Abstract
Objective
In this consensus statement, an international panel of experts deliver their opinions on key questions regarding the contribution of the human microbiome to carcinogenesis.
Design
International experts in oncology and/or microbiome research were approached by personal communication to form a panel. A structured, iterative, methodology based around a 1-day roundtable discussion was employed to derive expert consensus on key questions in microbiome-oncology research.
Results
Some 18 experts convened for the roundtable discussion and five key questions were identified regarding: (1) the relevance of dysbiosis/an altered gut microbiome to carcinogenesis; (2) potential mechanisms of microbiota-induced carcinogenesis; (3) conceptual frameworks describing how the human microbiome may drive carcinogenesis; (4) causation versus association; and (5) future directions for research in the field.
The panel considered that, despite mechanistic and supporting evidence from animal and human studies, there is currently no direct evidence that the human commensal microbiome is a key determinant in the aetiopathogenesis of cancer. The panel cited the lack of large longitudinal, cohort studies as a principal deciding factor and agreed that this should be a future research priority. However, while acknowledging gaps in the evidence, expert opinion was that the microbiome, alongside environmental factors and an epigenetically/genetically vulnerable host, represents one apex of a tripartite, multidirectional interactome that drives carcinogenesis.
Conclusion
Data from longitudinal cohort studies are needed to confirm the role of the human microbiome as a key driver in the aetiopathogenesis of cancer.
Keywords: microbiome, cancer, colorectal, oncology, consensus
Significance of this study.
What is already known on this subject?
Putative pathogens such as human papilloma virus and Helicobacter pylori have been demonstrated to be aetiological agents in a variety of cancers.
Humans possess anatomic niche-specific microbiomes which are involved in key aspects of host physiology and pathology.
Data from in vitro, animal and human studies suggest that the human microbiome may be involved in the aetiopathogenesis of cancer.
What are the new findings?
This is the first published expert consensus on the role of the microbiome in cancer.
Experts considered that there is currently no direct evidence that the human commensal microbiome is a key determinant in the aetiopathogenesis of cancer.
However, expert opinion was that the microbiome is one apex of a tripartite, multidirectional interactome alongside environmental factors and an epigenetically/genetically vulnerable host that combine to cause cancer.
How might it impact on clinical practice in the foreseeable future?
Clinicians should appreciate the current status of microbiome research with regards to the aetiology of cancer as it seems likely that manipulation of the microbiome may offer a therapeutic strategy for the prevention or treatment of cancer.
Introduction
The last decade has seen a surge in the number of studies of the gut microbiome and cancer, facilitated by next-generation sequencing platforms, ‘omics’ technologies and advanced bioinformatics approaches. In contrast to the human genome which is inherited, largely static and which consists of approximately 20 000 genes, the human microbiome is acquired, changes dynamically throughout life and contains approximately 10 to 20 million non-redundant genes.1 The human microbiota was once viewed as comprising either pathogens, causing infectious disease, or passive bystanders. In appreciating the beneficial functions of commensal microorganisms, we have departed from this dichotomy to embrace a novel, subtler paradigm in which host microorganisms contribute to both physiology and pathology. Commensal microorganisms, by definition, do not harm their host. Similarly, pathogens are defined by their ability to cause disease. However, we now appreciate that there is a large and poorly defined middle ground occupied by ‘pathogens’ coexisting with a disease free-host (eg, Helicobacter pylori) and the so-called ‘commensals’ that may contribute to disease under certain conditions. This article is principally concerned with the latter cohort, which may be considered dynamic symbionts—a group of organisms whose symbiotic nature may vary along a spectrum from mutualism and commensalism to parasitism and amensalism. For simplicity, we will refer to this group of dynamic symbionts as the human microbiome (figure 1).
Figure 1.
Key terms in the microbiome literature.
Genetic and environmental factors do not fully explain individual cancer risk—why do some (largely genetically identical) individuals who have similar lifestyles and ages get cancer while others do not? The random distribution of DNA replicative errors is part of the explanation,2 but the significant interindividual variation in microbiome composition and function, coupled with known and emerging potentially carcinogenic microbial-host interactions, suggest that the human microbiome may also be an important cofactor driving cancer risk. Evidence supporting this hypothesis has begun to accumulate and, furthermore, the human microbiome appears to have importance in relation to cancer prognosis3 and treatment.4
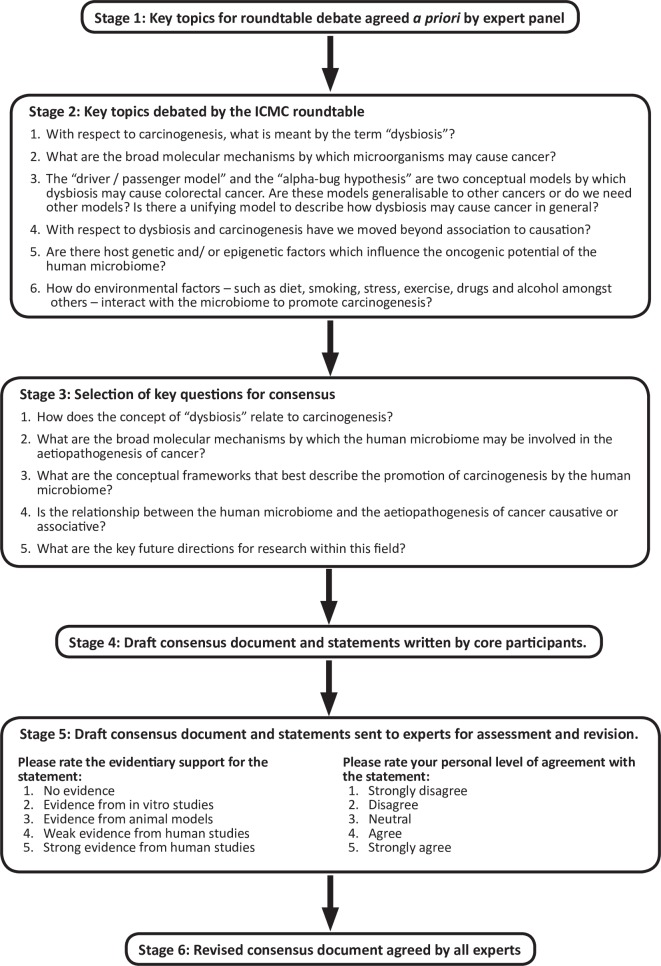
The International Cancer Microbiome Consortium (ICMC) was established in 2017 in recognition of the emerging significance of the human microbiome in oncology. The ICMC comprises academic and clinical experts in oncology and the microbiome and its aim is to promote education and international research collaboration. In its inaugural meeting, ICMC hosted a 1-day roundtable discussion to draw together current research and establish an expert consensus regarding the role of the human microbiome in carcinogenesis.5 Key topics (figure 2) were identified and debated before generating the written document, which was subsequently revised by experts to produce the final consensus.
