Abstract
Purpose
To characterize the global profile of circular RNAs (circRNAs) and their differential expression levels in homocysteine (Hcy)-treated ARPE-19 cells, a line of human retinal pigment epithelial (RPE) cells.
Materials and Methods
We treated ARPE-19 cells with and without Hcy to investigate the influence of Hcy on circRNA expression levels using dedicated human circRNA microarrays.
Results
A total of 12,233 circRNAs were identified out of them 54 were differentially expressed (17 were down-regulated, and 37 were up-regulated) with a fold change >2.0 (p < 0.05) in Hcy-treated versus untreated cells.
Conclusions
To our knowledge, this is the first report profiling circRNAs in human RPE cells post-Hcy treatment mimicking hyperhomocysteinemic (HHcy) conditions that negatively affect retinal biology and vision. These findings are of potential clinical significance as they will help understand Hcy metabolism and HHcy-mediated diseases and identify potential diagnostic and therapeutic targets for eye diseases that are caused by elevated Hcy concentrations.
Keywords: CircRNA, epigenetics, eye diseases, inflammation, retina
Introduction
Retinovascular disease, comprised primarily of diabetic retinopathy (DR) and retinal vein occlusion (RVO), is a major cause of vision loss in the working age population. Its cardinal features are vascular leakage, inflammation, thrombosis, and angiogenesis.1–5 Despite advances in anti-angiogenic treatment in ophthalmology, the retinovascular disease still causes significant morbidity. Recently, circRNAs have been shown to regulate RNA and protein expression in the retina.6 Circular RNAs (circRNAs) are a recently re-discovered class of endogenous RNAs that are generated from exons, introns, or both via a diverse number of cellular mechanisms. Rapid progress in high-throughput sequencing and bioinformatics has revealed that many circRNAs can modulate expression of microRNAs (miRNAs or miRs).7–10 More importantly, circRNAs fine-tune post-transcriptional expression via their interactions with miRNAs or with single or multiple RNA-binding proteins.8
In the eye, circRNAs can either promote ocular homeostasis and normal function or contribute to disease. For example, circRNA-ZNF609 can rescue vascular endothelial dysfunction in an oxygen-induced retinopathy model whereas CircHIIPK3 plays a negative role in DR.11,12 Identification of circRNAs that are expressed in retinal pigment epithelial (RPE) cells could shed light on their role in the eye, especially under hyperhomocysteinemic (HHcy) conditions. This could also assist in finding clinical biomarkers for treatment of retinal disorders. To the best of our knowledge, no study has examined circRNAs under HHcy conditions in the human eyes. This study sought to characterize the global profile of circRNAs in RPE cells under HHcy conditions. The RPE is an essential monolayer of cells immediately deep to the neurosensory retina which provides it with nutrition, allows the diffusion of oxygen to the outer retina, and phagocytoses outer photoreceptor segments to keep these cells functional. The proper function of the RPE is also critical to the maintenance of the blood-retina barrier (BRB). An intact BRB prevents the accumulation of serous fluid under or potentially within the outer retina, which are common structural complications of retinovascular disease and the most common cause of vision loss in both DR and RVO.13 Thus, a better understanding of RPE pathobiology is instrumental to better understanding of retinovascular disease in general. Diabetics are known to have increased levels of serum homocysteine (Hcy), which correlates with the level of DR severity.14 Moreover, HHcy conditions disrupt the BRB in retinal endothelial cells and disrupt RPE cell structure and function.4,15 Thus, HHcy-treated ARPE-19 cells are a model for studying the molecular and cellular effects of the retinovascular disease. Using a mouse model of hyperhomocysteinemia, we recently reported an abundant pool of circRNAs representing all cell types from the eye and opine that these elegant molecules constitute an inherent regulatory axis in the mammalian eyes and brain.16
Materials and methods
A fresh batch of ARPE-19 cells was obtained from American Type Culture Collection (ATCC, Manassas, VA, USA) and was handled as per standard practice.17 Cells were grown in sufficient quantities in Petri-dishes/cell culture flasks and treated with Hcy (150 μM/L) (Sigma). Triplicate samples of untreated were designated as Group 1 while triplicate samples in Hcy-treated as Group 2; all samples were kept in the incubator for 24 h at 37°C, 5% CO2. Total RNAs were isolated and purified using RNeasy Mini kit (Invitrogen Inc.). RNA quality and quantity were measured using a NanoDrop spectrophotometer (ND-1000; NanoDrop; Thermo Fisher Scientific), and RNA integrity was determined by gel electrophoresis. Arraystar human circRNA microarray V2.0 (Arraystar, Inc.) designed for identification of globally expressed human circRNAs was used for profiling of circRNAs that covers 13,617 human circRNAs curated from circRNA studies dealing with landmark publications allowing systematic