Introduction
The growing knowledge of the human genome sequence has spurred the interest in functional genomics.1 Functional genomics aims to understand the complex relationship between genotype and phenotype. Using high-throughput methods, functional genomics has expanded biomedical science beyond reductionist approaches and strive to gain an unbiased view of the regulation of gene transcription, translation and protein-protein interactions on a genomic scale. A more focused functional genomics approach determines the functional impact of genomic variation, such as mutations and polymorphisms associated with complex traits and diseases. The endeavor of functional genomics is fueling an explosion of new insights into novel biology and therapeutic strategies.2
The purpose of this Recent Highlights article is to provide a concise but comprehensive overview of recent studies in the journal of Arteriosclerosis, Thrombosis, and Vascular Biology on functional genomics applied to cardiovascular research and medicine, while also highlighting cutting edge functional genomic tools and their applications.
Transcriptome and proteome profiling for characterization of molecular signature and discovery of novel biology
Discovering the molecular signature associated with a cellular or disease state by transcriptomic, proteomic, and metabolomic profiling is one of the main quests in functional genomic studies. Integrating these “omics” data not only allows for an unbiased interpretation of biological processes, but also facilitates hypothesis generation and target prioritization for deeper mechanistic studies.
Molecular profiling of specific cell types in tissue specimen provides insights into cell-type specific mechanisms in disease pathogenesis. Macrophages are intricately involved in atherogenesis, but how macrophages in vulnerable plaques differ from those in stable plaques remains undetermined.3 Microarray analysis of macrophages laser captured from symptomatic and asymptomatic human atherosclerotic plaques reveal differentially expressed genes.3 The transcriptome feature of symptomatic plaque macrophages corresponded to the upregulation of 7 functional pathways, including inflammation, lipid metabolism, hypoxic response, cell proliferation, apoptosis, antigen presentation, and cellular energetics, providing the transcriptomic signature of human plaque macrophages.3 Proteomic profiling of vascular cells in the entire vascular beds including macro- and micro- blood vessels and capillary vascular beds allows the characterization of the systemic nature of vascular diseases. Serra et al. developed whole-body differential perfusion with increasing concentrations of detergent buffer to selectively solubilize distinct layers of vascular bed tissue, including endothelial glycocalyx, endothelial cells, and vascular smooth muscle cells (VSMC). By incorporating quantitative proteomic analysis, the method allows for the detection of system-wide proteomic change in vascular beds in vivo.4 Multi-omics profiling, either at bulk or single cell level, that also captures spatial organization and function of specific cell types in tissue specimen will undoubtedly provide a more complete picture of diverse tissues and organisms.5
RNA-sequencing is widely used to discover novel transcripts and identify differentially expressed transcripts for functional follow-up. Dnm3os, one of the top differentially expressed long non-coding RNAs (lncRNAs) upregulated in bone marrow-derived macrophages from type 2 diabetic db/db mice, has been discovered as a positive regulator of macrophage inflammatory responses.6 LncRNA MYOSLID was highly induced in VSMC by overexpression of MYOCD – the master regulator for VSMC differentiation, and has been found to promote VSMC differentiation while inhibiting proliferation.7 LncRNA MALAT1 was repressed by increased matrix stiffness in cultured human VSMC. Knockdown of MALAT1 limited stiffness-induced proliferation and migration of human VSMC, and knockout of Malat1 was protective in in vivo vascular injury response.8 Despite success, how to prioritize potential functional lncRNAs from RNA-seq screen for further workup remains the key challenge.9 Detailed discussion on the decision scheme for the prioritization pipeline in published work and efforts to potentially establish standardized pipelines for scalable hypothesis creation will facilitate efficient prioritization and discovery of novel functional transcripts.
Interpreting GWAS findings with functional genomic data for prioritization of causal variants and genes
Genome-wide association studies (GWAS) have successfully identified a large number of genetic loci associated with risks of complex traits and cardiovascular diseases.10 Determining the molecular functions of these loci will provide new insights into novel biology and therapeutic targets.1 The majority of genetic variants associated with complex diseases are found within noncoding regions of the genome, highlighting the necessity of understanding regulatory variation.
Consortium projects, such as ENCODE (Encyclopedia of DNA Elements),11 the Epigenomics Roadmap,12 and FANTOM (Functional Annotation of the Mammalian Genome),13 have focused on producing large catalogs of functional elements in the genome that are used to annotate the putative regulatory function of genetic variants. The GTEx (Genotype-Tissue Expression) project allows eQTL (expression quantitative trait locus) analysis that associates genotypes to the levels and directionality of gene expression in population samples. Indeed, disease- and trait-associated genetic variants are enriched in tissue-specific epigenetic marks,12 and eQTL variants in disease-relevant tissues and cell types are overrepresented among GWAS loci.14, 15
Candidate causal genes at each of the GWAS loci are generally prioritized based on proximity to the lead SNP (single nucleotide polymorphism) by fine mapping, overlap with epigenetic regulatory marks, eQTL data, and long-range chromatin interactions of variants with gene promoters by chromosome conformation capture-based techniques. In addition, statistical colocalization analysis to estimate whether the variants for GWAS and eQTL signals are shared provides a stronger hypothesis that the gene expression change indicated by the eQTL is causally related to the GWAS.16 Additional tools, such as DEPICT (Data-Driven Expression-Prioritized Integration for Complex Traits)17 and FUMA (Functional Mapping and Annotation of GWAS)18 are also being developed and applied to prioritize variants and genes from known GWAS loci for functional follow-up.