Figure 2.
A multistage, structured, iterative methodology was employed. ICMC, International Cancer Microbiome Consortium.
Methods
The aim of this consensus statement was to perform a qualitative assessment of the current evidence relevant to the hypothesis that the human microbiome plays a causative role in the aetiopathogenesis of cancer. Discussion and conclusions were weighted towards bacteria (as opposed to viruses or fungi) because bacteria accounted for most of the available data. Discussion was not restricted to the gut and all cancer types were considered, though again, the gut microbiome and colorectal cancer (CRC) feature prominently because of biases within the literature. Given that our aim was to convey current expert opinion on a novel area of science, as opposed to a specific clinical question, a structured, iterative, ‘pseudo-Delphi’ methodology was employed rather than a formal Delphi methodology. This structured approach is summarised in figure 2. The core participants (AJS, JLA, CAM, JMK and JRM) identified contributors based on their expertise in the microbiome, surgical and medical oncology (upper and lower GI cancer, urological cancers and oncogynaecology) and bioinformatics and invited them to contribute to the consensus by personal communication. Some 18 experts from Canada, China, Europe and the USA agreed to form the expert panel. In stage 1, panel members agreed, by email, on six key topics for discussion centred on the theme of the microbiome and carcinogenesis. In stage 2, panel members were assigned to each of these topics according to their expertise and led the debate and discussion on each topic during a 1-day face-to-face roundtable discussion which was recorded and minuted. In stage 3, the core participants reviewed the discussion and distilled it into five key questions to be addressed in the consensus statement. These key questions reflect the roundtable discussion rather than the a priori identified topics directly and were chosen because of their perceived interest to the academic and clinical community and their depth of discussion during the face-to-face meeting:
How does the concept of ‘dysbiosis’ relate to carcinogenesis?
What are the broad molecular mechanisms by which the human microbiome may be involved in the aetiopathogenesis of cancer?
What are the conceptual frameworks that best describe the promotion of carcinogenesis by the human microbiome?
Is the relationship between the human microbiome and the aetiopathogenesis of cancer causative or associative?
What are the key future directions for research in this field?
Extensive quotes from the discussion are available (see supplement) while selected quotes are included in figures alongside the discussion pertaining to each key question (figures 3–7). In stage 4, the core participants drew on the roundtable debate and suggested literature to draft statements and supporting discussion in response to each key question. Statements were distributed electronically to the expert panel who rated the strength of evidentiary support and their personal level of agreement with each statement according to Likert scales (figure 2, stage 5). Some 16 (out of the 18 present at the panel discussion) experts responded and statements were assigned the modal response (number given in brackets, see supplement for full responses). Panel members also had the opportunity to revise the content of the consensus statement. During stage 6, the expert panel reviewed and revised the manuscript to arrive at consensus over multiple iterations.
Figure 3.
Select quotes on dysbiosis and the microbiome.
Figure 4.
Select quotes on mechanisms of microbiome-induced carcinogenesis. HPV, human papilloma virus.
Figure 5.
Select quotes on conceptual frameworks describing microbiome-induced carcinogenesis.
Figure 6.
Select quotes on the potential causal relationship between the human microbiome and carcinogenesis.
Figure 7.
Select quotes regarding future directions in the field.
Results and Discussion
Question one: How does the concept of dysbiosis relate to carcinogenesis (table 1)?
Table 1 How does the concept of dysbiosis relate to carcinogenesis?
