysis buffer supplemented with ATP and centrifuged at 350 × g to pellet cell debris. The supernatant was transferred to a new tube and centrifuged at 100,000 × g at 37 °C for 1 h to pellet the F-actin. After the G actin–containing supernatant was transferred to a new tube, a depolymerization buffer was added to the pellet and incubated on ice for 1 h to depolymerize the F-actin. Following depolymerization, a 5× SDS sample buffer was added to the G-actin– and F-actin–containing samples and heated at 95 °C for 5 min to dissolve the proteins, which were then analyzed by Western blotting.
Subcellular fractionation
Nuclear and cytosolic fractions were separated using the Nuclear/Cytosol Fractionation Kit from BioVision (catalog no. K266-25; Milpitas, CA) per manufacturer's instructions. Briefly, cultured fibroblasts were harvested and dissolved in cytosol extraction buffer A, vortexed vigorously for 15 s, and ice-cold cytosol extraction buffer B added to each tube and vortexed for an additional 5 s. The suspension was centrifuged for 5 min at 16000 × g, and the supernatant (cytosolic extract) transferred immediately to a new tube. The pellet (nuclei) was resuspended in ice-cold Nuclear Extraction Buffer Mix and vortexed for 15 s every 10 min for a total of 40 min, followed by centrifugation at 16,000 × g for 10 min. The supernatant (nuclear extract) was transferred immediately to a new tube.
Immunoprecipitation
Fibroblasts were scraped off the culture dishes and lysed in radioimmune precipitation assay (RIPA) buffer (25 mm Tris, 150 mm NaCl, 0.1% SDS, 0.5% sodium deoxycholate, 1% Nonidet P-40, 0.5 mm EDTA) supplemented with protease inhibitor mixture (EMD Millipore, Billerica, MA). A 10-μl aliquot of either anti-SRF antibody (catalog no. 5147, Cell Signaling Technology, Danvers, MA) or IgG control (catalog no. 2729, Cell Signaling Technology) was added to 500 μg/500 μl of lysate and incubated at 4 °C overnight. After the incubation, 20 μl of pre-equilibrated protein A/G agarose beads (SC2003, Santa Cruz Biotechnology, Dallas, TX) was added to each sample and incubated with rotation at 4 °C for 2 h, followed by washing with RIPA buffer three times. Protein elution was done by heating the beads in 45 μl of 5× Laemmli sample buffer at 95 °C for 5 min, followed by centrifugation at 15,000 rpm for 15 s. The supernatant containing the eluted proteins was transferred to a new tube. Eluted proteins were probed for SRF and MRTF using Western blotting (see below).
Western blot analysis
Primary antibodies purchased from Cell Signaling Technology were anti-phospho-Smad2 (Ser-465/467, catalog no. 3108), anti-Smad2 (catalog no. 5339), anti-phospho-Smad3 (Ser-423/425, catalog no. 9520), anti-Smad3 (catalog no. 9523), anti-phospho-ERK1/2 (catalog no. 9102), anti-phospho-p38MAPK (Thr-180/Tyr-182, catalog no.9212), anti-p38MAPK (catalog no. 9212), anti-MRTF/MKL1 (catalog no. 14760), anti-SRF (catalog no. 5147), and anti-PARP (catalog no. 9542). Primary antibodies purchased from Santa Cruz Biotechnology were anti-CD44 (catalog no. sc-7051-R) and anti-GAPDH (catalog no. sc-25778). Polyclonal rabbit anti-α-smooth muscle actin (catalog no. ab5694) was purchased from Abcam (Cambridge, MA). Secondary antibodies including goat anti-rabbit or goat anti-mouse IgG conjugated with horseradish peroxidase (HRP) were obtained from Jackson ImmunoResearch Laboratories (West Grove, PA). At the end of treatments, cells were scraped off the culture plates and lysed in RIPA buffer supplemented with protease inhibitor cocktails (Millipore) and were then diluted in 4× NuPAGE LDS sample buffer (Life Technologies) and heated at 70 °C for 10 min. Equal amounts of proteins were loaded onto a 4–12% gradient polyacrylamide gel (Life Technologies) and separated by electrophoresis and transferred to a PVDF membrane (Immobilon-P, Millipore). The membranes were blocked in 5% nonfat milk dissolved in TBS with 0.05% Tween 20 (TBS-T) for 1 h at room temperature before probing with primary antibodies overnight at 4 °C, followed by incubation in the appropriate secondary antibody for 1 h at room temperature. The resulting signals were developed using an Enhanced Chemiluminescence (ECL) Western Blotting Detection Reagent kit (GE Healthcare). Digital records were obtained from each blot and the protein bands of interest were quantified using 1-D analysis software (Gel Logic, Carestream Molecular Imaging, Nutley, NJ). The membranes were stripped and reprobed for GAPDH as the loading control.