There are continued efforts to detect novel gene-trait association. Importantly, most GWAS have mainly focused on populations of European ancestry. The inclusion of multiethnic and admixed populations will provide additional power to detect associations for genetic variants segregating at low frequency in European populations.19 A recent GWAS in participants of Han ethnicity has identified 3 novel intragenic SNPs associated with CAD (coronary artery diseases) and has shown that knockdown of the host genes, SCML4 and THSD7A, affected endothelial function and monocyte adhesion.20 Furthermore, miRNAs21 and lncRNAs22 harboring GWAS variants have shown functional effects on trait-related cellular phenotypes, supporting their potential roles as the causal non-coding RNAs. In addition to human GWAS, QTL mapping in model organism using the inbred mice has successfully identified novel genetic variation and candidate genes regulating macrophage cholesterol metabolism,23 efferocytosis,24 apoptotisis,25 and atherosclerosis susceptibility.24, 25 QTL studies in mice have a high degree of concordance with mapped phenotype regions in the human genome but require far fewer resources and time, complementing observations in human populations.26
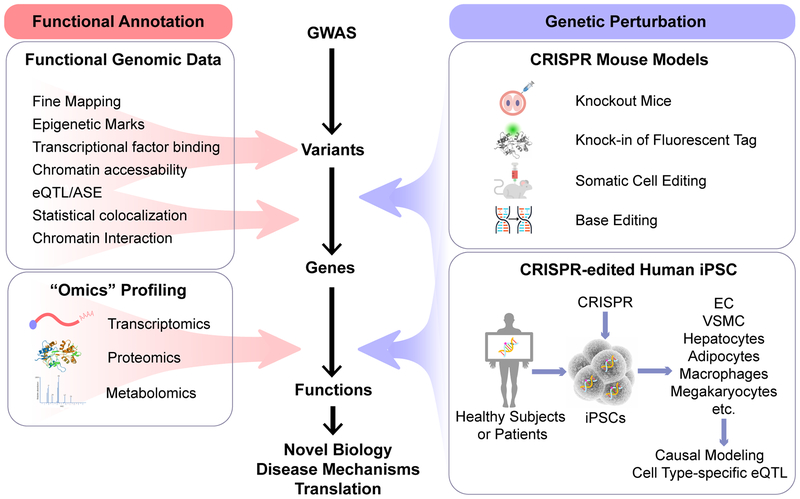
Although there is no guaranteed algorithm or community-curated standard to map GWAS variants to causal genes, fine mapping and other complementary functional genomic tools have provided first clue on the potential causal variants and genes, and the directionality of the effects. (Figure 1) Further validation in suitable experimental models is required to establish the causality and the functional mechanisms of the gene-to-disease association. The following section discusses examples of these efforts.
Figure 1. Interpreting GWAS findings with functional genomic tools for novel biology and translation.
Functional genomic data are essential for the functional annotation and prioritization of potential causal variants and genes. Perturbation of the candidate variants and genes in relevant tissues and cell types or model organisms establishes the causality and biological mechanisms of the variant-disease association. Integrating “omics” profiling facilitates hypothesis generation and mechanistic studies. ASE, allele-specific expression; CRISPR, clustered regularly interspaced short palindromic repeats; eQTL, expression quantitative trait loci; GWAS, genome-wide association studies; iPSC, induced pluripotent stem cells.
Interrogating the functional impact of candidate variants and genes
A noncoding variant can affect gene expression via epigenetic mechanisms, but may also serve as a proxy that links to other non-synonymous coding variants responsible for their causal effects on candidate gene expression. Perturbation of the genetic variants and gene-of-interest in relevant tissues and cell types or model organisms would therefore reveal the functional consequences of candidate variants and genes.
rs17514846, the lead SNP associated with CAD, is located within the intronic region of the FURIN gene.27 Leukocytes from individuals carrying the risk allele (A) of rs17514846 show higher FURIN expression. An analysis of isogenic THP-1 monocytic cell lines created by CRISPR (clustered regularly interspaced short palindromic repeats)-mediated genome editing further supported that isogenic cells with the A/A genotype for rs17514846 had higher FURIN abundance than the isogenic cells with the C/C genotype. Lentivirus-mediated overexpression of Furin in RAW264.7 cells promoted monocyte/macrophage migration and proliferation while inhibiting apoptosis, providing a biological mechanism for the association between genetic variants in FURIN and CAD risk.27 CAD-associated lead variant rs2487928 lies within the intronic regions of JCAD and is associated with JCAD expression in atherosclerotic arteries.28 JCAD protein localizes to endothelial cell junctions and disruption of JCAD inhibited angiogenesis in vitro and in vivo,29 At molecular level, JCAD interacts with large tumor suppressor kinase 2 and negatively regulates Hippo signaling leading to increased activity of Yes-associated protein, the transcriptional effector of the pathway.28 It is, however, unknown whether rs2487928 is the causal variant without perturbation experiment.