identification and profiling of circRNA transcriptomes under physiological and pathophysiological conditions. Each circRNA is represented by a circular splice junction probe which can reliably and accurately detect circRNA, even in the presence of its linear counterparts. The procedure employs a random primer-based labeling system that is coupled with RNase R pre-treatment to ensure specific and efficient labeling of circRNAs. RNA spike-in controls was included to monitor labeling and hybridization efficiencies. The circRNAs were annotated with predicted miRNA target sites to help unravel their functional roles as a natural miRNA sponge(s). Sample labeling and array hybridization were performed according to the protocol (Arraystar Inc.). Briefly, total RNAs were digested with RNase R (Epicentre, Inc.) to remove linear RNAs and enrich circRNAs. Then, enriched circRNAs were amplified and transcribed into fluorescent cRNAs utilizing a random priming method (Arraystar super RNA labeling kit). Labeled cRNAs were purified by RNeasy Mini Kit (Qiagen). Concentrations and specific activities of labeled cRNAs (pmol Cy3/μg cRNA) were measured by NanoDrop ND-1000. One microgram of each labeled cRNA was fragmented by adding 5 μl 10 × blocking agent and 1 μl of 25 × fragmentation buffer, then heated mixture at 60°C for 30 min. Finally, 25 μl 2 × hybridization buffer was added to dilute labeled cRNAs. Fifty microliter of hybridization solution was dispensed into gasket-slides and assembled to circRNA expression microarray-slides which were incubated for 17 h at 65° C in an Agilent hybridization oven. Hybridized arrays were washed, fixed and scanned using Agilent scanner G2505C.
For data analysis scanned images were imported into Agilent’s feature extraction software version 11.0.1.1 (Agilent Technologies, Inc.) for raw data extraction. Quantile normalization of raw data and subsequent data processing was performed using R software package. Following quantile normalization of raw data, low-intensity filtering was performed and circRNAs having the “P” or “M” flags (“all targets value”) in ≥3 out of 6 samples were retained for further analyses. Subsequently, samples were clustered hierarchically with cluster software version 2.0 (http://bonsai.hgc.jp/~mdehoon/software/cluster/software.htm) to evaluate the robustness of formed clusters, using correlation-centered metric and average linkage-clustering algorithm. When comparing two groups of profile differences (such as treated versus control), “fold change” (i.e., the ratio of group averages) between groups for each circRNA was computed. Statistical significance of difference was estimated by t-test. The circRNAs having fold changes ≥2.0 and p ≤ 0.05 were selected as significantly differentially expressed entities.
Results
RNAs were assessed for their qualities, quantities and then used for experiments (Figure 1a). Human circRNA microarrays were used to assess differences in circRNA expression profiles between Hcy-treated and the untreated ARPE-19 cells. A total of 12,233 circRNAs were identified. Differentially expressed circRNAs with statistical significance between Hcy-treated ARPE-19 cells were compared with untreated cells and then were identified through fold-change criteria. There was a homogeneous distribution of expression values in Hcy-treated and untreated ARPE-19 cells after comparing normalized distributions of intensities from all samples (Figure 1b). The majority of cRNAs showed similar levels of expression between the two groups (Figure 1c,d). A volcano plot for evaluation of differential expression as derived from expression profiling of circRNAs’ from Hcy-treated and untreated ARPE-19 cells did reveal a difference for several circRNA from one another between Group 2 (Hcy-treated) versus Group 1 (untreated cells, Figure 1c). A Scatter-plot of cRNAs’ averaged normalized value variation between treated and untreated cells displays the circRNAs that were found to be more than 2.0-fold changes as shown above the top green line and below the bottom green line (Figure 1d). The heat map and hierarchical clustering of Group 2 (Hcy-treated) versus Group 1 (untreated) were also created and that showed distinguishable circRNA expression patterns among samples (Figure 2a,b). The circRNAs showing fold changes ≥2.0 and p ≤ 0.05 were selected as significantly differentially expressed molecules. The microarray data demonstrate that expression of circRNAs in treated (Group 2) as compared to the untreated ARPE-19 cells were significantly different. A total of 54 significantly differentially expressed circRNAs were found. Among them, 17 were down-regulated (Table 1), and 37 were up-regulated (Table 2). Up-regulated circRNAs (~68%) were found to be more common than down-regulated circRNAs (~ 32 %). Differentially expressed circRNAs according to extent of changes between Hcy-treated versus the untreated cells are shown as down-regulated and up- regulated circRNAs and the nature of each circRNA’s origin is color-coded in Figure 2c while their chromosomal distributions are illustrated with respect to the abundance of total circRNAs between Group 1 versus Group 2 in Figure D.
Figure 1.