Immunofluorescence microscopy
Fibroblasts were grown in 35-mm cell culture dishes for immunofluorescence assays. For staining of α-SMA, the cells were fixed and permeabilized in a cold acetone:methanol mixture (1:1) for 10 min. For staining of other targeted proteins, the cells were fixed in 4% paraformaldehyde/PBS for 20 min and permeabilized with 0.1% Triton X-100/PBS for 3 min. After fixation and permeabilization, cells were blocked in 2% goat serum plus 1% BSA in PBS for 30 min at room temperature, incubated with primary antibody at 4 °C overnight, then incubated with secondary antibody conjugated with Alexa Fluor 568 or 488 (Invitrogen/Life Technologies) at room temperature for 2 h. Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI). Primary antibodies were anti–α-smooth muscle actin (catalog no. ab5694, Abcam), anti-MRTF/Mkl1 (catalog no. ab49311, Abcam), and anti-SRF (catalog no. 5147, Cell Signaling Technology). F-actin was visualized using phalloidin conjugated with Alexa Fluor 488 (catalog no. A12379, Invitrogen/Thermo Scientific). G-actin was labeled by DNase I conjugated with Alexa Fluor 594. Images were acquired using a Leica DM5500B upright microscope (Leica Microsystems, GmbH, Wetzlar, Germany) with a Retiga SRV Cooled CCD camera (QImaging, Surrey, BC Canada), and ImagePro Plus software (Media Cybernetics, Rockville, MD).
Quantitation of HA by FACE
The HA content in the cell layer and conditioned media of mouse skin fibroblast cultures was measured as described previously (38). Briefly, after proteolytic digestion and ethanol precipitation, HA in the samples was digested down to disaccharides using hyaluronidase S.D. (2.5 milliunits/μl; 100741-1A, Seikagaku America, Inc.) and labeled with 2-aminoacridone (Invitrogen) at 6.25 mm in 42.5% Me2SO, 7.5% glacial acetic acid, and 0.625 m sodium cyanoborohydride (1.25 μl/cm2 of tissue culture surface area). The labeled HA disaccharides were electrophoresed in a Bio-Rad mini-PROTEAN Tetra system using a gel composition of 20% acrylamide (37.5:1; Bio-Rad), 40 mm Tris acetate (pH 7.0), 2.5% glycerol, 10% ammonium persulfate, and 0.1% TEMED. After electrophoresis at 500 V (constant voltage) for 50 min at 4 °C, gels were imaged on a UV transilluminator at 365 nm using a CCD camera. The HA disaccharide band was quantified using Gel-Pro Analyzer® version 3.0 (Media Cybernetics, Rockville, MD).
Statistical analysis
Statistical analyses were performed using a two-sided Student's t test. p < 0.05 was considered statistically significant.
Author contributions
Y. W. conceptualization; Y. W. and E. V. M. data curation; Y. W. and E. V. M. formal analysis; Y. W. and J. A. M. validation; Y. W. investigation; Y. W. methodology; Y. W. writing-original draft; J. A. M. and E. V. M. project administration; J. A. M. and E. V. M. writing-review and editing; E. V. M. supervision; E. V. M. funding acquisition.