rs9390459 is a synonymous variant in STXBP5 and the lead SNP associated with altered plasma von Willebrand factor concentration.30 However, functional genomic data do not support rs9390459 as a non-coding variant that affect DNA epigenetic modification, transcriptional factor binding, distal interaction, or eQTL. Instead, a non-synonymous SNP rs1039084, in high LD (linkage disequilibrium) with the lead SNP rs9390459, is the causal variant for a decreased thrombotic phenotype.30 Zhu et al. knocked the rs1039084 minor allele of human STXBP5 into the orthologous mouse Stxbp5 locus by CRISPR and found that mice carrying the minor allele showed lower plasma von Willebrand factor concentration, prolonged bleeding and decreased thrombosis, phenotypes consistent with human minor allele carriers.30 Morris et al. also suggested that non-synonymous variant rs1051338, in high LD with the CAD lead SNP in LIPA gene, is associated with lower LIPA expression and enzymatic activity in lysosomes of human monocyte-derived macrophages,31 but causal modeling has not been performed.32
rs12740374 has previously been identified as the causal variant responsible for the association of the SORT1 locus with low-density lipoprotein cholesterol and CAD.33 rs12740374 is a noncoding SNP that lies ~120 kb away from the promoter of the SORT1 gene.33 The minor allele of rs12740374 creates a binding site for CCAAT-enhancer-binding protein transcription factors, resulting in liver-specific transcriptional activation of SORT1.33 Wang et al. further demonstrated a direct causal link between rs12740374 and SORT1 by perturbing the site of the SNP and evoking an alteration of SORT1 expression.34 CRISPR-targeting of the rs12740374 minor allele sequence in the liver of a locus-humanized transgenic mouse model results in a profound reduction of hepatic SORT1 protein expression, supporting the causality of the variant.34
Much progress has been made but it remains challenging to conclusively confirm the causal variants, causal genes, and the biological mechanisms of gene-to-disease association. Integrating state-of-the-art tools, such as human induced pluripotent stem cell (iPSC) technology and CRISPR genome editing techniques, will maximize the value of functional genomic data and accelerate its translation.
Human induced pluripotent stem cell differentiation for functional interrogation of GWAS variants and candidate genes
The differentiation of human iPSC to cell types of relevance to cardiovascular biology and diseases offers a powerful platform with enormous potential for disease modeling, drug screening and cell therapeutics. The combination of human iPSC technology with genome editing technique allows the generation of isogenic cell lines that differ in single genetic changes for causal modeling of candidate variants and genes, offering a new tool linking genotypes to phenotypes in the study of human cell biology and disease. (Figure 1)
We have previously generated iPSC-derived macrophages with knockout of the LIPA gene in human macrophages.35 Macrophages with loss-of-function of LIPA showed expected phenotype of impaired hydrolysis of cholesteryl ester and lysosomal cholesteryl ester accumulation, but the expression of ATP-binding cassette transporter ABCA1 and cholesterol efflux capacity were not affected.35 Impaired ABCA1 expression and function were, however, observed in skin fibroblasts derived from Cholesteryl Ester Storage Disease patients with loss-of-function mutations of LIPA,35 suggesting potential cell type-specific differences in LIPA metabolic phenotypes and the value of iPSC-derived cells in the study of human cell biology.
Since iPSC differentiation provides an unlimited source of subject genotype-specific cells, iPSC-derived hepatocytes-like cells36, 37 and adipocytes37 from a cohort of healthy subjects have been used to create a hepatocyte- and adipocyte-specific eQTL database that is complementary to traditional eQTL studies using bulk tissues. The iPSC-hepatocyte cohort successfully identify a large number of eQTL genes shared with the GTEx liver cohort, as well as many new eQTL genes. The iPSC-hepatocytes, however, poorly model the strong eQTL for rs12740374 genotype and SORT1 expression observed in both whole-liver samples and primary human hepatocytes.34 This suggests that iPSC-derived cells can be useful for establishing causal SNP-gene relationships for many, but not all loci. The eQTL database of iPSC-derived cells provides a useful resource for planning experiments to model these relationships.34
ATVB has recently published an “ATVB in Focus” series on iPSC-derived cell types relevant to atherothrombosis. The series includes 6 review articles on the differentiation, characterization and application of iPSC-derived endothelial cells,38, 39 VSMC,40 macrophages,41 megakaryocytes and platelets,42 and hepatocytes.43 We therefore refer to these contributions to guide further reading.
The expanding CRISPR toolbox for genome engineering
The advent of CRISPR genome editing techniques has greatly revolutionized functional genomic research. (Figure 1) CRIPSR/Cas9 (CRISPR Associated Protein 9) system permits rapid generation of novel animal models with a specific genetic background in one-step by co-injecting Cas9 mRNA, guide RNA, and DNA repair templates to zygotes.44 The microinjection can be done in C57BL/6 zygote, a strain commonly used in the studies of atherosclerosis and metabolic diseases,45 eliminating the need for extensive backcrossing.
Wang et al. generated intermedin knockout mice using CRIPSR and found that intermedin induced quiescent endothelial cells to proliferate, resulting in continuous vascular lumen expanding and a more effective blood perfusion.46 Because of founder mosaicism in CRISPR-derived mouse model, it is recommended that each F1 mouse is treated as the start of an individual mutant line. This study has also demonstrated that multiple mutant lines have the same phenotypes.46 Mouse models with fluorescent tagging of endogenous proteins can be useful to spatiotemporally track protein expression, localization, and binding activity in vivo. Lyu et al. generated a mouse model with CRISPR-mediated knock-in of epitope tags (carboxyl-terminal 3×FLAG or 3×HA) at the C-terminal end of the Myocd locus,47 which allows for detection of nuclear localization of Myocd and associated DNA binding.47
Somatic editing with CRISPR has also been established. Jarrett et al. developed a new method for generating an atherosclerosis mouse model through somatic deletion of Ldlr in liver of adult mice by single injection of adeno-associated viral vectors expressing Cas9 and a guide RNA targeting the Ldlr gene.48 The edited mice were compared with germline Ldlr−/− mice, and mice with overexpression of PCSK9 (proprotein convertase subtilisin/kexin type 9) that promotes degradation of low-density lipoprotein receptor in the liver,49, 50 models currently used to study atherosclerosis. Notable sexual dimorphism was observed, wherein somatic editing of Ldlr was superior for Ldlr removal in male mice, PCSK9 overexpression was more effective in female mice.48 Both models provide useful alternatives to the use of Ldlr−/− mice in atherosclerosis research.