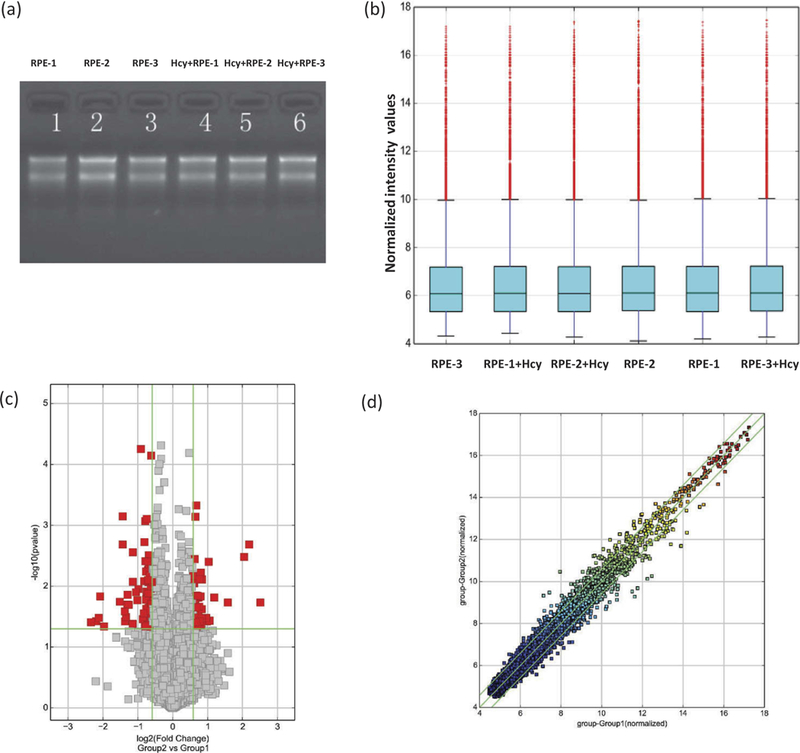
Assessment of RNAs’ integrity, genomic DNA contamination, and cRNAs’ differential expressions. (a) Denaturing agarose gel electrophoresis of samples: Lane 1, 2, and 3 represent total RNAs from untreated, and 4, 5, and 6 from Hcy-treated ARPE-19 cells. (b) A box plot showing a homogeneous distribution expression values in Hcy-treated and untreated ARPE-19 cells after comparing the distributions of intensities from all samples after normalization. (c) Volcano plot for the evaluation of differential expression as derived from the expression profiling of circRNAs’ from Hcy-treated and untreated ARPE-19 cells. Respective difference of each circRNA from one another between Group 2 (Hcy-treated) versus Group 1 (untreated cells). Gray points represent circRNAs that show no statistical significance while the red points represent the differentially expressed circRNAs with statistical significance. The vertical lines correspond to 2.0-fold (log2 scaled) up and down, respectively, and the horizontal line represents a p-value of 0.05 (−log10 scaled). (d) The scatter-plot of cRNAs’ averaged normalized value variation between Group 1; untreated (x-axis) and Group 2; Hcy-treated RPE cells (y-axis). The circRNAs above the top green line and below the bottom green line indicate more than 2.0-fold changes in circRNAs. Middle green line refers to no difference between the groups.
Figure 2.
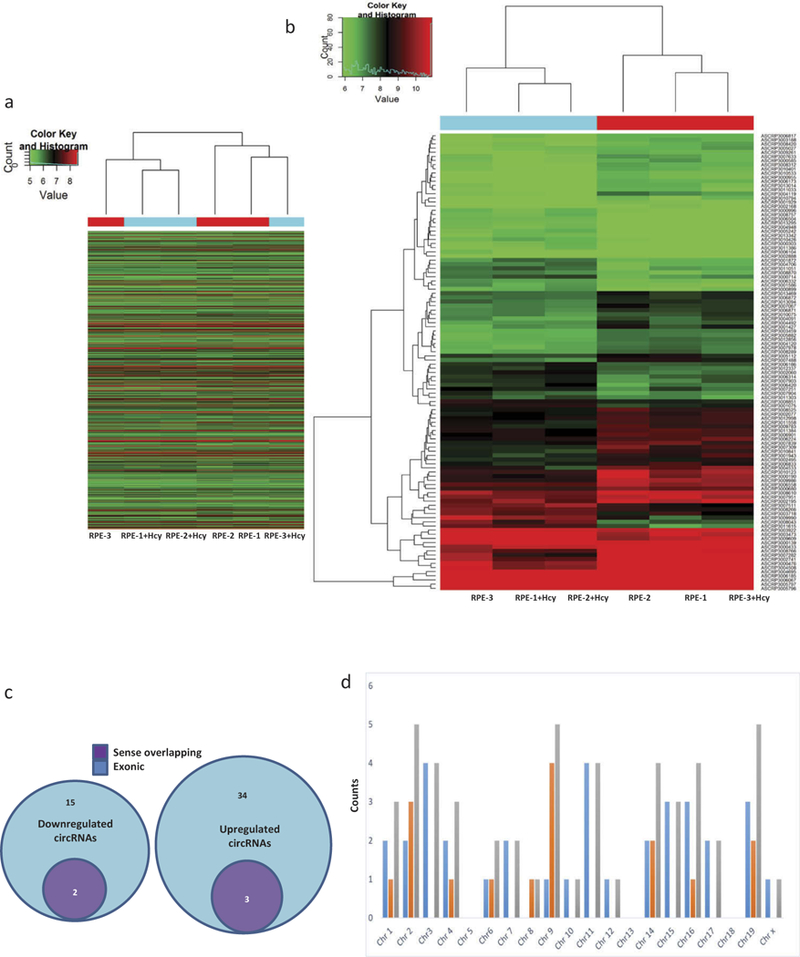
(a) Heat map of Group 2 (Hcy-treated) versus Group 1 (untreated) showing expression of all target values of circRNAs, their respective up and down-regulated patterns between ARPE-19 cells. Each column in the map represents a sample, and each row represents a circRNA. The red strip represents high relative expression, and green strip represents low relative expression. (b) Measurement of circRNAs’ relative expression activities employing the hierarchical cluster analysis for the respective up and down-regulated data sets between the Group 2 (Hcy-treated) versus Group-1 (untreated) ARPE-19 cells. Red represents high relative expression and green low relative expression. (c) Differentially expressed circRNAs according to the extent of changes between Hcy-treated and untreated ARPE-19 cells depicting down-regulated and up-regulated circRNAs. Nature of each circRNA’s origin is color-coded. (d) Chromosomal distributions of differentially expressed circRNAs with color codes illustrate their expression profiles with respect to the abundance of total circRNAs between Group 1 versus Group 2.