Supplementary Material
Acknowledgments
We sincerely thank Dr. Vincent C. Hascall for support and mentorship. We thank Dr. Veronique Orian-Rousseau at Karlsruhe Institute of Technology for generously providing us with CD44s expression plasmid. We sincerely thank Valbona Cali and Dr. Ronald Midura for assistance in generating the FACE data, and Dr. Judy Drazba and the team at Lerner Research Institute's Digital Imaging Core for assistance with cell imaging.
This work was supported by National Institutes of Health Grants P01 HL107147, NIH1K12HL141952, and S10OD019972. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S4.
- MF
- myofibroblasts
- α-SMA
- α-smooth muscle actin
- SRF
- serum response factor
- MRTF
- myocardin-related transcription factor
- G-actin
- globular actin
- F-actin
- filamentous actin
- MAPK
- mitogen-activated protein kinase
- HA
- hyaluronan
- EGFR
- epidermal growth factor receptor
- 4-MU
- 4-methylumbelliferone
- rTGF-β1
- recombinant TGF-β1
- FACE
- fluorophore-assisted carbohydrate electrophoresis
- RIPA
- radioimmune precipitation assay buffer
- DAPI
- 4′,6-diamidino-2-phenylindole
- scram
- scrambled
- qPCR
- quantitative PCR.
References
- 1. Hinz B. (2016) The role of myofibroblasts in wound healing. Curr. Res. Transl. Med. 64, 171–177 10.1016/j.retram.2016.09.003 [DOI] [PubMed] [Google Scholar]
- 2. Stunova A., and Vistejnova L. (2018) Dermal fibroblasts-A heterogeneous population with regulatory function in wound healing. Cytokine Growth Factor Rev. 39, 137–150 10.1016/j.cytogfr.2018.01.003 [DOI] [PubMed] [Google Scholar]
- 3. Andrews J. P., Marttala J., Macarak E., Rosenbloom J., and Uitto J. (2016) Keloids: The paradigm of skin fibrosis—pathomechanisms and treatment. Matrix Biol. 51, 37–46 10.1016/j.matbio.2016.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. van Caam A., Vonk M., van den Hoogen F., van Lent P., and van der Kraan P. (2018) Unraveling SSc pathophysiology; the myofibroblast. Front. Immunol. 9, 2452 10.3389/fimmu.2018.02452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ebrahimi H., Naderian M., and Sohrabpour A. A. (2016) New concepts on pathogenesis and diagnosis of liver fibrosis; a review article. Middle East J. Dig. Dis. 8, 166–178 10.15171/mejdd.2016.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lederer D. J., and Martinez F. J. (2018) Idiopathic pulmonary fibrosis. N. Engl. J. Med. 378, 1811–1823 10.1056/NEJMra1705751 [DOI] [PubMed] [Google Scholar]
- 7. Gabbiani G. (2003) The myofibroblast in wound healing and fibrocontractive diseases. J Pathol. 200, 500–503 10.1002/path.1427 [DOI] [PubMed] [Google Scholar]
- 8. Hinz B., Dugina V., Ballestrem C., Wehrle-Haller B., and Chaponnier C. (2003) α-Smooth muscle actin is crucial for focal adhesion maturation in myofibroblasts. Mol. Biol. Cell 14, 2508–2519 10.1091/mbc.e02-11-0729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Strauch A. R., and Hariharan S. (2013) Dynamic interplay of smooth muscle α-actin gene-regulatory proteins reflects the biological complexity of myofibroblast differentiation. Biology (Basel) 2, 555–586 10.3390/biology2020555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Olson E. N., and Nordheim A. (2010) Linking actin dynamics and gene transcription to drive cellular motile functions. Nat. Rev. Mol. Cell Biol. 11, 353–365 10.1038/nrm2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Miralles F., Posern G., Zaromytidou A. I., and Treisman R. (2003) Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell 113, 329–342 10.1016/S0092-8674(03)00278-2 [DOI] [PubMed] [Google Scholar]
- 12. Penn J. W., Grobbelaar A. O., and Rolfe K. J. (2012) The role of the TGF-β family in wound healing, burns and scarring: A review. Int. J. Burns Trauma 2, 18–28 [PMC free article] [PubMed] [Google Scholar]