The CRISPR toolbox is expanding. By fusing catalytically inactive Cas9 (dCas9) protein to cytosine deaminase domain, this “base editor” can convert cytosine bases to thymine bases in the genomic DNA near the guide RNA target site, allowing precise knock-in of specific nucleotide changes without inducing double-strand breaks.51 Chadwick et al. introduced site-specific nonsense mutations into the PCSK9 gene in the liver of adult mice, resulting in substantially decreased plasma PCSK9 protein concentrations by >50% and reduced plasma cholesterol concentrations by ~30%, demonstrating the ability to precisely introduce therapeutically relevant nucleotide variants into the genome in somatic tissues of adult mammals.52 By fusing different transcription regulators to the dCas9 protein, CRISPRi and CRISPRa can induce transcriptional silencing and activation, respectively.53 CRISPRi allows for precise perturbation of lncRNA transcription without eliminating the lncRNA genes, therefore well suited for the study of lncRNA function. Chowdhury et al. used CRISPRi to knockdown a natural antisense transcripts from the tie1 locus – tie1 antisense (tie1AS), and found that knockdown of tie1AS led to increased tie1 mRNA abundance and tie1-mediated vascular phenotypes in zebrafish.54
In addition to targeted genome editing, CRISPR has been harnessed to achieve large-scale functional screens. The ultimate aim for such screens is to identify novel genes and genetic variants that influence a specific phenotype in an unbiased fashion with genome-wide coverage. The application of CRISPR screen has been extensively covered in other review articles,55 and we expect CRISPR screens that annotate gene function at scale to accelerate many research projects in the field of cardiovascular science.
Summary
Novel genetic loci associated with cardiovascular diseases will continue to emerge through GWAS in diverse populations. The post-GWAS functional studies will dedicate to delineating the causal genetic variants and genes, and the underlying biological mechanisms. Novel functional genomic technologies at single-cell and single-molecule level will further reveal cell type-specific regulatory mechanisms.56 The promise of functional genomics is to understand the function of all components of the genome, and ultimately, translate genetic findings toward clinical applications in the diagnosis and treatment of both rare and common cardiovascular diseases.
Acknowledgement
The authors would like to thank Shuang Wu, PhD for his assistance in graphic design.
Sources of Funding
The authors’ research work has received funding from the National Center for Advancing Translational Sciences (NCATS) and National Institutes of Health (NIH) through Grant Number UL1TR001873, the NIH R00HL130574 (to Dr. Zhang), and R01HL139748 (to Dr. Lu). The content in this manuscript is solely the responsibility of the authors and does not necessarily represent the official views of the NCATS and the NIH.
Footnotes
Disclosures
None.
References
- 1.Nurnberg ST, Zhang H, Hand NJ, Bauer RC, Saleheen D, Reilly MP and Rader DJ. From Loci to Biology: Functional Genomics of Genome-Wide Association for Coronary Disease. Circulation research. 2016;118:586–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cappola TP and Margulies KB. Functional genomics applied to cardiovascular medicine. Circulation. 2011;124:87–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chai JT, Ruparelia N, Goel A, Kyriakou T, Biasiolli L, Edgar L, Handa A, Farrall M, Watkins H and Choudhury RP. Differential Gene Expression in Macrophages From Human Atherosclerotic Plaques Shows Convergence on Pathways Implicated by Genome-Wide Association Study Risk Variants. Arteriosclerosis, thrombosis, and vascular biology. 2018;38:2718–2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Serra A, Gallart-Palau X, Park JE, Lim GGY, Lim KL, Ho HH, Tam JP and Sze SK. Vascular Bed Molecular Profiling by Differential Systemic Decellularization In Vivo. Arteriosclerosis, thrombosis, and vascular biology. 2018;38:2396–2409. [DOI] [PubMed] [Google Scholar]