| Statement | Evidentiary support | Expert agreement |
| With respect to carcinogenesis, ‘dysbiosis’ should be considered a persistent departure of the host microbiome from the health-associated, homeostatic state, towards a cancer promoting and/or sustaining phenotype. | Weak evidence from human studies (9) | Agree (10) |
| At the present time, there is no accepted quantitative definition of a ‘normal’ microbiome. | Strong evidence from human studies (7) | Agree (8) |
When considering the influence of the microbiome on disease states,6 the term dysbiosis is frequently invoked within the microbiome literature and is mostly based on ‘relative abundance’ inferred from 16S rRNA gene sequencing data. However, the term is often loosely applied, poorly defined and has drawn criticism from the scientific community.7 We therefore sought to address the concept of dysbiosis specifically within the context of carcinogenesis. Dysbiosis has been defined as an abnormality—in composition and/or function—of the host symbiotic microbial ecosystem that exceeds its restitutive capacity and has negative effects on the host.8 However, dysbiosis is likely host specific and disease specific—a microbiome may be dysbiotic in one individual, but not in another and/or may promote one pathology, but not another. Therefore, dysbiosis is not an absolute and cannot be defined outside of the context of the host and the disease in question. The broad hypothesis that the expert consensus explored is that abnormalities in the host microbiome composition and/or function—or dysbiosis—are associated with the aetiopathogenesis of cancer. But how can we elucidate these abnormalities when even consensus regarding what comprises a ‘normal’ microbiome remains elusive? For example, although there can be said to be a ‘core microbiome’ at the phylum level (consisting primarily of Bacteroidetes and Firmicutes), there is significant difference between apparently healthy individuals at lower taxonomic levels.9 We stress ‘apparently’ because the long lag time associated with the development of most cancers makes it difficult to be sure that an individual is as ‘healthy’ as they seem without long-term follow-up. Interindividual variation in microbiome composition far exceeds variation associated with a given disease state. Such compositional variation may arise from the degree of functional redundancy displayed by the microbiome. For example, numerous microorganisms can produce short chain fatty acids (SCFA, eg, butyrate)—harbouring any of these organisms may be considered normal. Conversely, some functions are more conserved and may even be strain specific. 16S rRNA taxonomic profiling cannot resolve this detail and instead researchers are considering whether there is a core metagenome or metabolome.10
We suggest that such a functional approach is of more utility in discussing normality and dysbiosis. With respect to the aetiopathogenesis of cancer, we propose that dysbiosis should be considered a persistent departure of the host microbiome from the health-associated homeostatic state (consisting of mutualists and commensals), towards a cancer promoting and/or sustaining phenotype (parasitism or amensalism). This dysbiosis is specific to the individual and thus can only be defined by prospective longitudinal analysis. For example, based on the current literature, the health-associated gut microbiome can be said to have several core features. It is diverse, resilient to short-term environmental pressures and has sufficient plasticity to adapt to the benefit of the host in the face of longer term pressures.11 The health-associated microbiome should synergise with the host to drive beneficial immune responses12 and metabolic mutualism.13 Finally, the microbiome should have a tumour-suppressant effect on the host.14 15 Departure from these core features can be considered dysbiotic and may have the potential to incite or sustain cancer. It should be noted that the health-associated microbiomes of other niches will have different core features.
Question two: What are the broad molecular mechanisms by which the human microbiome may be involved in the aetiopathogenesis of cancer (table 2)?
Table 2 Whatare the broad molecular mechanisms by which the human microbiome may be involved in the aetiopathogeneis of cancer?
| Statement: the mechanisms by which the microbiome may initiate and/or drive carcinogenesis can be classified into: | Evidentiary support | Expert agreement |
| Genomic integration | Strong evidence from human studies (9) | Agree (8) |
| Genotoxicity | Evidence from animal studies (6) | Agree (9) |
| Inflammation | Strong evidence from human studies (7) | Agree (8) |
| Immunity | Evidence from animal studies (6)/weak evidence from human studies (6) | Agree (9) |
| Metabolism | Strong evidence from human studies (7) | Agree (13) |
At the molecular level, the mechanisms by which microorganisms can contribute to carcinogenesis are multiple and varied and have been extensively discussed elsewhere.16 The mechanisms may broadly be categorised into genomic integration, genotoxicity, inflammation, immunity and metabolism. Many of these mechanisms are likely not evolved to cause pathologies in the host but rather do so as collateral damage, as the organism optimises its chances of survival. Ultimately, all mechanisms converge on a final common pathway of prolonged host cell survival, enhanced replicative capacity and dedifferentiation.
Genomic integration
Integration of microbial DNA into the host genome is a virulence mechanism for multiple viruses that are causally associated with the development of cancer. Cervical cancer caused by human papilloma virus (HPV-16 and HPV-18) is a classic example. Insertion of two HPV genes, E6 and E7, into the host genome in cervical cells confers a survival advantage as their protein derivatives bind to and inactivate tumour-suppressor gene products (p53 and pRB).17 Note that this mechanism relies on transcription of viral proteins which exerts a carcinogenic effect on the host. This mechanism contrasts with insertional mutagenesis which describes the deregulation of host gene expression caused by insertion of the exogenous genetic material. If the host genes in question are tumour suppressor genes or oncogenes, cellular transformation to an oncogenic phenotype can ensue. Insertional mutagenesis is commonly associated with retroviruses and has been proposed as contributing to the oncogenicity of human T-cell lymphotrophic virus.18
Genotoxicity
Genotoxicity describes structural DNA damage, for example, strand breaks, adducts, deletions and rearrangements. If host DNA damage does not lead to cell death, it may affect tumour suppressor genes or oncogenes with carcinogenic effects. Cytolethal distending toxin (CDT) and colibactin are two well-characterised genotoxins. CDT is produced by Escherichia coli and Campylobacter jejuni (among others) and induces double-strand DNA breaks via its DNAse activity.19 CDT-deficient strains have attenuated carcinogenic potential in murine CRC models.20 Members of the Enterobacteriaceae family can also produce colibactin which induces DNA strand breaks and has been associated with human CRC.21 Aside from specific toxins, bacterial metabolites may also exert genotoxic effects. Reactive oxygen species (produced by Porphyromonas sp) and hydrogen sulphide (produced by Bilophila and Fusobacterium) are two examples that have been associated with colorectal neoplasia.22 23
Inflammation