- 13. Hinz B. (2016) Myofibroblasts. Exp. Eye Res. 142, 56–70 10.1016/j.exer.2015.07.009 [DOI] [PubMed] [Google Scholar]
- 14. Akhurst R. J., and Hata A. (2012) Targeting the TGFβ signalling pathway in disease. Nat. Rev. Drug Discov. 11, 790–811 10.1038/nrd3810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kim S. I., and Choi M. E. (2012) TGF-β-activated kinase-1: New insights into the mechanism of TGF-β signaling and kidney disease. Kidney Res. Clin. Pract. 31, 94–105 10.1016/j.krcp.2012.04.322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Stambe C., Nikolic-Paterson D. J., Hill P. A., Dowling J., and Atkins R. C. (2004) p38 Mitogen-activated protein kinase activation and cell localization in human glomerulonephritis: Correlation with renal injury. J. Am. Soc. Nephrol. 15, 326–336 10.1097/01.ASN.0000108520.63445.E0 [DOI] [PubMed] [Google Scholar]
- 17. Meyer-ter-Vehn T., Han H., Grehn F., and Schlunck G. (2011) Extracellular matrix elasticity modulates TGF-β–induced p38 activation and myofibroblast transdifferentiation in human tenon fibroblasts. Invest. Ophthalmol. Vis. Sci. 52, 9149–9155 10.1167/iovs.10-6679 [DOI] [PubMed] [Google Scholar]
- 18. Molkentin J. D., Bugg D., Ghearing N., Dorn L. E., Kim P., Sargent M. A., Gunaje J., Otsu K., and Davis J. (2017) Fibroblast-specific genetic manipulation of p38 mitogen–activated protein kinase in vivo reveals its central regulatory role in fibrosis. Circulation 136, 549–561 10.1161/CIRCULATIONAHA.116.026238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ponta H., Sherman L., and Herrlich P. A. (2003) CD44: From adhesion molecules to signalling regulators. Nat. Rev. Mol. Cell Biol. 4, 33–45 10.1038/nrm1004 [DOI] [PubMed] [Google Scholar]
- 20. Jordan A. R., Racine R. R., Hennig M. J., and Lokeshwar V. B. (2015) The role of CD44 in disease pathophysiology and targeted treatment. Front. Immunol. 6, 182 10.3389/fimmu.2015.00182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Orian-Rousseau V. (2015) CD44 acts as a signaling platform controlling tumor progression and metastasis. Front. Immunol. 6, 154 10.3389/fimmu.2015.00182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Knudson W., Chow G., and Knudson C. B. (2002) CD44-mediated uptake and degradation of hyaluronan. Matrix Biol. 21, 15–23 10.1016/S0945-053X(01)00186-X [DOI] [PubMed] [Google Scholar]
- 23. Maytin E. V. (2016) Hyaluronan: More than just a wrinkle filler. Glycobiology 26, 553–559 10.1093/glycob/cww033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Webber J., Jenkins R. H., Meran S., Phillips A., and Steadman R. (2009) Modulation of TGFβ1-dependent myofibroblast differentiation by hyaluronan. Am. J. Pathol. 175, 148–160 10.2353/ajpath.2009.080837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Webber J., Meran S., Steadman R., and Phillips A. (2009) Hyaluronan orchestrates transforming growth factor-β1–dependent maintenance of myofibroblast phenotype. J. Biol. Chem. 284, 9083–9092 10.1074/jbc.M806989200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tammi R. H., and Tammi M. I. (2009) Hyaluronan accumulation in wounded epidermis: A mediator of keratinocyte activation. J. Invest. Dermatol. 129, 1858–1860 10.1038/jid.2009.91 [DOI] [PubMed] [Google Scholar]
- 27. Passi A., Sadeghi P., Kawamura H., Anand S., Sato N., White L. E., Hascall V. C., and Maytin E. V. (2004) Hyaluronan suppresses epidermal differentiation in organotypic cultures of rat keratinocytes. Exp. Cell Res. 296, 123–134 10.1016/j.yexcr.2004.01.031 [DOI] [PubMed] [Google Scholar]