- 5.Moffitt JR, Bambah-Mukku D, Eichhorn SW, Vaughn E, Shekhar K, Perez JD, Rubinstein ND, Hao J, Regev A, Dulac C and Zhuang X. Molecular, spatial, and functional single-cell profiling of the hypothalamic preoptic region. Science. 2018;362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Das S, Reddy MA, Senapati P, Stapleton K, Lanting L, Wang M, Amaram V, Ganguly R, Zhang L, Devaraj S, Schones DE and Natarajan R. Diabetes Mellitus-Induced Long Noncoding RNA Dnm3os Regulates Macrophage Functions and Inflammation via Nuclear Mechanisms. Arteriosclerosis, thrombosis, and vascular biology. 2018;38:1806–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao J, Zhang W, Lin M, Wu W, Jiang P, Tou E, Xue M, Richards A, Jourd’heuil D, Asif A, Zheng D, Singer HA, Miano JM and Long X. MYOSLID Is a Novel Serum Response Factor-Dependent Long Noncoding RNA That Amplifies the Vascular Smooth Muscle Differentiation Program. Arteriosclerosis, thrombosis, and vascular biology. 2016;36:2088–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu CK, Xu T, Assoian RK and Rader DJ. Mining the Stiffness-Sensitive Transcriptome in Human Vascular Smooth Muscle Cells Identifies Long Noncoding RNA Stiffness Regulators. Arteriosclerosis, thrombosis, and vascular biology. 2018;38:164–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freedman JE and Miano JM. Challenges and Opportunities in Linking Long Noncoding RNAs to Cardiovascular, Lung, and Blood Diseases. Arteriosclerosis, thrombosis, and vascular biology. 2017;37:21–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lappalainen T, Scott AJ, Brandt M and Hall IM. Genomic Analysis in the Age of Human Genome Sequencing. Cell. 2019;177:70–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, Ziller MJ, Amin V, Whitaker JW, Schultz MD, Ward LD, Sarkar A, Quon G, Sandstrom RS, Eaton ML, Wu YC, Pfenning AR, Wang X, Claussnitzer M, Liu Y, Coarfa C, Harris RA, Shoresh N, Epstein CB, Gjoneska E, Leung D, Xie W, Hawkins RD, Lister R, Hong C, Gascard P, Mungall AJ, Moore R, Chuah E, Tam A, Canfield TK, Hansen RS, Kaul R, Sabo PJ, Bansal MS, Carles A, Dixon JR, Farh KH, Feizi S, Karlic R, Kim AR, Kulkarni A, Li D, Lowdon R, Elliott G, Mercer TR, Neph SJ, Onuchic V, Polak P, Rajagopal N, Ray P, Sallari RC, Siebenthall KT, Sinnott-Armstrong NA, Stevens M, Thurman RE, Wu J, Zhang B, Zhou X, Beaudet AE, Boyer LA, De Jager PL, Farnham PJ, Fisher SJ, Haussler D, Jones SJ, Li W, Marra MA, McManus MT, Sunyaev S, Thomson JA, Tlsty TD, Tsai LH, Wang W, Waterland RA, Zhang MQ, Chadwick LH, Bernstein BE, Costello JF, Ecker JR, Hirst M, Meissner A, Milosavljevic A, Ren B, Stamatoyannopoulos JA, Wang T and Kellis M. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forrest AR, Kawaji H, Rehli M, Baillie JK, de Hoon MJ, Haberle V, Lassmann T, Kulakovskiy IV, Lizio M, Itoh M, Andersson R, Mungall CJ, Meehan TF, Schmeier S, Bertin N, Jorgensen M, Dimont E, Arner E, Schmidl C, Schaefer U, Medvedeva YA, Plessy C, Vitezic M, Severin J, Semple C, Ishizu Y, Young RS, Francescatto M, Alam I, Albanese D, Altschuler GM, Arakawa T, Archer JA, Arner P, Babina M, Rennie S, Balwierz PJ, Beckhouse AG, Pradhan-Bhatt S, Blake JA, Blumenthal A, Bodega B, Bonetti A, Briggs J, Brombacher F, Burroughs AM, Califano A, Cannistraci CV, Carbajo D, Chen Y, Chierici M, Ciani Y, Clevers HC, Dalla E, Davis CA, Detmar M, Diehl AD, Dohi T, Drablos F, Edge AS, Edinger M, Ekwall K, Endoh M, Enomoto H, Fagiolini M, Fairbairn L, Fang H, Farach-Carson MC, Faulkner GJ, Favorov AV, Fisher ME, Frith MC, Fujita R, Fukuda S, Furlanello C, Furino M, Furusawa J, Geijtenbeek TB, Gibson AP, Gingeras T, Goldowitz D, Gough J, Guhl S, Guler R, Gustincich S, Ha TJ, Hamaguchi M, Hara M, Harbers M, Harshbarger J, Hasegawa A, Hasegawa Y, Hashimoto T, Herlyn M, Hitchens KJ, Ho Sui SJ, Hofmann OM, Hoof I, Hori F, Huminiecki L, Iida K, Ikawa T, Jankovic BR, Jia H, Joshi A, Jurman G, Kaczkowski B, Kai C, Kaida K, Kaiho A, Kajiyama K, Kanamori-Katayama M, Kasianov AS, Kasukawa T, Katayama S, Kato S, Kawaguchi S, Kawamoto H, Kawamura YI, Kawashima T, Kempfle JS, Kenna TJ, Kere J, Khachigian LM, Kitamura T, Klinken SP, Knox AJ, Kojima M, Kojima S, Kondo N, Koseki H, Koyasu S, Krampitz S, Kubosaki A, Kwon AT, Laros JF, Lee W, Lennartsson A, Li K, Lilje B, Lipovich L, Mackay-Sim A, Manabe R, Mar JC, Marchand B, Mathelier A, Mejhert N, Meynert A, Mizuno Y, de Lima Morais DA, Morikawa H, Morimoto M, Moro K, Motakis E, Motohashi H, Mummery CL, Murata M, Nagao-Sato S, Nakachi Y, Nakahara F, Nakamura T, Nakamura Y, Nakazato K, van Nimwegen E, Ninomiya N, Nishiyori H, Noma S, Noma S, Noazaki T, Ogishima S, Ohkura N, Ohimiya H, Ohno H, Ohshima M, Okada-Hatakeyama M, Okazaki Y, Orlando V, Ovchinnikov DA, Pain A, Passier R, Patrikakis M, Persson H, Piazza S, Prendergast JG, Rackham OJ, Ramilowski JA, Rashid M, Ravasi T, Rizzu P, Roncador M, Roy S, Rye MB, Saijyo E, Sajantila A, Saka A, Sakaguchi S, Sakai M, Sato H, Savvi S, Saxena A, Schneider C, Schultes EA, Schulze-Tanzil GG, Schwegmann A, Sengstag T, Sheng G, Shimoji H, Shimoni Y, Shin JW, Simon C, Sugiyama D, Sugiyama T, Suzuki M, Suzuki N, Swoboda RK, t Hoen PA, Tagami M, Takahashi N, Takai J, Tanaka H, Tatsukawa H, Tatum Z, Thompson M, Toyodo H, Toyoda T, Valen E, van de Wetering M, van den Berg LM, Verado R, Vijayan D, Vorontsov IE, Wasserman WW, Watanabe S, Wells CA, Winteringham LN, Wolvetang E, Wood EJ, Yamaguchi Y, Yamamoto M, Yoneda M, Yonekura Y, Yoshida S, Zabierowski SE, Zhang PG, Zhao X, Zucchelli S, Summers KM, Suzuki H, Daub CO, Kawai J, Heutink P, Hide W, Freeman TC, Lenhard B, Bajic VB, Taylor MS, Makeev VJ, Sandelin A, Hume DA, Carninci P and Hayashizaki Y. A promoter-level mammalian expression atlas. Nature. 