Table 1.
Biological information regarding down-regulated circRNAs in Hcy-treated versus untreated ARPE-19 cells.
| CircRNA ID | Fold change | FDR | p Value | circRNA type | Chr | Best transcript | Gene symbol |
|---|---|---|---|---|---|---|---|
| has_circRNA_102686 | 2.86 | 0.9 | 0.02 | Exonic | chr2 | NM_016441 | CRIM1 |
| has_circRNA_087856 | 4.59 | 1.00 | 0.04 | Exonic | chr9 | NM_002874 | RAD23B |
| has_circRNA_404514 | 3.63 | 1.00 | 0.44 | Exonic | chr1 | NM_024646 | ZYG11B |
| has_circRNA_104851 | 2.47 | 0.92 | 0.02 | Exonic | chr9 | NM_002874 | RAD23B |
| has_circRNA_104852 | 3.08 | 1.00 | 0.07 | Exonic | chr9 | NM_002874 | RAD23B |
| has_circRNA_104700 | 2.22 | 1.00 | 0.04 | Exonic | chr8 | NM_005607 | PTK2 |
| hsa_circRNA_077755 | 2.32 | 1.00 | 0.09 | Exonic | chr6 | uc003pyr.3 | GJA1 |
| hsa_circRNA_063089 | 2.01 | 1.00 | 0.06 | Exonic | chr22 | NM_002473 | MYH9 |
| hsa_circRNA_104854 | 4.34 | 1.00 | 0.03 | Exonic | chr9 | NM_002874 | RAD23B |
| hsa_circRNA_013055 | 2.58 | 1.00 | 0.04 | Exonic | chr1 | NM_005274 | GNG5 |
| hsa_circRNA_002178 | 2.40 | 0.92 | 0.02 | Sense overlapping | chr14 | NR_002312 | RPPH1 |
| hsa_circRNA_001846 | 2.53 | 1.00 | 0.03 | Sense overlapping | chr14 | NR_002312 | RPPH1 |
| hsa_circRNA_102688 | 2.45 | 0.88 | 0.01 | Exonic | chr2 | NM_016441 | CRIM1 |
| hsa_circRNA_102403 | 2.21 | 0.76 | 0.01 | Exonic | chr19 | NM_018959 | DAZAP1 |
| hsa_circRNA_101828 | 2.39 | 1.00 | 0.05 | Exonic | chr16 | NM_025187 | C16orf70 |
| hsa_circRNA_102402 | 2.70 | 0.53 | 0.00 | Exonic | chr19 | NM_018959 | DAZAP1 |
| hsa_circRNA_103670 | 2.08 | 1.00 | 0.41 | Exonic | chr4 | NM_144571 | CNOT6L |
CircRNA types: Different classes of circRNAs as: “exonic”, “intronic”, and sense overlapping. P value: p value calculated from paired t-test. FDR: FDR is calculated from Benjamini Hochberg FDR. Fold Change: The absolute ratio (no log scale) of normalized intensities between two conditions. Chr: chromosome; Best transcript, is transcribed from the same gene position with circular RNA, the sequence information is most similar to circular RNA. Expression values for circRNAs that are known to be down-regulated more than three folds indicated in bold.