- 28. Simpson R. M., Wells A., Thomas D., Stephens P., Steadman R., and Phillips A. (2010) Aging fibroblasts resist phenotypic maturation because of impaired hyaluronan-dependent CD44/epidermal growth factor receptor signaling. Am. J. Pathol. 176, 1215–1228 10.2353/ajpath.2010.090802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Meran S., Luo D. D., Simpson R., Martin J., Wells A., Steadman R., and Phillips A. O. (2011) Hyaluronan facilitates transforming growth factor-β1–dependent proliferation via CD44 and epidermal growth factor receptor interaction. J. Biol. Chem. 286, 17618–17630 10.1074/jbc.M111.226563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li Y., Jiang D., Liang J., Meltzer E. B., Gray A., Miura R., Wogensen L., Yamaguchi Y., and Noble P. W. (2011) Severe lung fibrosis requires an invasive fibroblast phenotype regulated by hyaluronan and CD44. J. Exp. Med. 208, 1459–1471 10.1084/jem.20102510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ansorge H. L., Beredjiklian P. K., and Soslowsky L. J. (2009) CD44 deficiency improves healing tendon mechanics and increases matrix and cytokine expression in a mouse patellar tendon injury model. J. Orthop. Res. 27, 1386–1391 10.1002/jor.20891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Porsch H., Mehić M., Olofsson B., Heldin P., and Heldin C. H. (2014) Platelet-derived growth factor β-receptor, transforming growth factor β type I receptor, and CD44 protein modulate each other's signaling and stability. J. Biol. Chem. 289, 19747–19757 10.1074/jbc.M114.547273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Velasco J., Li J., DiPietro L., Stepp M. A., Sandy J. D., and Plaas A. (2011) Adamts5 deletion blocks murine dermal repair through CD44-mediated aggrecan accumulation and modulation of transforming growth factor β1 (TGFβ1) signaling. J. Biol. Chem. 286, 26016–26027 10.1074/jbc.M110.208694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Brown T. A., Bouchard T., St. John T., Wayner E., and Carter W. G. (1991) Human keratinocytes express a new CD44 core protein (CD44E) as a heparan-sulfate intrinsic membrane proteoglycan with additional exons. J. Cell Biol. 113, 207–221 10.1083/jcb.113.1.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Afify A., Purnell P., and Nguyen L. (2009) Role of CD44s and CD44v6 on human breast cancer cell adhesion, migration, and invasion. Exp. Mol. Pathol. 86, 95–100 10.1016/j.yexmp.2008.12.003 [DOI] [PubMed] [Google Scholar]
- 36. Midgley A. C., Rogers M., Hallett M. B., Clayton A., Bowen T., Phillips A. O., and Steadman R. (2013) Transforming growth factor-β1 (TGF-β1)-stimulated fibroblast to myofibroblast differentiation is mediated by hyaluronan (HA)-facilitated epidermal growth factor receptor (EGFR) and CD44 co-localization in lipid rafts. J. Biol. Chem. 288, 14824–14838 10.1074/jbc.M113.451336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schmitt M., Metzger M., Gradl D., Davidson G., and Orian-Rousseau V. (2015) CD44 functions in Wnt signaling by regulating LRP6 localization and activation. Cell Death Differ. 22, 677–689 10.1038/cdd.2014.156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang Y., Lauer M. E., Anand S., Mack J. A., and Maytin E. V. (2014) Hyaluronan synthase 2 protects skin fibroblasts against apoptosis induced by environmental stress. J. Biol. Chem. 289, 32253–32265 10.1074/jbc.M114.578377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Crider B. J., Risinger G. M. Jr., Haaksma C. J., Howard E. W., and Tomasek J. J. (2011) Myocardin-related transcription factors A and B are key regulators of TGF-β1–induced fibroblast to myofibroblast differentiation. J. Invest. Dermatol. 