2014;507:462–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foroughi Asl H, Talukdar HA, Kindt AS, Jain RK, Ermel R, Ruusalepp A, Nguyen KD, Dobrin R, Reilly DF, Schunkert H, Samani NJ, Braenne I, Erdmann J, Melander O, Qi J, Ivert T, Skogsberg J, Schadt EE, Michoel T and Bjorkegren JL. Expression quantitative trait Loci acting across multiple tissues are enriched in inherited risk for coronary artery disease. Circulation Cardiovascular genetics. 2015;8:305–15. [DOI] [PubMed] [Google Scholar]
- 15.Zhong H, Beaulaurier J, Lum PY, Molony C, Yang X, MacNeil DJ, Weingarth DT, Zhang B, Greenawalt D, Dobrin R, Hao K, Woo S, Fabre-Suver C, Qian S, Tota MR, Keller MP, Kendziorski CM, Yandell BS, Castro V, Attie AD, Kaplan LM and Schadt EE. Liver and Adipose Expression Associated SNPs Are Enriched for Association to Type 2 Diabetes. PLOS Genetics. 2010;6:e1000932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lappalainen T Functional genomics bridges the gap between quantitative genetics and molecular biology. Genome Res. 2015;25:1427–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pers TH, Karjalainen JM, Chan Y, Westra HJ, Wood AR, Yang J, Lui JC, Vedantam S, Gustafsson S, Esko T, Frayling T, Speliotes EK, Boehnke M, Raychaudhuri S, Fehrmann RS, Hirschhorn JN and Franke L. Biological interpretation of genome-wide association studies using predicted gene functions. Nat Commun. 2015;6:5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Watanabe K, Taskesen E, van Bochoven A and Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017;8:1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin AR, Gignoux CR, Walters RK, Wojcik GL, Neale BM, Gravel S, Daly MJ, Bustamante CD and Kenny EE. Human Demographic History Impacts Genetic Risk Prediction across Diverse Populations. The American Journal of Human Genetics. 2017;100:635–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y, Wang DW, Chen Y, Chen C, Guo J, Zhang S, Sun Z, Ding H, Yao Y, Zhou L, Xu K, Song C, Yang F, Zhao B, Yan H, Wang WJ, Wu C, Lu X, Yang X, Dong J, Zheng G, Tian S, Cui Y, Jin L, Liu G, Cui H, Wang S, Jiang F, Wang C, Erdmann J, Zeng L, Huang S, Zhong J, Ma Y, Chen W, Sun J, Lei W, Chen S, Rao S, Gu D, Schunkert H and Tian XL. Genome-Wide Association and Functional Studies Identify SCML4 and THSD7A as Novel Susceptibility Genes for Coronary Artery Disease. Arteriosclerosis, thrombosis, and vascular biology. 2018;38:964–975. [DOI] [PubMed] [Google Scholar]
- 21.Wagschal A, Najafi-Shoushtari SH, Wang L, Goedeke L, Sinha S, deLemos AS, Black JC, Ramirez CM, Li Y, Tewhey R, Hatoum I, Shah N, Lu Y, Kristo F, Psychogios N, Vrbanac V, Lu YC, Hla T, de Cabo R, Tsang JS, Schadt E, Sabeti PC, Kathiresan S, Cohen DE, Whetstine J, Chung RT, Fernandez-Hernando C, Kaplan LM, Bernards A, Gerszten RE and Naar AM. Genome-wide identification of microRNAs regulating cholesterol and triglyceride homeostasis. Nature medicine. 2015;21:1290–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang H, Xue C, Wang Y, Shi J, Zhang X, Li W, Nunez S, Foulkes AS, Lin J, Hinkle CC, Yang W, Morrisey EE, Rader DJ, Li M and Reilly MP. Deep RNA Sequencing Uncovers a Repertoire of Human Macrophage Long Intergenic Noncoding RNAs Modulated by Macrophage Activation and Associated With Cardiometabolic Diseases. Journal of the American Heart Association. 2017;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hai Q, Ritchey B, Robinet P, Alzayed AM, Brubaker G, Zhang J and Smith JD. Quantitative Trait Locus Mapping of Macrophage Cholesterol Metabolism and CRISPR/Cas9 Editing Implicate an ACAT1 Truncation as a Causal Modifier Variant. Arteriosclerosis, thrombosis, and vascular biology. 2018;38:83–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kayashima Y, Makhanova N and Maeda N. DBA/2J Haplotype on Distal Chromosome 2 Reduces Mertk Expression, Restricts Efferocytosis, and Increases Susceptibility to Atherosclerosis. Arteriosclerosis, thrombosis, and vascular biology. 2017;37:e82–e91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Erbilgin A, Seldin MM, Wu X, Mehrabian M, Zhou Z, Qi H, Dabirian KS, Sevag Packard RR, Hsieh W, Bensinger SJ, Sinha S and Lusis AJ. Transcription Factor Zhx2 Deficiency Reduces Atherosclerosis and Promotes Macrophage Apoptosis in Mice. Arteriosclerosis, thrombosis, and vascular biology. 2018;38:2016–2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Flint J, Valdar W, Shifman S and Mott R. Strategies for mapping and cloning quantitative trait genes in rodents. Nature reviews Genetics. 2005;6:271–86. [DOI] [PubMed] [Google Scholar]