Table 2.
Biological information regarding up-regulated circRNAs in Hcy-treated and untreated ARPE-19 cells.
| CircRNA ID | Fold change | FDR | p Value | circRNA type | Chr | Best transcript | Gene symbol |
|---|---|---|---|---|---|---|---|
| hsa_circRNA_406494 | 2.11 | 1.00 | 0.33 | Exonic | chr4 | NM_133636 | HELQ |
| hsa_circRNA_405036 | 2.77 | 1.00 | 0.42 | Exonic | chr12 | NM_172240 | POC1B |
| hsa_circRNA_404798 | 2.08 | 1.00 | 0.57 | Exonic | chr10 | NM_015221 | DNMBP |
| hsa_circRNA_102620 | 2.04 | 0.75 | 0.00 | Exonic | chr2 | NM_145693 | LPIN1 |
| hsa_circRNA_101504 | 2.39 | 1.00 | 0.09 | Exonic | chr15 | NM_005313 | PDIA3 |
| hsa_circRNA_407344 | 2.07 | 1.00 | 0.33 | Exonic | chrX | NM_080612 | GAB3 |
| hsa_circRNA_060876 | 2.16 | 1.00 | 0.24 | Exonic | chr20 | NM_006045 | ATP9A |
| hsa_circRNA_092493 | 2.45 | 1.00 | 0.24 | Sense overlapping | chr2 | NM_003387 | WIPF1 |
| hsa_circRNA_103649 | 2.17 | 1.00 | 0.29 | Exonic | chr4 | NM_018475 | TMEM165 |
| hsa_circRNA_082335 | 2.10 | 1.00 | 0.11 | Exonic | chr7 | NM_014997 | KLHDC10 |
| hsa_circRNA_103348 | 2.07 | 1.00 | 0.04 | Exonic | chr3 | NM_012235 | SCAP |
| hsa_circRNA_101693 | 3.08 | 1.00 | 0.25 | Exonic | chr16 | NM_020677 | NMRAL1 |
| hsa_circRNA_063411 | 2.61 | 1.00 | 0.22 | Exonic | chr22 | NM_015088 | TNRC6B |
| hsa_circRNA_039626 | 2.07 | 1.00 | 0.06 | Exonic | chr16 | NM_016284 | CNOT1 |
| hsa_circRNA_033628 | 2.33 | 1.00 | 0.51 | Exonic | chr14 | NM_001311 | CRIP1 |
| hsa_circRNA_101744 | 2.24 | 1.00 | 0.31 | Exonic | chr16 | NM_020314 | C16orf62 |
| hsa_circRNA_404655 | 2.35 | 1.00 | 0.50 | Sense overlapping | chr1 | TCONS_00001278 | XLOC 000566 |
| hsa_circRNA_104940 | 2.07 | 1.00 | 0.41 | Exonic | chr9 | NM_005085 | NUP214 |
| hsa_circRNA_048764 | 2.33 | 1.00 | 0.57 | Exonic | chr19 | NM_015414 | RPL36 |
| hsa_circRNA_103461 | 2.98 | 1.00 | 0.13 | Exonic | chr3 | NM_007283 | MGLL |
| hsa_circRNA_101437 | 2.07 | 1.00 | 0.17 | Exonic | chr14 | NM_004434 | EML1 |
| hsa_circRNA_007237 | 2.44 | 1.00 | 0.22 | Sense overlapping | chr3 | NR_126560 | KCNMB2-AS1 |
| hsa_circRNA_404853 | 2.16 | 1.00 | 0.10 | Exonic | chr11 | NM_017508 | SOX6 |
| hsa_circRNA_050649 | 2.19 | 1.00 | 0.60 | Exonic | chr19 | ENST00000004982 | HSPB6 |
| hsa_circRNA_050648 | 2.04 | 1.00 | 0.53 | Exonic | chr19 | ENST00000004982 | HSPB6 |
| hsa_circRNA_101460 | 2.01 | 1.00 | 0.07 | Exonic | chr15 | NM_014608 | CYFIP1 |
| hsa_circRNA_100311 | 2.06 | 1.00 | 0.24 | Exonic | chr1 | NM_006699 | MAN1A2 |
| hsa_circRNA_100832 | 2.28 | 0.88 | 0.02 | Exonic | chr11 | NM_013402 | FADS1 |
| hsa_circRNA_402755 | 2.19 | 1.00 | 0.09 | Exonic | chr22 | NM_025225 | PNPLA3 |
| hsa_circRNA_000367 | 4.11 | 0.75 | 0.00 | Exonic | chr11 | NM_170601 | SIAE |
| hsa_circRNA_405330 | 5.70 | 0.91 | 0.02 | Exonic | chr15 | NM_005313 | PDIA3 |
| hsa_circRNA_402964 | 2.57 | 1.00 | 0.17 | Exonic | chr3 | NM_000532 | PCCB |
| hsa_circRNA_000735 | 2.12 | 1.00 | 0.48 | Exonic | chr17 | NM_002558 | P2RX1 |
| hsa_circRNA_403691 | 2.42 | 1.00 | 0.17 | Exonic | chr6 | NR_125845 | LOC101927768 |
| hsa_circRNA_102225 | 1.16 | 1.00 | 0.54 | Exonic | chr17 | NM_017921 | NPLOC4 |
| hsa_circRNA_082319 | 2.49 | 1.00 | 0.36 | Exonic | chr7 | NM_016478 | ZC3HC1 |
| hsa_circRNA_100834 | 2.07 | 1.00 | 0.17 | Exonic | chr11 | NM_004265 | FADS2 |
CircRNA types: Different classes of circRNAs as: “exonic”, “intronic”, and sense overlapping. P value: p value calculated from paired t-test. FDR: FDR is calculated from Benjamini Hochberg FDR. Fold Change: The absolute ratio (no log scale) of normalized intensities between two conditions. Chr: chromosome; Best transcript, is transcribed from the same gene position with circular RNA, the sequence information is most similar to circular RNA. Expression values for circRNAs that are known to be up-regulated more than three folds indicated in bold.