131, 2378–2385 10.1038/jid.2011.219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Velasquez L. S., Sutherland L. B., Liu Z., Grinnell F., Kamm K. E., Schneider J. W., Olson E. N., and Small E. M. (2013) Activation of MRTF-A–dependent gene expression with a small molecule promotes myofibroblast differentiation and wound healing. Proc. Natl. Acad. Sci. U.S.A. 110, 16850–16855 10.1073/pnas.1316764110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mack C. P., and Hinson J. S. (2005) Regulation of smooth muscle differentiation by the myocardin family of serum response factor co-factors. J. Thromb. Haemost. 3, 1976–1984 10.1111/j.1538-7836.2005.01316.x [DOI] [PubMed] [Google Scholar]
- 42. Luchsinger L. L., Patenaude C. A., Smith B. D., and Layne M. D. (2011) Myocardin-related transcription factor-A complexes activate type I collagen expression in lung fibroblasts. J. Biol. Chem. 286, 44116–44125 10.1074/jbc.M111.276931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shiwen X., Stratton R., Nikitorowicz-Buniak J., Ahmed-Abdi B., Ponticos M., Denton C., Abraham D., Takahashi A., Suki B., Layne M. D., Lafyatis R., and Smith B. D. (2015) A role of myocardin related transcription factor-A (MRTF-A) in scleroderma related fibrosis. PLoS One 10, e0126015 10.1371/journal.pone.0126015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Small E. M. (2012) The actin-MRTF-SRF gene regulatory axis and myofibroblast differentiation. J. Cardiovasc. Transl. Res. 5, 794–804 10.1007/s12265-012-9397-0 [DOI] [PubMed] [Google Scholar]
- 45. Gasparics Á., and Sebe A. (2018) MRTFs-master regulators of EMT. Dev. Dyn. 247, 396–404 10.1002/dvdy.24544 [DOI] [PubMed] [Google Scholar]
- 46. Muehlich S., Wang R., Lee S. M., Lewis T. C., Dai C., and Prywes R. (2008) Serum-induced phosphorylation of the serum response factor coactivator MKL1 by the extracellular signal-regulated kinase 1/2 pathway inhibits its nuclear localization. Mol. Cell. Biol. 28, 6302–6313 10.1128/MCB.00427-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ronkina N., Lafera J., Kotlyarov A., and Gaestel M. (2016) Stress-dependent phosphorylation of myocardin-related transcription factor A (MRTF-A) by the p38(MAPK)/MK2 axis. Sci. Rep. 6, 31219 10.1038/srep31219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sebe A., Masszi A., Zulys M., Yeung T., Speight P., Rotstein O. D., Nakano H., Mucsi I., Szászi K., and Kapus A. (2008) Rac, PAK and p38 regulate cell contact-dependent nuclear translocation of myocardin-related transcription factor. FEBS Lett. 582, 291–298 10.1016/j.febslet.2007.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ghatak S., Markwald R. R., Hascall V. C., Dowling W., Lottes R. G., Baatz J. E., Beeson G., Beeson C. C., Perrella M. A., Thannickal V. J., and Misra S. (2017) Transforming growth factor β1 (TGFβ1) regulates CD44V6 expression and activity through extracellular signal-regulated kinase (ERK)-induced EGR1 in pulmonary fibrogenic fibroblasts. J. Biol. Chem. 292, 10465–10489 10.1074/jbc.M116.752451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Midgley A. C., Oltean S., Hascall V., Woods E. L., Steadman R., Phillips A. O., and Meran S. (2017) Nuclear hyaluronidase 2 drives alternative splicing of CD44 pre-mRNA to determine profibrotic or antifibrotic cell phenotype. Sci. Signal. 10, eaao1822 10.1126/scisignal.aao1822 [DOI] [PubMed] [Google Scholar]