- 27.Zhao G, Yang W, Wu J, Chen B, Yang X, Chen J, McVey DG, Andreadi C, Gong P, Webb TR, Samani NJ and Ye S. Influence of a Coronary Artery Disease-Associated Genetic Variant on FURIN Expression and Effect of Furin on Macrophage Behavior. Arteriosclerosis, thrombosis, and vascular biology. 2018;38:1837–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones PD, Kaiser MA, Ghaderi Najafabadi M, Koplev S, Zhao Y, Douglas G, Kyriakou T, Andrews S, Rajmohan R, Watkins H, Channon KM, Ye S, Yang X, Bjorkegren JLM, Samani NJ and Webb TR. JCAD, a Gene at the 10p11 Coronary Artery Disease Locus, Regulates Hippo Signaling in Endothelial Cells. Arteriosclerosis, thrombosis, and vascular biology. 2018;38:1711–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hara T, Monguchi T, Iwamoto N, Akashi M, Mori K, Oshita T, Okano M, Toh R, Irino Y, Shinohara M, Yamashita Y, Shioi G, Furuse M, Ishida T and Hirata KI. Targeted Disruption of JCAD (Junctional Protein Associated With Coronary Artery Disease)/KIAA1462, a Coronary Artery Disease-Associated Gene Product, Inhibits Angiogenic Processes In Vitro and In Vivo. Arteriosclerosis, thrombosis, and vascular biology. 2017;37:1667–1673. [DOI] [PubMed] [Google Scholar]
- 30.Zhu QM, Ko KA, Ture S, Mastrangelo MA, Chen MH, Johnson AD, O’Donnell CJ, Morrell CN, Miano JM and Lowenstein CJ. Novel Thrombotic Function of a Human SNP in STXBP5 Revealed by CRISPR/Cas9 Gene Editing in Mice. Arteriosclerosis, thrombosis, and vascular biology. 2017;37:264–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morris GE, Braund PS, Moore JS, Samani NJ, Codd V and Webb TR. Coronary Artery Disease-Associated LIPA Coding Variant rs1051338 Reduces Lysosomal Acid Lipase Levels and Activity in Lysosomes. Arteriosclerosis, thrombosis, and vascular biology. 2017;37:1050–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang H and Reilly MP. LIPA Variants in Genome-Wide Association Studies of Coronary Artery Diseases: Loss-of-Function or Gain-of-Function? Arteriosclerosis, thrombosis, and vascular biology. 2017;37:1015–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Musunuru K, Strong A, Frank-Kamenetsky M, Lee NE, Ahfeldt T, Sachs KV, Li X, Li H, Kuperwasser N, Ruda VM, Pirruccello JP, Muchmore B, Prokunina-Olsson L, Hall JL, Schadt EE, Morales CR, Lund-Katz S, Phillips MC, Wong J, Cantley W, Racie T, Ejebe KG, Orho-Melander M, Melander O, Koteliansky V, Fitzgerald K, Krauss RM, Cowan CA, Kathiresan S and Rader DJ. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature. 2010;466:714–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang X, Raghavan A, Peters DT, Pashos EE, Rader DJ and Musunuru K. Interrogation of the Atherosclerosis-Associated SORT1 (Sortilin 1) Locus With Primary Human Hepatocytes, Induced Pluripotent Stem Cell-Hepatocytes, and Locus-Humanized Mice. Arteriosclerosis, thrombosis, and vascular biology. 2018;38:76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bowden KL, Bilbey NJ, Bilawchuk LM, Boadu E, Sidhu R, Ory DS, Du H, Chan T and Francis GA. Lysosomal acid lipase deficiency impairs regulation of ABCA1 gene and formation of high density lipoproteins in cholesteryl ester storage disease. J Biol Chem. 2011;286:30624–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pashos EE, Park Y, Wang X, Raghavan A, Yang W, Abbey D, Peters DT, Arbelaez J, Hernandez M, Kuperwasser N, Li W, Lian Z, Liu Y, Lv W, Lytle-Gabbin SL, Marchadier DH, Rogov P, Shi J, Slovik KJ, Stylianou IM, Wang L, Yan R, Zhang X, Kathiresan S, Duncan SA, Mikkelsen TS, Morrisey EE, Rader DJ, Brown CD and Musunuru K. Large, Diverse Population Cohorts of hiPSCs and Derived Hepatocyte-like Cells Reveal Functional Genetic Variation at Blood Lipid-Associated Loci. Cell Stem Cell. 2017;20:558–570.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Warren CR, O’Sullivan JF, Friesen M, Becker CE, Zhang X, Liu P, Wakabayashi Y, Morningstar JE, Shi X and Choi J. Induced pluripotent stem cell differentiation enables functional validation of GWAS variants in metabolic disease. Cell Stem Cell. 2017;20:547–557.e7. [DOI] [PubMed] [Google Scholar]
- 38.Lin Y, Gil CH and Yoder MC. Differentiation, Evaluation, and Application of Human Induced Pluripotent Stem Cell-Derived Endothelial Cells. Arteriosclerosis, thrombosis, and vascular biology. 2017;37:2014–2025. [DOI] [PubMed] [Google Scholar]