Discussion
The circRNAs are a novel class of endogenous molecules that are characterized by covalently closed continuous loops without 5′ to 3′ polarities and polyadenylated tails. They serve crucial roles as novel regulators in a swath of biological processes. However, their abundance, expression profiles, and clinical significance in HHcy-induced retinal associated disorders are unknown. They are generated from exonic or intronic sequences and seem to be conserved across species. Interestingly, circRNAs show tissue/developmental stage-specific expression patterns. Because they are more stable than their linear counterparts owing to their higher nuclease stability, they carry an enormous advantage from a diagnostic standpoint as a novel and promising class of clinical biomarkers. Further, they have been shown to act as miRNA sponges, thereby lifting the suppressive effect of miRNAs on transcription. Recent findings have demonstrated that they interact with disease-associated miRNAs, suggesting their importance in disease pathogenesis.9,10 Dysregulated circRNAs are associated with several human diseases of the nervous and cardiovascular diseases and with cancer.18–23 Mounting evidence suggests that circRNAs serve important roles in cellular metabolism and regulatory processes including development, proliferation, differentiation, and apoptosis.24,25 Most circRNAs have been identified in the brain.9,26–30 They are known to accumulate with aging and appear that they may be important in aging/age-related diseases.31 Increased abundance of circRNAs might impact age-related decline in neural functions as their levels are dynamically modulated in neurons both during differentiation and following bursts of electrical activity. One study showed that 283 circRNAs were altered after transient middle cerebral artery occlusion emphasizing their roles in regulating biological and molecular functions like cell metabolism, cell communication, and binding to proteins, ions, and nucleic acids. Interestingly, ciRS-7 contains more than 70 selectively miRNA target sites and functions as a miR-7 sponge to regulate expression of human EGFR, IRS2, SNCA.32
One of the major limitations of this study is the use of only a single cell line and the lack of serum measurements of circRNA levels in vivo. While the RPE is vital to survival and function of photoreceptors, the profile of circRNAs in the cells of the neurosensory retina or retinal endothelial cells, which form the inner BRB, is still unknown and requires further study. While DR clinically behaves like a retinovascular disease, it is well known to be a retina-wide neuropathy as well. Thus, the profile of circRNAs from photoreceptors, Muller cells, and glial cells is likely abnormal in DR. Second, since the goal of this work is ultimately to generate novel biomarkers for retinovascular disease, clinical samples from the eye are cumbersome to obtain and pose some risk in routine clinical practice. Serum biomarkers are preferable. While we did not measure any serum circRNAs, this study has at least established plausible candidates for serum biomarkers. A study on clinical samples for precisely this purpose is currently underway in our laboratory and would be reported on its completion in due course of time.
Vascular dysfunction is a hallmark of ischemic, neoplastic, and inflammatory diseases contributing to disease progression. We previously reported that MMPs induce cerebrovascular dysfunction and that hyperhomocysteinemia mediates vascular remodeling in various body systems.14,33 It is known that circRNA-ZNF 609 ameliorates vascular endothelial dysfunction in oxygen-induced retinopathy and circHIIPK3 plays a role in DR.11,12 In other studies, several circRNAs have been shown to be differentially expressed in patients with DR and hsa_circRNA_103410 is a known regulator of miR-126, which inhibits VEGF and MMP-9.34 circRNAs may be involved in paracrine signaling or cell-to-cell cross-talk in the retina. Thus, identification and profiling of circRNAs would lead to better understanding of their mechanisms of action that can pave the way for early diagnosis and superior therapeutics for numerous retinal diseases that presently have no cure. The clinical significance of circRNAs in HHcy conditions in general and their role in retinovascular diseases are not known. This study is a first step in shedding light on this complex topic.