- 51. Shatirishvili M., Burk A. S., Franz C. M., Pace G., Kastilan T., Breuhahn K., Hinterseer E., Dierich A., Bakiri L., Wagner E. F., Ponta H., Hartmann T. N., Tanaka M., and Orian-Rousseau V. (2016) Epidermal-specific deletion of CD44 reveals a function in keratinocytes in response to mechanical stress. Cell Death Dis. 7, e2461 10.1038/cddis.2016.342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Govindaraju P., Todd L., Shetye S., Monslow J., and Pure E. (2018) CD44-dependent inflammation, fibrogenesis, and collagenolysis regulates extracellular matrix remodeling and tensile strength during cutaneous wound healing. Matrix Biol. 75–76, 314–330 10.1016/j.matbio.2018.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yonemura S., Hirao M., Doi Y., Takahashi N., Kondo T., Tsukita S., and Tsukita S. (1998) Ezrin/radixin/moesin (ERM) proteins bind to a positively charged amino acid cluster in the juxta-membrane cytoplasmic domain of CD44, CD43, and ICAM-2. J. Cell Biol. 140, 885–895 10.1083/jcb.140.4.885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bennett V., and Healy J. (2009) Membrane domains based on ankyrin and spectrin associated with cell-cell interactions. Cold Spring Harb. Perspect. Biol. 1, a003012 10.1101/cshperspect.a003012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bourguignon L. Y. (2008) Hyaluronan-mediated CD44 activation of RhoGTPase signaling and cytoskeleton function promotes tumor progression. Semin. Cancer Biol. 18, 251–259 10.1016/j.semcancer.2008.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bourguignon L. Y. (2014) Matrix hyaluronan-activated CD44 signaling promotes keratinocyte activities and improves abnormal epidermal functions. Am. J. Pathol. 184, 1912–1919 10.1016/j.ajpath.2014.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bourguignon L. Y., Gilad E., Rothman K., and Peyrollier K. (2005) Hyaluronan-CD44 interaction with IQGAP1 promotes Cdc42 and ERK signaling, leading to actin binding, Elk-1/estrogen receptor transcriptional activation, and ovarian cancer progression. J. Biol. Chem. 280, 11961–11972 10.1074/jbc.M411985200 [DOI] [PubMed] [Google Scholar]
- 58. Bourguignon L. Y., Peyrollier K., Gilad E., and Brightman A. (2007) Hyaluronan-CD44 interaction with neural Wiskott-Aldrich syndrome protein (N-WASP) promotes actin polymerization and ErbB2 activation leading to β-catenin nuclear translocation, transcriptional up-regulation, and cell migration in ovarian tumor cells. J. Biol. Chem. 282, 1265–1280 10.1074/jbc.M604672200 [DOI] [PubMed] [Google Scholar]
- 59. Bourguignon L. Y., Ramez M., Gilad E., Singleton P. A., Man M. Q., Crumrine D. A., Elias P. M., and Feingold K. R. (2006) Hyaluronan-CD44 interaction stimulates keratinocyte differentiation, lamellar body formation/secretion, and permeability barrier homeostasis. J. Invest. Dermatol. 126, 1356–1365 10.1038/sj.jid.5700260 [DOI] [PubMed] [Google Scholar]
- 60. Bourguignon L. Y., Singleton P. A., and Diedrich F. (2004) Hyaluronan-CD44 interaction with Rac1-dependent protein kinase N-γ promotes phospholipase Cγ1 activation, Ca2+ signaling, and cortactin-cytoskeleton function leading to keratinocyte adhesion and differentiation. J. Biol. Chem. 279, 29654–29669 10.1074/jbc.M403608200 [DOI] [PubMed] [Google Scholar]
- 61. Bourguignon L. Y., Wong G., Earle C., Krueger K., and Spevak C. C. (2010) Hyaluronan-CD44 interaction promotes c-Src-mediated twist signaling, microRNA-10b expression, and RhoA/RhoC up-regulation, leading to Rho-kinase-associated cytoskeleton activation and breast tumor cell invasion. J. Biol. Chem. 285, 36721–36735 10.1074/jbc.M110.162305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Konopka A., Zeug A., Skupien A., Kaza B., Mueller F., Chwedorowicz A., Ponimaskin E., Wilczynski G. M., and Dzwonek J. (2016) Cleavage of hyaluronan and CD44 adhesion molecule regulate astrocyte morphology via Rac1 signalling. PLoS One 11, e0155053 10.1371/journal.pone.0155053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Roszkowska M., Skupien A., Wójtowicz T., Konopka A., Gorlewicz A., Kisiel M., Bekisz M., Ruszczycki B., Dolezyczek H., Rejmak E., Knapska E., Mozrzymas J. W., Wlodarczyk J., Wilczynski G. M., and Dzwonek J. (2016) CD44: A novel synaptic cell adhesion molecule regulating structural and functional plasticity of dendritic spines. Mol. Biol. Cell 27, 4055–4066 10.1091/mbc.e16-06-0423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Shimizu Y., Van Seventer G. A., Siraganian R., Wahl L., and Shaw S. (1989) Dual role of the CD44 molecule in T cell adhesion and activation. J. Immunol. 143, 2457–2463 [PubMed] [Google Scholar]