Inflammation is a central feature of carcinogenesis regardless of the aetiological agent and is the principal oncogenic mechanism underlying numerous well-defined, causal, microbial associations with cancer.24 Microbial virulence factors induce chronic inflammation of host tissue, stimulating cellular proliferation which can ultimately become deregulated and, combined with failure of apoptosis, result in a malignant phenotype.25 These effects may be mediated by microbial interaction with specific host cell intracellular signalling pathways. For example, Fusobacterium nucleatum (F. nucleatum; associated with CRC) can induce activation of the nuclear factor-kB (NF-kB) pathway26 while both F. nucleatum and Bacteroides fragilis possess virulence factors which negatively regulate E-cadherin, activating WNT/β-catenin signalling and driving cell proliferation.27 28 Enterotoxigenic B. fragilis (ETBF) secretes B. fragilis toxin and stimulates an TH-17/IL-17-dependent colitis and promotes tumourigenesis.29 In addition to direct, niche-organ-specific effects, there is also evidence that microbe-associated molecular patterns can induce proinflammatory effects in remote organs via their interactions with host pattern recognition receptors, such as toll-like (TLR) and nucleotide-binding oligomerization domain-like (NOD) receptors. Interaction between bacterial lipopolysaccharide and TLR4 results in the downstream activation of cell survival pathways and has been cited as a mechanism by which the intestinal microbiome may contribute to carcinogenesis outside the GI tract.30
Immunity
The interactions between the human microbiota and the immune system are well documented and have significant implications for a wide range of pathologies from atopy and autoimmunity to cancer.31 32 The host immune system plays a key role in preventing carcinogenesis by inducing cell death in abnormal host cells with neoplastic potential. The microbiome may interfere with this process at multiple levels. HIV has tropism towards CD4+ T lymphocytes, impairing the host’s ability to detect potentially neoplastic cells and increasing the rate of carcinogenesis. F. nucleatum expresses the Fap2 cell surface protein which interacts with T and Natural Killer cells to supress antitumour cytotoxicity.33 Although the microbiota may suppress host immunity, in the healthy state, it is hypothesised that microbiota-immune crosstalk facilitates the maintenance of an immune ‘tone’ promoting basal anticancer immunosurveillance. Various mechanisms are proposed including broadening the T cell receptor repertoire and enhancing the intensity of immune responses.34
Metabolism
In addition to the immune system, metabolism is the second key level at which the host and the microbiota interact.13 The human microbiome is replete with genes which influence the metabolism of dietary vitamins and nutrients, xenobiotics and host-derived compounds such as bile acids. Bacterial metabolism appears to be a crucial cofactor that underpins the observed associations between diet and various cancers.35 Pertinent to CRC, gut bacterial fermentation of dietary fibre to SCFA, such as butyrate, is believed to play an important role in suppressing oncogenesis via its anti-inflammatory and antiproliferative effects.15 In contrast, bacterial metabolism of bile acids and proteins can result in the formation of carcinogenic aromatic amines and sulphides.35 The microbiome also plays a substantial xenometabolic role which includes the formation of ultimate carcinogenic end products, acetaldehyde production from alcohol for example.36 However, the effects of microbial metabolites are likely to be context dependent on host factors. For example, microbial butyrate production has been suggested to induce CRC in mice deficient in the MSH2 gene (which codes a protein playing a key role in DNA mismatch repair) by driving hyperproliferation of colonocytes.37
Question three: What are the conceptual frameworks that best describe the promotion of carcinogenesis by the human microbiome (table 3)?
Table 3 Whatare the conceptual frameworks that best describe the promotion of carcinogenesis by rhe human microbiome?
| Statement | Evidentiary support | Expert agreement |
| With respect to the pathogenesis of CRC, ‘the driver–passenger’ model40 accounts for key observations from mechanistic studies and investigations of the on-tumour versus off-tumour microbiome. | Weak evidence from human studies (9) | Agree (8) |
| The role of the microbiome in the aetiopathogenesis of cancer can be conceptualised as one apex of a tripartite, multidirectional interactome alongside the environment and an epigenetically/genetically vulnerable host. | Weak evidence from human studies (7) | Strongly agree (9) |
Traditional carcinogens are generally conceptualised as initiating agents, causing DNA damage and inciting carcinogenesis. This conceptual framework can also be applied to well-established microbial carcinogens which induce DNA damage either directly via genomic integration (eg, HPV) or indirectly via immunosuppression (eg, HIV) and/or chronic inflammation (eg, HCV). However, our understanding of how the dynamic symbionts of the human microbiome may contribute to carcinogenesis is less well established and are largely drawn from the investigation of CRC. Based on in vitro and murine in vivo data, Sears and Pardoll proposed the ‘alpha-bug hypothesis’ in which key members of the host microbiome possess specific virulence traits that are both directly oncogenic and that also remodel the microbial community towards an oncogenic phenotype.38 They cite ETBF as a candidate ‘alpha-bug’, causing colonic epithelial cell damage directly via B. fragilis toxin and indirectly by modifying mucosal immune function and intracellular signalling events.27 29 In support of this hypothesis, investigators have demonstrated that high abundance of ETBF in colonic tissues is associated with early-stage carcinogenic lesions.39 However, numerous studies of the on-tumour microbiome show large variation and fail to find consistent overabundance of putative alpha bugs in carcinoma tissue. In recognition of this observation, Tjalsma et al proposed a refinement to the alpha-bug hypothesis dubbed the ‘driver–passenger model’.40 They agreed that certain bacterial drivers (akin to alpha bugs) are responsible for the initial epithelial DNA damage that incites oncogenesis. However, they propose that the oncogenic process creates a unique tumour microenvironment that favours the proliferation of opportunistic passenger bacteria that gradually outcompete the driver species. Thus, the colorectal tumour microbiome undergoes a temporal evolution correlating with tumour stage. This hypothesis is supported by observational data regarding bacterial communities present on colorectal tumours of varying stages.41 42 The ‘driver–passenger model’ does not exclude bacterial passengers from playing an active role in cancer progression. Fusobacterium sp have been consistently found to colonise CRC tissue in numerous studies and represent an archetypal passenger bacterium.43 44 Colorectal tumours are rich in amino acids, an essential substrate for Fusobacterium sp,45 and express ligands for Fusobacterium sp cell-surface receptors of which may explain their overabundance.46 Fusobacterium sp may also promote tumour progression via interactions with intracellular signalling pathways and immune cells.26 28
Although these models account for key observations from mechanistic studies and investigations of the on-tumour microbiome, they perhaps fall short of addressing the aetiopathogenesis of cancer at the macrolevel. For example, how do environmental exposures, such as diet, contribute to cancer risk? What is the impact of host genetic factors? Are these independent risk factors for cancer development or do they act in consort with the microbiome? In consideration of these questions, we propose a broad framework for modelling the role of the microbiome in the development of cancer based on the concept of the interactome. In this model, we consider carcinogenesis to be the ultimate outcome from harmful, tripartite, multidirectional interactions between the microbiome, the environment and the epigenetically/genetically vulnerable host. The relative contributions of these three apices will vary among cancer subtypes. In the case of CRC, for example, it seems likely that the microbiota is necessary, but not sufficient for carcinogenesis; a hypothesis supported by chemical (environment)47 or genetic (vulnerable host)48 rodent models of CRC that display significantly attenuated or no tumour development in germ-free settings. Bacterial driver functions (such as toxin, hydrogen sulphide or (reduced) butyrate production) are the key transducers of environmental cues (such as diet) that can incite carcinogenesis in a vulnerable host (eg, specific TLR genetic polymorphisms). Progression of a neoplasm may depend on continued exposure to environmental stimuli, maladaptive/adaptive changes in microbiome function and/or host response.