- 39.Carcamo-Orive I, Huang NF, Quertermous T and Knowles JW. Induced pluripotent stem cell–derived endothelial cells in insulin resistance and metabolic syndrome. Arteriosclerosis, thrombosis, and vascular biology. 2017;37:2038–2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maguire EM, Xiao Q and Xu Q. Differentiation and Application of Induced Pluripotent Stem Cell-Derived Vascular Smooth Muscle Cells. Arteriosclerosis, thrombosis, and vascular biology. 2017;37:2026–2037. [DOI] [PubMed] [Google Scholar]
- 41.Zhang H and Reilly MP. Human Induced Pluripotent Stem Cell-Derived Macrophages for Unraveling Human Macrophage Biology. Arteriosclerosis, thrombosis, and vascular biology. 2017;37:2000–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Borst S, Sim X, Poncz M, French DL and Gadue P. Induced Pluripotent Stem Cell-Derived Megakaryocytes and Platelets for Disease Modeling and Future Clinical Applications. Arteriosclerosis, thrombosis, and vascular biology. 2017;37:2007–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pournasr B and Duncan SA. Modeling Inborn Errors of Hepatic Metabolism Using Induced Pluripotent Stem Cells. Arteriosclerosis, thrombosis, and vascular biology. 2017;37:1994–1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miano JM, Zhu QM and Lowenstein CJ. A CRISPR Path to Engineering New Genetic Mouse Models for Cardiovascular Research. Arteriosclerosis, thrombosis, and vascular biology. 2016;36:1058–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Daugherty A, Tall AR, Daemen M, Falk E, Fisher EA, Garcia-Cardena G, Lusis AJ, Owens AP, 3rd, Rosenfeld ME and Virmani R. Recommendation on Design, Execution, and Reporting of Animal Atherosclerosis Studies: A Scientific Statement From the American Heart Association. Arteriosclerosis, thrombosis, and vascular biology. 2017;37:e131–e157. [DOI] [PubMed] [Google Scholar]
- 46.Wang LJ, Xiao F, Kong LM, Wang DN, Li HY, Wei YG, Tan C, Zhao H, Zhang T, Cao GQ, Zhang K, Wei YQ, Yang HS and Zhang W. Intermedin Enlarges the Vascular Lumen by Inducing the Quiescent Endothelial Cell Proliferation. Arteriosclerosis, thrombosis, and vascular biology. 2018;38:398–413. [DOI] [PubMed] [Google Scholar]
- 47.Lyu Q, Dhagia V, Han Y, Guo B, Wines-Samuelson ME, Christie CK, Yin Q, Slivano OJ, Herring P, Long X, Gupte SA and Miano JM. CRISPR-Cas9-Mediated Epitope Tagging Provides Accurate and Versatile Assessment of Myocardin-Brief Report. Arteriosclerosis, thrombosis, and vascular biology. 2018;38:2184–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jarrett KE, Lee C, De Giorgi M, Hurley A, Gillard BK, Doerfler AM, Li A, Pownall HJ, Bao G and Lagor WR. Somatic Editing of Ldlr With Adeno-Associated Viral-CRISPR Is an Efficient Tool for Atherosclerosis Research. Arteriosclerosis, thrombosis, and vascular biology. 2018;38:1997–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vozenilek AE, Navratil AR, Green JM, Coleman DT, Blackburn CMR, Finney AC, Pearson BH, Chrast R, Finck BN, Klein RL, Orr AW and Woolard MD. Macrophage-Associated Lipin-1 Enzymatic Activity Contributes to Modified Low-Density Lipoprotein-Induced Proinflammatory Signaling and Atherosclerosis. Arteriosclerosis, thrombosis, and vascular biology. 2018;38:324–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roche-Molina M, Sanz-Rosa D, Cruz FM, Garcia-Prieto J, Lopez S, Abia R, Muriana FJ, Fuster V, Ibanez B and Bernal JA. Induction of sustained hypercholesterolemia by single adeno-associated virus-mediated gene transfer of mutant hPCSK9. Arteriosclerosis, thrombosis, and vascular biology. 2015;35:50–9. [DOI] [PubMed] [Google Scholar]
- 51.Komor AC, Kim YB, Packer MS, Zuris JA and Liu DR. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chadwick AC, Wang X and Musunuru K. In Vivo Base Editing of PCSK9 (Proprotein Convertase Subtilisin/Kexin Type 9) as a Therapeutic Alternative to Genome Editing. Arteriosclerosis, thrombosis, and vascular biology. 2017;37:1741–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chadwick AC and Musunuru K. CRISPR-Cas9 Genome Editing for Treatment of Atherogenic Dyslipidemia. Arteriosclerosis, thrombosis, and vascular biology. 2018;38:12–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chowdhury TA, Koceja C, Eisa-Beygi S, Kleinstiver BP, Kumar SN, Lin CW, Li K, Prabhudesai S, Joung JK and Ramchandran R. Temporal and Spatial Post-Transcriptional Regulation of Zebrafish tie1 mRNA by Long Noncoding RNA During Brain Vascular Assembly. Arteriosclerosis, thrombosis, and vascular biology. 2018;38:1562–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Doench JG. Am I ready for CRISPR? A user’s guide to genetic screens. Nature reviews Genetics. 2018;19:67–80. [DOI] [PubMed] [Google Scholar]
- 56.Shema E, Bernstein BE and Buenrostro JD. Single-cell and single-molecule epigenomics to uncover genome regulation at unprecedented resolution. Nat Genet. 2019;51:19–25. [DOI] [PubMed] [Google Scholar]