- 65. Merzak A., Koocheckpour S., and Pilkington G. J. (1994) CD44 mediates human glioma cell adhesion and invasion in vitro. Cancer Res. 54, 3988–3992 [PubMed] [Google Scholar]
- 66. Morrison H., Sherman L. S., Legg J., Banine F., Isacke C., Haipek C. A., Gutmann D. H., Ponta H., and Herrlich P. (2001) The NF2 tumor suppressor gene product, merlin, mediates contact inhibition of growth through interactions with CD44. Genes Dev. 15, 968–980 10.1101/gad.189601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Busche S., Descot A., Julien S., Genth H., and Posern G. (2008) Epithelial cell-cell contacts regulate SRF-mediated transcription via Rac-actin-MAL signalling. J. Cell Sci. 121, 1025–1035 10.1242/jcs.014456 [DOI] [PubMed] [Google Scholar]
- 68. Fan L., Sebe A., Péterfi Z., Masszi A., Thirone A. C., Rotstein O. D., Nakano H., McCulloch C. A., Szászi K., Mucsi I., and Kapus A. (2007) Cell contact-dependent regulation of epithelial-myofibroblast transition via the rho-rho kinase-phospho-myosin pathway. Mol. Biol. Cell 18, 1083–1097 10.1091/mbc.e06-07-0602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Larsson C. (2006) Protein kinase C and the regulation of the actin cytoskeleton. Cell Signal. 18, 276–284 10.1016/j.cellsig.2005.07.010 [DOI] [PubMed] [Google Scholar]
- 70. Legg J. W., Lewis C. A., Parsons M., Ng T., and Isacke C. M. (2002) A novel PKC-regulated mechanism controls CD44 ezrin association and directional cell motility. Nat. Cell Biol. 4, 399–407 10.1038/ncb797 [DOI] [PubMed] [Google Scholar]
- 71. Nagano O., Murakami D., Hartmann D., De Strooper B., Saftig P., Iwatsubo T., Nakajima M., Shinohara M., and Saya H. (2004) Cell-matrix interaction via CD44 is independently regulated by different metalloproteinases activated in response to extracellular Ca2+ influx and PKC activation. J. Cell Biol. 165, 893–902 10.1083/jcb.200310024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Zhang P., Goodrich C., Fu C., and Dong C. (2014) Melanoma up-regulates ICAM-1 expression on endothelial cells through engagement of tumor CD44 with endothelial E-selectin and activation of a PKCα-p38-SP-1 pathway. FASEB J. 28, 4591–4609 10.1096/fj.11-202747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Bourguignon L. Y., Spevak C. C., Wong G., Xia W., and Gilad E. (2009) Hyaluronan-CD44 interaction with protein kinase Cϵ promotes oncogenic signaling by the stem cell marker Nanog and the production of microRNA-21, leading to down-regulation of the tumor suppressor protein PDCD4, anti-apoptosis, and chemotherapy resistance in breast tumor cells. J. Biol. Chem. 284, 26533–26546 10.1074/jbc.M109.027466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Evanko S. P., Potter-Perigo S., Petty L. J., Workman G. A., and Wight T. N. (2015) Hyaluronan controls the deposition of fibronectin and collagen and modulates TGF-β1 induction of lung myofibroblasts. Matrix Biol. 42, 74–92 10.1016/j.matbio.2014.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zöller M. (2011) CD44: can a cancer-initiating cell profit from an abundantly expressed molecule? Nat. Rev. Cancer 11, 254–267 10.1038/nrc3023 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.