Question four: Is the relationship between the human microbiome and the aetiopathogenesis of cancer causative or associative (table 4)?
Table 4 Isthe relationship between the human microbiome and the aetiopathogenesis of cancer causative or associative?
| Statement | Evidentiary support | Expert agreement |
| At the single-organism level, the role of microorganisms as aetiological agents in carcinogenesis is well established. | Strong evidence from human studies (10) | Strongly agree (8) |
| There are plausible mechanisms by which the human microbiome may cause cancer. | Evidence from animal studies (8) | Strongly agree (10) |
| There is a causal relationship between the human microbiome and the aetiopathogenesis of some cancers. | Weak evidence from human studies (8) | Agree (10) |
Despite a re-evaluation that considered modern sequence-based bacterial identification,49 Koch’s postulates cannot be applied to prove the causation of cancer by the host microbiome because it is not an ‘infectious agent’. The development of cancer is determined by the cumulative effects of a wide variety of risk factors and protective factors on an individual. Despite this complexity, at the single organism level, the role of microorganisms as aetiological agents (or risk factors) in carcinogenesis is well established. Numerous viruses, including human herpesvirus 8 select virotypes of HPV, hepatitis B and C, Epstein-Barr virus and HIV are classified as class 1 carcinogens by the International Agency for Research on Cancer (Lyon, France). H. pylori, schistosomes and liver flukes are also known to be carcinogenic. Such causal relationships are founded on Bradford Hill’s criteria which can be broadly categorised as: direct evidence from studies that a relationship between an exposure and an outcome is causal; mechanistic evidence that connects the exposure and outcome and parallel evidence that supports the causal relationship suggested in a study.50 For example, direct evidence for the causal relationship between H. pylori and gastric cancer (controversial for some time) comes from Uemura et al’s landmark prospective cohort study demonstrating that gastric cancer developed in approximately 3% of individuals infected with H. pylori compared with none of the uninfected individuals.51 This finding has been supported by numerous studies demonstrating mechanisms by which H. pylori contributes to gastric carcinogenesis including chronic inflammation and direct genotoxic effects mediated by a variety of defined virulence factors and effectors (eg, CagA, cagPAI and VacA).52 Parallel evidence comes from randomised trials demonstrating a reduction in the incidence of gastric cancer following H. pylori eradication.53 It is notable that not all individuals infected with H. pylori develop gastric cancer. Infection alone is not sufficient to cause disease; there are other factors which govern an individual’s susceptibility.
In contrast, a causal relationship between the human symbiotic microbiome and the development of cancer is not firmly established. The nature of the microbiome presents several challenges for proving causation. The microbiome—including the virome, mycobiome and other organisms—is orders of magnitude more complex than a discrete risk factor such as smoking or colonisation by a single organism. The microbiome is dynamic, changing with age, environmental exposures (eg, diet, antibiotics and some surgeries) and the development of cancer itself. While a causal relationship has been posited for numerous cancer types, the role of the gut microbiome in colorectal carcinogenesis has received the most attention and offers the strongest evidence. We have already summarised the mechanistic evidence supporting causation. Parallel evidence comes from murine studies in which germ-free mice chemically or genetically predisposed to intestinal neoplasia, develop fewer tumours compared with conventionally housed counterparts, while carcinogenesis can be potentiated by exposing mice to specific bacteria or stool from patients with CRC.26 54 Human studies have demonstrated that the CRC microbiome differs from that of nearby normal tissue with reduced diversity and altered community structure with lower relative abundance of potentially protective taxa (eg, butyrate-producing Roseburia) and increased abundance of taxa with potentially procarcinogenic phenotypes (eg, Fusobacterium, Bacteroides, Campylobacter, Escherichia and Porphyromonas).43 44 55–57 However, it is currently unclear whether this altered community structure is a risk factor for CRC development or rather evolves secondarily to the unique tumour microenvironment. Studies have shown a linear relationship between microbiome composition and tumour stage, moving through the adenoma-carcinoma sequence, and also with other histological tumour features such as lymphovascular and perineural invasion.41 42 These findings are at least consistent with causation but could also be argued to reflect changes in the tumour microenvironment. However, uninvolved, normal colonic mucosa from patients with CRC also demonstrates taxonomic differences (eg, abundance of sulfidogneic Bilophila wadswothia) compared with healthy mucosa from controls suggesting a microenvironment-independent underlying field change.58 Nonetheless, direct evidence supporting causation in a human population is lacking as current studies have been cross sectional with single-time-point sampling—there are no prospective, longitudinal data from large cohort studies. The evidence for other cancer types and microbiome niches is generally weaker than that for the gut microbiome and CRC and is therefore also unproven.
Question five: What are the key directions for future research to develop our understanding of the role of the microbiome in carcinogenesis (table 5)?
Table 5 Whatare the key directions for future research to develop our understanding of the role of the microbiome in carcinogenesis?
| Statement: key areas for future development with respect to the investigation of the microbiome and carcinogenesis are: | Expert agreement |
| Large, international cohort studies | Strongly agree (10) |
| Prospective longitudinal sampling | Strongly agree (14) |
| More focus on interventional, rather than purely observational studies | Strongly agree (6)/agree (6) |
| Integration of microbiome analysis with other oncological research projects | Strongly agree (12) |
| Standardisation and transparency in reporting microbiome research | Strongly agree (10) |
With the aims of making a more definitive statement regarding causality and generating true translational impact, the consortium suggested several key directions for future work on the microbiome and cancer.