Acknowledgments
Members of the laboratory are gratefully acknowledged for their continuous support and encouragement.
Funding
This work was supported by grants (HL-74815, HL-107640, HL-139047, and AR071789) from the National Institute of Health (NIH).
Footnotes
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- 1.Leizaola-Fernández C, Suárez-Tatá L, Quiroz-Mercado H, Colina- Luquez J, Fromow-Guerra J, Jiménez-Sierra JM, Guerrero-Naranjo JL, Morales-Cantón V. Vitrectomy with complete posterior hyaloid removal for ischemic central retinal vein occlusion: series of cases. BMC Ophthalmol. 2005; 5(10):1–5. doi: 10.1186/1471-2415-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Markand S, Saul A, Roon P, Prasad P, Martin P, Rozen R, Ganapathy V, Smith SB. Retinal ganglion cell loss and mild vasculopathy in methylene tetrahydrofolate reductase (Mthfr)-deficient mice: a model of mild hyperhomocysteinemia. Invest Ophthalmol Vis Sci. 2015;56 (4):2684–95. doi: 10.1167/iovs.14-16190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stitt AW, Curtis TM, Chen M, Medina RJ, McKay GJ, Jenkins A, Gardiner TA, Lyons TJ, Hammes HP, Simo R, et al. The progress in understanding and treatment of diabetic retinopathy. Prog Retin Eye Res. 2016;51(2):156–86. doi: 10.1016/j.preteyeres.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 4.Ibrahim AS, Mander S, Hussein KA, Elsherbiny NM, Smith SB, Al-Shabrawey M, Tawfik A. Hyperhomocysteinemia disrupts retinal pigment epithelial structure and function with features of age-related macular degeneration. Oncotarget. 2016;7 ,(8):8532–45. doi: 10.18632/oncotarget.7384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Homme RP, Singh M, Majumder A, George AK, Nair K, Sandhu HS, Tyagi N, Lominadze D, Tyagi SC. Remodeling of retinal architecture in diabetic retinopathy: disruption of ocular physiology and visual functions by inflammatory gene products and pyroptosis. Front Physiol. 2018;9(1268):1–18. doi: 10.3389/fphys.2018.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Han J, Gao L, Dong J, Bai J, Zhang M, Zheng J. The expression profile of developmental stage-dependent circular RNA in the immature rat retina. Mol Vis. 2017;23:457–69. [PMC free article] [PubMed] [Google Scholar]
- 7.Glazar P, Papavasileiou P, Rajewsky N. CircBase: a database for circular RNAs. Rna. 2014;20(11):1666–70. doi: 10.1261/rna.043687.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–38. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 9.Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9 (9):1–15, e1003777. doi: 10.1371/annotation/f782282b-eefa-4c8d-985c-b1484e845855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barrett SP, Salzman J. Circular RNAs: analysis, expression and potential functions. Development. 2016;143(11):1838–47. doi: 10.1242/dev.128074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu C, Yao MD, Li CP, Shan K, Yang H, Wang JJ, Liu B, Li XM, Yao J, Jiang Q, et al. Silencing of circular RNA-ZNF609 ameliorates vascular endothelial dysfunction. Theranostics. 2017;7(11):2863–77. doi: 10.7150/thno.19353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shan K, Liu C, Liu BH, Chen X, Dong R, Liu X, Zhang YY, Liu B, Zhang SJ, Wang JJ, et al. Circular noncoding RNA HIPK3 mediates retinal vascular dysfunction in diabetes mellitus. Circulation. 2017;136(17):1629–42. doi: 10.1161/CIRCULATIONAHA.117.029004. [DOI] [PubMed] [Google Scholar]
- 13.Gardner TW, Davila JR. The neurovascular unit and the pathophysiologic basis of diabetic retinopathy. Graefes Arch Clin Exp Ophthalmol. 2017;255(1):1–6. doi: 10.1007/s00417-016-3548-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldstein M, Leibovitch I, Yeffimov I, Gavendo S, Sela BA, Loewenstein A. Hyperhomocysteinemia in patients with diabetes mellitus with and without diabetic retinopathy. Eye (Lond). 2004;18(5):460–65. doi: 10.1038/sj.eye.6700702. [DOI] [PubMed] [Google Scholar]
- 15.Mohamed R, Sharma I, Ibrahim AS, Saleh H, Elsherbiny NM, Fulzele S, Elmasry K, Smith SB, Al-Shabrawey M, Tawfik A. Hyperhomocysteinemia alters retinal endothelial cells barrier function and angiogenic potential via activation of oxidative stress. Sci Rep. 2017;7(1):1–11, 11952. doi: 10.1038/s41598-016-0028-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singh M, George AK, Homme RP, Majumder A, Laha A, Sandhu HS, Tyagi SC. Circular RNAs profiling in the cystathionine-beta-synthase mutant mouse reveals novel gene targets for hyperhomocysteinemia induced ocular disorders. Exp Eye Res. 2018;174(9):80–92. doi: 10.1016/j.exer.2018.05.026. [DOI] [PubMed] [Google Scholar]