Large, longitudinal international cohort studies
Proving causation in cancer is difficult because of the lag between exposure and phenotype and the complex interactive nature of potential causative factors. However, these difficulties can be overcome by conducting large, international, longitudinal cohort studies as exemplified by the European Prospective Investigation into Cancer and Nutrition (EPIC)59 and Framingham60 studies. The execution of such studies within the microbiome field is not without challenge61 but is necessary to move from testing association to causation.
Increased focus on interventional studies
Observational studies have provided valuable knowledge regarding microbiome composition and function in cancer but if we are to translate this to patient benefit, more emphasis must be placed on interventional research. We need to develop our understanding of how manipulation of the microbiome can impact cancer. For example, early studies have suggested that antibiotic use may increase cancer risk.62
Integration with other oncology research
One way in which to deliver the above three goals is to consider the integration of microbiome research into oncology studies and trials. It is relatively common for clinical trials to have a basic/translational aspect that frequently delves into mechanistic aspects of disease. Given the potential involvement of the host microbiome in the aetiopathogenesis of cancer, cancer diagnosis as well as the efficacy and toxicity of chemotherapy, immunotherapy and radiotherapy, microbiome analysis would be a pertinent angle of inquiry in many oncology trials. Primary or secondary cancer prevention studies with interventions such as dietary change are prime examples.
Standardisation and transparency in reporting microbiome research
Microbiome science has captured the imaginations of scientists, clinicians and the public alike but often blinds consumers with complex ‘omics’ methodologies while simultaneously failing to deliver on its translational potential. This has precipitated criticism of the entire field63. Reporting guidelines have become de rigueur in many aspects of basic and clinic research and the panel would welcome the application of similar frameworks to microbiome research. Transparent reporting of experimental and analytical methods alongside detailed metadata is essential to facilitate reproducibility.
Conclusion
In this consensus statement, we have reflected on current expert opinion regarding the crucial question of whether the human microbiome plays a causative role in the aetiopathogenesis of cancer, with a focus on CRC. While cancer causation by specific microorganisms (such as H. pylori) is beyond doubt, proving causation for the human microbiome in the broader sense is more challenging. There are plausible mechanisms and ample parallel or supporting evidence from in vitro, murine and cross-sectional human studies. However, direct evidence from large longitudinal cohort studies is lacking and should be a key focus in the future. Until then, the expert panel considers that the case in support of a causative role for the human microbiome in the aetiopathogenesis of cancer remains unproven, but a majority nevertheless agrees with the hypothesis. The panel considers the microbiome to be one aspect of an interactome with an epigenetically/genetically vulnerable host and the environment. However, the host genotype is largely fixed and evidence suggests that the microbiome may be a key player in modulating/mediating the response to environmental pressures. In this context, the microbiome presents a very exciting opportunity to better understand cancer and how it might be prevented.
gutjnl-2019-318556supp001.pdf (127.8KB, pdf)
Acknowledgments
The authors would like to thank Jessica Younes, Science liaison at Winclove Probiotics, and Alexander Swidsinki, Charité Universitätsmedizin Berlin, for their contributions to the roundtable discussion.
Footnotes
Contributors: AJS, JA, CAM, JMK and JM planned the consensus statement and organised the ICMC roundtable discussion, proposing broad topics for discussion. AJS, JA, CAM, JMK, JM, DC, CJ, RB, JA, SJO, HRG, JT, JY, DJH, HV, JB and DWR contributed to active discussion during the roundtable event. AJS, JA, CAM, JMK and JM drafted the manuscript. DC, CJ, RB, JA, SJO, HRG, JT, JY, DJH, HV, JB, PWO and DWR revised the manuscipt. JMK and JM act as guarantors of the content of the manuscript.
Funding: The roundtable discussion was funded by Cancer Research UK (Award C42378/A25637). The Division of Integrative Systems Medicine and Digestive Disease at Imperial College London receives financial support from the National Institute of Health Research (NIHR) Imperial Biomedical Research Centre (BRC) based at Imperial College Healthcare NHS Trust and Imperial College London. This article is independent research funded by the NIHR BRC, and the views expressed in this publication are those of the authors and not necessarily those of the NHS, NIHR, or the Department of Health.
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Correction notice: This article has been corrected since it published Online First. The ninth author’s name has been corrected.
Patient consent for publication: Not required.
References
- 1. Lloyd-Price J, Mahurkar A, Rahnavard G, et al. Strains, functions and dynamics in the expanded Human Microbiome Project. Nature 2017;550:61–6. 10.1038/nature23889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tomasetti C, Li L, Vogelstein B, et al. somatic mutations, cancer etiology, and cancer prevention. Science 2017;355:1330–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mima K, Nishihara R, Qian ZR, et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut 2016;65:1973–80. 10.1136/gutjnl-2015-310101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zitvogel L, Ma Y, Raoult D, et al. The microbiome in cancer immunotherapy: Diagnostic tools and therapeutic strategies. Science 2018;359:1366–70. 10.1126/science.aar6918 [DOI] [PubMed] [Google Scholar]
- 5. Scott AJ, Merrifield CA, Alexander JL, et al. Highlights from the Inaugural International Cancer Microbiome Consortium Meeting (ICMC), 5–6 September 2017, London, UK. Ecancermedicalscience;11:791 10.3332/ecancer.2017.791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carding S, Verbeke K, Vipond DT, et al. Dysbiosis of the gut microbiota in disease. Microb Ecol Health Dis 2015;26:26191 10.3402/mehd.v26.26191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hooks KB, O’Malley MA. Dysbiosis and Its Discontents. MBio 2017;8 10.1128/mBio.01492-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Levy M, Kolodziejczyk AA, Thaiss CA, et al. Dysbiosis and the immune system. Nat Rev Immunol 2017;17:219–32. 10.1038/nri.2017.7 [DOI] [PubMed] [Google Scholar]
- 9. Human Microbiome Project C. Structure, function and diversity of the healthy human microbiome. Nature 2012;486:207–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zierer J, Jackson MA, Kastenmüller G, et al. The fecal metabolome as a functional readout of the gut microbiome. Nat Genet 2018;50:790–5. 10.1038/s41588-018-0135-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lozupone CA, Stombaugh JI, Gordon JI, et al. Diversity, stability and resilience of the human gut microbiota. Nature 2012;489:220–30. 10.1038/nature11550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell 2014;157:121–41. 10.1016/j.cell.2014.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nicholson JK, Holmes E, Kinross J, et al. Host-gut microbiota metabolic interactions. Science 2012;336:1262–7. 10.1126/science.1223813 [DOI] [PubMed] [Google Scholar]
- 14. Bhatt AP, Redinbo MR, Bultman SJ. The role of the microbiome in cancer development and therapy. CA Cancer J Clin 2017;67:326–44. 10.3322/caac.21398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ríos-Covián D, Ruas-Madiedo P, Margolles A, et al. Intestinal Short Chain Fatty Acids and their Link with Diet and Human Health. Front Microbiol 2016;7:185 10.3389/fmicb.2016.00185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schwabe RF, Jobin C. The microbiome and cancer. Nat Rev Cancer 2013;13:800–12. 10.1038/nrc3610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Münger K, Baldwin A, Edwards KM, et al. Mechanisms of human papillomavirus-induced oncogenesis. J Virol 2004;78:11451–60. 10.1128/JVI.78.21.11451-11460.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cook L, Melamed A, Yaguchi H, et al. The impact of HTLV-1 on the cellular genome. Curr Opin Virol 2017;26:125–31. 10.1016/j.coviro.2017.07.013 [DOI] [PubMed] [Google Scholar]
- 19. Nesić D, Hsu Y, Stebbins CE. Assembly and function of a bacterial genotoxin. Nature 2004;429:429–33. 10.1038/nature02532 [DOI] [PubMed] [Google Scholar]
- 20. He Z, Gharaibeh RZ, Newsome RC, et al. Campylobacter jejuni promotes colorectal tumorigenesis through the action of cytolethal distending toxin. Gut 2019;68:289–300. 10.1136/gutjnl-2018-317200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Buc E, Dubois D, Sauvanet P, et al. High prevalence of mucosa-associated E. coli producing cyclomodulin and genotoxin in colon cancer. PLoS One 2013;8:e56964 10.1371/journal.pone.0056964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Attene-Ramos MS, Wagner ED, Plewa MJ, et al. Evidence that hydrogen sulfide is a genotoxic agent. Mol Cancer Res 2006;4:9–14. 10.1158/1541-7786.MCR-05-0126 [DOI] [PubMed] [Google Scholar]
- 23. Huycke MM, Gaskins HR. Commensal bacteria, redox stress, and colorectal cancer: mechanisms and models. Exp Biol Med 2004;229:586–97. 10.1177/153537020422900702 [DOI] [PubMed] [Google Scholar]
- 24. Francescone R, Hou V, Microbiome GSI. inflammation, and cancer. Cancer J 2014;20:181–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pagano JS, Blaser M, Buendia MA, et al. Infectious agents and cancer: criteria for a causal relation. Semin Cancer Biol 2004;14:453–71. 10.1016/j.semcancer.2004.06.009 [DOI] [PubMed] [Google Scholar]
- 26. Kostic AD, Chun E, Robertson L, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 2013;14:207–15. 10.1016/j.chom.2013.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wu S, Rhee KJ, Zhang M, et al. Bacteroides fragilis toxin stimulates intestinal epithelial cell shedding and gamma-secretase-dependent E-cadherin cleavage. J Cell Sci 2007;120:1944–52. 10.1242/jcs.03455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rubinstein MR, Wang X, Liu W, et al. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe 2013;14:195–206. 10.1016/j.chom.2013.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wu S, Rhee KJ, Albesiano E, et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat Med 2009;15:1016–22. 10.1038/nm.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dapito DH, Mencin A, Gwak GY, et al. Promotion of hepatocellular carcinoma by the intestinal microbiota and TLR4. Cancer Cell 2012;21:504–16. 10.1016/j.ccr.2012.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science 2012;336:1268–73. 10.1126/science.1223490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Routy B, Le Chatelier E, Derosa L, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 2018;359:91–7. 10.1126/science.aan3706 [DOI] [PubMed] [Google Scholar]
- 33. Gur C, Ibrahim Y, Isaacson B, et al. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity 2015;42:344–55. 10.1016/j.immuni.2015.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zitvogel L, Ayyoub M, Routy B, et al. Microbiome and Anticancer Immunosurveillance. Cell 2016;165:276–87. 10.1016/j.cell.2016.03.001 [DOI] [PubMed] [Google Scholar]