- 17.Singh M, Tyagi SC. Homocysteine mediates transcriptional changes of the inflammatory pathway signature genes in human retinal pigment epithelial cells. Int J Ophthalmol. 2017;10(5):696–704. doi: 10.18240/ijo.2017.05.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song X, Zhang N, Han P, Moon BS, Lai RK, Wang K, Lu W. Circular RNA profile in gliomas revealed by identification tool UROBORUS. Nucleic Acids Res. 2016;44(9):1–12, e87. doi: 10.1093/nar/gkv1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shang X, Li G, Liu H, Li T, Liu J, Zhao Q, Wang C. Comprehensive circular RNA profiling reveals that hsa_-circ_0005075, a new circular RNA biomarker, is involved in hepatocellular crcinoma development. Medicine (Baltimore). 2016;95(22):1–7, e3811. doi: 10.1097/MD.0000000000003811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen S, Li T, Zhao Q, Xiao B, Guo J. Using circular RNA hsa_circ_0000190 as a new biomarker in the diagnosis of gastric cancer. Clin Chim Acta. 2017;466(3):167–71. doi: 10.1016/j.cca.2017.01.025. [DOI] [PubMed] [Google Scholar]
- 21.Li P, Chen H, Chen S, Mo X, T L, Xiao B, Yu R, Guo J. Circular RNA 0000096 affects cell growth and migration in gastric cancer. Br J Cancer. 2017;116(5):626–33. doi: 10.1038/bjc.2016.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cui X, Niu W, Kong L, He M, Jiang K, Chen S, Zhong A, Li W, Lu J, Zhang L. hsa_circRNA_103636: potential novel diagnostic and therapeutic biomarker in major depressive disorder. Biomark Med. 2016;10(9):943–52. doi: 10.2217/bmm-2016-0130. [DOI] [PubMed] [Google Scholar]
- 23.Wang K, Long B, Liu F, Wang JX, Liu CY, Zhao B, Zhou LY, Sun T, Wang M, Yu T, et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J. 2016;37(33):2602–11. doi: 10.1093/eurheartj/ehv713. [DOI] [PubMed] [Google Scholar]
- 24.Wang YH, Yu XH, Luo SS, Han H. Comprehensive circular RNA profiling reveals that circular RNA100783 is involved in chronic CD28-associated CD8(+)T cell ageing. Immun Ageing. 2015;12(17):1–10. doi: 10.1186/s12979-015-0028-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang J, Song YX, Wang ZN. Non-coding RNAs in gastric cancer. Gene. 2015;560(1):1–8. doi: 10.1016/j.gene.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 26.Rybak-Wolf A, Stottmeister C, Glazar P, Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss R, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 2015;58 (5):870–85. doi: 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 27.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. Rna. 2013;19(2):141–57. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade M, et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell. 2017;66(1):22–37.e9. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Veno MT, Hansen TB, Veno ST, Clausen BH, Grebing M, Finsen B, Holm IE, Kjems J. Spatio-temporal regulation of circular RNA expression during porcine embryonic brain development. Genome Biol. 2015;16(245):1–17. doi: 10.1186/s13059-014-0572-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mehta SL, Pandi G, Vemuganti R. Circular RNA expression profiles alter significantly in mouse brain after transient focal ischemia. Stroke. 2017;48(9):2541–48. doi: 10.1161/STROKEAHA.117.017469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gruner H, Cortes-Lopez M, Cooper DA, Bauer M, Miura P. CircRNA accumulation in the aging mouse brain. Sci Rep. 2016;6(12):1–14, 38907. doi: 10.1038/s41598-016-0001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–88. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 33.Familtseva A, Jeremic N, Kunkel GH, Tyagi SC. Toll-like receptor 4 mediates vascular remodeling in hyperhomocysteinemia. Mol Cell Biochem. 2017;433(1–2):177–94. doi: 10.1007/s11010-017-3026-9. [DOI] [PubMed] [Google Scholar]
- 34.Gu Y, Ke G, Wang L, Zhou E, Zhu K, Wei Y. Altered expression profile of circular RNAs in the serum of patients with diabetic retinopathy revealed by microarray. Ophthalmic Res. 2017;58(3):176–84. doi: 10.1159/000479156. [DOI] [PubMed] [Google Scholar]