- 35. O’Keefe SJ, Li JV, Lahti L, et al. Fat, fibre and cancer risk in African Americans and rural Africans. Nat Commun 2015;6:6342 10.1038/ncomms7342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Seitz HK, Stickel F. Molecular mechanisms of alcohol-mediated carcinogenesis. Nat Rev Cancer 2007;7:599–612. 10.1038/nrc2191 [DOI] [PubMed] [Google Scholar]
- 37. Belcheva A, Irrazabal T, Robertson SJ, et al. Gut microbial metabolism drives transformation of MSH2-deficient colon epithelial cells. Cell 2014;158:288–99. 10.1016/j.cell.2014.04.051 [DOI] [PubMed] [Google Scholar]
- 38. Sears CL, Pardoll DM. Perspective: alpha-bugs, their microbial partners, and the link to colon cancer. J Infect Dis 2011;203:306–11. 10.1093/jinfdis/jiq061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Purcell RV, Pearson J, Aitchison A, et al. Colonization with enterotoxigenic Bacteroides fragilis is associated with early-stage colorectal neoplasia. PLoS One 2017;12:e0171602 10.1371/journal.pone.0171602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tjalsma H, Boleij A, Marchesi JR, et al. A bacterial driver-passenger model for colorectal cancer: beyond the usual suspects. Nat Rev Microbiol 2012;10:575–82. 10.1038/nrmicro2819 [DOI] [PubMed] [Google Scholar]
- 41. Kinross J, Mirnezami R, Alexander J, et al. A prospective analysis of mucosal microbiome-metabonome interactions in colorectal cancer using a combined MAS 1HNMR and metataxonomic strategy. Sci Rep 2017;7:8979 10.1038/s41598-017-08150-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Flanagan L, Schmid J, Ebert M, et al. Fusobacterium nucleatum associates with stages of colorectal neoplasia development, colorectal cancer and disease outcome. Eur J Clin Microbiol Infect Dis 2014;33:1381–90. 10.1007/s10096-014-2081-3 [DOI] [PubMed] [Google Scholar]
- 43. Marchesi JR, Dutilh BE, Hall N, et al. Towards the human colorectal cancer microbiome. PLoS One 2011;6:e20447 10.1371/journal.pone.0020447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Castellarin M, Warren RL, Freeman JD, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res 2012;22:299–306. 10.1101/gr.126516.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dai ZL, Wu G, Zhu WY. Amino acid metabolism in intestinal bacteria: links between gut ecology and host health. Front Biosci 2011;16:1768–86. 10.2741/3820 [DOI] [PubMed] [Google Scholar]
- 46. Abed J, Emgård JE, Zamir G, et al. Fap2 Mediates Fusobacterium nucleatum Colorectal Adenocarcinoma Enrichment by Binding to Tumor-Expressed Gal-GalNAc. Cell Host Microbe 2016;20:215–25. 10.1016/j.chom.2016.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Uronis JM, Mühlbauer M, Herfarth HH, et al. Modulation of the intestinal microbiota alters colitis-associated colorectal cancer susceptibility. PLoS One 2009;4:e6026 10.1371/journal.pone.0006026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kado S, Uchida K, Funabashi H, et al. Intestinal microflora are necessary for development of spontaneous adenocarcinoma of the large intestine in T-cell receptor beta chain and p53 double-knockout mice. Cancer Res 2001;61:2395–8. [PubMed] [Google Scholar]
- 49. Fredricks DN, Relman DA. Sequence-based identification of microbial pathogens: a reconsideration of Koch’s postulates. Clin Microbiol Rev 1996;9:18–33. 10.1128/CMR.9.1.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Howick J, Glasziou P, Aronson JK. The evolution of evidence hierarchies: what can Bradford Hill’s ’guidelines for causation' contribute? J R Soc Med 2009;102:186–94. 10.1258/jrsm.2009.090020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Uemura N, Okamoto S, Yamamoto S, et al. Helicobacter pylori infection and the development of gastric cancer. N Engl J Med 2001;345:784–9. 10.1056/NEJMoa001999 [DOI] [PubMed] [Google Scholar]
- 52. Ishaq S, Nunn L. Helicobacter pylori and gastric cancer: a state of the art review. Gastroenterol Hepatol Bed Bench 2015;8:S6–S14. [PMC free article] [PubMed] [Google Scholar]
- 53. Fuccio L, Zagari RM, Eusebi LH, et al. Meta-analysis: can Helicobacter pylori eradication treatment reduce the risk for gastric cancer? Ann Intern Med 2009;151:121–8. 10.7326/0003-4819-151-2-200907210-00009 [DOI] [PubMed] [Google Scholar]
- 54. Wong SH, Zhao L, Zhang X, et al. Gavage of Fecal Samples From Patients With Colorectal Cancer Promotes Intestinal Carcinogenesis in Germ-Free and Conventional Mice. Gastroenterology 2017;153:1621–33. 10.1053/j.gastro.2017.08.022 [DOI] [PubMed] [Google Scholar]
- 55. Yu J, Feng Q, Wong SH, et al. Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer. Gut 2017;66:70–8. 10.1136/gutjnl-2015-309800 [DOI] [PubMed] [Google Scholar]
- 56. Kostic AD, Gevers D, Pedamallu CS, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res 2012;22:292–8. 10.1101/gr.126573.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Flemer B, Lynch DB, Brown JM, et al. Tumour-associated and non-tumour-associated microbiota in colorectal cancer. Gut 2017;66:633–43. 10.1136/gutjnl-2015-309595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yazici C, Wolf PG, Kim H, et al. Race-dependent association of sulfidogenic bacteria with colorectal cancer. Gut 2017;66:1983–94. 10.1136/gutjnl-2016-313321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Riboli E, Kaaks R. The EPIC Project: rationale and study design. European Prospective Investigation into Cancer and Nutrition. Int J Epidemiol 1997;26(Suppl 1):6S–14. 10.1093/ije/26.suppl_1.S6 [DOI] [PubMed] [Google Scholar]
- 60. Mahmood SS, Levy D, Vasan RS, et al. The Framingham Heart Study and the epidemiology of cardiovascular disease: a historical perspective. Lancet 2014;383:999–1008. 10.1016/S0140-6736(13)61752-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Vandeputte D, Tito RY, Vanleeuwen R, et al. Practical considerations for large-scale gut microbiome studies. FEMS Microbiol Rev 2017;41:S154–S67. 10.1093/femsre/fux027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Cao Y, Wu K, Mehta R, et al. Long-term use of antibiotics and risk of colorectal adenoma. Gut 2018;67:672–8. 10.1136/gutjnl-2016-313413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hanage WP. Microbiology: Microbiome science needs a healthy dose of scepticism. Nature 2014;512:247–8. 10.1038/512247a [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
gutjnl-2019-318556supp001.pdf (127.8KB, pdf)